Abstract
Deubiquitinating enzyme dysregulation has been linked to the development of a variety of human malignancies, including breast cancer. However, the exact involvement of the deubiquitinating enzyme USP39 in the progression of breast cancer is yet unknown. Cell viability and colony formation analysis was used to assess the effects of USP39 knockdown on breast cancer cells in this study. The interaction between USP39 and FOXM1 was investigated using co-immunoprecipitation (co-IP) and in vitro deubiquitination analysis. The expression of USP39 and FOXM1 in breast cancer tissues was studied using the TCGA database. According to our findings, USP39 deubiquitinates and stabilizes FOXM1, promoting breast cancer cell proliferation, colony formation, and tumor growth in vivo. Furthermore, elevated USP39 expression lowers FOXM1 ubiquitination, resulting in increased transcriptional activity. In addition, the high expression of USP39 reduces the ubiquitination of FOXM1, thereby enhancing the transcriptional activity of FOXM1 and regulating the expression of downstream genes Cdc25b and Plk1. USP39 is positively correlated with the expression level of FOXM1 in breast cancer cells. In general, our research revealed the USP39-FOXM1 axis as a critical driver of breast cancer cell proliferation and provided a theoretical foundation for targeting the USP39-FOXM1 axis for pancreatic cancer treatment.
Keywords: Breast cancer, ubiquitin-specific protease 39, transcription factor FOXM1, cell proliferation
Introduction
USP39 belongs to the family of ubiquitin-specific proteins and contains an N-terminal RS-like ZNF domain and a C-terminal ubiquitin hydrolase domain [1]. Recent studies have shown that USP39 is highly expressed in a variety of malignant tumors and is involved in a variety of biological functions such as cell proliferation [2,3], cell cycle progression [4], cell mitosis [5], cell migration and invasion [6,7], DNA damage repair [8], pre-messenger RNA (mRNA) splicing [9,10], cell apoptosis [11], cell regulation [6,12], and drug tolerance [8,13]. Exome sequencing studies involving breast cancer patients have shown that USP39 regulates cancer-related tumor suppressors, including CHEK2, and has a tumor-inducing effect [14]. Interference with USP39 could inhibit the growth of breast cancer cells [15]. RNAi-mediated USP39 significantly reduced the proliferation and colony formation ability of McF-7 cells, and decreased expression of USP39 could induce G0/G1 phase arrest and cell apoptosis [15]. However, the role and potential molecular mechanism of USP39 in breast cancer tumorigenesis have not been reported so far.
FOXM1 is a kind of typical hyperplasia related transcription factors [16]. It is a member of the Forkhead box (FOX) transcription factor family, which has 44 members in humans and shares a forkhead DNA binding motif [17,18]. As far as gene function is concerned, FOXM1 has been shown to be an oncogene that regulates growth and proliferation [19], and plays a particularly important role in stem cell differentiation [20,21], DNA damage repair [22,23], cell mitosis [24,25], drug tolerance [25,26], and cell metabolism [27,28]. The FOXM1 protein is known to regulate the cell cycle [29], mainly by regulating the transcriptome to promote mitosis and ensuring chromosomal stability [30-32]. The significant upregulation of certain cancers and cancer-related genes controlled by FOXM1 was found to occur in addition to its important role in the normal cell cycle [33,34]. A significant amount of FOXM1 is expressed in most human tumors, as well as liver, prostate, brain, breast, lung, colon, and pancreatic cancers [35-37]. Recent studies have also shown that the expression of FOXM1 promotes the growth, proliferation, invasion and angiogenesis of breast, gliomas, pancreas, colon, and lung cancer cells [38-40]. In addition, we found that FOXM1 has a strict regulatory effect on the proliferation and tumorigenicity of breast cancer cells [41,42]. Despite the fact that FOXM1 is overexpressed in breast cancer tumors, the molecular mechanism underlying its overexpression is unclear [43]. In addition, studies have confirmed that the E3 ubiquitin ligase APC/Cdh1 regulates the degradation of FOXM1 [44-46], However, deubiquitinase may play a role in FOXM1 protein stability. FOXM1 may be potentially stabilized by targeting the enzymes that prevent its destruction. Therefore, we tried to delineate the regulation of FOXM1 stability by removing ubiquitin, thereby protecting FOXM1 from degradation. USP39 is known to regulate FOXM1 expression [47,48], but the underlying molecular mechanism remains unknown.
DUB Ubiquitin-Specific Protease (USP) 39 is a member of the USP family, which contains 56 members united by a highly conserved domain [49]. This study demonstrates the relationship between FOXM1 and USP39. We observed that USP39 regulated the abundance of FOXM1. FOXM1 is protected from proteosomal degradation by USP39 binding to and reducing its ubiquitination levels. Moreover, we showed that USP39 expression could alter FOXM1 transcriptional activity, which affected the proliferation of cells and the development of tumors. Furthermore, we found that FOXM1 and USP39 were specifically upregulated in breast cancer. The depletion of USP39 can reduce the level of FOXM1, and the reduced level of FOXM1 inhibits the expression of USP39, so my conclusion is that FOXM1 is able to regulate the expression of USP39 based on a feedback loop. These findings suggest that USP39 regulates cell proliferation by maintaining the stability of FOXM1, interfering with USP39 can inhibit the proliferation of breast cancer, interfering with USP39 while overexpressing FOXM1 can alleviate the inhibition of breast cancer proliferation, FOXM1 and USP39 have a synergistic effect, so FOXM1 and USP39 as a potential drug target for breast cancer.
Materials and methods
Cell lines and cell culture
MCF10A, SUM185, BT474, SUM190, BT549, MDA-MB-231, MDA-MB-453, MCF-7, ZR-75-30, MDA-MB-468, SUM159 and HEK 293T, Cells were purchased from ATCC (Manassas, USA). Cells were grown in DMEM cell culture medium (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA) at 37°C in a 5% CO2 incubator.
Construction of plasmids
pCMV-FOXM1, pCMV-flag-FOXM1, and p6×FOXM1 Binding-Luc reporter plasmid were obtained from Dr. Yongjun Tan labarary [50]. pCMV-USP39, pCMV-C-myc-USP39, pCMV-Cdh1, and pCMV-HA-Ub were purchased from GenePharma (Shanghai, China).
Purification and expression of recombinant proteins
We transformed pHis-FoxM1, pHis-M1-234, pHis-GFP, pET-15b-GST, and pET-15b-GST-M1-234 into Rosetta/DE3 E. Coli cells and PCR was performed on selected colonies to identify positive colonies. To induce protein expression, cells were grown in LB medium at 37°C for 12 hours at 28°C, then added 1 mM IPTG for another 12 h. FOXM1N, GFP, or FOXM1 recombinant proteins were purified using Ni-SepharoseTM 6 Fast Flow (GE) according to the manufacturer’s instructions. The GST or GST-M1-234 (1-234 aa) recombinant protein was purified through the use of Glutathione SepharoseTM 4B (GE) following the manufacturer’s instructions.
Cell viability assays
The cells were plated in 96-well plates (4*103 cells in each well) and were incubated with 10% fetal bovine serum for 24 hours at 37°C in a humidified atmosphere containing 5% CO2. After transfection with SiUSP39#1, SiUSP39#2, and pCMV-USP39 expression plasmids, cell count assay was performed at 24 h, 48 h, and 72 h, respectively. MDA-MB-231 and MCF-7 cells were counted in order to analyze the effect of interference and high levels of USP39 on cell proliferation. Three repetitions of the tests were conducted.
Colony formation assays
A 6-well plate was seeded with 400 cells per dish at 37°C for 24 hours, then transfected with siUSP39#1, siUSP39#2, and pCMV-USP39 expression vectors. Fresh medium was replenished every three days. A 14-day incubation followed by paraformaldehyde fixation and three PBS rinses was followed by crystal violet staining for 10 minutes and three PBS washes were completed. Crystal violet-stained cell monoclonal photographs were taken with a camera.
Lentiviral transductions
MCF-7 or MDA-MB-231 cells were treated by LV-shUSP39 (shUSP39-1, sh-USP39-2; OBiO Technology, Shanghai, China) and LV-FOXM1 infection. 48 h later, the cells were cultured in DMEM containing 2 μg/mL purinomycin (Thermo Fisher Scientfic) for another 2 weeks to select the cells containing the constructs. For ectopic expression of USP39 or FOXM1, MDA-MB-231 or McF-7 cells were infected with lentiviral constructs expressing shUSP39 or full-length FOXM1. The shRNA interference sequence are as follows: sh-negative control, 5’-UUC UCC GAA CGU GUC ACG UTT-3’; sh-USP39-1, 5’-GCA UAU GAU GGU ACC ACU UTT-3’; sh-USP39-2, 5’-CCU UCA GGC UCU AUC UAA UTT-3’.
RNA interference
Transfection of FOXM1 siRNA (SantaCruz sc-43769) and USP39 siRNA (siRNA#1: CCTTCCAGACAACTATGAGAT, siUSP39#2: GCCGGGTATTGTGGGACTGAA) were carried out according to the manufacturer’s instructions using Lipofectamine 2000 (Invitrogen).
Quantitative real-time PCR (qPCR)
Total RNA (Omega) was extracted according to the reagent manufacturer’s instructions. Total RNA (1.0 μg) was reverse transcribed into 10 μl cDNA (Promega) using the Revert Aid First Strand Kit. Detection was performed using the qPCR kit SYBR Green (Toyobo) with the following sense (S) and antisense (AS) primers: hUSP39-S, 5’-TGA CCT CAT TGC CAA CAT CGT-3’ and hUSP39-AS, 5’-TTG CCT GTCCC ATG ATG AAGC-3’; hFOXM1-S, 5’-GCT TGC CAG AGT CCT TTT TGC-3’ and hFOXM1-AS, 5’-CCA CCT GAG TTC TCG TCA ATG C-3’; hCDC25B-S, 5’-CAG CGA CTT GCT GCT CAA AA-3’ and hCDC25BAS, 5’-CGA CAG GGA GGA TTC GGA TG-3’; hPLK1-S, 5’-CCG CAA TTA CAT GAG CGA GC-3’ and hPLK1-AS, 5’-AGG AGA CTC AGG CGG TAT GT-3’; hGAPDH-S, 5’-GGA GCG AGA TCC CTC CAA AAT-3’ and hGAPDH-AS, 5’-GGC TGT TGT CAT ACT TCT CAT GG-3’. Realplex2 qPCR system (Eppendorf) was used for qPCR. GAPDH is used to standardize relative mRNA levels.
Protein extraction and western blotting
Cells were washed twice in pre-chilled PBS and then lysed in cell lysate containing protease inhibitors (150 mM NaCl, 20 mM Tris pH 7.4, 0.1 mM EDTA, 1% Nonidet P-40). TissueLyser (Qiagen) was used to homogenize tumor tissue using tissue lysate containing 150 mM NaCl, 50 mM Tris-HCl pH 8.0, 2 mM EDTA pH 8.0, 20% glycerol, 1% Nonidet P-40 and 10 mM NaF, plus protease inhibitors. The BCA protein assay reagent was used to determine the protein concentration in supernatants after centrifugation (Thermo Fisher Scientific).
Lysates were combined with sample buffer containing beta-mercaptoethanol and heated to 95°C for 5 minutes. SDS-PAGE was used to separate the lysates, which were then transferred to a PVDF membrane for western blotting. The following antibodies and dilutions were used for Western blotting: rabbit anti-FOXM1 (1:1000, ABclonal, A2493), rabbit anti-FOXM1 (C-20) (1:5000, SantaCruz sc-502, recognizing FOXM1 C-terminus), rabbit anti-FOXM1 (K-19) (1:5000, SantaCruz sc-500, recognizing FOXM1 N-terminus), rabbit anti-USP39 (1:1000, Proteintech 23865-1-AP) mouse anti-flag (1:5000, Abmart #M22000), mouse anti-C-Myc (1:500, SantaCruz sc-40), mouse anti-GFP (1:500, Beyotime AG281), rabbit anti-GST (1:1000, Abcam ab19256), rabbit anti-CDC25B (1:1000, SantaCruz sc-326), rabbit anti-PLK1 (1:1000, CUSABIO, CSB-PA004668), mouse anti-Cdh1 (1:1000, SantaCruz sc-56312), rabbit anti-HA (1:1000, Proteintech 51064-2-AP), and mouse anti-β-actin (1:10000, Abcam ab49900). Signals from primary antibody were obtained via HRP conjugated anti-rabbit IgG (1:5000, ABclonal AS014) and anti-mouse IgG (1:5000; ABclonal AS003) magnification, and detected by the Kodak 4000 MM Imaging System using Super Signal West Femto Maximum Sensitivity Substrate (Thermo) (Kodak). Membranes were sliced horizontally to allow various antibodies to be detected. ImageJ software was used to quantify the bands of Western blotting.
Confocal imaging
1×104 Cells were inoculated in a 35 mm glass plate (MatTek) and fixed with 4% paraformaldehyde 24 hours later. Non-specific binding sites were sealed with skim milk powder. Then the cells were incubated with mouse anti-FoxM1 (1:500, SantaCruz SC-376471) and rabbit anti-USP39 (1:300, Proteintech 23865-1-AP), followed by the incubation of a goat anti-rabbit IgG-FITC (1:500, Santa Cruz sc-2012) or goat anti-mouse IgG-PE (1:500, SantaCruz sc-3738) secondary antibody. The FluoView FV1000 confocal imaging system (Olympus) was used to image the cells. Fluorescent FITC or CFL were excited by 488 nm or 559 nm laser, respectively.
Luciferase activity assays
Luciferase reporter gene vector (1 μg) and gene expression vector (1 μg) were transfected into 293T cells. The loading control for each transfection was the pRL-CMV plasmid (20 ng). Following the manufacturer’s instructions, Thirty-six hours after transfection, cells from each group were harvested and luciferase activity was assayed using a dual-luciferase assay system (Promega).
In vitro de-ubiquitination assays
DUB activity assay kit was purchased from Cayman chemical (NO. 701490). DUB activity assays were performed according to the kit protocol. Fluorescence between 355 nm and 365 nm and a wavelength of 455-465 nm are measured every minute for 20-60 minutes in a plate reader.
Pull-down and Co-IP analysis
Cells were collected and cell lysates were prepared. Lyse for 30 min using IP cell lysis buffer containing 100 mM NaCl, 50 mM Tris-Cl, 2.5 mM EDTA, 1% NP-40, 2.5 mM EGTA and 5% glycerol. The lysate was obtained by centrifuging for 10 min at 4°C at 12,000 rpm. The lysate of 500 µg proteins was incubated at 4°C for 2 h with 20 µl of beads (Ni-beads or GST Agarose beads, GE). In certain cases, GST, GST-M1-234, His-GFP, His-FOXM1, or His-FOXM1N1-234 (FOXM1N) was added to the reactions. Beads were then washed three times in IP buffer and analyzed by Western blotting or Mass spectrometric detection.
Co-IP involves incubating 500 µg protein lysates with 20 µL of Protein A/G PLUS-Agarose beads (SantaCruz sc-2003) and 2 µg anti-FOXM1 (Abcam, ab234077) or anti-IgG (CST#2729S) antibody at 4°C for 4 h. The beads were washed five times in IP buffer and western blotting was performed.
Immunohistochemistry
Tumor tissue was fixed with 4% paraformaldehyde and embedded in paraffin. Tumor sections (4 μm) were deparaffinized and hydrated, followed by endogenous peroxidase quenching, antigen retrieval, and nonspecific binding site blocking. Sections were then incubated with goat anti-CyclinD1 (1:200, ABclonal A11022) or goat anti-CDC25B (1:200, ABclonal A9758) followed by a horseradish peroxidase-conjugated anti-goat secondary antibody. Color was developed using 3, 3’ diaminobenzidine and finally photographed using a TE2000 microscope (Nikon).
Nude mice xenograft tumor formation experiment
Animal experiments were performed in accordance with institutional animal use guidelines, care, and ethics, and were approved by the Hubei Provincial Laboratory Animal Center, China (protocol number SYXK [Hubei] 2018-0071). BALB/c nude mice (female, 5 weeks old) were purchased from Suzhou Xishan Biotechnology Co., Ltd. (Suzhou, China). To investigate the effects of USP39 and FOXM1 on tumor formation and growth in vivo, independent experiments were performed. The mice were randomly divided into four groups (four mice in each group), which were shNC, shUSP39, shFOXM1 and shUSP39+LvFOXM1 groups, respectively. 1×106 MDA-MB-231 cells in each group were injected into the right armpit, and the mice were routinely observed. Tumor formation, size of transplanted tumors and mouse body weight were measured every two days, and tumor samples were collected on day 39 after injection of cancer cells. Tumors were measured with vernier calipers, and tumor volume (V) was calculated as follows: V = length × width2 ×1/2.
Statistical analysis
To determine the mean and standard deviation between samples, we utilize the Microsoft Excel application or Prism 7. In a nutshell, the measurement data is represented as Mean SD, and the comparison between the two groups of data is performed using an unpaired Student’s t test. For each sample value between the control and experimental groups, perform a two-way analysis of variance or T test. Data comparison among multiple groups was performed by one-way ANOVA. *, P≤0.05; **, P≤0.01; ***, P≤0.001; ##, P≤0.01. P≤0.05 is thought to represent a statistically significant difference.
Results
USP39 regulates FOXM1 protein level
FOXM1 is a known transcription factor that promotes the proliferation of various cancer cells [51]. FOXM1 is related to a variety of tumors and has been found to be highly expressed in a wide range of clinical tumor sampless; its high expression was related to genome instability [31]. Inhibiting FOXM1 ubiquitination and maintaining its stability regulated breast tumor progression [52]. According to previous studies, ubiquitination plays a key role in the cell proliferation signaling pathways [53,54]. Through mass spectrometry, ubiquitin-specific protease 39 (USP39) was identified as a potential regulator of FOXM1 stability (Supplementary Figure 1). Several studies have reported that ubiquitination mediates FOXM1 levels and regulates FOXM1 functions through the ubiquitin-proteasome system [55,56]. Researchers have confirmed that USP39 does not have deubiquitinating enzyme activity [1,5]; it has been also confirmed that USP39 can regulate stability of CHK2, PD-L1, and STAT1 [8,57,58]. However, the functions of USP39 and FOXM1 function in breast cancer have not been clarified. We interfered with the expression of the ubiquitin-specific protease USP39 in the breast cancer cell line MDA-MB-231. The results showed that silencing with USP39 significantly inhibited FOXM1 levels, but had no effect on FOXM1 mRNA levels (Figure 1A and 1B). In addition, upon treating MDA-MB-231 cells with the proteasome inhibitor MG132, the reduced level of FOXM1 was rescued (Figure 1C). To further determine whether USP39 regulates the stability of FOXM1, we treated cells with cycloheximide (CHX) and tested the stability of FOXM1. As shown in Figure 1D, when USP39 was present in MDA-MB-231 cells, the stability of FOXM1 was enhanced, and when USP39 was depleted, the stability of FOXM1 was significantly reduced. Together, these findings suggest USP39 regulates FOXM1 protein levels and stability via the proteasome, and that USP39 may function as a DUB of FOXM1.
Figure 1.

USP39 regulates FOXM1 protein levels. A. After siUSP39 was transfected for 24 h, 48 h and 72 h, qPCR was used to detect the changes of FOXM1 mRNA level in MDA-MB-231 cells. Relative mRNA levels were normalized to GAPDH. The levels of SiCTL group were referred as one. Values represented the mean ± SD of three independent experiments. (*, P≤0.05). B. After siUSP39 was transfected for 24 h, 48 h and 72 h, Western Blotting was used to detect the changes of FOXM1 protein level in MDA-MB-231 cells. C. MDA-MB-231 cells interfered with by siUSP39 and treated with the proteasome inhibitor MG-132, Western Blotting was used to detect changes in the protein levels of FOXM1 and USP39. D. MDA-MB-231 cells in the siUSP39 silence group and the control group were treated with cycloheximide (CHX) for 0 h, 0.5 h, 1 h, 2 h, 4 h, and 8 h, and the cells were collected, respectively, and Western Blotting detects changes in FOXM1 and USP39 protein levels.
USP39 maintains FOXM1 protein stability by competitively binding to FOXM1
For further confirmation of how USP39 affects FOXM1, we tested the expression and protein binding levels of USP39 and FOXM1 in breast cancer. We analyzed the expression levels of USP39 and FOXM1 in various tumors in the TCGA database. As shown in Supplementary Figure 2A and 2B, USP39 and FOXM1 were found to be highly expressed in various tumors (except thymoma), including breast cancer tumor tissues. To test whether USP39 binds to FOXM1 in order to further prove how USP39 regulates FOXM1 levels, we measured its binding to USP39. As shown in Figure 2A, Flag-tagged FOXM1 was able to pulldown USP39 and C-myc-tagged USP39 was able to pull down FOXM1 in 293T cells, as observed during immunoprecipitation assays. At the same time, the CoIP experiment also confirmed that endogenous USP39 and FOXM1 interact in breast cancer MDA-MB-231 cells (Supplementary Figure 3). Further, through cell immunostaining detection, it was found that FOXM1 and USP39 were co-localized in the nucleus (Figure 2B), which was consistent with the localization of FOXM1 in the nucleus to perform its functions [59]. To further confirm the direct binding of FOXM1 and USP39, prokaryotic E. coli DH5a cells were used to express and purify GST-M1-234 protein. The pull-down experiment revealed that GST-Sepharose beads could pull down the USP39 protein through the GST-tagged M1-234 in the cellular protein mixture (Supplementary Figure 4). These results confirmed that FOXM1 interacts with USP39 and does so via the N-terminus of FOXM1.
Figure 2.
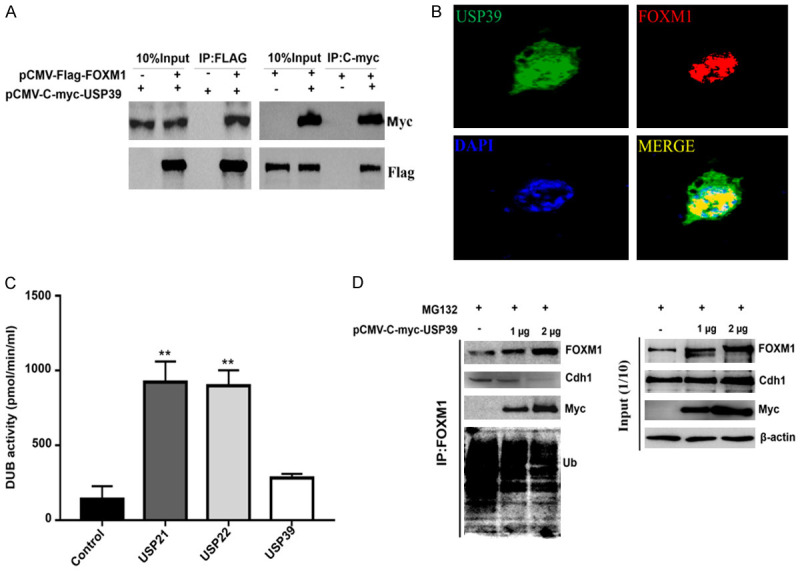
FOXM1 interacts with USP39. A. Flag-FOXM1 and C-myc-USP39 eukaryotic vector were co-transfected into 293T cells. After culturing for 48 hours, the cells were collected, and the interaction between FOXM1 and USP39 was detected by Co-IP. B. The co-localization of FOXM1 and USP39 was detected by cellular immunostaining. C. The USP21, USP22 and USP39 proteins were purified in vitro, and the deubiquitinating enzyme activity detection kit was used to detect the activity of USP39, USP21 and USP22 deubiquitinating enzymes. Values represented the mean ± SD of three independent experiments. (**, P≤0.01). D. pCMV-Myc-USP39 plasmids were transfected into MDA-MD-231 cells, cultured for 36 hours and then treated with 5 mM proteasome inhibitor, and then collected after culturing for 12 hours Cell sample. The CoIP experiment detects the interaction between USP39, FOXM1 and APC/Cdh1, and Western Blotting detects the level of FOXM1 ubiquitination, USP39, Cdh1 and FOXM1 protein levels.
Previous studies have confirmed that USP39 has no deubiquitinating enzyme activity [1,5], while other researchers have confirmed that USP39 can regulate the stability of CHK2, PD-L1, and STAT1 [8,57,58]. However, whether USP39 exhibits deubiquitinating enzyme activity and the mechanism that USP39 uses to regulate FOXM1 stability have not been clarified. Therefore, USP21, USP22, and USP39 proteins were purified in vitro, and the deubiquitinating enzyme activity detection kit was used to detect the activities of USP39, USP21, and USP22 deubiquitinating enzymes. The results showed that USP39 had lower enzymatic activity, but this was not significant compared with the control group; USP21 and USP22 had significant deubiquitinating enzyme activity (Figure 2C). It is known that FOXM1 is degraded by APC/Cdh1 ubiquitination [44,45,55]. Hence, it was of interest to analyze whether USP39 affected the ubiquitination level of FOXM1 by APC/Cdh1. CoIP experiment results showed that APC/Cdh1 significantly enhanced FOXM1 ubiquitination, whereas high expression of USP39 reduced the ubiquitination level of FOXM1 by APC/Cdh1, and weakened the binding ability between Cdh1 and FOXM1 (Figure 2D and Supplementary Figure 5). In addition, with the increase in the expression of USP39, the ubiquitination level of FOXM1 was significantly reduced, and the binding of APC/Cdh1 to FOXM1 also gradually decreased, and the results of endogenous and exogenous proteins were consistent (Figure 2D and Supplementary Figure 5). These data confirmed that USP39 blocks the ubiquitination of FOXM1 by APC/Cdh1 by competitively binding to FOXM1, thereby maintaining the stability of FOXM1.
USP39 promotes the transcriptional activity of FOXM1
The transcription factor FOXM1 acts as a checkpoint in regulating the cell cycle [24], therefore we tested whether USP39 was able to activate FOXM1 transcriptional activity in co-transfection experiments. When FOXM1 and USP39 were co-transfected, FXM1-mediated stimulation of the FOXM1 binding promoter (a promoter containing the artificial 6xFOXM1 binding sequence) was observed (Figure 3A). On the other hand, siRNA against USP39 abolished FOXM1 transcription on FOXM1 binding promoter (Figure 3B).
Figure 3.

USP39 affects FOXM1 transcriptional activity and typical downstream target gene expression. A. The breast cancer MDA-MB-231 cell line was co-transfected with pCMV-USP39 and pCMV-FOXM1, and the luciferase kit was used to detect the transcriptional activity of the FOXM1 binding motif promoter (6XFOXM1pro-luc). Values represented the mean ± SD of three independent experiments. (**, P≤0.01). B. The luciferase reporter gene vector containing the FOXM1 binding motif promoter (6XFOXM1pro-luc) was used to co-transfect with pCMV-FOXM1 vector and SiUSP39. Cell samples were collected after 48 hours. The luciferase kit detects USP39 silence or the effect of high expression on the transcriptional activity of FOXM1. Values represented the mean ± SD of three independent experiments. (**, P≤0.01; ***, P≤0.001). C and D. SiUSP39 and pCMV-FOXM1 transfected cells, qPCR and Western Blotting were used to detect the effect of silence or high expression of USP39 on the expression of typical downstream target genes CDC25B and PLK1 of FOXM1. Values represented the mean ± SD of three independent experiments. (*, P≤0.05; ##, P≤0.01).
In order to test whether USP39 affects the expression of FOXM1 target genes in breast cancer cells, The following two genes are typical examples of the many targets regulated by the FOXM1 transcription factor: proliferation-related CDC25B and PLK1 [29,60]. USP39, CDC25B and PLK1 mRNA and protein levels were determined using qPCR and Western blotting. According to the results, the mRNA and protein levels of CDC25B and PLK1 increased in the pCMV-USP39 group, While CDC25B and PLK1 were down-regulated in the siUSP39 group (Figure 3C, 3D), confirming that USP39 is involved in regulating the transcription of FOXM1 target genes.
FOXM1 is a key mediator that regulates tumor cell proliferation
USP39 may modulate cell proliferation by deubiquitinating and stabilizing FOXM1. As an experiment to verify this hypothesis, cells with high expression of USP39, interference with USP39, interference with FOXM1, or combined interference of USP39 and FOXM1 were used to detect analyze the proliferation of MDA-MB-231 cells. As shown in Figure 4A-C, interference with USP39 caused a significant decrease in cell proliferation and a sharp decrease in the number of clones formed. As shown in Figure 4D-F, high expression of USP39 promoted cell proliferation, and the number of clone formation increased dramatically. In addition, when FOXM1 was depleted, USP39 knockdown had no further effect on cell proliferation and clone formation (Figure 4G, 4H). Taken together, USP39 regulates tumor cell proliferation and clone formation through FOXM1.
Figure 4.

The effect of high expression and silence of USP39 on tumor cell proliferation and monoclonal formation. A. SiUSP39#1 and SiUSP39#2 were transfected into MDA-MB-231 cells respectively. After 48 hours, Western Blotting detected the effect of interference with USP39. B. Cell counting experiment to detect cell proliferation. MDA-MB-231 cells were transfected with SiUSP39#1 and SiUSP39#2, and photographed with a microscope at 24 h, 48 h, 72 h, and 96 h respectively, and the cell count was counted. Values represented the mean ± SD of three independent experiments. (**, P≤0.01). C. Monoclonal formation test to detect tumorigenic ability, MDA-MB-231 cells were transfected with SiUSP39#1 and SiUSP39#2, 14 days later, 10 minutes after crystal violet staining, washed with PBS 3 times, stereomicroscope photographed, statistical interference with USP39. Effect on the cloning ability of MDA-MB-231 cells. The colonies were imaged and the graph represented the number ± SD of colonies/well. (**, P≤0.01). D. The pCMV-USP39 vector was transfected into MCF-7 cells. After 48 hours, Western Blotting was used to detect the effect of high expression of USP39. E. Cell counting experiment to detect cell proliferation. MCF-7 was transfected with pCMV-USP39 vector, and photographed under microscope at 24 h, 48 h, 72 h, and 96 h, respectively, to count the number of cells. Values represented the mean ± SD. (*, P≤0.05). F. Monoclonal formation test to detect tumorigenesis ability, MCF-7 was transfected with pCMV-USP39 vector, 14 days later, crystal violet stained for 10 min, washed with PBS 3 times, stereomicroscope photographed, and counted the number of MCF-7 cell clones formed. The colonies were imaged and the graph represented the number ± SD of colonies/well. (**, P≤0.01). G. Cell counting experiment to detect cell proliferation. MDA-MB-231 was transfected with SiUSP39 and SiFOXM1, and the cells were photographed with a microscope at 48 h to count the number of cells. Values represented the mean ± SD. (*, P≤0.05). H. Monoclonal formation test to detect tumorigenic ability, MDA-MB-231 was transfected with SiUSP39 and SiFOXM1, 14 days later, 10 minutes after crystal violet staining, washed 3 times with PBS, stereomicroscope photographed, and counted MCF-7 cell clone formation number. The colonies were imaged and the graph represented the number ± SD of colonies/well. (**, P≤0.01).
USP39 is positively correlated with FOXM1 expression in clinical breast cancer samples
Various cancer samples were analyzed for gene expression, it was found that the expression of FOXM1 was elevated in cancer cells, and its level has been proposed as an indicator of cancer diagnosis and prognosis [61]. Conditional knockout of FOXM1 inhibited the occurrence and progression of a variety of solid tumors such as breast cancer, liver cancer, lung cancer and colorectal cancer [62-65]. However, the mechanism underlying FOXM1 stabilization in breast cancer remains unclear. We were interested in studying the relationship between USP39 and FOXM1 in breast cancer. We examined the expression of USP39 and FOXM1 in various breast cancer cell lines. As shown in Figure 5A, USP39 and FOXM1 were highly expressed in breast cancer MDA-MB-231, MDA-MB-468, and SUM159 cells lines. The expression of USP39 and FOXM1 in the breast cancer cell line MCF7 was found to be lower than that in other breast cancer cell lines. At the same time, we examined the expression of USP39 and FOXM1 in breast normal cell line (MCF10A cell) and different breast cancer cell lines (Luminal A, Luminal B, Basal, Claudin-low and HER2). Compared with normal breast cell line (MCF10A cell), USP39 and FOXM1 proteins were highly expressed in breast cancer cell lines (Luminal A, Luminal B, Basal, Claudin-low and HER2), and the expression trend was positively correlated. In addition, USP39 and FOXM1 were highly expressed in breast Claudin-low cell type (MDA-MB-231) and lower in breast cancer Luminal A cell type (MCF7) (Supplementary Figure 6).
Figure 5.

The expressions of FOXM1 and USP39 in TCGA clinical samples were significantly up-regulated and positively correlated. A. FOXM1 and USP39 protein levels were detected for breast cancer cell lines, and FOXM1 and USP39 protein levels were positively correlated in different cell lines. B. TCGA breast cancer tumor clinical sample bank conducts statistical analysis of USP39 and FOXM1 expression levels, and conducts correlation analysis of tumor tissue expression. C-F. Analyze the relationship between FOXM1 and USP39 expression and prognosis in breast cancer patients through the Kaplan Meier Plotter database (http://kmplot.com).
We analyzed the expression levels of USP39 and FOXM1 in breast cancer tumor tissues. As shown in Supplementary Figure 2B, USP39 and FOXM1 were observed to be highly expressed in breast cancer tumor tissues. TCGA database analysis showed that the expression of USP39 and FOXM1 proteins in breast cancer was positively correlated (Figure 5B). In addition, we further explored the key roles of FOXM1 and USP39 in the survival of patients with breast cancer. The Kaplan-Meier plotter tool was used through the public data set (http://kmplot.com/analysis/index.php?p = service&cancer = breast). Kaplan-Meier curve and log-rank test analysis showed that the increase in FOXM1 and USP39 mRNA levels was significantly correlated with overall survival (OS), distant metastasis-free survival (DMFS), recurrence-free survival (RFS), and overall breast cancer post-progressive survival (PPS) (P<0.05) (Figure 5C-F). In other words, breast cancer patients with high levels of FOXM1 and USP39 mRNAs are expected to have low OS, DMFS, FP, and PPS.
USP39 is the target gene of FOXM1
To further examine the relationship between FOXM1 and USP39, we expressed or interfered with FOXM1 in MDA-MB-231 and MCF7 cells respectively. QPCR and western blotting detected the effect of high expression or interference with FOXM1 on the mRNA and protein levels of USP39. It was found that high expression of FOXM1 promoted the mRNA and protein levels of USP39, while silencing FOXM1 inhibited the mRNA and protein levels of USP39 (Figure 6A, 6B). In addition, we also found that FOXM1 high or low expression of FOXM1 affected the transcription level of USP39 (data not shown). To further confirm that FOXM1 regulates the USP39 promoter, luciferase reporter assay was carried out and the results showed that high expression of FOXM1 could activate the USP39 promoter (Figure 6C). These results together indicated that the FOXM1 transcription factor controls USP39 transcription.
Figure 6.

USP39 is the target gene of FOXM1. A. Gradient transfection of pCMV-FOXM1 expression plasmid or siFOXM1 interference sequence, 36 hours later, cells were collected, cDNA was obtained by reverse transcription kit, and the mRNA expression of FOXM1 and USP39 was detected by qPCR. Relative mRNA levels were normalized to GAPDH. Values represented the mean ± SD (**, P≤0.01, ***, P≤0.001). B. Gradient transfection of pCMV-FOXM1 expression plasmid and siFOXM1 interference sequence. After 48 hours, cells were collected to obtain cell lysate. Total protein was extracted. Western Blotting was used to detect the protein levels of FOXM1 and USP39. C. After the breast cancer MDA-MB-231 cell line was transfected with pCMV-FOXM1, the luciferase kit detects the activity of pUSP39pro-luc. Values represented the mean ± SD (*, P≤0.05, ***, P≤0.001).
Knockdown of USP39 inhibits xenograft tumor growth in vivo
In order to study the function of USP39 and FOXM1 on the growth of xenografts in vivo, we implanted MDA-MB-231 (shNC, shUSP39, shFOXM1 and shUSP39+FOXM1) cells subcutaneously in nude mice to establish a xenograft tumor model. As shown in Figure 7A-C, all mice developed tumors, while tumor volumes were significantly lower in the shUSP39 and shFOXM1 groups than in the shNC group. Compared with the shUSP39 and shFOXM1 groups, the tumors of the mice in the shUSP39+FOXM1 group were significantly increased, indicating that the high expression of FOXM1 alleviated the inhibitory effect of shUSP39 on tumors. There was no significant change in the body weight of mice in each group (Figure 7D). Immunohistochemical staining revealed significantly lower levels of FOXM1 target genes and proliferation markers CyclinD1 and CDC25B in xenografts after USP39 or FOXM1 knockout compared with control. There was no significant difference in the expression of CyclinD1 and CDC25B in tumors of mice in the shUSP39+LV-FOXM1 group (Figure 7E). These data suggest that USP39 is essential for tumor growth in vivo by regulating FOXM1.
Figure 7.
Knockdown of USP39 or FOXM1 inhibits xenograft tumor growth in vivo. A. The tumor length and width were measured every two days from the first day of transplanted tumor to the end of the animal experiment. Tumor volume (V) was calculated as: V = length × width2 ×1/2. Data points for tumor volume represent the mean in mm3 ± SD. **, P≤0.01, ***, P≤0.001. B. Body weight was measured every two days from the first day of transplanted tumor to the end of the animal experiment. Data points for mouse body weight represent the mean in mm3 ± SD. C. Representative mice on day 39 after injection of each group of breast cancer cell lines into the forelimbs of immunodeficient mice were imaged. D. Tumors from shNC (n=4), shUSP39 (n=4), shFOXM1 (n=4), shUSP39+Lv-FOXM1 (n=4) groups were harvested. Insets show representative collected tumors. E. Tumor tissue sections from shNC (n=4), shUSP39 (n=4), shFOXM1 (n=4), shUSP39+Lv-FOXM1 (n=4) groups were immunostained with anti-CyclinD1 or anti-CDC25B antibodies. Images were taken with an inverted microscope (200×, scale bar 50 µm).
Discussion
In conclusion, we demonstrated that USP39 regulates ubiquitination and stabilizes FOXM1 through competitive binding with APC/Cdh1, thereby regulating breast cancer cell proliferation. USP39 and FOXM1 were found to be upregulated and positively correlated in breast cancer, indicating that USP39 has a potential prognostic value in breast cancer. In addition, USP39 was found to stabilize FOXM1 in breast cancer cells, and FOXM1 was observed to regulate the expression of USP39 to form a closed-loop control of USP39/FOXM1 (Figure 8).
Figure 8.

USP39 facilitates breast cancer cell proliferation through stabilization of FOXM1. E3 ubiquitin ligase APC/Cdh1 regulates the degradation of FOXM1, USP39 and APC/Cdh1 compete with FOXM1 to activate the expression of downstream genes Cdc25b, Plk1 and Usp39 to maintain the stability of FOXM1 protein and up-regulate the transcriptional activity of FOXM1.
We identified USP39 as a stabilizing factor for the transcription factor FOXM1. USP39 stabilizes FOXM1, high expression of USP39 increases the abundance of FOXM1, and subsequently upregulates the FOXM1 transcription network. This activates breast cancer cell proliferation. FOXM1 and USP39 are highly expressed in clinical samples of breast cancer. However, they are negatively correlated with survival. High expression of FOXM1 increased the expression of USP39. This indicates that FOXM1 is a USP39 transcription factor. Therefore, we envisage that USP39 and FOXM1 are expressed in an interdependent manner and can be used as molecular markers for breast cancer diagnosis.
FoxM1 is a member of the Fox transcription factor family. This transcription factor family has more than 50 members, which are involved in the regulation of differentiation, cell cycle progression, cell proliferation, angiogenesis, metabolism, apoptosis, stem cell pluripotency [19,64,66], and other physiological processes. Studies have shown that the balance of phosphorylation-dephosphorylation and ubiquitination-deubiquitination is important for maintaining FOXM1 activity in cells [46,67,68]. In a cell cycle-dependent manner, FoxM1 transcriptional activity is controlled by temporally regulated phosphorylation and dephosphorylation events. Dephosphorylation of FoxM1 occurs simultaneously with the withdrawal of mitosis. Recent studies have shown that ubiquitination plays a significant role in regulating FOXM1 protein levels and functions [52,69,70]. Previous studies have confirmed that USP39 has no deubiquitinating enzyme activity, while recent studies have revealed that USP39 can regulate the protein stability of CHK2, PD-L1, SP1, ZEB1 and STAT1. Thus far, whether USP39 has deubiquitination enzyme activity, whether it has protein stability, and what is the mechanism of deubiquitination. It has been found that USP39 promotes tumor malignant phenotype and drug resistance through effective splicing of oncogenes [7,9,10,13,71]. USP39 was observed to maintain the stability of oncogenes through deubiquitination enzyme activity [8,57,58,72,73]. In addition, USP39 is also known to promote tumor progression by regulating oncogenes or tumor suppressor genes [3,6,12,74]. In this study, we discovered a new mechanism involving USP39 that leads to the inhibition of ubiquitination by competitive APC/Cdh1 binding to FOXM1 (Figures 2D and 8). Stabilization of FoxM1 leads to the stimulation of tumor cell proliferation [75], cell differentiation [51], cell cycle progression [52], and drug resistance [76], as well as can induce chromosomal instability [31], and expression of DNA repair genes [77].
FOXM1 has been shown to play an important role in the maintenance and development of monoclonal cells in several studies [78-80]. Silencing USP39 for 72 hours leads to statistically significant downregulation of FOXM1 mRNA levels (Figure 1A), USP39 may also have some functions in FOXM1 mRNA splicing [48]. Loss of FOXM1 affects cell proliferation and the process of monoclonal formation. Therefore, we also studied the role of the USP39-FOXM1 axis in cell proliferation and monoclonal formation. As reported previously, we found that depleting USP39 inhibits the proliferation of breast cancer cells by causing FOXM1 to degrade (Figures 1C, 4B). However, knocking out FOXM1 in USP39-depleted cells had no further effect on cell proliferation or monoclonal formation (Figure 4G, 4H). A high expression of USP39 activates FOXM1 transcription and promotes FOXM1 target genes like CDC-25B and PLK1 (Figure 3A, 3C and 3D). In addition, we found that FOXM1 can regulate the expression of USP39 (Figure 6). According to these findings, USP39 regulates breast cancer cell proliferation by deubiquitinating FOXM1.
In previous studies, FOXM1 and USP39 were found to be upregulated in cancer cells, leading to tumor cell proliferation and cycle progression [47,48]. However, the cancer-promoting mechanism of USP39 remains unclear. We believe that the upregulation of USP39 may be a that mechanism underlies for the upregulated of FOXM1 in breast cancer. In this study, we found that the levels of USP39 and FOXM1 were up-regulated and positively correlated in breast cancer cell lines and GEO database clinical cancer samples. In addition, USP39 may compete with APC/cdh1 to combine with FOXM1 to promote the proliferation of breast cancer cells. Previous studies have confirmed the correlation between breast cancer and APC/Cdh1. Therefore, Cdh1 may be an important part of tumor suppression and can be regarded as a new biomarker for breast cancer [81]. Han et al found that the interaction between the APC/C co-activator Cdh1 regulates breast tumorigenesis [82]. Myatt SS et al. analysis of FOXM1 wild-type and mutant results showed that facilitates translocation to the cytoplasm, SUMOylation suppresses FOXM1 activity, and maintain APC/Cdh1-mediated FOXM1 ubiquitination and degradation, Ubiquitination and degradation of FOXM1-SUMOylation due to the APC/Cdh1 complex [70]. It can be seen that APC/Cdh1 can be used as potential sites for breast cancer treatment intervention. Therefore, the FOXM1 gene plays a central role in the cancer-promoting response. APC/Cdh1 promotes decreased FOXM1 expression, FOXM1 ubiquitination increases the cellular response to anticancer drugs, indicating that FOXM1 ubiquitination negative regulation of the expression and activity. FOXM1 regulates gene expression mainly in response to DNA damage repair and cell cycle. The Ubiquitination of FOXM1 mediates its degradation and nucleocytoplasmic transport, which will eventually reduce its transcriptional activity. Therefore, Ubiquitination of FOXM1 may act as a molecular switch that ultimately controls cell proliferation and cell cycle.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 81870173 to C.L.), the Natural Science Foundation of Hubei Province (Grant Number 2020CFB206 to Z.Z.), the Hubei Provincial Department of Education (Grant Number Q20202805 to Z.Z.), Hubei Provincial Department of Education “Hundred Schools and Hundred Counties” (Grant Number BXLBX0806 to Z.Z.), and the Doctoral Foundation of HuBei University of Science & Technology Science (Grant Number BK202028 and 2021TNB02 to Z.Z.).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hadjivassiliou H, Rosenberg OS, Guthrie C. The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing. RNA. 2014;20:656–669. doi: 10.1261/rna.042838.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraile JM, Manchado E, Lujambio A, Quesada V, Campos-Iglesias D, Webb TR, Lowe SW, López-Otín C, Freije JM. USP39 deubiquitinase is essential for kras oncogene-driven cancer. J Biol Chem. 2017;292:4164–4175. doi: 10.1074/jbc.M116.762757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan J, Li X, Zhang G, Cheng W, Wang W, Lei Y, Ma Q, Song G. USP39 mediates p21-dependent proliferation and neoplasia of colon cancer cells by regulating the p53/p21/CDC2/cyclin B1 axis. Mol Carcinog. 2021;60:265–278. doi: 10.1002/mc.23290. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Yao X, Li M, Xi Y, Zhao L. USP39 regulates the cell cycle, survival, and growth of human leukemia cells. Biosci Rep. 2019;39:BSR20190040. doi: 10.1042/BSR20190040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008;7:2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J, Zhang G, Li X, Ma Q, Cheng W, Wang W, Zhang B, Hu T, Song G. Knocking down USP39 inhibits the growth and metastasis of non-small-cell lung cancer cells through activating the p53 pathway. Int J Mol Sci. 2020;21:8949. doi: 10.3390/ijms21238949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Ma W, Hu W, Di Q, Zhao X, Ma X, Chen X, Sun P, Wu H, Wu Z, Chen W. Ubiquitin-specific peptidase 39 promotes human glioma cells migration and invasion by facilitating ADAM9 mRNA maturation. Mol Oncol. 2022;16:388–404. doi: 10.1002/1878-0261.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Chen Y, Geng G, Li L, Yin P, Nowsheen S, Li Y, Wu C, Liu J, Zhao F, Kim W, Zhou Q, Huang J, Guo G, Zhang C, Tu X, Gao X, Lou Z, Luo K, Qiao H, Yuan J. USP39 regulates DNA damage response and chemo-radiation resistance by deubiquitinating and stabilizing CHK2. Cancer Lett. 2019;449:114–124. doi: 10.1016/j.canlet.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Wang Z, Li J, Qin J, Song J, Li Y, Zhao L, Zhang X, Guo H, Shao C, Kong B, Liu Z. Splicing factor USP39 promotes ovarian cancer malignancy through maintaining efficient splicing of oncogenic HMGA2. Cell Death Dis. 2021;12:294. doi: 10.1038/s41419-021-03581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding K, Ji J, Zhang X, Huang B, Chen A, Zhang D, Li X, Wang X, Wang J. RNA splicing factor USP39 promotes glioma progression by inducing TAZ mRNA maturation. Oncogene. 2019;38:6414–6428. doi: 10.1038/s41388-019-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan Z, Han K, Lin S, Hu H, Shen Z, Min D. Knockdown of ubiquitin-specific peptidase 39 inhibited the growth of osteosarcoma cells and induced apoptosis in vitro. Biol Res. 2017;50:15. doi: 10.1186/s40659-017-0121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai J, Liu T, Huang P, Yan W, Guo C, Xiong L, Liu A. USP39, a direct target of microRNA-133a, promotes progression of pancreatic cancer via the AKT pathway. Biochem Biophys Res Commun. 2017;486:184–190. doi: 10.1016/j.bbrc.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Chen T, Li X, Yan W, Lou Y, Liu Z, Chen H, Cui Z. USP39 promotes ovarian cancer malignant phenotypes and carboplatin chemoresistance. Int J Oncol. 2019;55:277–288. doi: 10.3892/ijo.2019.4818. [DOI] [PubMed] [Google Scholar]
- 14.Kuligina ES, Sokolenko AP, Bizin IV, Romanko AA, Zagorodnev KA, Anisimova MO, Krylova DD, Anisimova EI, Mantseva MA, Varma AK, Hasan SK, Ni VI, Koloskov AV, Suspitsin EN, Venina AR, Aleksakhina SN, Sokolova TN, Milanović AM, Schürmann P, Prokofyeva DS, Bermisheva MA, Khusnutdinova EK, Bogdanova N, Dörk T, Imyanitov EN. Exome sequencing study of Russian breast cancer patients suggests a predisposing role for USP39. Breast Cancer Res Treat. 2020;179:731–742. doi: 10.1007/s10549-019-05492-6. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Ji X, Liu X, Yao R, Chi J, Liu S, Wang Y, Cao W, Zhou Q. Lentivirus-mediated inhibition of USP39 suppresses the growth of breast cancer cells in vitro. Oncol Rep. 2013;30:2871–2877. doi: 10.3892/or.2013.2798. [DOI] [PubMed] [Google Scholar]
- 16.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 17.Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA, Costa RH. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo T, Ando M, Nagai N, Tomisato W, Srirat T, Liu B, Mise-Omata S, Ikeda M, Chikuma S, Nishimasu H, Nureki O, Ohmura M, Hayakawa N, Hishiki T, Uchibori R, Ozawa K, Yoshimura A. The NOTCH-FOXM1 axis plays a key role in mitochondrial biogenesis in the induction of human stem cell memory-like CAR-T cells. Cancer Res. 2020;80:471–483. doi: 10.1158/0008-5472.CAN-19-1196. [DOI] [PubMed] [Google Scholar]
- 21.Molinuevo R, Freije A, de Pedro I, Stoll SW, Elder JT, Gandarillas A. FOXM1 allows human keratinocytes to bypass the oncogene-induced differentiation checkpoint in response to gain of MYC or loss of p53. Oncogene. 2017;36:956–965. doi: 10.1038/onc.2016.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Wang Z, Na L, Dong D, Wang W, Zhao C. FZD5 contributes to TNBC proliferation, DNA damage repair and stemness. Cell Death Dis. 2020;11:1060. doi: 10.1038/s41419-020-03282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranski OA, Kalinichenko VV, Adami GR. Increased FOXM1 expression can stimulate DNA repair in normal hepatocytes in vivo but also increases nuclear foci associated with senescence. Cell Prolif. 2015;48:105–115. doi: 10.1111/cpr.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, Samejima K, Xie L, Paulson JR, Earnshaw WC, Cortez D, Meyer T, Cimprich KA. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018;361:806–810. doi: 10.1126/science.aap9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson MB, Sun H, Robichaux J, Pfeifer M, McDermott U, Travers J, Diao L, Xi Y, Tong P, Shen L, Hofstad M, Kawakami M, Le X, Liu X, Fan Y, Poteete A, Hu L, Negrao MV, Tran H, Dmitrovsky E, Peng D, Gibbons DL, Wang J, Heymach JV. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12:eaaz4589. doi: 10.1126/scitranslmed.aaz4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, Shin K, Seo YJ, Choi YS, Lee JI, Lee J, Joo KM, Nam DH. FoxM1 promotes stemness and radio-resistance of glioblastoma by regulating the master stem cell regulator Sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyuno T, Kohno T, Konno T, Yamaguchi H, Kyuno D, Imamura M, Kimura Y, Kojima T, Takemasa I. Glucose-dependent FOXM1 promotes epithelial-to-mesenchymal transition via cellular metabolism and targeting snail in human pancreatic cancer. Pancreas. 2020;49:273–280. doi: 10.1097/MPA.0000000000001485. [DOI] [PubMed] [Google Scholar]
- 28.Saavedra-García P, Nichols K, Mahmud Z, Fan LY, Lam EW. Unravelling the role of fatty acid metabolism in cancer through the FOXO3-FOXM1 axis. Mol Cell Endocrinol. 2018;462:82–92. doi: 10.1016/j.mce.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Muller GA, Quaas M, Fischer M, Han N, Stutchbury B, Sharrocks AD, Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227–236. doi: 10.1128/MCB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiler SME, Pinna F, Wolf T, Lutz T, Geldiyev A, Sticht C, Knaub M, Thomann S, Bissinger M, Wan S, Rossler S, Becker D, Gretz N, Lang H, Bergmann F, Ustiyan V, Kalin TV, Singer S, Lee JS, Marquardt JU, Schirmacher P, Kalinichenko VV, Breuhahn K. Induction of chromosome instability by activation of yes-associated protein and forkhead box M1 in liver cancer. Gastroenterology. 2017;152:2037–2051. e2022. doi: 10.1053/j.gastro.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Laoukili J, Kooistra MR, Brás A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong A, Huang S. FoxM1 and Wnt/beta-catenin signaling in glioma stem cells. Cancer Res. 2012;72:5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kambach DM, Sodi VL, Lelkes PI, Azizkhan-Clifford J, Reginato MJ. ErbB2, FoxM1 and 14-3-3zeta prime breast cancer cells for invasion in response to ionizing radiation. Oncogene. 2014;33:589–598. doi: 10.1038/onc.2012.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, He K, Yan S, Yang Y, Gao X, Zhang M, Xia Z, Huang Z, Huang S, Zhang N. Metadherin/astrocyte elevated gene-1 positively regulates the stability and function of forkhead box M1 during tumorigenesis. Neuro Oncol. 2017;19:352–363. doi: 10.1093/neuonc/now229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. doi: 10.1016/B978-0-12-407190-2.00016-2. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Chen H, Yu L, Shan L, Xie L, Hu J, Chen T, Tan Y. Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther. 2013;20:117–124. doi: 10.1038/cgt.2012.94. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, Jiang G, Lu M, Zhang Z, Yin J, Zeng S, Chen X, Deng M, Jia X, Gu Y, Chen D, Zheng G, He Z. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966–983. doi: 10.1038/s41418-019-0389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song X, Fiati Kenston SS, Zhao J, Yang D, Gu Y. Roles of FoxM1 in cell regulation and breast cancer targeting therapy. Med Oncol. 2017;34:41. doi: 10.1007/s12032-017-0888-3. [DOI] [PubMed] [Google Scholar]
- 44.Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008;7:2720–2726. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- 45.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008;28:5162–5171. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Li L, Xu S, Liu Z, Zhou C, Li Z, Liu Y, Wu W, Huang Y, Kuang M, Fan S, Li H, Li X, Song G, Wu WS, Chen J, Hou Y. A Cdh1-FoxM1-Apc axis controls muscle development and regeneration. Cell Death Dis. 2020;11:180. doi: 10.1038/s41419-020-2375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W, Ding Y. USP39 regulates the growth of SMMC-7721 cells via FoxM1. Exp Ther Med. 2017;13:1506–1513. doi: 10.3892/etm.2017.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W, Guan W, Zhou J, Wu Y, Qiu Y, Ding Y. USP39 promotes the growth of human hepatocellular carcinoma in vitro and in vivo. Oncol Rep. 2015;34:823–832. doi: 10.3892/or.2015.4065. [DOI] [PubMed] [Google Scholar]
- 49.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Bu H, Yu J, Chen Y, Pei C, Yu L, Huang X, Tan G, Tan Y. The cell-penetrating FOXM1 N-terminus (M1-138) demonstrates potent inhibitory effects on cancer cells by targeting FOXM1 and FOXM1-interacting factor SMAD3. Theranostics. 2019;9:2882–2896. doi: 10.7150/thno.32693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng Y, Yu C, Liu Y, Hu C, Ma R, Lu X, Ji P, Chen J, Mizukawa B, Huang Y, Licht JD, Qian Z. FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML. Nat Commun. 2020;11:928. doi: 10.1038/s41467-020-14590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arceci A, Bonacci T, Wang X, Stewart K, Damrauer JS, Hoadley KA, Emanuele MJ. FOXM1 deubiquitination by USP21 regulates cell cycle progression and paclitaxel sensitivity in basal-like breast cancer. Cell Rep. 2019;26:3076–3086. e6. doi: 10.1016/j.celrep.2019.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai J, Culley MK, Zhao Y, Zhao J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell. 2018;9:754–769. doi: 10.1007/s13238-017-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou T, Lin Z. The involvement of ubiquitination machinery in cell cycle regulation and cancer progression. Int J Mol Sci. 2021;22:5754. doi: 10.3390/ijms22115754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manzione MG, Rombouts J, Steklov M, Pasquali L, Sablina A, Gelens L, Qian J, Bollen M. Co-regulation of the antagonistic repoman: Aurora-B pair in proliferating cells. Mol Biol Cell. 2020;31:419–438. doi: 10.1091/mbc.E19-12-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gartel AL. A new target for proteasome inhibitors: FoxM1. Expert Opin Investig Drugs. 2010;19:235–242. doi: 10.1517/13543780903563364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Y, Guo J, Sun T, Fu Y, Zheng H, Dong C, Xiong S. USP39 Serves as a deubiquitinase to stabilize STAT1 and sustains type I IFN-induced antiviral immunity. J Immunol. 2020;205:3167–3178. doi: 10.4049/jimmunol.1901384. [DOI] [PubMed] [Google Scholar]
- 59.Zanin R, Pegoraro S, Ros G, Ciani Y, Piazza S, Bossi F, Bulla R, Zennaro C, Tonon F, Lazarevic D, Stupka E, Sgarra R, Manfioletti G. HMGA1 promotes breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. J Exp Clin Cancer Res. 2019;38:313. doi: 10.1186/s13046-019-1307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koo CY, Muir KW, Lam EW. FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Ros S, Wright AJ, D’Santos P, Hu DE, Hesketh RL, Lubling Y, Georgopoulou D, Lerda G, Couturier DL, Razavi P, Pelossof R, Batra AS, Mannion E, Lewis DY, Martin A, Baird RD, Oliveira M, de Boo LW, Linn SC, Scaltriti M, Rueda OM, Bruna A, Caldas C, Brindle KM. Metabolic imaging detects resistance to PI3Kalpha inhibition mediated by persistent FOXM1 expression in ER(+) breast cancer. Cancer Cell. 2020;38:516–533. e9. doi: 10.1016/j.ccell.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 66.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, Huang X, Tan Y. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38:8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen YJ, Dominguez-Brauer C, Wang Z, Asara JM, Costa RH, Tyner AL, Lau LF, Raychaudhuri P. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J Biol Chem. 2009;284:30695–30707. doi: 10.1074/jbc.M109.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y, Li Y, Xue J, Gong A, Yu G, Zhou A, Lin K, Zhang S, Zhang N, Gottardi CJ, Huang S. Wnt-induced deubiquitination FoxM1 ensures nucleus beta-catenin transactivation. EMBO J. 2016;35:668–684. doi: 10.15252/embj.201592810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, Man EP, Khongkow P, Tsang JW, Khoo US, Medema RH, Freire R, Lam EW. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2016;35:1433–1444. doi: 10.1038/onc.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Myatt SS, Kongsema M, Man CW, Kelly DJ, Gomes AR, Khongkow P, Karunarathna U, Zona S, Langer JK, Dunsby CW, Coombes RC, French PM, Brosens JJ, Lam EW. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene. 2014;33:4316–4329. doi: 10.1038/onc.2013.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Y, Geng H, Liu G, Ji Q, Cheng X, Li X, Liu W, Thorne RF, Zhang R, Liu X. The deubiquitinase USP39 promotes ESCC tumorigenesis through pre-mRNA splicing of the mTORC2 component rictor. Front Oncol. 2021;11:667495. doi: 10.3389/fonc.2021.667495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, Yuan J, Song C, Lei Y, Xu J, Zhang G, Wang W, Song G. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021;28:2315–2332. doi: 10.1038/s41418-021-00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong X, Liu Z, Zhang E, Zhang P, Wang Y, Hang J, Li Q. USP39 promotes tumorigenesis by stabilizing and deubiquitinating SP1 protein in hepatocellular carcinoma. Cell Signal. 2021;85:110068. doi: 10.1016/j.cellsig.2021.110068. [DOI] [PubMed] [Google Scholar]
- 74.Yuan X, Sun X, Shi X, Wang H, Wu G, Jiang C, Yu D, Zhang W, Xue B, Ding Y. USP39 promotes colorectal cancer growth and metastasis through the Wnt/beta-catenin pathway. Oncol Rep. 2017;37:2398–2404. doi: 10.3892/or.2017.5454. [DOI] [PubMed] [Google Scholar]
- 75.Liu J, Guo S, Li Q, Yang L, Xia Z, Zhang L, Huang Z, Zhang N. Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J Neurooncol. 2013;111:245–255. doi: 10.1007/s11060-012-1018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang N, Wang C, Wang J, Wang Z, Huang D, Yan M, Kamran M, Liu Q, Xu B. Aurora kinase A stabilizes FOXM1 to enhance paclitaxel resistance in triple-negative breast cancer. J Cell Mol Med. 2019;23:6442–6453. doi: 10.1111/jcmm.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo W, Gao F, Li S, Liu L. FoxM1 promotes cell proliferation, invasion, and stem cell properties in nasopharyngeal carcinoma. Front Oncol. 2018;8:483. doi: 10.3389/fonc.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujita T, Liu W, Doihara H, Date H, Wan Y. Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin Cancer Res. 2008;14:1966–1975. doi: 10.1158/1078-0432.CCR-07-1585. [DOI] [PubMed] [Google Scholar]
- 82.Han T, Jiang S, Zheng H, Yin Q, Xie M, Little MR, Yin X, Chen M, Song SJ, Beg AA, Pandolfi PP, Wan L. Interplay between c-Src and the APC/C co-activator Cdh1 regulates mammary tumorigenesis. Nat Commun. 2019;10:3716. doi: 10.1038/s41467-019-11618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



