Abstract
The expression, in Escherichia coli, of variants of the Erwinia chrysanthemi secretion genes outB and outS under the Ptac promoter is toxic to the cells. During attempts to clone E. chrysanthemi genes able to suppress this toxicity, I identified two genes, sotA and sotB, whose products are able to reduce the isopropyl-β-d-thiogalactopyranoside (IPTG) induction of the E. coli lac promoter. SotA and SotB belong to two different families of the major facilitator superfamily. SotA is a member of the sugar efflux transporter family, while SotB belongs to the multidrug efflux family. The results presented here suggest that SotA and SotB are sugar efflux pumps. SotA reduces the intracellular concentration of IPTG, lactose, and arabinose. SotB reduces the concentration of IPTG, lactose, and melibiose. Expression of sotA and sotB is not regulated by their substrates, but sotA is activated by the cyclic AMP receptor protein (CRP), while sotB is repressed by CRP. Lactose is weakly toxic for E. chrysanthemi. This toxicity is increased in a sotB mutant which cannot efficiently efflux lactose. This first evidence for a physiological role of sugar efflux proteins suggests that their function could be to reduce the intracellular concentration of toxic sugars or sugar metabolites.
Active efflux has been shown to be the cause of the drug resistance of many bacteria. Some of the proteins performing this efflux can deal with one type or a narrow range of structurally related compounds, like the Escherichia coli tetracycline exporter TetB (20). Other proteins are able to extrude from the cell a broad variety of unrelated compounds (antibiotics, quaternary amines, and basic dyes). The resistance conferred by these proteins is called multidrug resistance (MDR). The activity of these pumps gives bacterial pathogens a significant resistance to widely used antibiotics. The gram-negative MDR proteins belong to four different families of transport protein (23, 26). (i) The first is the major facilitator superfamily (MFS). This large group of proteins, which contains membrane transport proteins from bacteria to higher eukaryotes, is involved in the symport, antiport, or uniport of various substrates (25). It includes the well-studied MDR transporters QacA of Staphylococcus aureus and Bmr of Bacillus subtilis. These proteins contain 12 or 14 transmembrane segments. (ii) The second is the resistance nodulation division family (RND). Proteins of this family contain 12 transmembrane helices. The E. coli AcrB and the Pseudomonas aeruginosa MexB proteins belong to this group (19, 27). (iii) The third, the Smr family, exemplified by the S. aureus Smr protein, contains proteins smaller than those of the former groups, which possess four transmembrane segments (11). (iv) The fourth, the recently described MATE family, is exemplified by the Vibrio parahaemolyticus NorM protein (6).
The specificity of these MDR proteins is generally very broad, and a large spectrum of compounds can be effluxed (10). However, their substrates have been sought among antibiotics and drugs (basic dyes, sodium dodecyl sulfate [SDS], quaternary ammonium compounds, carbonyl cyanide m-chlorophenylhydrazone, etc.). Recent experiments have shown that other types of molecules, such as sugars, can be pumped out of bacteria. The CmlA efflux pump, which can export a wide variety of antibiotics from different families, can efflux the galactoside isopropyl-β-d-thiogalactopyranoside (IPTG), a gratuitous inducer of genes controlled by the LacI repressor (3). Induction-based suppression of toxic effects has led to the discovery of two E. coli genes, belonging to two different families, encoding sugar efflux proteins. The first of these genes, ydeA, is a member of the MFS with 12 transmembrane segments (5, 7). Its expression prevents accumulation of arabinose and IPTG in the cytoplasm. The second one, setA (yabM), belongs to a new subfamily of the MFS, the SET (sugar efflux transporter) family (18), which includes two other E. coli proteins (YieO and YicK), a protein from Yersinia pestis, and one from Deinococcus radiodurans. SetA is able to prevent the accumulation of lactose and IPTG in E. coli cells. Thus, sugars and glycosides can both be substrates of efflux pumps.
During a search for Erwinia chrysanthemi genes whose products are able to suppress the toxic effect due to the expression of the E. chrysanthemi outS and outB genes cloned under the Ptac promoter, I identified two genes able to reduce the intracellular IPTG concentration of the bacteria. This leads to a reduction in the toxicity resulting from the accumulation of OutS and OutB in the bacteria. I also showed that their products are able to prevent the accumulation of several sugars in the bacteria.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and drug resistance assays.
The strains and plasmids used in this study are listed in Table 1. E. chrysanthemi and E. coli strains were grown at 30 and 37°C, respectively, unless otherwise stated. The media used were Luria broth (LB) or M63 minimal medium supplemented with a carbon source (0.2%), and for cloning experiments, the following antibiotics were used at the concentrations indicated: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; and tetracycline, 10 μg/ml. The resistance of E. coli strains containing cloned genes to drugs was tested by spotting 10 μl of a 105-fold dilution of an overnight culture on LB plus ampicillin plates containing various dilutions of the tested compound. The ability of the strain to form single colonies was recorded after 24 h.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. chrysanthemi | ||
| 3937 | Wild type | Laboratory collection |
| A350 | lmrT(Con) lacZ | 15 |
| A2507 | lmrT(Con) lacZ crp::Cm | 29 |
| A3145 | lmrT(Con) lacZ sotA::uidA-kan | This work |
| A3501 | lmrT(Con) lacZ sotB::uidA-kan | This work |
| E. coli | ||
| NM522 | supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 rK− mK− F′ proAB lacIq ΔlacZM15 | Stratagene |
| P4X | Hfr metB relA1 spoT1 | Laboratory collection |
| NA1 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR uxaB::Mud(Aprlac) | 14 |
| 1701 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR (rbs-lac)op6.16 | J. Beckwith |
| 9665 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR trp::(malK-lac)1 | 1 |
| Plasmids | ||
| pACT3 | Cmr Ptac ori p15A | 9 |
| pEXT20 | Apr Ptac | 9 |
| pACTPB | pACT3 expressing pelB | This work |
| pABSC3 | pACT3 expressing pelBsp-outB | G. Condemine and V. E. Shevchik |
| pABCT2 | pACT3 expressing outB | G. Condemine and V. E. Shevchik |
| pACTS1 | pACT3 expressing pelBsp-outS | 32 |
| pBAD18 | Apr PBAD | 12 |
| pBADuid | uidA-kan cloned in the pBAD18 polylinker | This work |
| pBR322 | Tetr Apr | 4 |
| pBRS1 | pBR322 expressing sotA | This work |
| pBRS6 | pBR322 expressing sotB | This work |
| pSU9 | Cmr | 2 |
| pS1A | pSU9 expressing sotA | This work |
| pS6E | pSU9 expressing sotB | This work |
| pUC18 | Apr | Biolabs |
| pUCS1 | pUC18 expressing sotA | This work |
| pUCS6 | pUC18 expressing sotB | This work |
| pULB110 | Tetr Apr (Mu 3A) | 34 |
Genetic techniques.
Transduction with phage φEC2 was performed as described by Résibois et al. (28). Genetic mapping was performed with pULB110 as described by van Gijsegem and Toussaint (34). Marker exchange recombinations were obtained after growth in a low-phosphate medium as described by Roeder and Collmer (30).
Enzyme assays.
β-Glucuronidase assays were performed with toluenized cells grown to the exponential phase with p-nitrophenol-β-d-glucuronate as the substrate (24). β-Galactosidase assays were performed with toluenized cells grown to the exponential phase with o-nitrophenol-β-d-galactose as the substrate (21).
Recombinant DNA techniques.
Preparations of chromosome and plasmid DNA, restriction digestions, ligations, DNA electrophoresis, transformations, and electroporations were carried out as described by Sambrook et al. (31). Sequencing was performed by Genome Express SA (Grenoble, France).
SDS-PAGE and Western blotting.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (17). After electrophoresis, proteins were electroblotted onto nitrocellulose in a semidry apparatus at 1 mA/cm2 for 1 h in transfer buffer containing 40 mM glycine, 50 mM Tris, 0.4% SDS, and 10% methanol. The nitrocellulose was then saturated and incubated with antibodies and developed with the ECL enhanced chemiluminescence detection kit (Pharmacia-Amersham Biotech).
Nucleotide sequence accession number.
The complete DNA sequences of sotA and sotB have been deposited in the EMBL database under accession no. AJ249180 and AJ249181, respectively.
RESULTS
Isolation of two genes suppressing the toxicity of PelBsp-OutS and PelBsp-OutB.
OutS and OutB are two components of the E. chrysanthemi Out secretion machinery, involved in the secretion of pectinases in the external medium (8). OutS, an outer membrane lipoprotein, is a chaperone of the secretin OutD (32, 33). OutB is an inner membrane protein that interacts with OutD (G. Condemine and V. E. Shevchik, unpublished data). We have constructed variants of outS and outB in which the region coding for their N-terminal anchors has been replaced by the region coding for the PelB signal sequence (32; G. Condemine and V. E. Shevchik, unpublished data). These constructs have been cloned under the Ptac promoter of plasmid pACT3. When E. coli cells containing these plasmids were grown in the absence of IPTG at 30°C, the resulting proteins (PelBsp-OutS and PelBsp-OutB) were correctly synthesized, processed, and accumulated in the periplasm. When grown at 37°C in the presence of IPTG, the cells were unable to form colonies on plates and were lysed in liquid medium. To identify proteins interacting with OutS and OutB, I tried to isolate E. chrysanthemi genes which, expressed on a multicopy plasmid, could suppress the toxic effect due to the overexpression of PelBsp-OutS and PelBsp-OutB. An E. chrysanthemi gene library constructed in plasmid pUC18 was introduced into E. coli strain NM522 containing plasmid pACTS1 (pelBsp-outS). A total of 6 × 104 transformants were spread onto LB-ampicillin-chloramphenicol-IPTG plates that were incubated overnight at 37°C. Six transformants were able to grow on this medium. Restriction mapping of the six plasmids suppressing the toxicity of PelBsp-OutS showed that they fall into two groups. Five contained a common region, and one had a totally different restriction map. One plasmid of each type was retained for further studies. The two genes suppressing toxicity were named sotA and sotB (for suppressor of Out toxicity). The plasmids bearing sotA and sotB were introduced into NM522 containing plasmid pABSC3 (pelBsp-outB). Both transformants were able to grow at 37°C in the presence of IPTG, indicating that the sotA and sotB genes are also able to suppress the toxicity induced by PelBsp-OutB.
sotA and sotB modify IPTG induction of the Ptac promoter.
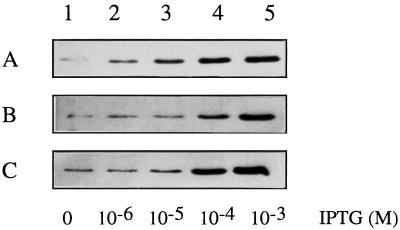
To determine the mode of action of SotA and SotB, I estimated the amount of PelBsp-OutS and PelBsp-OutB in cells expressing, or not, SotA and SotB in the absence or presence of IPTG. While in the absence of IPTG no significant variation was observed, the amounts of both PelBsp-OutS and PelBsp-OutB were reduced with either SotA or SotB. This effect was more pronounced when SotA was present (Fig. 1). To test whether the effect of sotA and sotB was specific to PelBsp-OutS and PelBsp-OutB, I quantified the products of two other genes (outB and pelB) cloned under the Ptac promoter of plasmid pACT3 and expressed under the same conditions. Overexpression of wild-type OutB or PelB from pACT3 is not toxic for the bacteria. The amount of OutB or PelB was also reduced when produced with SotA or SotB (Fig. 1). This reduction could result from a decrease in the copy number of plasmid pACT3, from a reduced induction of the Ptac promoter, or from an increased degradation of mRNAs or proteins. I first checked that the copy number of pACT3 was unchanged in the presence of sotA or sotB (data not shown). To test the other hypotheses, we expressed OutB under its own promoter in plasmid pBR322 and under the tac promoter of plasmid pEXT20. pEXT20 is a pBR322 derivative that contains the expression cassette comprising the Ptac promoter of plasmid pACT3. While expression of OutB from pBR322 was not modified by the presence of SotA and SotB, even in the presence of IPTG, expression from pEXT20 was reduced (data not shown). This result indicates that SotA and SotB do not modify the stability of RNAs or proteins, but they do influence the IPTG induction of the Ptac promoter of pEXT20 or pACT3. Induction of pelB cloned into pACT3 was tested by using increasing amounts of IPTG. Induction of PelB synthesis was observable with concentrations of IPTG as low as 10−6 M in the control strain. A 10−5 M concentration of IPTG was required to induce PelB synthesis when sotB was present, and 10−4 M IPTG was required when sotA was present (Fig. 2). Thus, SotA and SotB could decrease the sensitivity of Ptac to IPTG or reduce the intracellular concentration of IPTG.
FIG. 1.
Effect of the presence of sotA and sotB on the expression of genes cloned into plasmid pACT3. E. coli NM522 cells containing plasmids pACTS1 (pelBsp-outS) (A), pABSC3 (pelBsp-outB) (B), pABTC2 (outB) (C), and pACTPB (pelB) (D) were transformed with plasmids pUC18, pUCS1 (sotA), and pUCS6 (sotB). Cells were grown for 3 h in the absence or presence of 10−4 M IPTG. Aliquots were loaded onto SDS-PAGE gel and immunoblotted with anti-OutS (A), anti-OutB (B and C), and anti-PelB (D) antibodies.
FIG. 2.
Effect of the presence of sotA and sotB on the IPTG induction of pelB cloned into pACT3. E. coli NM522 cells containing pACTPB (pelB) and pUC18 (A), pUCS1 (sotA) (B), or pUCS6 (sotB) (C) were grown in LB medium with 0 (lane 1), 10−6 (lane 2), 10−5 (lane 3), 10−4 (lane 4), or 10−3 M IPTG (lane 5). Aliquots were loaded onto an SDS-PAGE gel (10% polyacrylamide) and immunoblotted with anti-PelB antibody.
The products of sotA and sotB are putative membrane transport proteins.
The DNA fragment containing the sotA gene, reduced to 1.6 kb by subcloning, was sequenced. It contains one putative open reading frame of 1,179 nucleotides, encoding a protein of 42,433 Da with 12 putative transmembrane domains. A similarity search in data banks revealed that it belongs to the recently identified sugar efflux transporter family, a subgroup of the MFS of membrane transporters. A strong homology exists with the proteins SetB (YeiO) (68% identity), YicK (61% identity), and SetA (YabM) (53% identity) of E. coli. SetA and SetB can efflux lactose and/or IPTG from E. coli cells.
The sotB gene has been localized by subcloning on a 2.1-kb DNA fragment which has been sequenced. It contains one putative complete open reading frame of 1,185 nucleotides, encoding a protein of 41,960 Da with 12 putative transmembrane domains. SotB belongs to another subfamily of the MFS which contains MDR proteins. SotB presents homology with the chloramphenicol resistance protein of Streptomyces lividans (CmlR) which exports the drug, the AraJ protein of E. coli, and the YdeA proteins of E. coli and Haemophilus influenzae. The highest identity was observed with YdeA of E. coli (61%). The E. coli YdeA protein is able to export arabinose and IPTG from the bacteria.
SotA and SotB can export several sugars.
The reduced induction of IPTG of the Ptac promoter in the presence of SotA and SotB and the homology of these proteins with sugar exporters suggested that SotA and SotB are sugar efflux pumps. To test this hypothesis and to determine the substrates of these pumps, we measured whether the presence of SotA and SotB on a plasmid in E. coli modifies the induction of sugar utilization operons by their substrate. Plasmids bearing sotA and sotB were introduced into E. coli P4X, and lacZ induction was monitored. A reduction in the induction level of the lacZ gene by IPTG was observed in the presence of SotA or SotB (Fig. 3A and B). However, the concentration at which this effect was observable was 10-fold lower with SotB than with SotA (3 × 10−6 M versus 4 × 10−5 M). The lac operon is also inducible by lactose and melibiose (21). While induction of lacZ by these compounds was barely modified in the presence of SotA, it was clearly diminished by SotB (Fig. 3C, D, and E). Efflux of galacturonate, ribose, and maltose was tested by measuring the induction of an uxaB-lacZ fusion, an rbs-lacZ fusion, and a malK-lacZ fusion in the presence of sotA and sotB. No modification of the induction rate of these fusions was observed, indicating that these sugars are not substrates of SotA or SotB. Efflux of arabinose was tested by measuring the induction of β-glucuronidase encoded by the uidA gene, which was cloned under the Para promoter of plasmid pBAD18. The presence of SotA, but not of SotB, was able to prevent, very efficiently, induction by arabinose of uidA expression (Fig. 3F).
FIG. 3.
Effect of the presence of sotA and sotB on the induction of the lac and ara promoter in E. coli by various compounds. Strains P4X (induction by IPTG, lactose, or melibiose) containing plasmid pBR322 (□), pBRS1 (sotA) (◊), or pBRS6 (sotB) (○), or NM522/pBADuid (induction by arabinose) carrying plasmid pSU9 (□), pS1A (sotA) (◊), or pS6E (sotB) (○) were grown in LB medium to an optical density at 600 nm of 0.5. Inducer was added at the concentration indicated, and the β-galactosidase (β-gal) or β-glucuronidase (β-glu) activity of the culture was measured over time. (A) IPTG (4 × 10−5 M). (B) IPTG (3 × 10−6 M). (C) Lactose (20 mg/liter). (D) Lactose (5 mg/liter). (E) Melibiose (40 mg/liter). (F) Arabinose (4 mg/liter).
The multidrug transporter CmlA is able to confer resistance to a wide range of drugs and also to efflux IPTG (3). To test whether SotA and SotB could also efflux drugs, I compared the resistance of strain NM522, with or without sotA and sotB, to 19 antibiotics and drugs (tetracycline, chloramphenicol, rifampin, nalidixic acid, novobiocin, kanamycin, spectinomycin, erythromycin, fusidic acid, trimethoprim, gentamicin, vancomycin, kasugamycin, crystal violet, SDS, ethidium bromide, oleic acid, coumaric acid, and ferrulic acid). The presence of SotA and SotB did not change the MICs of these compounds. Thus, SotA and SotB seem to efflux specifically sugars.
Analysis of sotA and sotB transcription.
sotA and sotB mutants were constructed by reverse genetics. A uidA-kan cassette was inserted into the unique SmaI site of sotA and into the unique BamHI site of sotB. Constructs in which the uidA gene was in the same orientation as sotA or sotB were retained, introduced into E. chrysanthemi, and recombined into the chromosome. The operon fusions obtained were used to study sotA and sotB transcription. Synthesis of multidrug transporters is often induced by one of the drugs they expel (22). Thus, we measured the expression of the sotA-uidA and sotB-uidA fusions in the presence of various sugars (Table 2). Expression of sotA was not induced by its substrates, IPTG and arabinose. Among the other sugars tested, only galacturonate gave a 1.5-fold induction. The presence of a crp mutation reduced by twofold the expression of sotA. The expression of sotB was not induced by lactose or IPTG, but increased fourfold in the presence of glucose. It was also induced to a lesser level by other metabolizable sugars, such as galacturonate, arabinose, and melibiose (Table 2). The fusion was expressed at an eightfold-higher level in a crp mutant. This suggested a repression of sotB by cyclic AMP receptor protein (CRP). A CRP binding site, which contains 9 out of the 10 nucleotides of the consensus (TGTGAN6TCGCA), can be found at position 533 to 548, 65 bases upstream of the sotB initiation codon.
TABLE 2.
Expression of sotA-uidA and sotB-uidA fusions in the presence of various sugars
| Growth conditiona | Additional mutation | β-Glucuronidase activityb
|
|
|---|---|---|---|
| sotA-uidA | sotB-uidA | ||
| Glycerol | —c | 141 ± 5 | 16 ± 2 |
| Glycerol + lactose | — | 135 ± 7 | 18 ± 2 |
| Glycerol + IPTG | — | 141 ± 14 | 12 ± 2 |
| Glycerol + melibiose | — | 140 ± 9 | 49 ± 14 |
| Glycerol + arabinose | — | 104 ± 8 | 38 ± 12 |
| Glycerol + galacturonate | — | 216 ± 8 | 48 ± 6 |
| Glycerol + rhamnose | — | 147 ± 9 | 15 ± 2 |
| Glycerol + maltose | — | 152 ± 6 | 13 ± 1 |
| Glucose | — | 99 ± 7 | 62 ± 6 |
| Glucose | crp | 72 ± 6 | 125 ± 6 |
Bacteria were grown in M63 medium with 0.2% glycerol or glucose as the carbon source and 0.2% of the sugars tested or 1 mM IPTG.
β-Glucuronidase activity is expressed in nanomoles of p-nitrophenol formed per minute per milligram (dry weight) of bacteria. Each value is the average of four determinations.
—, none.
Analysis of E. chrysanthemi sotA and sotB mutants.
The presence of lactose in the growth medium was detrimental to the growth of E. chrysanthemi A350, the strain used in this study (Fig. 4). It has been designed for the construction of lacZ gene fusion (15). Lactose uptake has been increased by the derepression of the expression of the broad-substrate-specificity transporter lmrT (involved in lactose, melibiose, and raffinose entry), and the lacZ gene has been inactivated. Thus, this strain is unable to metabolize lactose. The inhibitory effect of lactose on the growth of the sotA and sotB mutant was compared with that observed with A350. While the growth of the sotA mutant was equivalent to that of A350, the sotB mutant showed reduced growth, indicating that the presence of an efficient lactose efflux pump, SotB, is sufficient to reduce, at least partially, the lactose toxicity toward E. chrysanthemi.
FIG. 4.
Growth of strain A350 and of sotA and sotB mutants in the presence of lactose. M63 medium containing 0.2% glycerol and various concentrations of lactose was inoculated at a 1:100 dilution with a culture of strain A350, A3156, or A3501. After 16 h of growth, the optical density at 600 nm (OD600) was measured.
sotA and sotB have been mapped on the E. chrysanthemi chromosome. Chromosomal mobilization mediated by the plasmid pULB110 was used for conjugation with various polyauxotrophic recipient strains. Both genes were localized between the his-1 and gal-1 markers (16). sotA and sotB are cotransducible by phage φEC2 with a frequency of 63%, indicating that they are probably located within 20 kb of each other.
DISCUSSION
The multicopy suppression strategy I used, which involved looking for genes whose products could suppress the toxicity of PelBsp-OutS or PelBsp-OutB, was aimed at finding proteins that interact with OutS and OutB. This approach led to the identification of two genes that are not related to the Out secretion system, but whose products are able to reduce the intracellular concentration of the lac gene inducer, IPTG. The presence of sotA or sotB reduces the expression of any gene cloned under the Ptac promoter of plasmid pACT3 by lowering the intracellular concentration of IPTG. The results presented here suggest that SotA and SotB are sugar efflux proteins. The other known sugar efflux proteins have been isolated fortuitously by similar cloning strategies: cmlA and setA reduce induction by IPTG of toxic genes cloned under the Ptac promoter (3, 18), and ydeA prevents the arabinose-dependent expression of a toxic protein expressed under the control of the arabinose PBAD promoter (5, 7). Thus, the use of a multicopy suppression strategy when the toxic protein synthesis is under the control of an inducible promoter often leads to the identification of genes able to expel the inducer. New sugar efflux transporters could be identified, by the same strategy, by cloning toxic genes in other sugar-inducible promoter-based expression vectors.
Up to now, no physiological function has been assigned to sugar efflux pumps. The results presented here show that one of their functions could be to remove toxic sugars from the cytoplasm. Although the cause of lactose toxicity in E. chrysanthemi is not known, it is clear that the absence of an efflux system able to reduce the intracellular concentration of this sugar has a detrimental effect on the growth of the bacteria. Accumulation of nonmetabolizable sugars or of sugar metabolites, such as sugar phosphates, can have growth-inhibitory effects. The sugar efflux transporters could participate in the regulation of the intracellular concentration of these toxic compounds. The presence of three SET members in E. coli could indicate that this strategy of detoxification is widely used by bacteria. Huber et al. (13) have shown that when E. coli cells are grown in the presence of lactose, a large proportion of the β-galactosidase products are found in the culture medium. Sugar efflux pumps could help maintain an intracellular concentration of sugar within a range compatible with the biochemical and regulatory metabolism of the bacteria. Recently, amino acid efflux pumps have been described (35). It would be interesting to investigate whether the intracellular concentrations of other compounds could also be regulated by efflux pumps.
Initially, SotA and SotB may seem redundant, but the specificity of the two transporters appears different. IPTG, melibiose (6-O-α-d-galactopyranosyl-d-glucose), and lactose [β-d-galactose (1-4)-α-glucose] are apparently good substrates for SotB. Thus, SotB can be considered as an exporter of β-galactosides. The specificity of SotA seems to be wider, since it can apparently efflux IPTG, lactose, and arabinose, a pentose. The differences in substrate specificity may contribute as well to differences in the respective roles of SotA and SotB. Screening of other sugar efflux activities by SotA and SotB will show if they have a narrow specificity, recognizing only one type of glycoside, or if they are multisugar efflux pumps, with a specificity as broad as that of some multidrug efflux pumps. The absence of a specific regulation of sotA and sotB expression by lactose or arabinose and their control by the global regulator CRP, the homology of SotB with MDR proteins, suggest that they could have a broad specificity, contributing to the efflux of many sugars from the cytoplasm. However, in contrast to CmlA, they do not seem to be able to export drugs and antibiotics.
ACKNOWLEDGMENTS
I thank M. Heyde for strains, S. Reverchon for providing the E. chrysanthemi gene library, and N. Cotte-Pattat and Valerie James for reading the manuscript.
This work was supported by grants from the Centre National de la Recherche Scientifique and from the Ministère de l'Éducation Nationale de la Recherche et de la Technologie.
REFERENCES
- 1.Alonzo S, Heyde M, Laloi P, Portalier R. Analysis of the effect exerted by extracellular pH on the maltose regulon in Escherichia coli K-12. Microbiology. 1998;144:3317–3325. doi: 10.1099/00221287-144-12-3317. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Bohn C, Bouloc P. The Escherichia coli cmlA gene encodes the multidrug efflux pump Cmr/MdfA and is responsible for isopropyl-β-d-thiogalactopyranoside exclusion and spectinomycin sensitivity. J Bacteriol. 1998;180:6072–6075. doi: 10.1128/jb.180.22.6072-6075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heineker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 5.Bost S, Silva F, Belin D. Transcription activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose PBAD promoter. J Bacteriol. 1999;181:2185–2191. doi: 10.1128/jb.181.7.2185-2191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown M H, Paulsen I T, Skurray R A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 7.Carolé S, Pichoff S, Bouché J-P. Escherichia coli gene ydeA encodes a major facilitator pump which exports l-arabinose and isopropyl-β-d-thiogalactopyranoside. J Bacteriol. 1999;181:5123–5125. doi: 10.1128/jb.181.16.5123-5125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condemine G, Dorel C, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Some of the out genes involved in the secretion of pectate lyases in Erwinia chrysanthemi are regulated by kdgR. Mol Microbiol. 1992;6:3199–3211. doi: 10.1111/j.1365-2958.1992.tb01775.x. [DOI] [PubMed] [Google Scholar]
- 9.Dykxhoorn D M, St. Pierre R, Furlong C E. A set of compatible tac promoter expression vectors. Gene. 1996;177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- 10.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinary broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grinius L, Dreguniene G, Goldberg E B, Lio C H, Projan S J. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid. 1992;27:119–129. doi: 10.1016/0147-619x(92)90012-y. [DOI] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber R E, Lytton J, Fung E B. Efflux of β-galactosidase products from Escherichia coli. J Bacteriol. 1980;141:528–533. doi: 10.1128/jb.141.2.528-533.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Isolation of fusions between the lac genes and several genes of the exu regulon: analysis of their regulation, determination of the transcription direction of the uxaC-uxaA operon, in Escherichia coli K-12. Mol Gen Genet. 1981;182:279–287. doi: 10.1007/BF00269671. [DOI] [PubMed] [Google Scholar]
- 15.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Lactose catabolism in Erwinia chrysanthemi. J Bacteriol. 1985;169:1223–1231. doi: 10.1128/jb.169.3.1223-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugouvieux-Cotte-Pattat N, Reverchon S, Robert-Baudouy J. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol Microbiol. 1989;3:1587–1597. doi: 10.1111/j.1365-2958.1989.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Liu J Y, Miller P F, Gosink M, Olson E R. The identification of a new family of sugar efflux pumps in Escherichia coli. Mol Microbiol. 1999;31:1845–1851. doi: 10.1046/j.1365-2958.1999.01321.x. [DOI] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon G N, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 20.McMurry L, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Neyfakh A A. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–313. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novel G, Didier-Fichet M L, Stoeber F. Inducibility of β-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J Bacteriol. 1974;120:89–95. doi: 10.1128/jb.120.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Résibois A, Cole M, Faelen M, Schoonejans E, Toussaint A. phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology. 1982;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 29.Reverchon S, Expert D, Robert-Baudouy J, Nasser W. The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J Bacteriol. 1997;179:3500–3508. doi: 10.1128/jb.179.11.3500-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roeder D L, Collmer A. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J Bacteriol. 1985;164:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Shevchik V E, Condemine G. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology. 1998;144:3219–3228. doi: 10.1099/00221287-144-11-3219. [DOI] [PubMed] [Google Scholar]
- 33.Shevchik V E, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;11:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Gijsegem F, Toussaint A. In vivo cloning of Erwinia carotovora genes involved in the catabolism of hexuronates. J Bacteriol. 1983;154:1227–1235. doi: 10.1128/jb.154.3.1227-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zakataeva N P, Aleshin V V, Tokmakova I L, Troshin P V, Livshits V A. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 1999;452:228–232. doi: 10.1016/s0014-5793(99)00625-0. [DOI] [PubMed] [Google Scholar]