Abstract
The cellulosome of Clostridium thermocellum is a multiprotein complex with endo- and exocellulase, xylanase, β-glucanase, and acetyl xylan esterase activities. XynY and XynZ, components of the cellulosome, are composed of several domains including xylanase domains and domains of unknown function (UDs). Database searches revealed that the C- and N-terminal UDs of XynY and XynZ, respectively, have sequence homology with the sequence of a feruloyl esterase of strain PC-2 of the anaerobic fungus Orpinomyces. Purified cellulosomes from C. thermocellum were found to hydrolyze FAXX (O-{5-O-[(E)-feruloyl]-α-l-arabinofuranosyl}-(1→3)-O-β-d-xylopyranosyl-(1→4)-d-xylopyranose) and FAX3 (5-O-[(E)-feruloyl]-[O-β-d-xylopyranosyl-(1→2)]-O-α-l-arabinofuranosyl-[1→3]}-O-β-d-xylopyranosyl-(1→4)-d-xylopyranose), yielding ferulic acid as a product, indicating that they have feruloyl esterase activity. Nucleotide sequences corresponding to the UDs of XynY and XynZ were cloned into Escherichia coli, and the expressed proteins hydrolyzed FAXX and FAX3. The recombinant feruloyl esterase domain of XynZ alone (FAEXynZ) and with the adjacent cellulose binding domain (FAE-CBDXynZ) were characterized. FAE-CBDXynZ had a molecular mass of 45 kDa that corresponded to the expected product of the 1,203-bp gene. Km and Vmax values for FAX3 were 5 mM and 12.5 U/mg, respectively, at pH 6.0 and 60°C. PAX3, a substrate similar to FAX3 but with a p-coumaroyl group instead of a feruloyl moiety was hydrolyzed at a rate 10 times slower. The recombinant enzyme was active between pH 3 to 10 with an optimum between pH 4 to 7 and at temperatures up to 70°C. Treatment of Coastal Bermuda grass with the enzyme released mainly ferulic acid and a lower amount of p-coumaric acid. FAEXynZ had similar properties. Removal of the 40 C-terminal amino acids, residues 247 to 286, of FAEXynZ resulted in protein without activity. Feruloyl esterases are believed to aid in a release of lignin from hemicellulose and may be involved in lignin solubilization. The presence of feruloyl esterase in the C. thermocellum cellulosome together with its other hydrolytic activities demonstrates a powerful enzymatic potential of this organelle in plant cell wall decomposition.
Plant cell wall material composed mainly of cellulose, hemicelluloses, and lignin is one of the largest sources of renewable energy on earth. Arabinoxylan is one of the main hemicelluloses. Its backbone structure is a chain of β(1→4)-linked xylose moieties to which are attached side chains, including arabinose, acetate, and methyl-glucuronic acid (7, 40). The arabinose has ester-linked ferulic acid and p-coumaric acid. Ferulic acid links hemicellulose to lignin (39). Since feruloyl esterases hydrolyze the bond between the arabinose and ferulic acid, they may release the covalently bound lignin from hemicelluloses and aid in the degradation of plant cell walls. Feruloyl esterases have been found in bacteria and fungi (44).
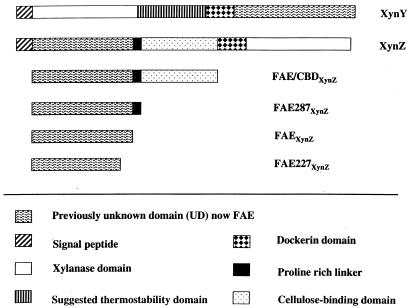
The cellulosome first discovered in Clostridium thermocellum (2, 29) is a multiprotein complex with a molecular mass of about 3,000 kDa. Cellulosomes are produced by several anaerobic bacteria (4) and anaerobic fungi (17, 32). The core of the cellulosome is an enzymatically inactive cellulosome integrating polypeptide (CipA) functioning as a scaffold. CipA of C. thermocellum contains nine copies of a cohesin domain, a type II dockerin domain, and a cellulose binding domain (CBD). At present, 18 catalytic active subunits of the cellulosome have been sequenced. They have endoglucanase, cellobiohydrolase (exoglucanase), xylanase, chitinase, or β-glucanase (lichenase) activity (2). All enzymatically active subunits have multidomain structures that include at least a catalytic domain and a dockerin domain which binds to the cohesins of CipA. Other domains present in some of the catalytic subunits include CBDs, immunoglobulin-like domains, serine- and threonine- or proline-rich linkers, and domains of unknown functions (UDs). Examples of subunits having UDs are XynY (20) and XynZ (22) (see Fig. 2). Starting with the N terminus, XynY has xylanase (glycosyl hydrolase family 10), a domain characterized as a thermostability domain, a dockerin, and a UD. Also starting with the N terminus, XynZ has a UD, a proline-rich linker, a CBD (family VI), a dockerin, and xylanase (glycosyl hydrolase family 10).
FIG. 2.
Domain organization of XynY (20), XynZ (22), and constructs. FAE-CBDXynZ, comprising 400 amino acid residues, is a truncated form of XynZ including the FAE domain and the CBD; FAE287XynZ, comprising 287 amino acid residues, includes the FAE domain and a linker; FAEXynZ, comprising 266 amino acid residues, is the FAE domain without a linker; and FAE227XynZ, with 227 amino acid residues, is a truncated FAE domain.
In the present study, we show that UDs of XynY and XynZ have homology with a feruloyl esterase (FaeA) (D. L. Blum, X.-L. Li, H. Z. Chen, and L. G. Ljungdahl, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. K-153, 1999) from the anaerobic fungus Orpinomyces PC-2 (GenBank accession no. AF164351) and that these domains exhibit feruloyl esterase activity. Consequently, XynY and XynZ are bifunctional enzymes with feruloyl esterase and xylanase activities. The presence of feruloyl esterase in the cellulosome of C. thermocellum points toward an additional ability of this organelle to hydrolyze plant tissue.
(A preliminary report of this work was given at the Mie Bioforum in 1998 [5]).
MATERIALS AND METHODS
Bacterial strains, vectors, and culture media.
C. thermocellum JW20 was cultivated in prereduced liquid medium (33) at 60°C under an atmosphere of nitrogen. For isolation of cellulosomes and to obtain subfractions of C. thermocellum, 0.2% Avicel (a microcrystalline cellulose [0.2%, wt/vol], 2- to 20-μm particle size; Baker TLC) and 0.5% (wt/vol) cellobiose were used, respectively, as carbon sources. Escherichia coli strain BL21(DE3) (Stratagene, La Jolla, Calif.) and plasmid pRSET B (Invitrogen, La Jolla, Calif.) were used as the host strain and the vector for protein expression, respectively. Initial work was done with pRSET B, with which we obtained satisfactory results, but these were improved considerably using pET-21b (Novagen, Madison, Wis.). The work described uses these plasmids. Recombinant E. coli cells were selected for by growing in Luria-Bertani medium containing 100 μg of ampicillin per ml.
Amplification and cloning of sequences coding for different domains of XynY and XynZ.
Genomic DNA was isolated from C. thermocellum as previously described (24). To clone fragments of DNA corresponding to the UDs of XynY and XynZ and deletions of the UD of XynZ, PCR primers were designed (Table 1) and synthesized on an Applied Biosystems DNA synthesizer (PE Biosystems, Foster City, Calif.). To facilitate the insertion of DNA sequence into pET-21b or pRSET B, BamHI and HindIII sites were added to forward and reverse primers, respectively (Table 1). PCRs were carried out on a Perkin-Elmer 480 Thermocycler (Norwalk, Conn.) for 30 cycles, with each cycle at 95°C for 1 min, 48°C for 1 min, and 72°C for 3 min. PCR products and the plasmid were digested with BamHI and HindIII, purified with a Geneclean Spin Kit (BIO 101, Inc., Vista, Calif.), and ligated with T4 ligase. E. coli BL21(DE3) was transformed with the ligation mixture, and at least four colonies of each construct were picked for analyzing feruloyl esterase expression. The inserted sequences were sequenced to verify the lack of unwanted mutations.
TABLE 1.
Primer designs amplifying various regions of xynY and xynZ of C. thermocellum
| Name | Sequencea | Gene | Direction | Positionb |
|---|---|---|---|---|
| XYF1 | TAGGATCCCCTGTAGCAGAAAATCCTTC | xynY | Forward | 795–800 |
| XYR1 | GAGGAAGCTTTTACATGGAAGAAATATGGAAG | xynY | Reverse | 1071–1077 |
| XZF1 | TAGGATCCCTTGTCACAATAAGCAGTACA | xynZ | Forward | 20–26 |
| XZR1 | GAGGAAGCTTTTAGTTGTTGGCAACGCAATA | xynZ | Reverse | 242–247 |
| XZR2 | GAGGAAGCTTAGTTTCCATCCCTCGTCAA | xynZ | Reverse | 281–286 |
| XZR3 | GAGGAAGCTTAGTCATAATCTTCCGCTTC | xynZ | Reverse | 302–307 |
| XZR4 | GAGGAAGCTTAAACGCCAAAAGTGAACCAGTC | xynZ | Reverse | 414–421 |
Restriction sites BamHI and HindIII are underlined and double underlined, respectively.
Amino acid positions correspond to xylanase sequences in the data banks.
Isolation and analysis of cellulosomes and subfractions of C. thermocellum.
Cellulosomes produced by C. thermocellum were isolated from 10 liters of culture fluid after complete Avicel exhaustion by the affinity digestion method (38). They were further purified by gel filtration with a fast protein liquid chromatography system with a Superose 6 column (Pharmacia, Piscataway, N.J.). The buffer used was 50 mM Tris-HCl and 100 mM NaCl at a flow rate of 0.2 ml/min. Fractions of 0.5 ml each were collected and stored at 4°C for further analysis. To prepare subfractions, C. thermocellum was grown on 0.5% cellobiose in 200 ml of culture. Cells were recovered by centrifugation, resuspended in 50 mM Tris-HCl buffer (pH 7.5), and sonicated. The cultural medium was concentrated to 5 ml with a PM10 Diaflo ultrafiltration membrane (Amicon, Inc., Beverly, Mass.). To remove cellulosomes from the medium, 0.5 mg of Avicel was added and the suspension was stirred at 4°C for 4 h. Avicel with bound cellulosomes was recovered by centrifugation, and the cellulosomes were released from the Avicel by elution with distilled water (33). All fractions were tested for avicelase, xylanase, and feruloyl esterase activity.
Enzyme assays.
Unless otherwise noted, enzyme assays were performed at 60°C in 50 mM Na-citrate buffer, pH 6.0. One unit of enzyme activity was defined as the amount of enzyme that releases 1 μmol of product min−1, and specific activity was given in units per milligram of protein. Protein was determined by the method of Bradford (9). Feruloyl esterase activity was measured using a modified version of the assay described by Borneman et al. (7). The appropriately diluted protein sample (25 μl) was added to 400 μl of buffer containing 8 mM substrate. Samples were incubated at 60°C for 5 min, and the reaction was stopped by adding 25 μl of 20% formic acid. Release of ferulic acid was measured via high-performance liquid chromatography (HPLC) using a mobile phase of 10 mM Na formate (pH 3) and 30% (vol/vol) methanol. For routine assays, FAXX and FAX3 purified from wheat bran were used as substrates (7). Ethyl ferulate and ethyl-p-coumarate esters were gifts from D. E. Akin (U.S. Department of Agriculture, Athens, Ga.). The hydrolysis of these (10 mM) were determined similarly to that of FAXX, but the HPLC analyses were performed with 50% methanol. HPLC runs were done with a Hewlett-Packard 1100 Series instrument (Wilmington, Del.) equipped with an autosampler and diode array detector with a Hypersil octyldecyl silane (125 by 4 mm) column. Ferulic acid and p-coumaric acid were used as standards. To determine the amount of feruloyl and p-coumaroyl groups released from plant cell walls, wheat bran and Coastal Bermuda grass were ground in a Thomas Wiley mill (VWR Scientific Products, Atlanta, Ga.) to pass through a 250-μm screen. Plant samples of 10 mg each were incubated for 1 h in 400 μl of 50 mM Na-citrate buffer (pH 6.0) plus 25 μl of enzyme. After the addition of 25 μl of 20% formic acid to stop the reaction, the samples were centrifuged at 16,000 × g in a microcentrifuge and then assayed for ferulic and p-coumaric acid by HPLC.
Assays with p-nitrophenol substrates were performed in microtiter plate wells. Two hundred microliters of substrate at a concentration of 100 μM was preincubated in wells heated to 40°C. Enzyme (10 μl) was added to the reaction mixture, and the absorbance was followed continuously at a wavelength of 405 nm. p-Nitrophenol was used as a standard. Xylanase and cellulase activities were measured by determining the amount of reducing sugar released with dinitrosalicylate (37).
Enzyme purification.
Cultures of 1 liter of the recombinant E. coli containing pET-21b plus insert were grown in Luria broth containing 100 μg of ampicillin per ml until an optical density at 600 nm of 0.5 was reached and then grown an additional 4 to 6 h after induction with 1 mM isopropyl-β-d-thiogalactopyranoside, depending on the construct. Cells were harvested by centrifugation at 10,000 × g. They were resuspended at a concentration of 1 g per 3 ml of 50 mM Tris-HCl (pH 7.5) and lysed with a French press cell. Cell debris was removed by centrifugation at 100,000 × g. The cell extract was heat treated at 70°C for 30 min. Denatured protein was removed by centrifugation at 100,000 × g. The supernatant was concentrated to a volume of 2 ml with a Centricon 10 concentrator (Amicon, Inc., Beverly, Mass.) and then applied to a TSK 3000SW column (TosoHaas, Montgomeryville, Pa.), which was run with 50 mM Tris-HCl (pH 7.5) and 5% glycerol as solvent. The purified enzyme was stored at 4°C in the elution buffer and was stable for at least 1 month with minimal loss.
Enzyme stability experiments.
Purified enzyme at a concentration of 13 μg/ml in 50 mM Na-citrate (pH 6.0) was placed in a water bath at the appropriate temperature and incubated at intervals of 1 h. Enzyme aliquots (25 μl) were removed, and assays were performed in triplicate with FAX3 as a substrate as described above. FAE-CBDXynZ was tested at temperatures of 50, 60, and 70°C, while FAEXynZ was tested at 70, 80, and 90°C.
Other analytical procedures.
Enzyme purity was monitored with sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels which were run by the method of Laemmli (28). Proteins were stained with Coomassie blue. The isoelectric point of the protein was determined by running the enzyme on a precast Serva IEF gel (Novex, San Diego, Calif.). The gel was run at 12 W of constant power for 45 min.
RESULTS
Demonstration of feruloyl esterase activity in the cellulosome.
The initial indication of the presence of feruloyl esterase in the cellulosome of C. thermocellum was obtained from data bank search and sequence analysis using the Genetics Computer Group package (University of Wisconsin Biotechnology Center, Madison) and the VAX/VMS system of the Bioscience Computing Resource of The University of Georgia, Athens). This search showed that the catalytic domain of Orpinomyces strain PC-2 FaeA was over 30% identical to sequences coding for the UD of XynY and the UD of XynZ. (Fig. 1).
FIG. 1.
Alignment of sequence homologous to Orpinomyces PC-2 FaeA. Sequences are FaeA_Orpin (accession no. AF164351) (5); XynZ_Clotm, xylanase Z from C. thermocellum (accession no. M22624) (22); XynY_Clotm, xylanase Y from C. thermocellum (accession no. P51584) (20); Xyn1_Rumin, xylanase 1 from a Ruminococcus sp. (accession no. S58235) (1); YIEL_Ecoli, gene encoding an unknown 44-kDa protein from E. coli (accession no. P31471) (10); and DPP_Aspfu, dipeptidyl peptidase from A. fumigatus (accession no. L48074) (3).
To confirm the presence of feruloyl esterase in C. thermocellum and to localize this activity, medium and cell extracts of C. thermocellum grown on cellobiose were obtained. As shown in Table 2, feruloyl esterase, avicelase (cellulase activity measured with Avicel as a substrate), and xylanase activities are mainly extracellular. To verify if feruloyl esterase activity is attributed to the cellulosomes, the latter were removed from the culture medium by adsorption onto Avicel. After removal of the cellulosomes, only 0.82% of feruloyl esterase, 9.8% of avicelase, and 16.9% of xylanase activities remained in the culture medium. Almost all of the feruloyl esterase (98.7%), avicelase (80.5%), and xylanase (73.3%) activities were recovered in the cellulosomal fraction released from the Avicel with distilled water (Table 2). Thus, the majority of feruloyl esterase activity seemed to belong to the cellulosome. Distribution of other activities between cellulosome and cultural medium treated with Avicel is in accordance with the presence of some noncellulosomal (free) cellulases and xylanases in the C. thermocellum culture medium. Finally, cellulosomes were obtained from cultures grown on Avicel. They were purified by the affinity digestion method (38) and gel filtration chromatography with a Superose 6 column (11). The cellulosomes with a mass over 2.0 million Da contained the majority of the feruloyl esterase activity. No activity was found in fractions with protein of a molecular mass less than 200 kDa. These data strongly suggest that feruloyl esterase activity resides in the cellulosome.
TABLE 2.
Distribution of proteins and hydrolytic activity in cells and culture medium of C. thermocellum grown on 0.5% cellobiose
| Fraction | Protein
|
Feruloyl esterase
|
Avicelase
|
Xylanase
|
||||
|---|---|---|---|---|---|---|---|---|
| mg | % | U | % | U | % | U | % | |
| Cell extracta | 0.81 | 39.7 | 0.005 | 2.1 | 0.001 | 2.4 | 0.49 | 5.3 |
| Culture mediuma | 1.23 | 60.3 | 0.238 | 97.9 | 0.04 | 97.6 | 8.72 | 94.7 |
| Culture medium after Avicel treatment | 0.98 | 48.0 | 0.002 | 0.8 | 0.004 | 9.8 | 1.56 | 16.9 |
| Purified cellulosome | 0.176 | 8.6 | 0.24 | 98.7 | 0.033 | 80.5 | 6.75 | 73.3 |
The sum of the activity in the cell extract and cultural medium was considered to be 100%.
Expression of the UDs of XynY and XynZ in E. coli.
Nucleotides corresponding to regions of DNA encoding amino acids in XynZ (accession no. M22624) from residues 20 to 421 and in XynY (accession no. X83269) from residues 795 to 1,077 were overexpressed in E. coli using the pET and pRSET systems, respectively. The XynZ sequence referred to as FAE-CBDXynZ incorporates the family VI CBD (Fig. 2), while the XynY protein designated FAEXynY contains only the catalytic domain (Fig. 2). The cell extracts containing the expressed proteins each hydrolyzed FAXX with release of ferulic acid, suggesting that these proteins are feruloyl esterases. E. coli cells lacking the plasmids or containing plasmids without C. thermocellum DNA inserts did not hydrolyze FAXX. The expressed FAEXynY and FAE-CBDXynZ had molecular masses of 31 and 45 kDa, respectively, consistent with the sequence data. Since these proteins had similar sequences and function and the XynZ protein had higher expression levels than the XynY protein (data not shown), we focused on the XynZ protein in subsequent experiments.
Deletion analysis of FAE-CBDXynZ.
Constructs were made which corresponded to proteins with amino acids from the original XynZ sequence of residues 20 to 307 (FAE287XynZ), 20 to 286 (FAEXynZ) and 20 to 247 (FAE227XynZ) (Fig. 2). FAE287XynZ is missing the CBD but contains the proline-rich linker which separates the CBD from the feruloyl esterase domain, while FAEXynZ does not contain this linker. When these constructs were expressed in E. coli in the same manner as FAE-CBDXynZ, they both exhibited feruloyl esterase activity, whereas FAE227XynZ was expressed but inactive. The data suggest that neither the CBD nor the linker is necessary for activity, but that C-terminal amino acids in the sequence from residues 247 to 286 of the FAE domain are necessary for activity.
Purification and characterization of the FAE-CBDXynZ and FAEXynZ.
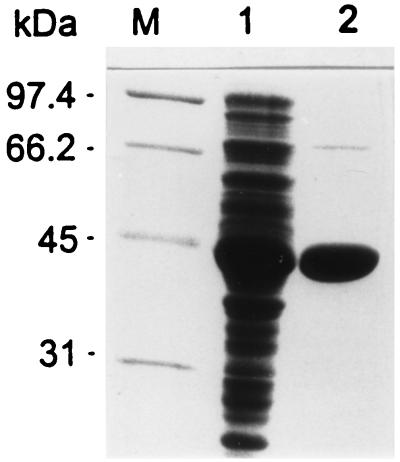
The FAE-CBDXynZ polypeptide was purified from E. coli cell extract by a single step of heat treatment at 70°C for 30 min. Over 200 mg of homogeneous FAE-CBDXnyZ (Fig. 3) was obtained from 2.5 g of crude proteins (Table 3). There was no evidence for aggregation of the esterase produced in E. coli or the presence of inclusion bodies.
FIG. 3.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of FAE-CBDXynZ overexpressed in E. coli. Lane M, low-range protein standards (Bio-Rad Laboratories, Richmond, Calif.), including phosphorylase B (97.4 kDa), serum albumin (66.2), ovalbumin (45 kDa), and carbonic anhydrase (31 kDa); lane 1, E. coli cell extract; lane 2, heat-treated cell extract (Table 3).
TABLE 3.
Purification of the recombinant FAE-CBDXynZ and FAEXynZ from E. coli
| Fraction and procedure | Protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| FAE-CBDXynZ cell extract | 2,597 | 3,250 | 1.3 | 100 |
| Heat treatment | 220 | 2,830 | 12.9 | 86 |
| FAEXynZ cell extract | 533 | 1,520 | 2.9 | 100 |
| Heat treatment | 213 | 1,630 | 7.7 | 107 |
| Gel filtration | 31 | 823 | 26.6 | 54 |
The purified protein had a Vmax of 12.5 μmol of ferulic acid released min−1 mg−1 and a Km of 5 mM with FAX3 as substrate. The enzyme had the highest activity towards FAXX but was almost as active toward FAX3 (Table 4). The protein was able to release low levels of ferulic acid from ethyl ferulic acid, ground wheat bran, and Coastal Bermuda grass, and p-coumaric acid from PAX3 and ethyl-p-coumarate. The protein lacked activity toward carboxymethyl cellulose, Avicel, p-nitrophenyl (pNP)-arabinopyranoside, pNP-glucopyranoside, pNP-xylopyranoside, and pNP-acetate. Isoelectric focusing indicated a pI of 5.8.
TABLE 4.
Substrate specificity of recombinant feruloyl esterase FAE-CBDXynZ
| Substrate | U/mga |
|---|---|
| FAXX | 12.5 |
| FAX3 | 11.8 |
| PAX3 | 1.4 |
| Ethyl ferulate | 0.066 |
| Ethyl-p-coumarate | 0.022 |
| Wheat bran | 0.06 |
| Coastal Bermuda grass | 0.1 |
The number of units per milligram is the amount of ferulic or p-coumaric acid formed per minute per milligram of enzyme. See Materials and Methods for details.
The FAEXynZ was also expressed and purified to homogeneity. The purification is shown in Table 3. The protein was expressed in a manner similar to that of FAE-CBDXynZ. In contrast to FAE-CBDXynZ, FAEXynZ obtained following heat treatment was not pure. An additional step involving gel filtration with a TSK 3000SW column resulted in a pure enzyme with a Vmax of 28.2 U mg−1 and a Km of 10.5 mM with FAX3 as substrate. FAEXynZ was inhibited by ferulic acid but not by xylose or arabinose. The FAEXynZ was most active between 50 and 60°C and had a high level of activity between pH 4 and 7. It and FAE-CBDXynZ were stable at 70°C for 6 h. At 80°C, FAEXynZ lost about 50% of the activity within 3 h and all activity after 1 h at 90°C. FAE-CBDXynZ bound weakly to acid-swollen cellulose, while the other constructs missing the CBD did not bind to acid-swollen cellulose.
DISCUSSION
The cellulosome of C. thermocellum is a remarkable extracellular organelle of about 3,000 kDa that also exists as a polycellulosome of a mass approaching 100,000 kDa (12). The first assessment of the number of different subunits in the cellulosome indicated the presence of 14 subunits (30). Later, Kohring et al. (26) studying C. thermocellum strain JW20 observed 26 subunits. At least 19 cellulosomal polypeptides have been cloned and sequenced (2). They include CipA and 18 catalytic subunits with various glycosyl hydrolase activities. It is now clear that the cellulosome is capable of hydrolyzing not only cellulose and the backbone of hemicelluloses but also substituents of xylan. Recently Hayashi et al. (23) sequenced two homologous xylanases, XynA and XynB, from C. thermocellum and demonstrated that they contain a NodB domain in addition to a family II xylanase domain. A NodB domain of a multidomain xylanase from Cellulomonas fimi deacetylates acetyl xylan (31). A Ruminococcus flavefaciens xylanase has acetyl xylan esterase activity (25). These xylanases clearly are bifunctional enzymes. Conceivably, the xylanases and the acetyl xylan esterases work together in the hydrolysis of the xylan. A synergistic effect between a separate xylanase and an acetyl xylan esterase has been demonstrated (6).
The situation may be similar regarding XynY and XynZ. As shown in this paper, both contain feruloyl esterase and xylanase domains. The xylanase domain of XynZ has been well studied and crystallized, and its three-dimensional structure has been solved (16, 42). The feruloyl esterase and xylanase may well work together in the two bifunctional enzymes. The feruloyl esterase domain and the xylanase domain may form a dumbbell-like structure, and arabino xylan may be hydrolyzed in a multicutting event involving the xylose chain as well as the ester linkage between the arabinosyl and feruloyl moieties. As with xylanase and acetyl xylan esterase, a synergistic effect has been shown between a separate xylanase and a feruloyl esterase (8). It has been proposed also that feruloyl esterases are responsible for the hydrolysis of bonds between lignin and hemicelluloses (40, 42). The anaerobic fungus Neocallimastix patriciarum solubilizes lignin (36) and a xylanolytic Butyrivibrio sp. has phenolic esterase activity (35).
Multifunctional enzymes—one gene with more than one catalytic domain—were discussed several years ago (43). Such enzymes with activities acting on the same substrate but different bonds may have the advantage that the substrate does not have to be released from the enzyme when undergoing two or more enzymatic reactions. Several bifunctional enzymes in addition to xylanase-acetyl xylan esterase and xylanase-feruloyl esterase are known. Also, trifunctional enzymes exist with separate catalytic domains in a single polypeptide. Examples include peroxidase-lipoxygenase from the sea whip coral Plexaura homomalla (27), xylanase-β-(1,3-1,4)-glucanase from R. flavefaciens (19), xylanase with two catalytic domains from N. patriciarum (21), methenyltetrahydrofolate cyclohydrolase-methylenetetrahydrofolate dehydrogenase from several clostridia (34), and the trifunctional C1-tetrahydrofolate synthase from yeast and liver (41).
The domains of xylanase and feruloyl esterase of XynZ and XynY are well separated in the polypeptides (Fig. 2). Separate expression of the domains in E. coli yielded active enzymes. As shown with the feruloyl esterase domain of XynZ, it is also active when the CBD and the linker regions are removed. Catalytic domains that are active without adjacent domains, such as CBD or dockerins, have been observed with many of the polypeptides from the C. thermocellum cellulosome. However, when amino acids 247 to 286 were removed from the C-terminal end of the feruloyl esterase domain of XynZ, the enzyme was inactive, indicating the requirement of this sequence for activity. An alignment of the feruloyl esterase domains of XynZ, XynY, and FaeA of Orpinomyces (Fig. 1) shows that these domains have substantial homology. This homology was not apparent with feruloyl esterases of Aspergillus niger and Aspergillus tubingensis (15), CinA and CinB from Butyrivibrio fibrisolvens (13, 14), and XylD from Pseudomonas fluorescens subsp. cellulosa (18). The sequence analysis implies that feruloyl esterases may be classified in families similar to cellulases and other glycohydrolyses. The Orpinomyces FaeA and the feruloyl esterase domains of XynZ and XynY have homology to a polypeptide of unknown function in E. coli (10) and to an unknown domain of the carboxy-terminal region of a xylanase from a Ruminococcus sp. (1). Feruloyl esterase activity is present in Ruminococcus spp. (40), whereas no feruloyl esterase activity has been demonstrated in E. coli. The E. coli gene may encode a dipeptidase instead, since homology exits between a dipeptidase of Aspergillus fumigatus and feruloyl esterase (Fig. 1) (3).
Removal of amino acids 247 to 286 (FAE227XynZ) resulted in an inactive protein. This was somewhat unexpected, since this region of the feruloyl xylan esterase has the least homology between the different enzymes (Fig. 1). It is possible that this region is important for the stability and configuration of the enzymes, but a short sequence of the Orpinomyces FaeA (PGGTHDFPVW; amino acids 437 to 446) has high homology to a C. thermocellum XynZ sequence (QGGGHDFNVW; amino acids 256 to 265). Similar sequences are also found in the other enzymes shown in Fig. 1. These sequence may be of importance for the catalytic activity of the feruloyl esterases.
ACKNOWLEDGMENTS
This work was funded by grant DE-FG02-93ER20127 from the Department of Energy (L.G.L.) and by Aureozyme, Inc. (X.-L.L.) and Georgia Research Alliance (X.-L.L.)
REFERENCES
- 1.Arakaki, C., R. Breves, A. Czihal, H. Baeumlein, B. Carillo, and J. Hofemeister. 1997. EMBL data library accession no. S58235.
- 2.Bayer E A, Shimon L J W, Shoham Y, Lamed R. Cellulosomes—structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 3.Beauvais A, Monod M, Debeaupui J P, Diaquin M, Kobayashi H, Latge J P. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J Biol Chem. 1997;272:6238–6244. doi: 10.1074/jbc.272.10.6238. [DOI] [PubMed] [Google Scholar]
- 4.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 5.Blum D L, Kataeva I, Li X-L, Ljungdahl L G. Phenolic acid esterase activity of Clostridium thermocellum cellulosome is attributed to previously unknown domains of XynY and XynZ. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Genetics, Biochemistry, and Ecology of Cellulose Degradation. Proceedings of Mie Bioforum 98. Tokyo, Japan: UNI Publishers Co.; 1998. p. 478. [Google Scholar]
- 6.Blum D L, Li X-L, Chen H, Ljungdahl L G. Characterization of an acetyl xylan esterase from the anaerobic fungus Orpinomyces sp. strain PC-2. Appl Environ Microbiol. 1999;65:3990–3995. doi: 10.1128/aem.65.9.3990-3995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman W S, Hartley R D, Himmelsbach D S, Ljungdahl L G. Assay for trans-p-coumaroyl esterase using a specific substrate from plant cell walls. Anal Biochem. 1990;190:129–133. doi: 10.1016/0003-2697(90)90145-y. [DOI] [PubMed] [Google Scholar]
- 8.Borneman W S, Ljungdahl L G, Hartley R D, Akin D E. Feruloyl and p-coumaroyl esterases from the anaerobic fungus Neocallimastix MC-2: properties and functions in plant cell wall degradation. In: Coughlan M P, Hazlewood G, editors. Hemicellulose and hemicellulases. Cambridge, United Kingdom: Portland Press; 1993. pp. 85–102. [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Burland V D, Plunkett III G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 11.Choi S K, Ljungdahl L G. Dissociation of the cellulosome of Clostridium thermocellum in the presence of ethylenediaminetetraacetic acid occurs with the formation of truncated polypeptides. Biochemistry. 1996;35:4897–4905. doi: 10.1021/bi9524629. [DOI] [PubMed] [Google Scholar]
- 12.Coughlan M P, Hon-nami K, Hon-nami H, Ljungdahl L G, Paulin J J, Rigsby W E. The cellulolytic enzyme complex of Clostridium thermocellum is very large. Biochem Biophys Res Commun. 1985;130:904–909. doi: 10.1016/0006-291x(85)90502-9. [DOI] [PubMed] [Google Scholar]
- 13.Dalrymple B D, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl esterase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 14.Dalrymple B D, Swadling Y, Cybinski D H, Xue G-P. Cloning of a gene encoding cinnamoyl ester hydrolase from the ruminal bacterium Butyrivibrio fibrisolvens E14 by a novel method. FEMS Microbiol Lett. 1996;143:115–120. doi: 10.1111/j.1574-6968.1996.tb08469.x. [DOI] [PubMed] [Google Scholar]
- 15.de Vries R P, Michelsen B, Poulsen C H, Kroon P A, van den Heuvel R H H, Faulds C B, Williamson G, van den Hombergh J P T W, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez R, Souchon H, Spinelli S, Dauter Z, Wilson K S, Chauvaux S, Béguin P, Alzari P M. A common protein fold and similar active site in two distinct families of β-glycanases. Nat Struct Biol. 1995;2:569–576. doi: 10.1038/nsb0795-569. [DOI] [PubMed] [Google Scholar]
- 17.Fanutti C, Ponyi T, Black G W, Hazlewood G P, Gilbert H J. The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic rumen fungi functions as a docking domain. J Biol Chem. 1995;270:29314–29322. doi: 10.1074/jbc.270.49.29314. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira L M A, Wood T M, Williamson G, Faulds C, Hazlewood G P, Black G W, Gilbert H J. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294:349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint H J, Martin J, McPherson C A, Daniel A S, Zhang J-X. A bifunctional enzyme, with separate xylanase and β-(1,3-1,4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J Bacteriol. 1993;175:2943–2951. doi: 10.1128/jb.175.10.2943-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontes C M G A, Hazlewood G P, Morag E, Hall J, Hirst B H, Gilbert H J. Evidence for a general role for noncatalytic thermostabilizing domains in xylananses from thermophilic bacteria. Biochem J. 1995;307:151–158. doi: 10.1042/bj3070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert H J, Hazlewood G P, Laurie J I, Orpin C G, Xue G P. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992;6:2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 22.Grépinet O, Chebrou M-C, Béguin P. Purification of Clostridium thermocellum xylanase Z expressed in Escherichia coli and identification of the corresponding product in the culture medium of C. thermocellum. J Bacteriol. 1988;170:4576–4581. doi: 10.1128/jb.170.10.4576-4581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi H, Takehara M, Hattori T, Kimura T, Karita S, Sakka K, Ohmiya K. Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum. Appl Microbiol Biotechnol. 1999;51:348–357. doi: 10.1007/s002530051401. [DOI] [PubMed] [Google Scholar]
- 24.Kataeva I, Li X-L, Chen H, Choi S-K, Ljungdahl L G. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J Bacteriol. 1999;181:5288–5295. doi: 10.1128/jb.181.17.5288-5295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirby J, Aurilia V, McCrae S I, Martin J C, Flint H J. Plant cell wall degrading enzyme complexes from the cellulolytic rumen bacterium Ruminococcus flavefaciens. Biochem Soc Trans. 1998;26:S169. doi: 10.1042/bst026s169. [DOI] [PubMed] [Google Scholar]
- 26.Kohring S, Wiegel J, Mayer F. Subunit composition and glycosidic activities of the cellulase complex from Clostridium thermocellum JW20. Appl Environ Microbiol. 1990;56:3798–3804. doi: 10.1128/aem.56.12.3798-3804.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koljak R, Boutaud O, Shieh B-H, Samel N, Brash A R. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lamed R, Setter E, Bayer E A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156:828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamed R, Setter E, Kenig R, Bayer E A. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng Symp. 1983;13:163–181. [Google Scholar]
- 31.Laurie J I, Clarke J H, Ciruela A, Faulds C B, Williamson G, Gilbert H J, Rixon J E, Millward-Sadler J, Hazlewood G P. The NodB domain of a multidomain xylanase from Cellulomonas fimi deacetylates acetylxylan. FEMS Lett. 1997;148:261–264. [Google Scholar]
- 32.Li X-L, Chen H, Ljungdahl L G. Two cellulases, CelA and CelC, from the polycentric anaerobic fungus Orpinomyces strain PC-2 contain N-terminal docking domains for a cellulase-hemicellulase complex. Appl Environ Microbiol. 1997;63:4721–4728. doi: 10.1128/aem.63.12.4721-4728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ljungdahl L G, Coughlan M P, Mayer F, Mori Y, Hon-nami K. Macrocellulase complexes and yellow affinity substance from Clostridium thermocellum. Methods Enzymol. 1988;160:483–500. [Google Scholar]
- 34.Ljungdahl L G, O'Brien W E, Moore M R, Liu M-T. Methylene-tetrahydrofolate dehydrogenase from Clostridium formicoaceticum and methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase (combined) from Clostridium thermoaceticum. Methods Enzymol. 1980;66:599–609. doi: 10.1016/0076-6879(80)66513-6. [DOI] [PubMed] [Google Scholar]
- 35.McSweeney C S, Dulieu A, Bunch R. Butyrivibrio spp. and other xylanolytic microorganisms from the rumen have cinnamoyl esterase activity. Anaerobe. 1998;4:57–65. doi: 10.1006/anae.1997.0130. [DOI] [PubMed] [Google Scholar]
- 36.McSweeney C S, Dulieu A, Katayama Y, Lowry J B. Solubilization of lignin by the ruminal anaerobic fungus Neocallimastix patriciarum. Appl Environ Microbiol. 1994;60:2985–2989. doi: 10.1128/aem.60.8.2985-2989.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller G L. Use of dinitrosaliclic acid reagent for determination of reducing sugars. Anal Chem. 1959;31:127–132. [Google Scholar]
- 38.Morag E, Bayer E A, Lamed R. Affinity digestion for the near-total recovery of purified cellulosome from Clostridium thermocellum. Enzyme Microb Technol. 1992;14:289–292. [Google Scholar]
- 39.Ralph J, Grabber J H, Hatfield R D. Lignin-ferulate cross-links in grasses: active incorporation of ferulate polysaccharides esters into ryegrass lignins. Carbohydr Res. 1995;275:167–178. [Google Scholar]
- 40.Selinger L B, Forsberg C W, Cheng K-J. The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe. 1996;2:263–284. doi: 10.1006/anae.1996.0036. [DOI] [PubMed] [Google Scholar]
- 41.Shannon K W, Rabinowitz J C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem. 1988;263:7717–7725. [PubMed] [Google Scholar]
- 42.Souchon H, Spinelli S, Béguin P, Alzari P M. Crystallization and preliminary diffraction analysis of the catalytic domain of xylanase Z from Clostridium thermocellum. J Mol Biol. 1994;235:1348–1350. doi: 10.1006/jmbi.1994.1089. [DOI] [PubMed] [Google Scholar]
- 43.Stark G R. Multifunctional proteins: one gene—more than one enzyme. Trends Biochem Sci. 1977;2:64–66. [Google Scholar]
- 44.Williamson G, Kroon P A, Faulds C B. Hairy plant polysaccharides: a close shave with microbial esterases. Microbiology. 1998;144:2011–2023. doi: 10.1099/00221287-144-8-2011. [DOI] [PubMed] [Google Scholar]