Abstract
Lung adenocarcinoma (LUAD) is a very heterogeneous cancer with a bad prognosis. Pyroptosis and ferroptosis are two newly discovered forms of regulated cell death, which can trigger inflammation-related immunosuppression in tumor microenvironments, thereby promoting tumor growth. So far, there has been no thorough systematic investigation of the predictive values of ferroptosis and pyroptosis-related genes in LUAD. Therefore, in this study, we conducted a combined analyses in the gene expression of ferroptosis and pyroptosis and identified four distinct subgroups: immobility, ferroptosis, pyroptosis, and mixed. The gene sets most closely associated to both ferroptosis and pyroptosis were utilized to build a risk prediction model based on their variations in survival and biological activities. More importantly, our conclusions from bioinformatics analyses were validated by external experiments in patients with LUAD. In conclusion, the establishment of LUAD subgroups based on the ferroptosis- and pyroptosis-related gene expression profile provided new insights into understanding the roles of programmed cell death in oncogenesis and might contribute to the development of individualized therapy.
Keywords: Lung adenocarcinoma, ferroptosis, pyroptosis, gene signatures, clinical outcomes
Introduction
Because of its poor prognosis, lung cancer is one of the leading causes of cancer-related mortality globally [1]. Non-small cell lung cancer (NSCLC) accounts for the vast majority of LUAD diagnoses, while the most common NSCLC histologic subtype accounts for around 40% of all lung cancer cases [2]. The two major clinical challenges with LUAD treatment are the late diagnosis and drug resistance [3]. Although multiple clinical trials have demonstrated encouraging effects in certain LUAD patients with immunotherapies based on immune checkpoint inhibitors (ICIs) [4], other LUAD patients who undergo ICI therapy fail to exhibit a significant improvement in OS, probably due to cancer cell resistance to apoptosis. Hence, searching for novel forms of cell death has emerged as a viable treatment strategy.
Apoptosis, necrosis, ferroptosis, parthanatos, oxeiptosis, oncosis, pyroptosis, and autophagy are a few of the several types of cell death that are involved in the pathophysiology and development of cancer. Of note, ferroptosis and pyroptosis have become the center of interest in recent years. Numerous studies have shown that the essential biological processes connected to the development of LUAD are ferroptosis and pyroptosis [7,8]. In contrast to apoptosis, necrosis, and autophagy, which are other types of programmed cell death [9], ferroptosis is iron-dependent, which is caused by an imbalance in cellular redox equilibrium and ultimately the excessive lipid peroxidation. Many studies have explored the functional mechanism and the potential of targeting ferroptosis for cancer treatment [10]. For example, Alvarez et al. have reported that decreasing the iron-sulfur cluster biosynthesis enzyme NFS1 in LUAD, in conjunction with limiting cysteine transport, induces ferroptosis in vitro and reduced xenograft tumor growth in vivo [11]. Zhang et al. have discovered that the translational control of SLC7A11 by RBMS1 modifies lung cancer ferroptosis [12]. Wang et al. found that the lung cancer stem cell factor SOX2 mediates ferroptosis resistance by overexpressing SLC7A11 [13]. Other newly identified regulators, such as erianin, may impede cell migration and cause Ca2+/CaM-dependent ferroptosis, potentially playing an anti-tumor role in lung cancer [15].
However, how ferroptosis affects the tumor immune microenvironment (TIME) remains elusive. It is reasonable to speculate the function of ferroptosis in TIME as TIME is connected to iron metabolism and homeostasis in vivo, whereas ferroptosis is necessary for tumor immunity [16]. Recent research has shown that CD8+ T lymphocytes activated by immunotherapy raise ferroptosis-specific lipid peroxidation in tumor cells, increasing the effectiveness of cancer immunotherapy [17]. It’s important to note that the expression of SLC7A11 and SLC3A2 is downregulated by IFN secreted by CD8+ T cells, which promotes ferroptosis and lipid peroxidation in addition to blocking tumor cells from absorbing cystine [18]. A connection between ferroptosis and the prognosis of non-small cell lung cancer was also found by Lai et al. [19]. LUAD and their activities are impacted by ferroptosis-related genes, although little is known about the processes behind these effects.
The synthesis of several proinflammatory mediators during pyroptosis, a lytic type of controlled cell death, is one of its defining features [20]. One of two signaling pathways-the GSDMD-dependent, which is regulated by caspase 1/4/5/11, or the GSDME-dependent, which is regulated by caspase 3-can initiate pyroptosis [21-24]. Gasdermin C and Gasdermin D are two important pyroptosis effectors that are overexpressed in several malignancies and are linked to tumor growth and the poor prognosis of patients [25,26]. Furthermore, Teng et al. discovered that the ROS/NF-κB/NLRP3/GSDMD signal axis was activated by PPVI, the caspase 1-mediated pyroptosis, which mediated the PPVI-induced suppression of NSCLC [27]. Furthermore, by triggering caspase 3/GSDME signaling, Zhang et al. discovered that the chemotherapy medicines paclitaxel and cisplatin differently promote pyroptosis in A549 lung cancer cells [28]. Both in vivo and in vitro, cucurbitacin B reduces NSCLC via causing TLR4/NLRP3/GSDMD-dependent pyroptosis [29]. Significantly, a unique gene signature for pyroptosis was discovered in LUAD to be used in prognostic prediction. The development of LUAD depends on the lncRNA KCNQ1OT1/miR-335-5p/NLRP1/NLRP7-mediated regulatory axis of pyroptosis [30]. Furthermore, Gasdermin E may promote tumor cell phagocytosis by macrophages, enhance the quantity and potency of CD8+ T lymphocytes, and boost the activity of natural killer (NK) cells, leading to tumor cell pyroptosis and generating a positive feedback loop [31].
The interaction among pyroptosis, ferroptosis, and TIME to influence their perspective function is intricate. CD8+ T lymphocytes could limit tumor growth by inducing ferroptosis and pyroptosis via different mechanisms. For instance, CD8+ T cells may release IFN-γ to inhibit SLC7A11 expression, leading to a buildup of lipid ROS and tumor cell ferroptosis [32]. The activation of ferroptosis, in turn, can further improve antitumor immunity. Additionally, in order to cause pyroptosis and activate the macrophage-derived cytokine IL-1, necessary for antitumor immunity, CD8+ T cells may produce GzmA (a GSDMB-cleaving enzyme) and GzmB (a GSDME-cleaving enzyme) [33]. At present, the impact of pyroptosis and ferroptosis on tumor biology is unknown. Therefore, it is critical to better understand the complex functions and the signaling pathways of pyroptosis and ferroptosis in order to develop pyroptosis and ferroptosis-related treatment strategies for LUAD.
In this study, we identified four important cell death subgroups of LUAD based on the consensus clustering of gene expression patterns associated with pyroptosis and ferroptosis. Each of these subgroups was correlated with unique immunological, mutational, and survival markers. The pyroptosis-ferroptosis score was also computed, and it has a greater capacity to predict the response to chemotherapy and immunotherapy in LUAD patients.
Materials and methods
LUAD datasets and data processing
The Cancer Genome Atlas (TCGA) RNA-sequencing data and comprehensive clinical information for 492 LUAD patients were retrieved via the GDC API. The FPKM (Fragments Per Kilobase per Million) expression data were converted to TPM (Transcripts Per Kilobase per Million) format and utilized as the training cohort. Complete patient clinical information from the GSE30219 dataset based on the Affymetrix HG-U133 Plus 2.0 Array platform, from the GSE72094 dataset based on the Rosetta/Merck Human RSTA Custom Affymetrix 2.0 platform, and from the GSE42127 dataset based on the Illumina HumanWG-6 v3.0 expression beadchip platform were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/) database and used as the test cohorts. In addition, two immunotherapy cohorts, GSE126044 and GSE135222, consisting of patients who underwent PD1 treatment were used for the prediction of immunotherapeutic responses. The IMvigor210 dataset was publiclyavailable, and the fully described software and packages downloaded at http://research-pub.gene.com/IMvigor210CoreBiologies were distributed under the Creative Commons 3.0 license. A total of 298 melanoma patients who received PD1 therapy and had full clinical data related to their diagnoses were included.
Somatic cell data collection
The TCGA-LUAD cohort’s somatic mutation data (in MAF format) were obtained from the UCSC Xena Data Center (https://xena.ucsc.edu). The maftools package in R was used to evaluate and illustrate the mutation type and frequency of the genes. In addition, copy number variants (CNV) in TCGA-LUAD patients were collected and analyzed; a score > 0.2 was defined as amplification, while the score < -0.2 was defined as deletion. The CNV summary map was visualized using the Circos package in R.
Pyroptosis and ferroptosis subgroups
The pyroptosis-related genes (n = 39) [34-36] and the ferroptosis-associated genes (n = 113) [37-39] were collated from published literatures as shown in Table S1. The ConsensusClusterPlus package was used to cluster the pyroptosis- and ferroptosis-related genes (set parameters were as follows: reps = 1000, pItem = 0.8, and pfeature = 1). Ward. D 2 and Pearson correlation were used as the clustering algorithm and distance measure, respectively, with k = 5. The median expression levels of co-expressed pyroptosis and ferroptosis genes were used to divide samples into the subgroups shown below: immobility, pyroptosis, ferroptosis, and mixed.
Development and verification of risk model based on subgroups
The risk model was built using samples from the TCGA database. The WGCNA tool in R was used to create a scale-free co-expression network, and the transcriptome was utilized to identify the most significant modules for the cell death subgroups. To increase matrix similarity, a soft threshold parameter of 12 (scale-free R2 = 0.91) was used, and a scale-free co-expression network was built. Hierarchical clustering of the weighted coefficient matrix was used to identify the modules further. The genes in the pink module most associated with subgroups were used for further analyses. Univariate analysis and the log-rank test were used to find the pertinent prognostic modular genes. The Cox proportional hazard model (iteration = 100) with LASSO penalty function was utilized for these possible prognostic genes, and the optimal gene model was chosen using R’s glmnet package. The best gene model was used to determine risk ratings using the following formula:
Risk Score = ∑ i Coefficient (mRNAi ) × Expression (mRNAi )
Moreover, the consistency C-index, proposed by Harrell et al. was calculated using the survcomp package in R [37-39], and the predictive powers of all the five datasets for scoring were validated; the higher the C-index, the more accurate the model’s prediction capacity.
Predicting chemotherapeutic and immunotherapeutic responses
The pRRophetic package in R and the Genomics of Drug Sensitivity in Cancer (GDSC) database were used to choose the five first-line therapeutic medicines utilized in the treatment of LUAD: cisplatin, gemcitabine, paclitaxel, docetaxel, and vinorelbine. The half-maximal inhibitory concentration (IC50) for each sample and the sensitivity of the high- and low-risk groups to chemotherapy were compared using ridge regression (RR). The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm and subclass mapping were used to forecast how patients in different risk categories will respond to anti-CTLA4 and anti-PD1 treatment. In addition, the potentil treatment targets included genes that substantially varied between the high- and low-risk groups. The CMap database (https://clue.io/) was used to look for prospective medications that may target these genes. As a result, in addition to choosing the anticipated drugs based on gene expression characteristics, the mode of action (MOA) of these pharmaceuticals targeting the relevant biochemical pathways was also determined. The compounds with concentration fraction < -96 were considered potential therapeutic drugs.
qRT-PCR
To verify the prediction accuracy, qRT-PCR was used to examine the expression levels of the critical genes such as ANLN, E2F7, ECT2, HMMR, and TK1 in thirty-six LUAD clinical samples and their paired adjacent normal lung tissues obtained from the Shanghai Pulmonary Hospital. The total RNA was extracted using the Trizol reagent, and its quality was confirmed by RNA gel electrophoresis. Then, 2 µg of the total RNA was transcribed into cDNA using a quantitative reverse transcription kit (Wuhan Seville biology company) followed by qPCR analysis. The preparation of qPCR reaction was according to the manufacturer’s instructions. Briefly, the fluorescence quantitative PCR kit SYBR Green qPCR Master Mix (Wuhan Seville biology company) was used, and the PCR reaction conditions were as follows: pre-denaturation at 95°C for 10 min; denaturation at 95°C for 10 seconds; annealing at 60°C for the 40 seconds, and final extension at 72°C for 40 seconds. Forty cycles were set for the qPCR reaction. The CT values were recorded, and the relative expressions were calculated using the quantitative 2-ΔΔCT method with GADPH as internal reference. The sequences of primers used in this study were listed in Table S2. This study was approved by Shanghai Pulmonary Hospital Ethics Committee (ethical lot number: K21-111Y).
Immunofluorescence staining
Formalin fixed, paraffin-embedded tissue blocks of LUAD and the corresponding paracancerous tissues were cut into 5-μm-thick sections. These sections were dewaxed, rehydrated, and incubated with primary antibodies against ANLN, EEF7, ECT2, HMMR, and TK1, followed by incubation with FITC-conjugated secondary antibodies. And DAPI (sigma) was used for nuclear counterstaining. The sections were examined under fluorescence microscope (Zeiss, Oberkochen, Germany) and photographed. The intensity of fluorescence staining of the sections was determined by Image J.
Bioinformatics and statistical analyses
The CIBERSORT package in R and LM22 characteristics were used to calculate the infiltration levels of 22 immune cell types in each patient in the TCGA cohort. The immunological and matrix scores for each sample were calculated using the ESTIMATE program. The GSVA program in R was used to assess the activity of immune-related pathways in each sample using the ssGSEA method. Gene markers for immune-related pathways are provided in Table S3. Next, the differentially expressed genes were obtained using the Limma package, and those with P.adjust < 0.05 and |Log2FC| > 1 were considered as significantly differentially expressed. The clusterprofiler software in R and the Metascape webtool (https://metascape.org/gp/index.html) were used to annotate gene functions. R program was used for all statistical analysis and mapping (version 4.04). For the above-mentioned two groups, the Kruskal-Wallis test was performed, and the Wilcoxon test was utilized for pairwise comparison. The Chi-Square test was used to compare proportional differences. To assess the survival curves for subgroups in each dataset, Kaplan-Meier curves were produced. To determine statistically significant differences, the logarithmic rank test was applied. The survivalROC package in R was used to conduct the time-dependent receiver operating characteristic (tROC) analysis, and the area under the curve (AUC) was determined to assess the predictive potential of variables. The rms package in R was used to plot nomograms and calibration curves. The Student’s t-test was performed to examine gene expression changes between tumor and normal tissues. Unless specified, two-tailed P < 0.05 was considered statistically significant for all tests.
Results
Identification of four distinct subgroups by combined analysis of ferroptosis and pyroptosis gene expression in LUAD
In total, 492 patients from the TCGA-LUAD cohort were included in our study. The LUAD tumors were classified based on the relative expression levels of ferroptosis- and pyroptosis-related genes. To select the co-regulated and biologically relevant genes in LUAD, we performed consensus clustering and identified two sets of stably co-expressed genes involved in the pyroptosis (N = 20) and ferroptosis (N = 66) pathways for cell death subgrouping as shown in Figures 1A, S1A-C. For each sample, the median values of pyroptosis and ferroptosis gene co-expression were computed and used to stratify four distinct subgroups: immobility (Pyroptosis ≤ 0, Ferroptosis ≤ 0), pyroptosis (Pyroptosis > 0, Ferroptosis ≤ 0), ferroptosis (Pyroptosis ≤ 0, Ferroptosis > 0), and mixed (Pyroptosis > 0, Ferroptosis > 0) (Figure 1B). Figure 1C showed the expression levels of pyroptosis and ferroptosis genes in each subgroup. Pyroptosis subgroup was the largest subgroup (148/492; 30.08%), followed by the ferroptosis (132/492; 26.83%), immobility (122/492; 24.80%), and mixed subgroup (90/492; 18.29%). As shown in Figure 1D, based on the survival analysis, the pyroptosis subgroup was associated with the best overall survival, while the ferroptosis and mixed subgroups exhibited the worst overall survival. The intersection of differentially expressed genes among these four subgroups was shown in Figure S1D and Table S4.
Figure 1.

Tumor stratification for LUAD based on relative expression levels of pyroptosis- and ferroptosis-related genes. A. The heat map depicted a consistent clustering solution generated for pyroptosis- and ferroptosis-related genes (k = 2) in 492 patients with LUAD. B. The scatter map showed the median expression levels of pyroptosis- (x-axis) and ferroptosis-related (y-axis) gene co-expression in each LUAD sample. The metabolic subgroups were allocated according to the relative expression levels of pyroptosis- and ferroptosis-associated genes. C. The heat map depicted the co-expression levels of pyroptosis- and ferroptosis-associated genes in each subgroup. D. Kaplan-Meier survival analysis for four subgroups of LUAD patients.
Association between these four subgroups and the immunophenotype
As shown in Figure 2A, the ferroptosis subgroup showed the highest tumor homogeneity and the lowest immune score as indicated by the ESTIMATE scores, while the pyroptosis subgroup was the opposite, exhibiting the lowest homogeneity and the highest immune score. We also examined the differential expression of immune checkpoint related genes, including CD274, HAVCR2, CTLA4, LAG3, IDO1, and PDCD1, and the immune response related genes, including CD8A, CXCL9, CXCL10, GZMA, GZMB, PRF1, IFNG, TNF, and TBX2 among the subgroups [41,42]. As shown in Figure 2B, their expressions were higher in the pyroptosis and mixed subgroups, while their expressions were the lowest in the ferroptosis subgroup. Next, the abundance of 22 immune cell types was quantified using a cyclic classification algorithm. Figure 2C showed the differences in levels of immune cell infiltration among the subgroups. CD8+ T cells and memory B cells showed higher infiltration in the pyroptosis subgroup. A greater infiltration of naive B cells and activated dendritic cells in the ferroptosis subgroup was observed.
Figure 2.
Systematic immune and functional enrichment analyses for the immune subgroups. (A-D) Box diagram of analyses for ESTIMATE score (A), immune checkpoint (B), immune infiltration (C), immune pathways (D) among the subgroups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (E) GO functional enrichment analysis, including significantly enriched biological processes, cellular components, molecular functions, and KEGG pathways. (F) Interaction network between the enrichment pathways.
Furthermore, we analyzed the differences in immunoreactive pathways among the subgroups (Figure 2D). Immunoreactive pathways were more active in the pyroptosis and mixed subgroups, while was the lowest in the ferroptosis subgroup, suggesting the significant differences in immunoactivities among the subgroups, which might attribute to the survival of patients. Moreover, through the enrichment analysis of the differentially expressed genes among subgroups, we found a significant enrichment in immune-related pathways, including the immunoregulatory interactions between a lymphoid and a non-lymphoid cell, immune system processes, and positive regulation of immune responses (Figure 2E). The interaction network among the enrichment pathways was shown in Figure 2F, among them, pathways related to lymphocyte activation, immune effector process, regulation of cytokine production, and leukocyte migration were highly enriched. Based on these data, we postulated that pyroptosis, a highly immunogenic type of cell death, might trigger local inflammation and attracted inflammatory cells into the area, which might serve as a potential therapeutic target to reverse TIME’s immunosuppression and trigger a systemic immune response to LUAD. In contrast, the ferroptosis subgroup showed the highest tumor homogeneity and the lowest immune score, which might attribute to its poor prognosis.
Association of the cell death subgroups with genomic changes
It has been well known that genetic alterations influence tumor immunity and immune invasion. Therefore, we analyzed the differences in copy number variations and somatic variations among the subgroups and found a higher tumor mutation burden (TMB) in ferroptosis and mixed subgroups, while a lower TMB in the pyroptosis and immobility subgroups (Figure 3A). In addition, the ferroptosis subgroup had the highest numbers of chromosomal amplifications and deletion events, while the pyroptosis subgroup had the least incidence (Figure 3B, 3C). The mutation landscape of the top 25 most commonly mutated genes in distinct subgroups was shown in Figure 3D. Furthermore, we evaluated the correlation between the expression of TP53 and TTN, the top two highest mutated genes, and the median expression of pyroptosis- and ferroptosis-related genes. As shown in Figure 3E-H, a positive correlation between TP53 and ferroptosis genes, a negative correlation between TTN and ferroptosis genes, and no significant correlations between either TP53 or TTN with pyroptosis genes were observed. These results suggested that the ferroptosis subgroup showed greater genomic alterations than the pyroptosis subgroup. Taken together, the tight association between ferroptosis and mutations may result in high tumor malignancy, thereby leading to worse prognosis of patients in ferroptosis subgroup.
Figure 3.
Tumor immune microenvironment patterns and immunogenic characteristics of LUAD associated with the cell death subgroups. A-C. Violin plots for death subgroups in individual LUAD sample were sectioned according to TMB, amplitudes, and deletions. D. Mutation landscape of the top 25 genes with the highest mutation frequency in different subgroups. E, F. Scatterplot plots delineated the association of the median TP53 gene expression with ferroptosis (left panel) and with pyroptosis (right panel). G, H. Scatterplot plots delineated the associations of the median TTN gene expression with ferroptosis (left panel) and with pyroptosis (right panel).
Construction of risk model based on the subgroups
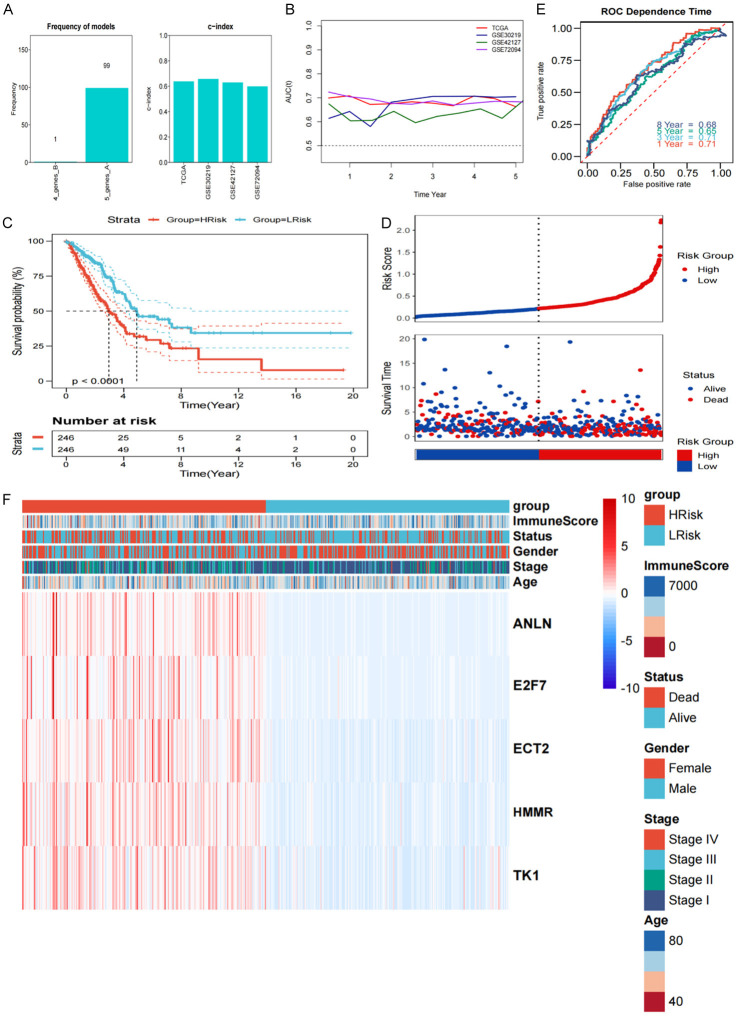
Given the significant differences in survival and biological functions among the subgroups, we further analyzed the differentially expressed genes. WGCNA was performed in the training cohort using the transcriptomic data for the differentially expressed genes and the subgroups. Using β = 12 as the best soft threshold, we ensured non-scale co-expression in the network graph (Figure S2A), and a total of eight non-gray module graphs were obtained (Figure S2B, S2C). Among these modules, the pink module (R = 0.43, P = 3e-21), with the highest correlation, was considered the most relevant for the cell death subgroups (Figure S2D, S2E). Furthermore, for the 189 genes in the pink module, univariate Cox analysis and the log-rank test were done, and 142 of them were shown to have substantial prognostic predicting capacity (P < 0.05). The detailed data was presented in Table S5. Furthermore, we performed a total of 100 iterations and included two gene sets for further analysis (Table S6). We identified a robust five gene signature: ANLN, E2F7, ECT2, HM, and TK1, as the frequency of the model was significantly higher (Figure 4A). As a result, the five-gene signature was best suited for the risk model’s creation. Accordingly, the risk score graphs (Figure S2F, S2G) were plotted, and the specific coefficient values were shown in Table S7. To evaluate the effectiveness of our risk score, we first calculated the C-index for predicting the OS, and the values for TCGA, GSE30219, GSE42127, and GSE72049 were 0.6383, 0.6580, 0.6302, and 0.5989, respectively. The tROC analysis showed that the AUC of the risk score plots for each point in the four cohorts for five years was greater than 0.6 (Figure 4B). Thus, the risk model had a high prediction accuracy for survival. According to KM analysis, the prognosis of patients in the high-risk category was poorer (P < 0.0001) (Figure 4C), and a poor survival was observed in the high-risk group (Figure 4D). ROC analysis showed that 1-, and 8-year AUC was 0.71 and 0.68, respectively, suggesting the better prognostic efficacy of risk score (Figure 4E). Consistently, similar results were observed in the three test cohorts (Figure S3). Figure 4F showed the expression of genes in different subgroups according to the model.
Figure 4.
Mining subgroups and construction of risk model by WGCNA. A. Model frequency was shown in the bar graph. B. The AUC curve of different datasets depicted changes annually. C. KM survival curves according to the risk scores obtained from TCGA data. D. The scatter plot for the survival and the risk score from our analysis. E. AUC for the risk model at 1, 3, 5, and 8 years. F. Gene expression values for different subgroups in the model.
Validation of the independent prognostic value of the risk model
According to the Chi-Square test, all variables except sex revealed significant differences between the high and low subgroups among the differences in each clinical characteristic across subgroups, as shown in Figure 5A. Risk score had higher prognostic accuracy than sex, age, and stage (Figure 5B). Following the incorporation of new clinical data, univariate and multivariate Cox regression analysis for the four cohorts revealed that the risk score was an independent prognostic predictor (Figure 5C, 5D). In addition, the subgroup analysis showed that the risk score of different clinical subgroups exhibited satisfactory predictive efficacy in four cohorts (Figure S4). Therefore, we constructed a nomogram to better visualize the predictive power of the model (Figure 5E), and the calibration curve, the indication of the good predictive accuracy of the nomogram, was presented in Figure 5F. Importantly, ROC analysis showed that the nomogram model had the best prediction accuracy as compared to other variables (Figure 5G).
Figure 5.
Effectiveness of the risk model. A. Pie chart showing the differences for each clinical variable among subgroups. B. The time-dependent AUC showed the prediction efficiency of risk score and clinical features (Risk score, Age, Gender, and Stage) for OS in training cohort. C, D. Univariate and multivariate forest plots constructed based on the risk model scores and other clinical variables from TCGA data. E. Nomograms based on risk model scores and other clinical variables. F. Rectification curves for nomograms. G. Prediction accuracy of AUC curves of the nomograms.
Differences in biological functions among the subgroups
We also evaluated the association between our risk score and numerous biological processes. As shown in Figure 6A, a high negative correlation was obsesrved between the mean risk score and the immune score (Pearson Correlation = -0.166, P < 0.01). The high-risk group exhibited higher tumor homogeneity and lower immune activity (Figure 6B), and most of the immune checkpoint and immunoreactivity characteristics were highly enriched in the high-risk group, in addition to TBX2 (Figure 6C). In supporting this, LUAD patients in the two groups showed differences in immune microenvironment (Figure 6D). The high-risk group showed higher M0/M1 macrophage activated-CD4 cells infiltration, while the low-risk group had higher infiltration of B cells, dendritic cells, mast cells, plasma cells, CD4 resting cells, and Tregs, suggesting active antigen presentation in the low-risk group. Although M0 and M1 macrophages are generally considered to produce antineoplastic proinflammatory cytokines, including ROS and nitric oxide (NO), to inhibit tumor growth and progression [41,42], M0/M1 macrophage infiltrations may also lead to a poor prognosis. For instance, a recent study showed that M1 macrophage recruitment was strongly associated with poor OS in medulloblastoma [44]. In our study, we further analyzed the balance between M1 and M2 macrophages in the high- and low-risk group, however, the difference was not statistically significant. A possible explanation for higher M0 and M1 macrophage infiltrations in the high-risk group is that pyroptosis and ferroptosis-related genes can regulate the tumor immune microenvironment. To test this, we analyzed the differences in immune response pathways between high- and low-risk groups and found that the MHC class I pathway was significantly upregulated in the high-risk group, while the HLA pathway and type II IFN response pathway were significantly enriched in the low-risk groups (Figure 6E). Furthermore, GSEA analysis revealed that cell cycle, DNA repair, and P53 signaling pathways were significantly enriched in the high-risk group, indicating active cell division and tumor malignancy, whereas asthma and T cell receptor pathways were enriched in the low-risk group, indicating the immune system’s active state (Figure 6F, 6G). Further enrichment analysis revealed that the up-regulated genes in high-risk groups were strongly related with cell division processes such as organelle fission, nuclear division, and chromosomal segregation. The down-regulated genes were associated with immune activity and protein production processes, including humoral immune response, antimicrobial humoral response, and protein processing (Figure 6H, 6I).
Figure 6.
Immunological and functional enrichment analyses for different risk groups. (A) Scatter plot of correlation between immune score and risk score; (B-E) Differences between high and low-risk groups in ESTIMATE score (B), immune checkpoint (C), immune infiltration (D), immune pathways (E). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; (F, G) GSEA analysis for up- and down-regulated genes in high- and low-risk groups; (H, I) GO pathway analysis for up- and down-regulated genes in high and low-risk groups.
Differences in genomic alterations among the subgroups
We examined at whether there were any differences in total mutation counts, nonsynonymous mutation counts, and synonymous mutation counts between the high- and low-risk groups (Figure 7A-C). There was a positive correlation between the risk score and the mutation load among the three mutation types; when the mutation load was higher, its correlation with all mutation counts was the lowest (R = 0.23, P = 2e-07), whereas its correlation with the synonymous mutation counts was the highest (R = 0.29, P = 0.89e-11) in the high-score group. The general topography of copy number differences in the high- and low-score groups was shown in Figure 7D and 7E. Figure 7F and 7G revealed that the high-score group had a considerable increase in amplifications and deletions. We also examined the mutations in the top 25 most mutated genes between the high- and low-score groups. As shown in Figure 7H, all the genes with significant differences had higher mutation rates in the high-score group. Figure 7I showed the landscape of mutations for the top 25 genes between high- and low-score groups.
Figure 7.
Differences in genomic mutations among different subgroups and the mutational landscape. A-C. Scatter plots based on the differences on all mutation counts, non-synonymous mutation counts, and synonymous mutation counts between the high- and low-risk groups. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; D, E. Overall landscape for copy number variations in the high- and low-risk score groups; F, G. Amplification and deletion variations were significantly greater in the high-risk score group; H. Mutation difference forest plot of the 25 genes with the highest mutation frequency in the high- and low-risk groups. All significantly different genes had higher mutation frequencies in the high-risk group (*P < 0.05; **P < 0.01; ***P < 0.001); I. Oncoplot of mutation landscape for the top 25 frequently mutated genes between high- and low-risk score groups.
Risk score as an indicator to guide treatment strategies
The sensitivity to five regularly used chemotherapy medications was compared between high- and low-score groups, and we discovered that the IC50 of these drugs was considerably greater in the low-score group, indicating a reduced sensitivity to chemotherapy (Figure 8A). The three external validation cohorts yielded similar findings. Because the differentially expressed genes between high- and low-score groups might be used as small chemical targets, we performed MOA analysis to identify 66 small molecule drugs targeting 39 molecular pathways (Figure S5A). Figure 8B showed a significant reduction in risk scores for patients who were responding to immunotherapy in the external dataset GSE126044 upon PD1 treatment. The survival analysis indicated that the overall survival was the worst in the NSCLC dataset of GSE135222 upon PD1 treatment in the high-score group (P = 0.0061) (Figure 8C). The risk score’s usefulness was further evaluated in the IMvigor210 cohort, which revealed that responders had lower scores (Figure 8D). Figure 8E demonstrated that patients with high scores had the worst overall survival. Consistently, the Chi-Square test also showed that the response rate to PD1 was higher in the low-score group (P = 0.013) (Figure 8F), and patients in the low-score group were sensitive to the PD1 treatment (FDR = 0.014) according to the subclass mapping algorithm (Figure 8G). TIDE algorithm also similarly showed that the low score group was more sensitive to PD1 treatment (P = 0.015) (Figure 8H). Consistent results were observed in the three validation cohorts (Figure S5B-D).
Figure 8.
Correlation between risk score and drug sensitivity. A. Differences in the IC50 values of five commonly used drugs between the high- and low-score groups; B. Differences in risk scores between responders and non-responders; C. The KM survival curve for the high- and low-score groups of patients in the GSE135222 data; D. Differences in risk scores between responders and non-responders in the IMvigor210 dataset; E. KM survival curve for the high- and low-score groups of patients in the IMvigor210 dataset; F-H. Different PD1 response rate based on different algorithms.
Validation of five hub genes in the clinical samples
To validate the accuracy and reliability of this signature, tumor samples and the paracancerous tissues from 36 newly diagnosed LUAD patients were collected and analyzed by qRT-PCR, and the differential expression of the five hub genes was compared by utilizing the paired t-test. The results showed that the expressions of ANLN, E2F7, ECT2, HMMR, and TK1 in LUAD were significantly up-regulated in tumor samples compared to those in paracancerous tissues (Figure 9A-E), which supported our conclusions from bioinformatics analyses. We use not only qRT-PCR to determine their transcription level but also histochemical staining to detect the protein expression of these five hub genes in these samples. A paired t-test confirmed that the fluorescence intensity of ANLN, E2F7, ECT2, HMMR, and TK1 in LUAD were significantly higher than that in normal adjacent tissues (Figure 10A-E), further demonstrating the reliability and accuracy of the hub genes we screened.
Figure 9.

Validation of the expression of the five-gene signature by qRT-PCR in LUAD and paracancerous tissues samples. (A) ANLN, (B) E2F7, (C) ECT2, (D) HMMR, and (E) TK1 (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Figure 10.

Histochemical staining to detect the expression levels of five hub genes. (A) ANLN, (B) E2F7, (C) ECT2, (D) HMMR, and (E) TK1 (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Discussion
Pyroptosis and ferroptosis are emerging as important forms of programmed cell death in normal and pathological conditions including cancer. Ferroptosis and pyroptosis can influence the TIME and suppress the occurrence and the development of cancer, thereby improving the prognosis of patients with cancer [45]. Ferroptosis and pyroptosis can also impact cancer therapy to affect the survival of patients [46]. The commonly used chemotherapeutic drugs and immune checkpoint inhibitors (ICIs) inhibit tumor progression by inducing apoptosis [47]; however, most tumors exhibit innate resistance to apoptosis [48]. Identifying new pathways of cell death induction will lead to the development of potentially new cancer therapy techniques. Therefore, our current studies on the non-apoptotic-related new cell death pathways will shed light on the dvelopment of therapeutic targets.
In this study, we stratified LUAD into four distinct subgroups: immobility, ferroptosis, pyroptosis, and mixed, based on the dual analysis of ferroptosis and pyroptosis-related gene expressions. The pyroptosis subgroup had the greatest patient survival, according to the survival study, while patients in the ferroptosis and mixed subgroups has the worst survival. Significant differences in the immunoreactivities among the subgroups were also observed; the ferroptosis subgroup showed the highest tumor homogeneity and the lowest immune score, while the pyroptosis subgroup showed the opposite features, which might attribute to the different survival among subgroups. Furthermore, there were differences in immunocyte infiltrations among subgroups; the infiltration of CD8+ T cells and memory B cells were higher in the pyroptosis subgroup, while naive B cells and activated dendritic cells were enriched in the ferroptosis subgroup. Thus, our findings provided new insights into the immunological features of different LUAD subgroups.
Previous research has shown the significance of genetic alterations in modulating tumor immunity and immune invasion patterns [49]. Thus, we analyzed the differences in copy number variations and somatic variations between different subgroups. Higher TMB was observed in the ferroptosis and mixed subgroups, while TMB in the pyroptosis and immobility subgroups was lower. The chromosome amplification and deletion events were most abundant in the ferroptosis subgroup, while lowest in the pyroptosis subgroup. In addition, a positive correlation between p53 and ferroptosis genes, a negative correlation between TTN and ferroptosis genes, and no significant correlation between TTN and pyroptosis genes were found. The tumour suppressor p53 and ferroptosis sensitivity are tightly related [50]. In p53 wildtype mice, p53 binds to the promoter of SLC7A11, an essential molecule for ferroptosis induction, and inhibits its transcription. But animals carrying numerous p53 mutations (K98R, K117R, K161R, and K162R) show a significant reduction in p53-dependent ferroptosis responses [51]. Ferroptosis is thought to be an innate mechanism for beginning tumour resistance given the high prevalence of p53 mutation in diverse malignancies [52]. Ferroptosis subgroup showed more genomic alterations as compared to the pyroptosis subgroup, and the significantly high association between ferroptosis and mutations may lead to malignant tumor and worse prognosis.
Given the differences in survival and biological functions among the subgroups, we analyzed the differential expression of ferroptosis and pyroptosis genes. Based on the WGCNA co-expression network, we selected the gene sets that were most relevant to pyroptosis and ferroptosis and finally identified ANLN, E2F7, ECT2, HMMR, and TK1 genes as the signature for the risk model. As a result, based on the median risk score, we divided the samples into high- and low-risk groups, and then we examined the variations between these groups. The development of the risk model should enable the prognostic prediction to be more precise.
Increasing evidence has shown that the immune system may influence cancer in a context-dependent way, either by promoting it or by inhibiting it. Accordingly, one of the most effective anticancer therapies, immune checkpoint therapy, has been developed. In our study, we found significant differences in the immune status of the low- and high-risk LUAD patients. M0/M1 macrophages and activated CD4 cells were found to show high infiltration in the high-risk group, while B cells, dendritic cells, mast cells, plasma cells, resting CD4 cells, and Tregs were enriched in the low-risk group, suggesting the active antigen presentation in the low-risk group. B cells can inhibit tumor cells, reduce the occult micrometastasis incidence, and prolong survival by limiting subsequent tumor spread [53]. Remark et al. have summarized some studies on the relationship between immune cells and survival outcomes in NSCLC and suggested that B cell density was a better prognostic marker [54]. We also observed that immune cell infiltration had a significant effect on LUAD survival outcomes. The degree of B cell infiltration reduced as the risk score went up, which is consistent with the high-risk patients’ shorter survival times. To examine the functional mechanisms underlying the gene signature, we performed gene enrichment analysis by utilizing GSEA. Compared to the low-risk group, the high-risk group showed significant enrichment in the activation of the cell cycle, DNA repair, and p53 signaling pathways, suggesting an active cell proliferation in the high-risk group.
We evaluated the effectiveness of employing risk score to forecast the chemotherapeutic response in order to more accurately assess the clinical viability of our risk model. The IC50 values of the five commonly used chemotherapeutic drugs were significantly high in the low-risk score group, suggesting that the patients in this group were less sensitivity to chemotherapy. In addition to chemotherapy, we also determined the relationship between patient characteristics and immunotherapeutic sensitivity. Our results suggested that patients in the low-risk score group were more sensitive to PD1 therapy, consistent with the findings that the characteristics of high-risk score group were significantly related to cell cycle, while those in the low-risk score group were mainly associated with cellular immunity. Collectively, our findings may provide rationales in selecting treatment options for LUAD patients.
In summary, our study has some advantages. First, the combined ferroptosis- and pyroptosis-related gene signatures were identified in the scRNA-seq data of LUAD, which were more accurate than the traditional biomarkers. Second, the risk model could systematically quantify individual LUAD patients from multiple perspectives, including function, immune infiltration, and genomic alterations. Hence, the risk model could perform bettr in predicting the responses to chemotherapy and immunotherapy. Finally, we used external experiments to validate the prognostic value of hub genes in clinical samples.
However, our study has some limitations. The data we used were from the TCGA database and the GEO datasets. All sample information in the four cohorts (TCGA-LUAD and three-GEO cohorts) that were used for construction and external validation of the prognostic signature were also from public databases. The ferroptosis-associated prognostic signature would be more reliable if validated in our center’s prospective clinical trial cohort.
Conclusion
In conclusion, we identified four LUAD subgroups based on the expressions of ferroptosis- and pyroptosis-related genes and constructed a risk model comprising of five ferroptosis- and pyroptosis-related genes. The findings of the predictive model indicated greater accuracy for prognostic prediction, which may guide the clinical treatment. Our study may provide insights into the identification of novel therapeutic targets for LAUD and contribute to the development of individualized therapies targeting unique tumor metabolic characteristics.
Acknowledgements
The authors hereby express their gratitude to all participants who supported the study. This work was supported by Natural Science Foundation of Jiangsu Province (Youth Found) (BK20200395) and National Natural Science Foundation of China (Youth Found) (82103309).
The study was approved by Shanghai Pulmonary Hospital Ethics Committee (K21-111Y). The patients consented to participate.
Disclosure of conflict of interest
None.
Abbreviations
- LUAD
Lung adenocarcinoma
- ROS
reactive oxygen specie
- TCGA
The Cancer Genome Atla
- FPKM
Fragments Per Kilobase per Million
- TPM
Transcripts Per Kilobase per Million
- CNV
copy number variants
- GDSC
Genomics of Drug Sensitivity in Cancer
- RR
ridge regression
- IC50
half-maximal inhibitory concentration
- TIDE
Tumor Immune Dysfunction and Exclusion
- MOA
mode of action
- tROC
time-dependent receiver operating characteristic
- AUC
area under the curve
- TMB
tumor mutation burden
- ROS
reactive oxygen species
- NO
nitric oxide
- TME
tumor microenvironment
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEGs
differentially expressed genes
- FDR
false discovery rate
Tables S1, S2, S5-S7 and Figures S1-S5
Table S3
Table S4
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bi K, Wei X, Qin X, Li B. BTK has potential to be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment remodeling: a study based on TCGA data mining. Front Oncol. 2020;10:424. doi: 10.3389/fonc.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spella M, Stathopoulos GT. Immune resistance in lung adenocarcinoma. Cancers (Basel) 2021;13:384. doi: 10.3390/cancers13030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naimi A, Mohammed RN, Raji A, Chupradit S, Yumashev AV, Suksatan W, Shalaby MN, Thangavelu L, Kamrava S, Shomali N, Sohrabi AD, Adili A, Noroozi-Aghideh A, Razeghian E. Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Commun Signal. 2022;20:44. doi: 10.1186/s12964-022-00854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Wang Y, Ren Y, Zhang Q, Yi P, Cheng C. Metabolic modulation of immune checkpoints and novel therapeutic strategies in cancer. Semin Cancer Biol. 2022 doi: 10.1016/j.semcancer.2022.02.010. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, He B, Cai Q, Zhang P, Peng X, Zhang Y, Xie H, Wang X. Combination of tumor mutation burden and immune infiltrates for the prognosis of lung adenocarcinoma. Int Immunopharmacol. 2021;98:107807. doi: 10.1016/j.intimp.2021.107807. [DOI] [PubMed] [Google Scholar]
- 7.Lin X, Zhou T, Hu S, Yang L, Yang Z, Pang H, Zhou X, Zhong R, Fang X, Yu Z, Hu K. Prognostic significance of pyroptosis-related factors in lung adenocarcinoma. J Thorac Dis. 2022;14:654–667. doi: 10.21037/jtd-22-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong R, He R, Liu B, Jiang W, Wang B, Li N, Geng Q. Ferroptosis: a new promising target for lung cancer therapy. Oxid Med Cell Longev. 2021;2021:8457521. doi: 10.1155/2021/8457521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Huang Y. Ferroptosis: an iron-dependent cell death form linking metabolism, diseases, immune cell and targeted therapy. Clin Transl Oncol. 2022;24:1–12. doi: 10.1007/s12094-021-02669-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, Possemato R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551:639–643. doi: 10.1038/nature24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Sun Y, Bai L, Zhi L, Yang Y, Zhao Q, Chen C, Qi Y, Gao W, He W, Wang L, Chen D, Fan S, Chen H, Piao H, Qiao Q, Xu Z, Zhang J, Zhao J, Zhang S, Yin Y, Peng C, Li X, Liu Q, Liu H, Wang Y. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11. J Clin Invest. 2021;131:e152067. doi: 10.1172/JCI152067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Chen Y, Wang X, Tian H, Wang Y, Jin J, Shan Z, Liu Y, Cai Z, Tong X, Luan Y, Tan X, Luan B, Ge X, Ji H, Jiang X, Wang P. Stem cell factor SOX2 confers ferroptosis resistance in lung cancer via upregulation of SLC7A11. Cancer Res. 2021;81:5217–5229. doi: 10.1158/0008-5472.CAN-21-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z, Jiang W, Mao M, Zhao J, Chen J, Cheng N. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin Transl Med. 2021;11:e390. doi: 10.1002/ctm2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P, Wu Q, Feng J, Yan L, Sun Y, Liu S, Xiang Y, Zhang M, Pan T, Chen X, Duan T, Zhai L, Zhai B, Wang W, Zhang R, Chen B, Han X, Li Y, Chen L, Liu Y, Huang X, Jin T, Zhang W, Luo H, Chen X, Li Y, Li Q, Li G, Zhang Q, Zhuo L, Yang Z, Tang H, Xie T, Ouyang X, Sui X. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduct Target Ther. 2020;5:51. doi: 10.1038/s41392-020-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Yu L, Ding J, Chen Y. Iron metabolism in cancer. Int J Mol Sci. 2018;20:95. doi: 10.3390/ijms20010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye L, Jin F, Kumar SK, Dai Y. The mechanisms and therapeutic targets of ferroptosis in cancer. Expert Opin Ther Targets. 2021;11:965–986. doi: 10.1080/14728222.2021.2011206. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, Xia H, Zhou J, Li G, Li J, Li W, Wei S, Vatan L, Zhang H, Szeliga W, Gu W, Liu R, Lawrence TS, Lamb C, Tanno Y, Cieslik M, Stone E, Georgiou G, Chan TA, Chinnaiyan A, Zou W. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai Y, Zhang Z, Li J, Li W, Huang Z, Zhang C, Li X, Zhao J. STYK1/NOK correlates with ferroptosis in non-small cell lung carcinoma. Biochem Biophys Res Commun. 2019;519:659–666. doi: 10.1016/j.bbrc.2019.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Yang F, Bettadapura SN, Smeltzer MS, Zhu H, Wang S. Pyroptosis and pyroptosis-inducing cancer drugs. Acta Pharmacol Sin. 2022 doi: 10.1038/s41401-022-00887-6. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agard NJ, Maltby D, Wells JA. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteomics. 2010;9:880–893. doi: 10.1074/mcp.M900528-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 24.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 25.Miguchi M, Hinoi T, Shimomura M, Adachi T, Saito Y, Niitsu H, Kochi M, Sada H, Sotomaru Y, Ikenoue T, Shigeyasu K, Tanakaya K, Kitadai Y, Sentani K, Oue N, Yasui W, Ohdan H. Gasdermin c is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated apc, promoting colorectal cancer proliferation. PLoS One. 2016;11:e166422. doi: 10.1371/journal.pone.0166422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T, Song Y. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol Rep. 2018;40:1971–1984. doi: 10.3892/or.2018.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng J, Mei Q, Zhou X, Tang Y, Xiong R, Qiu W, Pan R, Law BY, Wong VK, Yu C, Long H, Xiao X, Zhang F, Wu J, Qin D, Wu A. Polyphyllin VI induces caspase-1-mediated pyroptosis via the induction of ROS/NF-κB/NLRP3/GSDMD signal axis in non-small cell lung cancer. Cancers (Basel) 2020;12:193. doi: 10.3390/cancers12010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Li C, Wang Y, Xu L, He X, Zeng Q, Zeng C, Mai F, Hu B, Ouyang D. Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis. 2019;24:312–325. doi: 10.1007/s10495-019-01515-1. [DOI] [PubMed] [Google Scholar]
- 29.Yuan R, Zhao W, Wang Q, He J, Han S, Gao H, Feng Y, Yang S. Cucurbitacin B inhibits non-small cell lung cancer in vivo and in vitro by triggering TLR4/NLRP3/GSDMD-dependent pyroptosis. Pharmacol Res. 2021;170:105748. doi: 10.1016/j.phrs.2021.105748. [DOI] [PubMed] [Google Scholar]
- 30.Lin W, Chen Y, Wu B, Chen Y, Li Z. Identification of the pyroptosis-related prognostic gene signature and the associated regulation axis in lung adenocarcinoma. Cell Death Discov. 2021;7:161. doi: 10.1038/s41420-021-00557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, Junqueira C, Meza-Sosa KF, Mok TMY, Ansara J, Sengupta S, Yao Y, Wu H, Lieberman J. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockwell BR, Jiang X. A physiological function for ferroptosis in tumor suppression by the immune system. Cell Metab. 2019;30:14–15. doi: 10.1016/j.cmet.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13:110. doi: 10.1186/s13045-020-00946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karki R, Kanneganti T. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19:197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, Hu J. The role of pyroptosis in cancer: pro-cancer or pro-“host”? Cell Death Dis. 2019;10:650. doi: 10.1038/s41419-019-1883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Man SM, Kanneganti T. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, Bassik MC, Nomura DK, Dixon SJ, Olzmann JA. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schröder MS, Culhane AC, Quackenbush J, Haibe-Kains B. Survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics (Oxford, England) 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, Mcclanahan TK. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Santa F, Vitiello L, Torcinaro A, Ferraro E. The role of metabolic remodeling in macrophage polarization and its effect on skeletal muscle regeneration. Antioxid Redox Signal. 2019;30:1553–1598. doi: 10.1089/ars.2017.7420. [DOI] [PubMed] [Google Scholar]
- 44.Lee C, Lee J, Choi SA, Kim S, Wang K, Park S, Kim SH, Lee JY, Phi JH. M1 macrophage recruitment correlates with worse outcome in SHH Medulloblastomas. BMC Cancer. 2018;18:535. doi: 10.1186/s12885-018-4457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Wang Q, Zhang X, Guo Z, Cui X. Mechanisms of neoantigen-targeted induction of pyroptosis and ferroptosis: from basic research to clinical applications. Front Oncol. 2021;11:685377. doi: 10.3389/fonc.2021.685377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang B, Zhong W, Gu Y, Li Y. Emerging mechanisms and targeted therapy of pyroptosis in central nervous system trauma. Front Cell Dev Biol. 2022;10:832114. doi: 10.3389/fcell.2022.832114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W, Zhang L, Xiu Z, Guo J, Wang L, Zhou Y, Jiao Y, Sun M, Cai J. Combination of immune checkpoint inhibitors with chemotherapy in lung cancer. Onco Targets Ther. 2020;13:7229–7241. doi: 10.2147/OTT.S255491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors in cancer therapy: a focus on T-regulatory cells. Immunol Cell Biol. 2018;1:21–33. doi: 10.1111/imcb.1003. [DOI] [PubMed] [Google Scholar]
- 49.Cindy Yang SY, Lien SC, Wang BX, Clouthier DL, Hanna Y, Cirlan I, Zhu K, Bruce JP, El Ghamrasni S, Iafolla MAJ, Oliva M, Hansen AR, Spreafico A, Bedard PL, Lheureux S, Razak A, Speers V, Berman HK, Aleshin A, Haibe-Kains B, Brooks DG, Mcgaha TL, Butler MO, Bratman SV, Ohashi PS, Siu LL, Pugh TJ. Pan-cancer analysis of longitudinal metastatic tumors reveals genomic alterations and immune landscape dynamics associated with pembrolizumab sensitivity. Nat Commun. 2021;12:5137. doi: 10.1038/s41467-021-25432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang R, Kroemer G, Tang D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic Biol Med. 2019;133:162–168. doi: 10.1016/j.freeradbiomed.2018.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, Gu W. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 53.Bremnes RM, Al-Shibli K, Donnem T, Sirera R, Al-Saad S, Andersen S, Stenvold H, Camps C, Busund L. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol. 2011;6:824–833. doi: 10.1097/JTO.0b013e3182037b76. [DOI] [PubMed] [Google Scholar]
- 54.Remark R, Becker C, Gomez JE, Damotte D, Dieu-Nosjean M, Sautès-Fridman C, Fridman W, Powell CA, Altorki NK, Merad M, Gnjatic S. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191:377–390. doi: 10.1164/rccm.201409-1671PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.