Abstract
CCCTC‐binding factor (CTCF) is an eleven zinc finger (ZF), multivalent transcriptional regulator, that recognizes numerous motifs thanks to the deployment of distinct combinations of its ZFs. The great majority of the ~50,000 genomic locations bound by the CTCF protein in a given cell type is intergenic, and a fraction of these sites overlaps with transcriptional enhancers. Furthermore, a proportion of the regions bound by CTCF intersect genes and promoters. This suggests multiple ways in which CTCF may impact gene expression. At promoters, CTCF can directly affect transcription. At more distal sites, CTCF may orchestrate interactions between regulatory elements and help separate eu‐ and heterochromatic areas in the genome, exerting a chromatin barrier function. In this review, we outline how CTCF contributes to the regulation of the three‐dimensional structure of chromatin and the formation of chromatin domains. We discuss how CTCF binding and architectural functions are regulated. We examine the literature implicating CTCF in controlling gene expression in development and disease both by acting as an insulator and a factor facilitating regulatory elements to efficiently interact with each other in the nuclear space.
Keywords: enhancer, chromatin structure, CTCF, insulator, regulation of gene expression
Subject Categories: Chromatin, Transcription & Genomics
This review discusses how CTCF shapes the 3D chromatin structure and chromatin domains, how CTCF binding and architectural functions are regulated, and how CTCF controls gene expression in development and disease by acting as an insulator and a facilitator for regulatory element interaction in the nuclear space.
Glossary
- 3C
chromatin conformation capture
- 3D
three dimensional
- AID
auxin inducible degron
- CBS
CTCF Bound Site, genomic regions with ChIP‐seq signal indicating CTCF binding
- CRE
cis regulatory element
- CT
chromosome territory
- CTCF
CCCTC‐binding factor
- ES cells
embryonic stem cells
- FISH
fluorescence in situ hybridisation
- FRAP
fluorescence recovery after photobleaching
- G‐CIMP
CpG island methylator phenotype
- GIST
gastrointestinal stromal tumors
- Hi‐C
genome‐wide chromosome conformation capture
- ICR
imprinting control region
- IDH
isocitrate dehydrogenase
- IS
insulation score
- kb
kilo base pair
- Micro‐C
micrococcal nuclease‐assisted chromatin conformation capture
- PEI
promoter‐enhancer interactions
- PolII
RNA polymerase II
- RA
retinoic acid
- RBR
RNA‐binding region
- RT
residence time
- SDH
succinate dehydrogenase
- TAD
topologically associating domain
- TCGA
The Cancer Genome Atlas
- TF
transcription factor
- TSS
transcription start site
Introduction
Cell type‐specific gene expression is ensured by a concerted action of DNA cis regulatory elements (CRE) including promoters, enhancers, silencers and insulators. CREs bind transcription factors (TFs) thereby controlling the production of messenger RNAs. Enhancer activity is essential for context‐specific gene expression. Enhancer elements are frequently located at great genomic distances from their cognate promoters and one of the fundamental questions in the field is how, despite pronounced genomic separation, enhancers activate genes with specificity. Several lines of evidence suggest that the way the chromatin fibre is organized in the cell nucleus contributes to ensuring correct promoter–enhancer dialogues. The three‐dimensional organization of chromatin in the cell nucleus is non‐random (Misteli, 2020). Chromosomes occupy distinct territories (CT) and the radial position of a chromosome in the cell nucleus is related to its overall activity. Hi‐C, a genome‐wide chromosome conformation capture technology, allows to look deeply into the organization of the genome highlighting spatial segregation of CTs into A and B compartments that are grossly reminiscent of eu‐ and heterochromatin, respectively (Lieberman‐Aiden et al, 2009; Kalhor et al, 2011; Rao et al, 2014). At genomic distances within a mega base range, which typically separate promoters and enhancers, Hi‐C has revealed that chromatin is arranged into domains of strong self‐contact called topologically associating domains (TADs, Fig 1A and B; Nora et al, 2012; Dixon et al, 2012; Sexton et al, 2012) or contact domains (Rao et al, 2014). TADs are intricate in their structures and frequently feature smaller domains referred to as sub‐TADs. High‐resolution fluorescence in situ hybridization (FISH), which allows to visualize chromatin structure in 3D space at high resolution, confirmed spatial partitioning of genomes into domains of preferred self‐contact, which correspond to TADs (Wang et al, 2016; Bintu et al, 2018; Miron et al, 2020). FISH revealed that the positions of TAD boundaries vary substantially between cells (Bintu et al, 2018). Nonetheless, when averaged across hundreds of alleles, the positions of TAD boundaries inferred from FISH are congruent with the coordinates derived from Hi‐C (Bintu et al, 2018; Barth et al, 2020). Hence, TAD correspond to individual domains of chromatin organization and Hi‐C can be used to map them.
Figure 1. CTCF and cohesins build chromatin architecture.
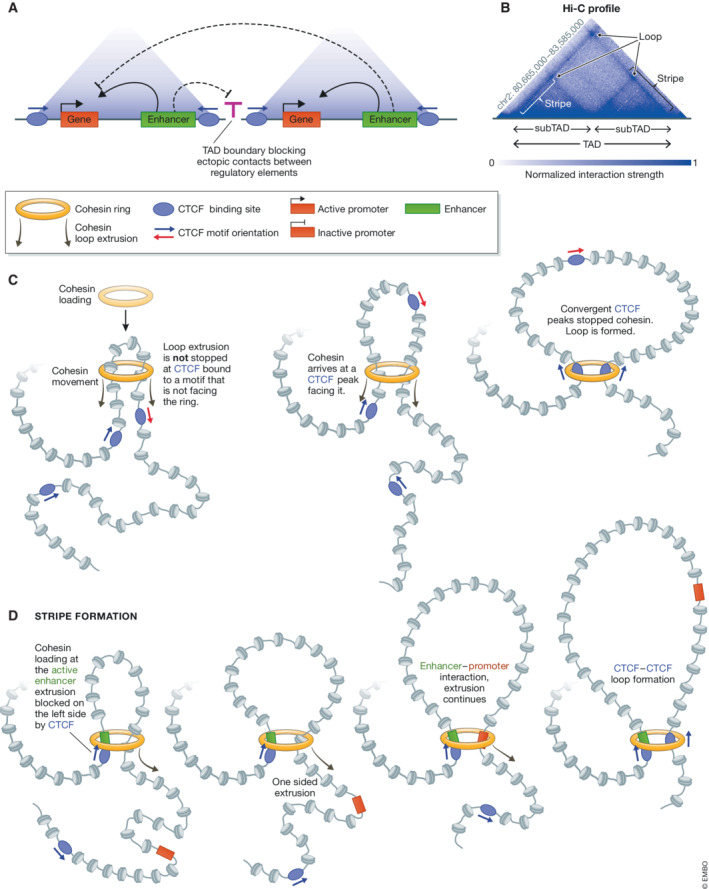
(A) Model of Topologically Associating Domains (TAD). TADs are regions of strong self‐contact. Promoter–enhancer interactions inside the domains are favoured while contacts with promoters and enhancers in adjacent domains are restrained. This is believed to help establish a functional organization of the genome. (B) Hi‐C profile illustrating TAD organization at an example locus in Neural Progenitor cells (data from Bonev et al, 2017). Increasing colour strength denotes enhanced interaction frequency. This in turn, can be interpreted as increased physical proximity in the three‐dimensional space of the cell nucleus. Triangles of Hi‐C signal reveal domains of enhanced interaction frequencies (TADs). Dots in the matrix (corner peaks) correspond to loops and reveal interactions between relatively short genomic intervals (here sub‐TAD boundaries). At some loci, TAD boundaries interact heavily with the entire TAD, which manifests itself as thin stripes of increased interaction frequency. (C) Loop extrusion model. Upon loading, cohesins (yellow ring) start translocating on chromatin (arrows) and their movement is accompanied by extrusion of an ever‐growing loop. Cohesins pass CTCF proteins bound to a motif which does not face them. Loop extrusion stops when cohesins encounter CTCF bound to a motif that is facing them (thick black arrow). (D) Model explaining the formation of architectural stripes. At genomic locations where cohesin loading occurs in the proximity of CTCF‐binding sites, including at active enhancers (green rectangle), CTCF bound to a motif oriented in a forward direction (en face) with respect to the loaded cohesin blocks loop extrusion immediately after loading. Loop extrusion proceeds fuelled by cohesin activity on the other side of the complex and allows the elements in the entire domain including promoters (red rectangle) to be “presented” to the fixed anchor overlapping the active enhancer (green rectangle). Depicted here is a single cohesin ring, it is unclear whether one or two cohesin rings extrude loops.
The observations that: (i) cognate and co‐regulated enhancer–promoter pairs tend to reside in the same TAD (Shen et al, 2012; de Laat & Duboule, 2013; Symmons et al, 2014), (ii) disruption of TAD boundaries can result in aberrant gene expression of loci in the merged domains (Guo et al, 2015; Lupiáñez et al, 2015; Franke et al, 2016), (iii) genomic intervals under the influence of a regulatory element largely coincide with TADs, as determined by serial insertions of a transcriptional sensor (Akhtar et al, 2013; Andrey et al, 2017; Despang et al, 2019) or of a well‐described promoter–enhancer pair (Zuin et al, 2022), and that (iv) co‐regulated enhancers and promoters are located in the same domain (Arner et al, 2015), led to the view that through structuring the genome TADs constitute functional units of genome organization (Akhtar et al, 2013; de Laat & Duboule, 2013; Kieffer‐Kwon et al, 2013; Symmons et al, 2014, 2016; Lupiáñez et al, 2015; Franke et al, 2016; Despang et al, 2019). These data together, echoed previous observations that chromatin is organized into functional units in metazoans as was initially appreciated in the fruit fly Drosophila melanogaster (Kellum & Schedl, 1991).
In vertebrates, the first chromatin boundaries limiting the action of regulatory elements were discovered at the chicken beta‐globin locus, where these elements either block aberrant gene silencing imposed by proximal heterochromatin (barrier function), or limit enhancer activity (insulator function; Recillas‐Targa et al, 1999; Chung et al, 1993). Likewise, in the mouse, insulators can shield promoters from being activated by an unrelated enhancer as in the case of the α/δ T‐cell receptor (Zhong & Krangel, 1997) and globin loci (Hanssen et al, 2017). Furthermore, insulators orchestrate allele‐specific expression as exemplified at the IGF2/H19‐imprinted gene locus. Deletion of an insulator can lead to inappropriate gene expression and morphological defects in metazoans (Hagstrom et al, 1996; Zhou et al, 1996; Zhou & Levine, 1999). In essence, the organization of chromatin into functional domains helps to maintain a proper DNA cis‐regulatory element (CRE) dialogue in the cell.
CCCTC‐binding factor (CTCF) is a conserved transcriptional regulator composed of 11 central zinc‐finger domains (ZFs) and peripheral, unstructured N‐ and C termini. CTCF binds a relatively long and complex motif which can be present in the DNA in a forward or reverse orientation. Hence, the motifs of two CTCF bound sites (CBS) can either be in tandem (the same direction), divergent or convergent (facing each other) orientation. As we will see below, the orientation of the motif within a CTCF peak with respect to other genomic features can have profound consequences on chromatin topology and activity (Fig 1).
CTCF was initially uncovered as a protein binding to the chicken Myc promoter, where it associates with a CCCTC‐sequence 180–230 bp upstream of the transcription start site (TSS; Lobanenkov et al, 1990; Klenova et al, 1993). Around that time, Rainer Renkawitz et al described Negative Protein 1 (NeP1), a transcriptional regulator cooperating with nuclear receptors in regulating the chicken lysozyme gene (Baniahmad et al, 1990). Later, the authors uncovered that NeP1 is identical to CTCF (Burcin et al, 1997). The subsequent discoveries that vertebrate insulators depend on CTCF (Bell et al, 1999; Recillas‐Targa et al, 2002; Cuddapah et al, 2009) and that CTCF contributes to the regulation of the CRE dialogue (Splinter et al, 2006; Majumder et al, 2008) genuinely transformed the research in the field of transcriptional regulation.
In this review, we outline how CTCF contributes to chromatin architecture. We discuss the function of CTCF as an insulator and recapitulate how CTCF‐bound regions may impact gene expression by integrating and shaping the locus‐specific regulatory landscape in the cell. We summarize recent findings linking CTCF to cell differentiation and disease with a special focus on cancer and neurological disorders.
CTCF and the cohesin complex build chromatin domains
In mammals, TAD boundaries are enriched in CBS (Dixon et al, 2012; Nora et al, 2012), which is consistent with the insulator role of CTCF (Bell et al, 1999; Recillas‐Targa et al, 2002). Cohesin complexes, composed of structural maintenance of chromosomes 1 and 3 and Rad21 (kleisin) and associated factors STAG1/2 and Pds5a/b, form TADs in an energy‐dependent fashion (Gassler et al, 2017; Haarhuis et al, 2017; Rao et al, 2017; Schwarzer et al, 2017; Wutz et al, 2017; Vian et al, 2018). CTCF interacts with cohesins (Rubio et al, 2008; Uuskula‐Reimand et al, 2016; Li et al, 2020), the two factors frequently co‐occupy genomic sites (Parelho et al, 2008; Rubio et al, 2008; Stedman et al, 2008; Wendt et al, 2008), and both CTCF and cohesins are required for TAD formation (Sofueva et al, 2013; Zuin et al, 2014; Nora et al, 2017; Wutz et al, 2017; preprint: Hsieh et al, 2021). Three essential features of TAD structures elucidated our understanding of the mechanisms driving domain and loop formation. First, CTCF‐bound motifs at TAD boundaries are directed primarily toward the interior of the TAD (that is in convergent orientation with respect to the interior of the TAD; de Wit et al, 2015; Vietri Rudan et al, 2015; Rao et al, 2014; Fig 1A). Second, at numerous loci, the two CTCF‐bound domain borders come together to form a loop (Fig 1B), connecting two CBS with convergent motifs (Fig 1A–D; Tang et al, 2015; Rao et al, 2014). Third, the fact that a relatively small protein (CTCF is ~5 nM in diameter) dictates formation of a large chromatin structure (TADs and loops are several hundreds of nanometres wide) in a way that is dependent on the orientation of its motif, collectively hinted at a one‐dimensional (1D) loop extrusion as the most likely mechanism underlying TAD formation (Nasmyth, 2001; Alipour & Marko, 2012; Dekker & Mirny, 2016). Computational simulations and experimental assessment of chromatin folding in cells with genetically engineered perturbations of CTCF motifs supported the loop extrusion hypothesis (Sanborn et al, 2015; Fudenberg et al, 2015; Guo et al, 2015; Fig 1C and D). In the loop extrusion model, upon loading, cohesins slide along the chromatin fibre and extrude a loop. This activity is stopped when they encounter CTCF that is bound to a motif that faces them. Recently, dedicated microfluidics devices coupled with fluorescence imaging, allowed for the direct visualization of cohesin‐mediated loop extrusion in‐vitro and in real‐time (Davidson et al, 2019; Kim et al, 2019; Golfier et al, 2020). It will be an exciting and ground‐breaking development to track chromatin loop extrusion in the living cell nucleus in real time.
CTCF can act as insulator and chromatin barrier element
Loss of CTCF binding at TAD boundaries can perturb their capacity to insulate contacts between domains leading to merging of adjacent TADs (Despang et al, 2019; Franke et al, 2016; Lupiáñez et al, 2015; Guo et al, 2015; Hanssen et al, 2017; Vian et al, 2018; Fig 2A). Functionally, TAD boundary deletion causes a spectrum of effects ranging from minor, as at the Sox9‐Kcnj2 locus (Despang et al, 2019) to substantial. For instance, deletion or inversion of TAD boundaries at the WNT6‐EPHA4‐PAX3 locus may lead to alterations in mouse digit number (Lupiáñez et al, 2015). At the murine alpha globin locus, removal of CBS that partition the domain into smaller sub‐TADs, leads to aberrant upregulation of genes otherwise silenced in the presence of the sub‐TAD boundary (Hansen et al, 2019). Essential cell identity genes are often demarcated by CTCF/cohesin loop anchors. Loop formation insulates these genes together with their regulatory neighbourhood, which helps to maintain the local chromatin environment and proper gene expression. Removal of the anchors of such loops leads to misexpression of key regulators of cell fate (Dowen et al, 2014) and can contribute to diseases including cancer (as will be discussed below).
Figure 2. Genome engineering reveals locus‐specific transcriptional and architectural consequences of TAD boundary deletion and insertion.
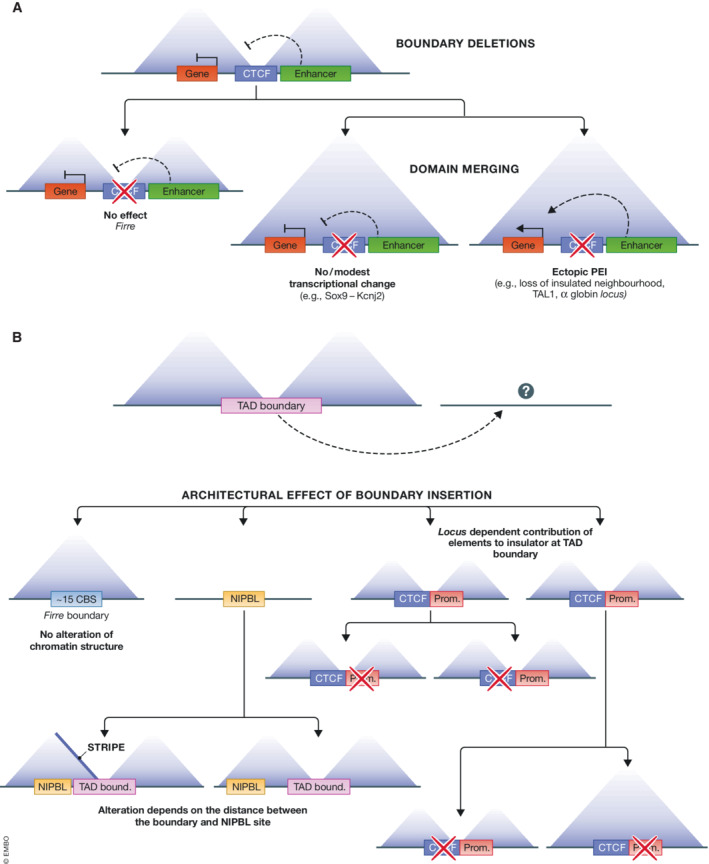
(A) TAD or a sub‐TAD boundary deletion may lead to no overt alteration of chromatin architecture as seen at the Firre locus (Barutcu et al, 2018), or to TAD and subTAD merging accompanied by either modest (Sox9/Kcjn2; Despang et al, 2019) or considerable transcriptional changes (e.g., loss of insulated neighbourhoods and oncogene activation; Hnisz et al, 2016), or aberrant activation of genes as, for example in the vicinity of otherwise insulated globin genes (Hanssen et al, 2017). (B) Ectopic insertion of a boundary element may lead to no change in the architecture of the recipient locus (Barutcu et al, 2018). When considering other CBS, boundaries can still be formed despite the deletion of the CBS. Depending on whether the ectopic boundary is inserted far or close to a Nipbl cohesin loader binding site, the boundary may form stripes (Redolfi et al, 2019). The contribution of distinct elements making up the boundary depends on the intrinsic features of the target locus. At one location, a boundary composed of a CBS site and a housekeeping gene promoter depends on both elements, while at another location CTCF appears less crucial for boundary formation (Zhang et al, 2020).
The auxin‐inducible degradation (AID) system allows to deplete a protein of interest efficiently and rapidly (Natsume et al, 2016). AID‐mediated acute removal of CTCF weakens TAD boundaries and disrupts CTCF–CTCF loops (Nora et al, 2017; Wutz et al, 2017; Hyle et al, 2019), establishing an essential role of CTCF in the formation of TADs. (It is worth noting here that despite a general effect, some borders are insensitive to the depletion of CTCF protein hinting at additional mechanisms driving the segmentation of the genome.) Acute removal of CTCF leads to transcriptional deregulation of numerous loci in embryonic stem (ES) cells, immortalized erythroid precursors, and B cell cancer cells (Nora et al, 2017; Hyle et al, 2019; Xu et al, 2021). Even in short time scales, loss of functional insulators could lead to global gene deregulation. The acute depletion of CTCF results in equal numbers of up and downregulated genes. Gene upregulation in the CTCF‐depleted cells can, to some degree, be explained by loss of boundary activity. In comparison with genes not affected by CTCF loss, loci with gained expression in the CTCF‐depleted cells are more frequently separated from nearby enhancers by a TAD boundary (Nora et al, 2017). Yet, despite prominent examples of gene deregulation in the absence of CTCF, the direct transcriptional effects of CTCF removal remain overall mild, as one would predict massive transcriptional changes upon genome‐wide abrogation of insulator activity (preprint: Hsieh et al, 2021; Luan et al, 2021). The lack of stronger effects may be due to only a limited dependence of gene expression on long‐range enhancer regulation. Likewise, epigenetic silencing of promoters could render them unresponsive to enhancers. Furthermore, promoter–enhancer specificity could be more often hardwired at the level of biochemical compatibility between CREs (Pachano et al, 2021) versus being regulated by insulators. Finally, a recent study revealed hundreds of promoter–enhancer interactions (PEI) that cross TAD boundaries suggesting that multiple enhancer–promoter pairs are perhaps unaffected by the insulating function of CTCF (preprint: Hsieh et al, 2021; see also below). Interestingly, prolonged depletion of CTCF leads to substantial gene deregulation (Nora et al, 2017) and is incompatible with cell differentiation (see below). Together, these results indicate that the functional perturbations elicited by the loss of CTCF are either caused by secondary effects subsequently to the initial deregulation of a handful of genes (Hyle et al, 2019; Xu et al, 2021), or that the direct effects of CTCF removal require time to unfold (preprint: Hsieh et al, 2021).
Chromatin boundaries may act as barrier elements that block spreading of heterochromatin, thereby inhibiting gene silencing. In essence, barrier activity allows to overcome chromatin position effects and shield genes to ensure stable expression despite repressive chromatin environments. Several paradigm insulators including 5'HS4 element at the chicken beta‐globin locus feature both insulator and barrier activities (Recillas‐Targa et al, 2002). Genome wide, CBS frequently coincide with zones of transition between open chromatin and histone 3 lysine 27 trimethylation (H3K27me3)‐enriched domains (Cuddapah et al, 2009). However, acute removal of CTCF in ES cells does not lead to spreading of H3K27me3 to adjacent domains (Nora et al, 2017), consistent with previous reports revealing uncoupling between CTCF and of 5'HS4 barrier activity at the chicken beta‐globin locus (Recillas‐Targa et al, 2002). Can CTCF nonetheless segregate regions of diverse chromatin activity? Hox gene clusters constitute paradigm loci for studying the interplay between trithorax and polycomb group proteins. The two complexes establish euchromatic H3K4me3 and heterochromatic H3K27me3 histone marks, respectively, thereby regulating spatiotemporal patterns of Hox gene expression, which are essential for axial patterning of the developing embryo. Exposure of ES cells to retinoic acid (RA) mimics cervical motor neuron development. At the HoxA locus, RA treatment of ES cells leads to upregulation of HoxA1‐6 rostral genes, leaving HoxA7‐13 silent (consistent with the pattern of expression during embryogenesis, HoxA7‐13 are normally expressed in caudal parts of the embryo). The boundary between HoxA1‐6 and HoxA7‐13 genes associates with CTCF; the removal of this CBS leads to spreading of histone marks related to open chromatin, loss of H3K27me3 and transcriptional activation of the HoxA7 gene (Narendra et al, 2015). The loss of the barrier role of CTCF is translated into gain of regulatory interactions at the HoxA locus and homeotic transformations of the embryo (Narendra et al, 2016).
Together, these experiments show essential roles for CTCF in the formation of TADs. TADs overall help maintain proper CRE dialogues in the cell. Through CTCF, TAD boundaries act as insulators and at some genomic locations as barrier elements. Yet, the contribution of TAD boundaries appears locus and context‐specific. It will be important to determine which CBS constitute genuine insulators and what properties of the domains and TAD sequences allow some CBS to exert enhancer‐blocking functions (Box: In need of answers). Likewise, it will be essential to determine how many barrier elements there are in the mammalian genome, and how they depend on CTCF.
Box: In need of answers
-
i
What mechanisms determine the choice of CBS for boundary function?
-
ii
How does RNA contribute to CTCF–CTCF loop formation and boundary activity, and can RNA affect the stability of CTCF dimers at the anchor of architectural loops?
-
iii
What mechanisms underlie the genome‐wide maturation of chromatin topology during embryonic stem cell differentiation?
-
iv
How does the local chromatin environment impact insulator activity and how do promoter‐enhancer interactions depend on CTCF?
-
v
How do post‐translational modifications and protein partners of CTCF contribute to its architectural functions?
Harnessing the unique properties of CTCF at gene promoters and enhancers
CBS frequently overlap promoters (Chen et al, 2012). Historically, the functions of CTCF were addressed at the P2 element, a CBS immediately downstream of the transcription start site (TSS) of Myc. The impact of CTCF at P2 is largely repressive (Filippova et al, 1996). Similarly, CBS within the silencer of the chicken lysozyme gene (LYZ; Arnold et al, 1996; Burcin et al, 1997), as well as CBS adjacent to thyroid hormone responsive element in the 3'UTR of the LYZ gene or the intronic CBS at the HLA‐DRB1 locus were suggested to inhibit gene expression based on in‐vitro reporter assays (Awad et al, 1999; Arnold et al, 2000). Can CTCF repress gene expression, and can it do so directly, by binding to promoters? It is hard to answer this question without an extensive effort in genome engineering, but acute CTCF depletion experiments may provide some insight into this matter.
Transcriptional activation corollary to acute CTCF loss in B cell leukemic cells is linked to gain in chromatin accessibility at promoters (Xu et al, 2021). This result is compatible with CTCF acting directly at the promoter to inhibit gene expression. CTCF can induce DNA bending (Arnold et al, 1996) which might impact binding of other proteins to DNA thereby silencing gene expression. Furthermore, CTCF can interact with a co‐repressor SIN3A, which can remodel chromatin and induce histone deacetylation leading to reporter gene downregulation (Lutz et al, 2000).
CTCF can also exert a stimulatory role when bound to gene promoters and the mechanisms of CTCF‐mediated positive impact on transcription might entail the regulation of both the local chromatin landscape and the large‐scale genome architecture. CTCF can co‐purify with the largest subunit of RNA Polymerase II (PolII; Chernukhin et al, 2007), promote PolII clustering in the nucleus (Lee et al, 2022) and recruit TBP‐associated core promoter factor TAF3 to upregulate gene expression (Liu et al, 2011). Diminished levels of CTCF lead to loss of a fraction of peaks of CTCF at promoters and transcriptional downregulation of genes implicated in oncogenesis (Aitken et al, 2018; see also below). Promoters of genes that are downregulated upon removal of CTCF frequently contain CBS and, at a subset of these promoters, CTCF binds only 60 bp upstream of the TSS. At these sites, the CTCF motif is predominantly oriented in the same direction as the gene promoter (Nora et al, 2017). The significance of this observation is unclear. Given that CTCF binding is related to well‐positioned nucleosome arrays, CTCF might impact gene expression by orchestrating nucleosome phasing (Nora et al, 2017). Alternatively, by limiting antisense transcription, CTCF might favour gene expression (Cho et al, 2005; Degner et al, 2011). Why only a small minority of CTCF‐bound promoters (10%, Nora et al, 2017) cause transcriptional upregulation in response to CTCF depletion remains unclear.
The positive role of CTCF at gene promoters likely relies, at least in part, on its architectural functions. Promoter–enhancer loops can connect convergently oriented CBS with distal CREs including enhancers and promoters (Rao et al, 2014; Tang et al, 2015). Hence, in addition to impacting nucleosome positioning, CBS within gene promoters might act as anchor or docking sites that facilitate PEI. For instance, a CBS located 2 kb upstream of the MYC promoter is conserved in multiple cancer cell lines. This CBS is required for the MYC promoter to receive input from various cell type‐specific super‐enhancers located in the genomic surroundings of the MYC gene (Schuijers et al, 2018). Many more genes harbour CTCF sites in their extended promoter region that could potentially exert enhancer docking functions (Schuijers et al, 2018) and modify the affinity of promoters to enhancers (Oh et al, 2021b). The extent to which this model describes the implication of CTCF in regulation of oncogene expression is, however, under debate (Hyle et al, 2019).
Promoters may display enhancer activity towards other genes (Dao et al, 2017; Diao et al, 2017). Artificial tethering of CTCF to the Vcan promoter facilitates its transcriptional upregulation during neural induction via establishing contacts with the promoter of the Tmem167 gene located 350 kb downstream of the Vcan TSS (Kubo et al, 2021). Likewise, CBS in the vicinity of enhancers may act to favour the interaction with their cognate promoter. At the human Sonic hedgehog (SHH) locus, CTCF sites flanking the limb‐specific enhancer ZRS are required for SHH expression; lack of these CBS is related to acheiropody, a congenital condition featuring limb truncation (Ushiki et al, 2021).
The mechanism of CTCF‐driven formation of PEI most likely depends on cohesin action. The cohesin loader Nipbl is enriched at active enhancers (Kagey et al, 2010; Kieffer‐Kwon et al, 2013; Liu et al, 2021) and proper cohesin dynamics are essential for promoter–enhancer transactions. Loss of cohesins diminishes PEIs (Lavagnolli et al, 2015; el Khattabi et al, 2019) affecting the capacity of enhancers to fully upregulate genes (Aljahani et al, 2022). Removal of cohesins primarily affects genes regulated by remote enhancers (Lavagnolli et al, 2015; Calderon et al, 2022). Likewise, failure to remove cohesin complexes from chromatin also affects PEI. Deletion of the Wapl cohesin unloader leads to exhaustion of the free cohesin pool in the nucleoplasm thereby blocking loading of new cohesin complexes at enhancers affecting timely formation of PEIs (Liu et al, 2021). At genomic sites where cohesins are loaded in close vicinity of CBS with a motif oriented en face of the newly loaded loop extruders, cohesin activity is immediately blocked on one site which leads to fixation of the cohesin loading enhancer at the anchor of the loop (Fig 1D). In this configuration, extrusion proceeds on the other side of the ring reeling in and presenting the entire TAD to the enhancer, in Hi‐C maps this is accompanied by formation of architectural stripes (Barrington et al, 2019; Vian et al, 2018; Fig 1B and D). Recently, using an orthogonal, crosslinking‐free method to score for chromatin structure, the Giorgetti lab has provided evidence that cohesin loading in the vicinity of a CBS leads to the formation of stripes (Redolfi et al, 2019).
CTCF removal leads to loss of numerous PEI (Thiecke et al, 2020; Kubo et al, 2021; Lee et al, 2022) and a reversible disruption of PolII‐enriched transcriptional condensates (Lee et al, 2022). Yet, as we have seen above, the immediate transcriptional consequences remain somewhat scarce and the acute degradation of CTCF does not lead to overt changes in transcription initiation or elongation genome‐wide (preprint: Hsieh et al, 2021; Luan et al, 2021). Likewise, although substantial data point to an essential role of TADs in the spatiotemporal regulation of gene expression (de Laat & Duboule, 2013), the relationship between the three‐dimensional structure of chromatin, TAD formation and transcriptional regulation appears complex. While activation of genes frequently coincides with formation of promoter–enhancer loops as detected by live cell microscopy (Chen et al, 2018), chromatin conformation capture assays (Sanyal et al, 2012; Kieffer‐Kwon et al, 2013; Mifsud et al, 2015; Bonev et al, 2017; Pekowska et al, 2018; Hua et al, 2021) and genome architecture mapping (Beagrie et al, 2017), these techniques show that promoter–enhancer loops can also be formed in the absence of enhancer activity (de Laat & Duboule, 2013; Ghavi‐Helm et al, 2014; Williamson et al, 2016; Phanstiel et al, 2017). At some loci, the distance between promoters and their cognate enhancers can be uncoupled from gene activation (Alexander et al, 2019; Benabdallah et al, 2019). Promoters might form only a transient interaction with condensates containing the mediator complex (Cho et al, 2018b). Such an extremely dynamic nature of PEI would hence require an ultra‐deep sequencing of Hi‐C libraries for detection (Bonev et al, 2017; Hua et al, 2021). Technological improvements, including the introduction of micro‐C which allows to detect substantially larger repertoires of PEIs due to a higher signal to noise ratio (preprint: Hsieh et al, 2021), indicate that PEI rely on physical contacts. Piggy‐Back‐mediated genetic engineering experiments allowed to place an enhancer at increasing genomic distances from its cognate promoter and, combined with Hi‐C, recently demonstrated a complex interplay between physical proximity, presence of other CREs in a TAD and functional interactions between regulatory elements stressing the need for context‐specific models of PEI control (Zuin et al, 2022).
While the data suggest that CTCF is implicated in the setup of PEI, recent reports show that a substantial fraction of PEIs appears immune to acute (3 h) removal of CTCF or cohesins, suggesting a “time‐buffering” model, where the established local chromatin environment would be sufficient to keep PEI for at least 3 h without these architectural factors (preprint: Hsieh et al, 2021). Micro‐C also revealed thousands of previously unappreciated PEIs. Remarkably, in mouse embryonic stem cells, 20% of the PEIs connect elements located in two adjacent TADs, and are hence not blocked by CTCF‐bound borders (preprint: Hsieh et al, 2021). This discovery is interesting in light of the observation that, in the context of an engineered locus, where a CTCF site is located between an enhancer and its cognate promoter, truncations in the enhancer element that diminish its strength (as established by reporter assays) also render the action of the enhancer more susceptible to insulation by CTCF (Zuin et al, 2022). It will be fascinating to assess more broadly whether inter‐TAD PEIs are functional and how the enhancer strength contributes to the capacity of PEIs to overcome the constrains imposed by CTCF.
Deletion of CTCF sites leads to increased heterogeneity of gene expression mouse T cells (Ren et al, 2017). Likewise, CTCF and cohesin depletion cripples the response of macrophages to inflammatory stimuli (Cuartero et al, 2018; Stik et al, 2020) and can reduce the impact of enhancers on a subset of promoters (Vos et al, 2021; Aljahani et al, 2022) perhaps by influencing the formation of transcriptional condensates (Lee et al, 2022) or by regulating the capacity of TFs to bind to chromatin (preprint: Hsieh et al, 2021). These data indicate that rather than strictly allowing, CTCF modulates PEIs. It remains an open question how PEIs are formed and how the specificity of these interactions is ensured. We predict that further technological developments will reveal an even larger complexity of PEIs, and a combination of live‐cell imaging and improved chromatin conformation capture assays will most likely be instrumental to address these questions (Brandão et al, 2021). (See also Concluding remarks.)
Regulatory element composition and genomic context impact strength of insulators
Insulation score (IS) is defined as the ratio between the number of interactions that cross a given genomic position and the number of interactions that are formed at both sides of the assayed position (Sofueva et al, 2013). Using advanced computational methods to increase the robustness of Hi‐C, allowed Gong et al to classify TAD boundaries based on their IS. Apart from showing that the strongest boundaries tend to have more pronounced CTCF binding (Gong et al, 2018), this analysis revealed that the cell‐type invariant TAD boundaries differ in strength between tissues (Gong et al, 2018) as also shown in developing cells (Pekowska et al, 2018). Hence, the IS constitutes a tuneable parameter of chromatin architecture.
The IS reflects the fraction of interactions built on both sides of a boundary to the ones that cross it. Cohesin loading and hence loop extrusion is not uniform across the genome (Vian et al, 2018; Hua et al, 2021) and it is unclear how the density of the loop‐extruding complexes impacts the IS. Recent data show that both boundary‐encoded, and locus intrinsic features shape the capacity of CBS to sustain insulator functions (Fig 2). When inserted into an unrelated genomic locus, both an artificial construct containing three CBS and a TAD boundary separating the HoxD locus into two domains retain their insulator functions at the ectopic site (Redolfi et al, 2019; Willemin et al, 2021). In contrast, the CTCF‐enriched TAD boundary intersecting the long‐noncoding RNA locus Firre, exerts no overt effect on chromatin structure at an ectopic locus (Barutcu et al, 2018). Hence, the capacity of the TAD boundaries to insulate regions from each other in some cases depends on the site where they are inserted.
TAD boundaries frequently coincide with CTCF‐binding sites and housekeeping gene promoters (Dixon et al, 2012). To what extent do these elements collaborate to regulate the IS? A boundary element containing both the CTCF‐binding site, and the transcription start site (TSS) of a housekeeping gene PARL, requires both elements to exert its function (Zhang et al, 2020). Yet, the strength of insulation depends not only on the CBS and PARL promoter but also on the local chromatin context of the region where the boundary was knocked in. Removal of the CBS in the inserted boundary reduced the IS; while the excision of the TSS from the boundary exerted a more pronounced effect on IS at one ectopic locus. In contrast, when inserted at another genomic position, the TSS and CBS of the same boundary worked in an additive fashion (Zhang et al, 2020). The presence of the CBS within the ectopic locus and the distance of the inserted boundary to the nearest transcribed gene contribute to the IS of the boundary at the insertion site (Zhang et al, 2020). Thus, is transcription sufficient to elicit boundary formation? In the absence of CTCF, cohesins accumulate at sites of convergent transcription which might in principle lead to enhanced contact insulation and boundary formation (Busslinger et al, 2017). When inserted into the coding sequence of a gene, the TSS of PARL gene alone can drive boundary formation (Zhang et al, 2020). The fact that a fraction of boundaries gained upon ES cell differentiation is devoid of CTCF (Pekowska et al, 2018), but overlaps activated promoters, as exemplified at the Zfp608 locus (Bonev et al, 2017), suggests an instructive role of promoters in chromatin organization during differentiation. Yet, a precocious transcriptional activation of the Zfp608 promoter by dCas9‐VP64 is insufficient to elicit insulation (Bonev et al, 2017), which means that additional factors recruited to the Zfp608 promoter during differentiation might be required for boundary formation. The inhibition of transcription can lead to diminished boundary strength (Rowley et al, 2017; Barutcu et al, 2019) but the effect appears relatively weak and numerous other reports show no impact of a transcriptional block on TAD structures in the fruit fly (Hug et al, 2017; Hsieh et al, 2020; Jiang et al, 2020). A transcriptional block does not affect the restoration of chromatin loops neither upon reintroduction of cohesins (Vian et al, 2018) nor upon entry into the G1 phase after cell division (Zhang et al, 2021). Interestingly, emergence of TAD boundaries in fruit fly oocytes depends on the TF Zelda that induces a global transcriptional onset (Hug et al, 2017). It will be important to define which factors regulate CTCF‐less boundaries in mammalian cells and whether these factors might also impact CTCF‐bound insulators.
Using a highly efficient flippase‐assisted recombination‐based genome engineering, Huang et al assessed a set of CTCF‐bound TAD boundary elements for their insulator activity by inserting them between the Sox2 promoter and a super‐enhancer active in mouse ES cells (Huang et al, 2021). Insulators exerted a remarkably weak effect on Sox2 expression, out of the 11 tested CBS, the most potent element reduced the expression of Sox2 by only 11%. Inclusion of an increasing number of tandemly oriented CBS enhanced the IS and diminished the activity of Sox2 gene by at most ~40%, and the transcriptional effect correlated with the extent of changes in chromatin structure measured by Hi‐C (Huang et al, 2021). Does this mean that insulators exert only a very modest impact on gene expression? Or rather, that insulator functions are a derivative of both sequence composition of the boundary and the local chromatin environment as shown by Barutcu et al (2018)? Sleeping‐beauty transposon‐assisted insertion of a fluorescent reporter construct containing a strong enhancer and a weak promoter separated by a well‐established insulator revealed that reporter activity strongly depends on the presence of additional CREs at the locus (Ribeiro‐Dos‐Santos et al, 2022). Likewise, older in‐vivo experiments in the fruit fly show that the strength of the Fab‐7 boundary element depends on the promoter–enhancer pair, and some enhancers are blocked by insulators more readily than others (Zhou et al, 1996). A similar observation was recently made when assessing the capacity of a CBS to interfere with PEI at an engineered locus (Zuin et al, 2022). Taken together, these data indicate that transcriptional output at a given locus is most likely corollary to the combined action of multiple elements that dynamically interact and signal to one another thereby collectively moulding the activity of the locus. This property of transcriptional regulatory systems likely underlies the difficulty to assess insulator functions of CTCF sites in chromatin reporter assays that largely rely on insertion of insulator elements in‐between a known enhancer–promoter pair. The low activity of insulators in genomic engineering experiments might in part be due to the disability of ectopic sites chosen for the assay to provide the necessary environment for proper insulator action. Large scale in‐situ assessments of the implications of CTCF sites for regulating promoter–enhancer transactions will be needed to critically assess this model.
Intrinsic and extrinsic factors regulate CTCF binding to its cognate sites
CTCF interacts with an array of motifs with marked differences in DNA sequence. It does so by deploying distinct combinations of its ZFs depending on the site (Filippova et al, 1996, 1998; Burcin et al, 1997; Awad et al, 1999; Kanduri et al, 2000; Quitschke et al, 2000; Renda et al, 2007). The ZFs of CTCF interact with multiple DNA bases simultaneously, ZFs 4‐7 bind to the major groove of DNA, the interaction involves only one strand of the DNA double‐helix (Hashimoto et al, 2017; Yin et al, 2017). By overexpressing ZF mutants of CTCF and using ChIP‐seq to localize the engineered CTCF molecules, Nakahashi et al showed that not all the ZFs contribute equally to CTCF DNA binding (Nakahashi et al, 2013). ZF 4‐7 appear essential for the pattern of CTCF distribution, their mutants occupy less than 20% of the wild‐type CBS. These central ZF were previously annotated as binding to the core DNA‐binding motif (Filippova et al, 1996; Renda et al, 2007; Ohlsson et al, 2010). Recently, combining the acute depletion of the wild‐type form of CTCF with controlled expression of ZF CTCF mutants, allowed Soochit et al to obtain largely improved conditions to study the impact of individual ZFs; the ZFs mutants that destabilize CTCF binding to the largest degree display highest CTCF‐DNA‐binding dynamics and lowest capacity to form loops (Soochit et al, 2021). Therefore, the stability of the association between CTCF and DNA can be linked to loop formation and the control of chromatin architecture. Residence time (RT) of CTCF on chromatin measured by Fluorescence Recovery After Photobleaching (FRAP) is within a range of minutes in mouse ES cells (Hansen et al, 2017). This value appears high when compared to more classical transcriptional regulators. Given the link between the RT of CTCF binding and loop formation (Soochit et al, 2021), it is possible that a long RT is a perquisite for CTCF's architectural functions. Live‐cell imaging and tracking of CTCF‐CTCF loop anchors recently revealed that a loop at the Fbn2 locus can persist for up to 30 minutes in ES cells (Gabriele et al, 2022). Loop stability might be even more pronounced; estimates based on the measurements of the stability of cohesin binding to chromatin indicate that loop structures may persist for hours depending on the post‐translational modifications of STAG factors (Wutz et al, 2020). Yet, it remains unclear and relatively understudied whether RT of CTCF differs between tissues and whether chromatin loop formation can be impacted by mechanisms that influence the RT of CTCF in physiological settings. Remarkably, activation of quiescent B cells by exposure to conditions mimicking immune responses leads to a marked reduction of the RT of CTCF (Kieffer‐Kwon et al, 2017). In the future, it will be instrumental to understand the extent by which the RT of CTCF differs between cell types and how it is related to its architectural functions.
DNA methylation anticorrelates with CTCF binding and insulator activity
DNA methylation anticorrelates with CTCF binding (Bell & Felsenfeld, 2000); differences in the methylation of CpGs islands correlate with cell type‐specific CTCF binding and insulator activity (Wang et al, 2012). The imprinting control region (ICR) that regulates allele‐specific expression of Igf2 and H19 binds to CTCF; the insulator function of ICR depends on DNA methylation that anti‐correlates with insulation (Bell & Felsenfeld, 2000; Hark et al, 2000; Kanduri et al, 2000; Szabó et al, 2000; Cui et al, 2001; Holmgren et al, 2001). An analogous situation has been described at the Gtl2 and Dlk1 loci, where the ICR is hemi‐methylated and binds to CTCF at the unmethylated allele (Wylie et al, 2000). CTCF binding at other imprinted loci (Hikichi et al, 2003; Fitzpatrick et al, 2007; Lin et al, 2011) including Rasgrf1, Myotonic Dystrophy 1 (MD1), is also sensitive to DNA methylation (Filippova et al, 2001; Yoon et al, 2005). Mutations in isocitrate dehydrogenase (IDH) or succinate dehydrogenas (SDH) cause DNA hyper‐methylation in glioblastomas and gastrointestinal stromal tumors (GIST), respectively. The increased DNA methylation affects CTCF binding and insulator function, which favours oncogene expression (Flavahan et al, 2016, 2019). While KIT‐mutant, PDGFRA‐mutant and SDH‐mutant GIST share enhancer landscapes, they differ in transcriptional programmes. The CBS hyper‐methylation and insulator dysfunction in SDH‐mutant GISTs largely explains the differences in gene expression and in the future may help to contribute to the development of personalized anti‐cancer therapies (Flavahan et al, 2019).
However, while unmethylated motifs are bound by CTCF preferentially (Stadler et al, 2011; Feldmann et al, 2013), DNA methylation does not block the association between CTCF and DNA in vivo (Stadler et al, 2011). Likewise, only 40% of tissue or cell type‐specific CBS can be related to differential DNA methylation and genome‐wide loss of DNA methylation does not lead to a massive unmasking of CTCF motifs and a marked gain in new CBS (Stadler et al, 2011; Wang et al, 2012). While DNA demethylation exerts overall weak effects on the profile of CTCF binding, induction of DNA methylation seems to have a more pronounced effect. Genetic removal of ten‐eleven translocation 1 and 2 (Tet1 and Tet2) dioxygenases, that convert 5‐methylcytosine into hydroxymethylated, formylated (5fC) or carboxylated intermediates, increases DNA methylation and causes the loss of a substantial fraction of CTCF peaks in ES cells (3,916 CBS were lost, while 7,232 CBS were maintained in the Tet1/2−/− cells). This effect, pronounced at regions with low CpG density, is possibly caused by nucleosome repositioning and occlusion of CTCF motifs rendering them inaccessible to CTCF (Wiehle et al, 2019). In general, sites with a low CpG content seem to bind CTCF less and appear particularly vulnerable to DNA methylation levels (Wiehle et al, 2019). In ES cells, CRISPR‐dCas9‐Dnmt3a‐mediated methylation of the CTCF‐binding site insulating Nlrp12 and H2Q10 loci from the expressed miR290 and Pou5f1 genes caused transcriptional upregulation of Nlrp12 and H2Q10 (Liu et al, 2016). However, it needs to be determined whether the effect is caused by DNA methylation or by a possible occlusion of the CBS by the dCas9 protein.
Histone modifications and chromatin openness impact CTCF binding
CBS were originally annotated by analysing DNAseI sensitive sites. CTCF‐binding motifs are depleted of nucleosomes (Teif et al, 2012; Carone et al, 2014; Liu et al, 2016) and CBS feature up to 20 well‐positioned nucleosomes around the CTCF motif (Fu et al, 2008). Chromatin openness could be one of the signatures tagging CTCF motifs for recognition. Yet, open regions that intersect CTCF peaks are closed upon CTCF removal (Xie et al, 2020). CTCF interacts with chromatin remodelling complexes including switch/sucrose nonfermentable complex (SWI/SNF) and the Imitation SWItch (ISWI) complex (Wiechens et al, 2016; Marino et al, 2019; Valletta et al, 2020). The removal of Snf2h, the ATPase subunit of the ISWI complex reduces CTCF chromatin binding and CTCF‐CTCF loops in ES cells (Wiechens et al, 2016; Barisic et al, 2019). The latter result is somewhat unexpected, as the deletion of Snf2h leads to a reduction not abrogation of CTCF binding (Barisic et al, 2019), and according to the targeted protein degradation experiments, removal of over 90% of CTCF is required to detect robust loop loss in ES cells (Nora et al, 2017). The more recent data suggest that even relatively subtle changes in strength of CTCF binding (as detected by ChIP‐seq) may translate to pronounced architectural effects. It will be interesting to assess how Snf2h loss impacts the dynamics of CTCF binding to chromatin in real time. CTCF can promote chromatin opening and incorporation of a histone variant H3.3 (Weth et al, 2014) suggesting that it can act upstream of the establishment of DNA accessibility. H2A.Z, a histone variant of H2A, promotes nucleosome unwrapping, and surprisingly, the removal of H2A.Z can enhance CTCF binding (Wen et al, 2020), suggesting a destabilizing role for these euchromatin‐enriched histone variants in CTCF binding.
CTCF‐binding sites implicated in chromatin topology are ultra‐stable
Acute depletion of CTCF protein does not eliminate CTCF from all CBS, as thousands of sites remain occupied even after the removal of more than 90% of CTCF (Hyle et al, 2019; Luan et al, 2021). The stably bound CBS are, on one hand, depleted by the destabilizing downstream motif, and, on the other hand, enriched in the A/T‐rich motif located ~200 bp from the CTCF motif. Yet, the deletion of the A/T sequence does not affect CTCF binding as assessed at a silent Myrip locus. This suggests that additional factors are at play in the regulation of the stability of CTCF binding to chromatin (Luan et al, 2021). Promoters (Hyle et al, 2019) and enhancers (Luan et al, 2021) retain CTCF best, and a substantial fraction of CTCF peaks that withstand the degradation of the bulk of CTCF protein overlap TAD borders and loop anchors. Remarkably, the sites that do not lose CTCF in the auxin‐treated cells also remain occupied by CTCF during mitosis (Luan et al, 2021). More recent data has however challenged these observations showing that virtually all CTCF binding is lost from the CBS upon a 3‐h depletion of the CTCF protein (preprint: Hsieh et al, 2021). It will be essential to determine what regulates the stability of CTCF binding to TAD borders and loop anchors.
RNA regulates CTCF binding to DNA
RNA modulates CTCF occupancy in the genome indirectly and directly. Local transcription of long non‐coding RNAs can induce nucleosome repositioning and occlusion of CBS thereby evicting CTCF (Lefevre et al, 2008). Furthermore, CTCF harbours an RNA‐binding region (RBR) and associates with RNA in vivo (Saldaña‐Meyer et al, 2014; Kung et al, 2015). The RNA species that interact with CTCF are frequently produced in the vicinity of CBS and can locally and directly impact the association of CTCF with DNA. RNA can stabilize CTCF binding to chromatin (Hansen et al, 2019; Saldaña‐Meyer et al, 2019) or disrupt it possibly by competing with DNA (Sun et al, 2013; Oh et al, 2021a). Global transcriptional inhibition blocks CTCF binding primarily at promoters, which suggests a positive role of RNA in CTCF binding at these locations (Hansen et al, 2019; Saldaña‐Meyer et al, 2019; Miyata et al, 2021). (See also below.)
Factors controlling architectural functions of CTCF
There are tens of thousands of CBS in the genome, yet only a small fraction of them participates in the formation of TAD boundaries and loops. The architectural functions of CBS are likely a result of the regulatory element composition of TAD boundaries. Furthermore, local transcriptional activity may impinge on CTCF binding to chromatin. Likewise, the unique features of the CTCF protein including the ability of CTCF to interact with cohesins, the particularly stable binding of CTCF to chromatin, and the capacity of CTCF to homo‐oligomerize and form clusters are most probably essential for its architectural roles.
The unstructured N‐terminal tail of CTCF underlies the dialogue between CTCF and cohesins (Li et al, 2020; Nora et al, 2020; Pugacheva et al, 2020) and stabilizes cohesin binding to chromatin by directly competing with the cohesin release factor Wapl (Li et al, 2020). Remarkably, tethering of the unstructured CTCF termini to an unrelated ectopic genomic region using artificial ZFs is not sufficient to efficiently retain cohesin and block loop extrusion (Pugacheva et al, 2020), suggesting a combined role with other portions of the CTCF molecule in cohesin retention.
The non‐coding‐RNA steroid receptor RNA activator (SRA) and DEAD‐box RNA helicase p68 (DDX5) associate with CTCF at the IGF2/H19 locus. Knockdown of DDX5 and SRA hampers the association between CTCF and cohesins at the IGF2/H19 locus impacting allele‐specific expression patterns of Igf2/H19 (Yao et al, 2010). RNA stimulates the interactions between CTCF molecules in vitro (Yusufzai et al, 2004) and in vivo (Saldaña‐Meyer et al, 2014; Hansen et al, 2019). CTCF oligomerization in the cell nucleus is illustrated by a formation of clusters of variable sizes (Gu et al, 2020; Hansen et al, 2020). Deletion of the region in CTCF which is essential for CTCF–RNA interaction diminishes CTCF cluster size (Hansen et al, 2019), which is consistent with older data showing that a p53 antisense transcript Wrap53 favours CTCF clustering (Saldaña‐Meyer et al, 2014). Likewise, mutation of the RBR, precluding the association of CTCF with RNA, reduces CTCF binding at CBS and dismantles chromatin loops (Hansen et al, 2019; Saldaña‐Meyer et al, 2019). Noteworthy, RBR‐dependent cluster formation seems to aid CTCF in finding its cognate sites (Hansen et al, 2020), leading to the proposal that RNA molecules could act as road signs attracting CTCF and modulating its binding and perhaps also its architectural functions. Other data show that while this model can stand, the interactions between CTCF and RNA are complex and impact CTCF binding in a locus‐specific manner. Global transcription inhibition stabilizes CTCF clusters (Gu et al, 2020), and several non‐coding RNAs have been shown to destabilize the interaction between CTCF and DNA, which leads to loss of CTCF–CTCF loops (Sun et al, 2013; Oh et al, 2021a). Together, these data indicate that the non‐coding RNA portfolio in the cell might constitute an additional regulatory layer acting to fine tune CTCF binding to its cognate sites thereby impacting chromatin topology.
CTCF can undergo various post‐translational modifications, but their functional significance needs to be deepened further. Poly(ADP)ribosylation (PARylation) was shown to mark insulator‐bound CTCF proteins in low throughput chromatin immunoprecipitation coupled with microarray experiments (Yu et al, 2004). Treatment of cells with PARP1 inhibitor Olaparib attenuates insulator activity as measured by reporter assays. CTCF interacts with Poly‐ADP‐ribose polymerase I (PARP1) thereby impacting circadian gene repositioning to the heterochromatic nuclear lamina (Zhao et al, 2015). It is unclear how to combine these observations to a model that explains the contribution of PARP1 and PARylation in CTCF‐anchored loop and TAD formation. In addition to PARP1, there are numerous other proteins that co‐immunoprecipitate with CTCF (Uuskula‐Reimand et al, 2016). Amongst them, Myc‐Associated Zinc Finger Protein MAZ that was recently identified as a regulator of insulator functions of CTCF. CTCF‐ and MAZ‐binding sites coincide in the genome. The depletion of MAZ destabilizes CTCF‐anchored loops and diminishes insulation of PEI by CTCF genome‐wide (Xiao et al, 2021; Ortabozkoyun et al, 2022).
CTCF‐mediated chromatin topology during development
The genomic coordinates of TADs are overall rather preserved in distinct cell types (Dixon et al, 2012; Rao et al, 2014); more domain boundaries are shared than cell type‐specific between largely transcriptionally divergent pluripotent and neural stem cells (Dixon et al, 2015; Bonev et al, 2017; Pekowska et al, 2018). There are several notable examples of loci that alter interaction patterns upon differentiation including, for instance, the Sox2 locus (Li et al, 2014) or the Hox loci (Montavon et al, 2011; de Laat & Duboule, 2013; Rodríguez‐Carballo et al, 2017). TAD boundaries and loops emerge early during development and increase their strengths stepwise during cell commitment accompanying loss of totipotency (Flyamer et al, 2017), exit from pluripotency and lineage commitment (Bonev et al, 2017; Pekowska et al, 2018). The consolidation of TADs detected by Hi‐C corresponds to the decrease in domain intermingling in super‐resolution microscopy experiments (Szabo et al, 2020). Chromatin topology featuring more loose TAD borders and infrequent loops is characteristic for ES cells. These features of nuclear structure can be reinstalled in differentiated cells upon reprogramming to pluripotency (Pekowska et al, 2018). Cell maturation also enhances architectural loop formation. Transcriptional activation at the globin genes in erythroid cells correlates with the strengthening of the CTCF–CTCF loop that demarcates the locus (Hua et al, 2021). Likewise, neuronal commitment of progenitor cells further consolidates TADs and loops (Bonev et al, 2017). Akin to cell differentiation, cell maturation is also accompanied by changes in CTCF‐mediated chromatin topology. Activation of naïve B cells leads to exit from the G0 phase and massive upregulation of gene expression. This phenomenon is associated with gain of CTCF–CTCF loops and induction of PEI (Kieffer‐Kwon et al, 2017). It is unclear whether loop and TAD boundary strengthening is related to differences in cohesin loading. In this light, the recent base pair resolution chromatin conformation capture experiments revealed that the increase in Nipbl binding correlates with transcriptional upregulation and loop strengthening at the activated loci suggesting that gain of loops may indeed be caused by increased cohesin loading upon activation of the locus (Hua et al, 2021).
What is the functional role of TAD consolidation during development? While this question is under investigation, CTCF seems to stabilize the acquired cell identity, its removal leads to increased spontaneous dedifferentiation of ES cells to totipotent‐like cells in cell culture (Olbrich et al, 2021; Zhu et al, 2021). Furthermore, removal of CTCF at the early stages of B‐cell to macrophage trans‐differentiation favours the transition (Stik et al, 2020). Therefore, chromatin structure might help in maintaining acquired cell identities.
Implication of CTCF in disease
Genomic rearrangements can reshuffle the relative positions and distances between regulatory elements leading to altered expression patterns underlying disease states. Loss of CBS and TAD boundary activity can result in severe phenotypical consequences including homeotic transformations (deletion of the boundaries at the HoxA and HoxC loci; Narendra et al, 2016), alteration in digit numbers when the boundaries at the WNT6/IHH/EPHA4/PAX3 are misplaced (Lupiáñez et al, 2015), or a plethora of phenotypes including cleft palate, delayed ossification, short snout and shortened long bones in the case of domain boundary inversions and ectopic insertions, which reshuffle promoter–enhancer contacts at the Sox9/Kcjn2 locus (Despang et al, 2019). Likewise, in addition to DNA mutations and epimutations in the CBS, altered protein sequences and expression levels of CTCF are related to several human disorders including cancer and neurological conditions (Fig 3). As we will see, the impact of CTCF is largely context dependent. However, common effects of truncating mutations and deletions of CTCF on cell proliferation reveal CTCF as a critical regulator of normal cell and organ homeostasis.
Figure 3. Mutations in CTCF related to neurological syndromes.
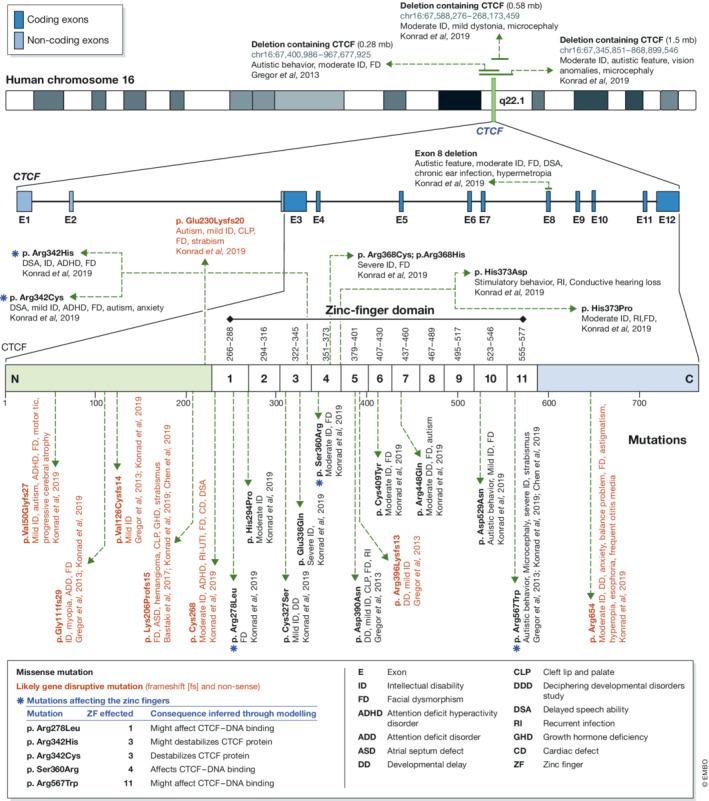
Multiple mutations including deletions have been reported for CTCF. These genetic perturbations are linked to numerous neurological manifestations. Genetic variants impacting CTCF binding sites associate with several disorders including neurological diseases. The predicted impact of the mutations in ZFs of CTCF on its 3D protein structure and the inferred possible effects on CTCF binding to chromatin.
Mutations of CTCF coding sequence in cancer and their functional role in oncogenesis
The DNA sequence encoding CTCF was first shown to be affected in breast and prostate tumours (Filippova et al, 1998). The analysis of several large Pan Cancer data sets including the TCGA data revealed a whole spectrum of CTCF mutations including deletions, amplifications and point mutations present in multiple tumours with varying frequency (Rubio‐Perez et al, 2015; Debaugny & Skok, 2020). Out of 273 detected mutations in CTCF (https://www.intogen.org/search?gene=CTCF accessed on the 19.03.2022), only 41 (15%) are synonymous while 165 are missense (60%) and 66 (24%) are truncating mutations; the genetic alterations localize throughout the coding sequence of CTCF and multiple mutations target the ZFs of CTCF (see also below; Rubio‐Perez et al, 2015).
CTCF acts as a tumour suppressor gene (Davoli et al, 2013; Gonzalez‐Perez et al, 2013; Rubio‐Perez et al, 2015). As we have noted above, most of the genetic alterations remove one allele of CTCF and were suggested to act as cancer drivers in breast, head and neck and uterine carcinomas (Gonzalez‐Perez et al, 2013) and endometrial cancers (Marshal et al, 2017). Prostate, ovarian and breast cancers frequently feature hemizygous deletions of CTCF (Filippova et al, 2002; Damaschke et al, 2020) and loss of one allele of CTCF in kidney and endometrial cancers correlates with poor patient survival (Kemp et al, 2014; Uhlen et al, 2017). What is the functional impact of the reduction of CTCF levels and how does CTCF prevent oncogenesis? Deletion of one copy of CTCF leads to loss of approximately 30% of CTCF protein in mouse embryonic fibroblasts (MEFs; Aitken et al, 2018). A diminished level of CTCF increases the susceptibility of the hemizygous animals to both spontaneous and radiation‐induced cancers (Kemp et al, 2014). The mechanism of the tumour suppressor action of CTCF might involve deregulated DNA methylation – the loss of CTCF binding can result in hypermethylation of CpGs (Kemp et al, 2014; Damaschke et al, 2020). Only a small fraction of up to a thousand CTCF peaks is lost in the Ctcf+/− MEFs, yet these sites seem particularly relevant as judged by the fact that they predominantly intersect gene promoters (Aitken et al, 2018). Hemizygous deletion of CTCF leads to downregulation of both the mRNA and protein levels of multiple genes linked to oncogenic pathways (Aitken et al, 2018). The affected loci frequently contain multiple CBS and display an altered chromatin signature at their promoter regions including diminished looping to putative enhancer elements (Aitken et al, 2018). Remarkably, similar sets of genes are deregulated in Ctcf+/− MEFs, spontaneously arising murine liver cancers as well as in human cancers with deleterious CTCF mutations (Aitken et al, 2018) which testifies the tumour suppressor action of CTCF. Further sustaining the view of a tumour suppressive action of CTCF, its loss leads to upregulation of the Programmed Cell Death Protein 1 (PD‐L1) which aids cancer cells to evade immune system surveillance (Martin et al, 2021; Oreskovic et al, 2022). Together, these data strongly argue for a tumour suppressive role of CTCF.
While the overexpression of CTCF blocks cell growth by slowing down the cell cycle (Rasko et al, 2001), some point mutations in CTCF that lead to gain of CTCF function can enhance cell survival by blocking apoptosis, as in the case of endometrial carcinomas (Marshal et al, 2017). Interestingly, adrenocortical carcinomas and testicular germ cell cancers frequently feature amplifications of the CTCF coding sequence (Debaugny & Skok, 2020), which might lead to CTCF overexpression. It will be important to determine whether and how the gain of CTCF might favour cancer development and whether the gain of CTCF copies constitutes a cause or a consequence of oncogenic pathway activation.
CTCF‐binding sites in cancer
Numerous mutations and epimutations in CBS can contribute to transcriptional deregulation in diseases. Cancers frequently feature the A•T>C•G or A•T>G•C substitutions within the CBS or sequences immediately adjacent to it (Katainen et al, 2015; Kaiser et al, 2016; Poulos et al, 2016; Guo et al, 2018), which likely leads to loss of insulator sites. Loss of CBS may elicit aberrant gene expression. For instance, the deletion of CTCF‐ bound insulators can lead to upregulation of TAL1 and LMO2 proto‐oncogenes in T‐cell acute lymphoblastic leukemia (Hnisz et al, 2016). In human Gliomas, mutations in the Isocitrate Dehydrogenase (IDH) gene functionally limit the action of Tet enzymes and induce CpG island methylator phenotypes (G‐CIMP). The resulting epimutations lead to loss of CTCF binding and disruption of TAD boundaries (Flavahan et al, 2016) accompanied by aberrant gene expression as exemplified at the locus coding PDGFRA, a glioma oncogene. A similar effect is observed in gastrointestinal stromal tumors where hypermethylation of CTCF motifs within the CBS at domain boundaries at KIT and FGF4 loci favours their expression (Flavahan et al, 2019).
It is unclear which insulator elements are most relevant in cancer. Cornell Non‐Coding Driver (CNCDriver) is a computational method that uses mutational imprints of distinct cancers to identify insulator drivers that affect the dialogue between promoters and enhancers thereby leading to cancer‐related gene deregulation (Liu et al, 2019). The cytokines of the TGFB family including TGFB1 are involved in metastasis of multiple cancer types (Padua & Massagué, 2009). CNCDriver uncovered a mutation in the CTCF motif at the CBS in the vicinity of the TGFB1 locus present in 17% of metastatic melanoma samples. This CBS might act as an insulator decreasing the expression of the TGFB1 gene (Liu et al, 2019).
As we saw above, in murine B cell tumours, the translocation of the immunoglobulin heavy chain (IGH) locus to the vicinity of the oncogene Myc brings along the IGH enhancer. At the resulting chromosome, the IGH enhancer can aberrantly upregulate Myc expression (Gostissa et al, 2009). This translocation also includes a cluster of CBSs that flank the IGH enhancer. The deletion of these CBS has two effects: downregulation of Myc expression due to inefficient Myc‐IgH enhancer communication in the absence of these CBS (which correlates with decreased proliferation of the cancer cells) and an ectopic activation of genes otherwise separated from the IGH enhancer by the CBSs (Vian et al, 2018). Hence, CBS impact enhancer activity at rearranged chromatin loci in cancer.
Interestingly some cancer‐related point mutations in CTCF affect the individual ZFs of CTCF thereby altering its genomic binding profile (Filippova et al, 2002). The cancer‐related mutations L309P, R339Q and R377H affecting the ZF2, ZF3 and ZF4/5, respectively cause variable impact on CTCF binding to its cognate sites as assessed by ChIP (preprint: Bailey et al, 2021). Remarkably, the residue R377 is a hotspot of cancer mutations that may affect CTCF binding to DNA (preprint: Bailey et al, 2021). The R377 is frequently mutated in uterine, skin and bowel cancer (cancerhotspots.org, accessed 31.05.2022). Efforts are on the way to establish the functional implications of ZF mutations for cancer growth and development, using CRISPR‐Cas9 driven mutation and ChIP‐seq experiments.
Role of CTCF in neuronal diseases
In humans, mutations in chromatin‐related factors often manifest themselves with neurological syndromes including intellectual impediments (Janowski et al, 2021). Patients with a heterozygous, four base pair deletion in the 5' end of the coding sequence of CTCF display developmental delay, intellectual disability and microcephaly (MIM#615502). This mutation shifts the reading frame of CTFC mRNA and introduces a premature stop codon thereby functionally knocking out one copy of the CTCF allele (Gregor et al, 2013). More recently, genetic analyses of 39 individuals with neurodevelopmental disorders (NDD) from mild to severe symptoms unveiled more CTCF variants related to neurological diseases (Konrad et al, 2019; Fig 3A) including mutations that might affect CTCF binding to DNA (Fig 3). Most of these variants reported in the study were de novo mutations, except for two types of familial CTCF mutations with mild effects of developmental delay.
Single Nucleotide Polymorphism (SNPs) at CBS have also been linked to neurological diseases (Table 1). The rs1990620 SNP is a risk variant linked to frontotemporal lobar degeneration, a complex disorder featuring a progressive decline in behaviour and dementia. The rs1990620 located upstream of the TSS of the transmembrane protein 106B (TMEM106B) gene increases CTCF binding. Overexpression of TMEM106B is neurotoxic and the genetic alteration correlates with a gain of chromatin contacts between the risk locus and the surrounding regulatory elements including the promoter of TMEM106B and a putative enhancer. This alteration of chromatin structure might lead to increased TMEM106B expression and decreased neuronal cell survival (Gallagher et al, 2017). Numerous other SNP risk variants of neurodegenerative diseases have been shown to intersect CBS (Gallagher et al, 2017) suggesting essential roles of CTCF in maintaining proper neuronal functions. Remarkably, schizophrenia disease risk variants often overlap anchors of CTCF–CTCF loops present in induced pluripotent stem cells‐derived neuronal cells (Rajarajan et al, 2018). These data suggest that changes in insulator activity contribute to transcriptional deregulation in this complex disease.
Table 1.
Single nucleotide polymorphism (SNP) impacting CTCF functionality and disease susceptibility.
| SNP | Locus | Functional impact | Disease | References |
|---|---|---|---|---|
| rs2535629 | ITIH3 (3p21.1) | Disrupts CTCF binding and regulates the expression of the SFMBT1 | Schizophrenia | Li et al (2022) |
| rs796364; rs281759 | 2q33.1 | Disrupts CTCF, RAD21 and FOXP2 binding leading to upregulation of TYW5 (schizophrenia associated factor in brain) | Schizophrenia | Li et al (2022) |
| rs1990620 | TMEM106B (7p21) | Increase in CTCF binding facilitates long range chromatin interactions perhaps leading to the upregulation of TMEM106B | Frontotemporal lobar degeneration | Gallagher et al (2017) |
| rs3825427 | UBAC2 gene (13q32.3) | Increase in UBAC2 expression by recruiting CTCF at the promoter | Noise induced hearing loss | Wan et al (2022) |
| rs34481144 | IFTIM3 (11p15.5) | Recruits CTCF to the promoter and downregulates the expression of IFTIM3 | Influenza disease | Allen et al (2017) |
| rs9820407 | CTNNB1 (3p22.1) | Increase in CTNNB1 expression possibly by CTCF mediated long range chromatin interaction | Osteoporosis | Wang et al (2021) |
Concluding remarks
CTCF is considered as one of the essential proteins implicated in the regulation of chromatin topology and gene expression. The recent data discussed here increase our understanding of the contribution of CTCF to the regulation of gene expression by directly acting at a promoter or by impacting promoter–enhancer contacts. Yet, a number of aspects of CTCF biology are still unclear (Box: In need of answers) and addressing these questions will be key to link chromatin topology to gene expression during development and in disease. Activation of regulatory elements increases their mobility (Gu et al, 2018) and association with mediator condensates (Cho et al, 2018b), which is attenuated when transcription is inhibited (Gu et al, 2018). How can one reconcile these observations? Gene activation correlates with a gain in intra‐TAD interactions (Kieffer‐Kwon et al, 2017; Pekowska et al, 2018) at least in part due to increased cohesin loading at activated promoters and enhancers (Hua et al, 2021). At sites where chromatin‐bound CTCF flanks an enhancer (Fig 1C), PEI are strengthened by CTCF and form architectural stripes (Vian et al, 2018). Recent nanoscopy and scanning electron microscopy data revealed that CTCF and active chromatin marks, including histone modifications typical for active enhancers, co‐localize on the surface of chromatin domains reminiscent of TADs (Miron et al, 2020). It is therefore possible that at loci forming architectural stripes, the enhancer is confined to the TAD surface and by fixing its position in space the PEI may be formed more efficiently.
The frequently observed, relatively subtle transcriptional effects accompanying TAD–TAD merging in the genome engineering experiments agree with the overall weak effect of cohesin and CTCF removal on gene expression (Nora et al, 2017; Rao et al, 2017). These data might be confounded by the fact that the gene expression redout comes from a population of cells. CBS contribute to the cell‐to‐cell variability in gene expression (Ren et al, 2017) particularly affecting transcriptional regulators implicated in the maintenance of cell identity (Wang et al, 2019). Corollary to this, the transcriptional effects of CTCF disruption might require time to fully manifest. Indeed, disruptions of individual CBS often perturb gene expression in long term.
Several groups described widespread immunity of promoters to the influence of new enhancers within the context of reshuffled TADs built at loci that underwent genomic rearrangements (Despang et al, 2019; Ghavi‐Helm et al, 2019; Laugsch et al, 2019). What else, apart from insulators, affects the responsiveness of promoters to enhancers? The sequence composition of regulatory elements likely plays an important role in this process (Zabidi et al, 2015; Arnold et al, 2017; Pachano et al, 2021). Furthermore, the combinatorial cis‐regulatory element landscape of each TAD impacts the promoter–enhancer dialogue by creating a competitive environment actively shaping the likelihood of establishing functional links between genes and enhancers (Lower et al, 2009; Furlong & Levine, 2018; Cho et al, 2018a; Hao et al, 2019; Oudelaar et al, 2019; Oh et al, 2021b; Zuin et al, 2022). TADs are in fact highly dynamic structures constantly built by cohesins and dismantled by the Wapl cohesin unloader (Haarhuis et al, 2017; Hansen et al, 2017; Bintu et al, 2018; Vian et al, 2018). Recent live‐cell imaging of loop dynamics in ES cells revealed that CTCF–CTCF loops are rather rare (3–6% of loci form loops at any given time in the cell population) and persist for up to 30 min (Gabriele et al, 2022). Depending on the composition of the cohesin complex, loops may have varying lifetimes (Wutz et al, 2020). It will be important to understand what regulates the composition of the cohesin complexes at a given genomic locus, how the cohesin composition impacts CTCF functions and whether it affects PEI. Likewise, understanding the mechanisms by which CTCF regulates locus‐specific gene expression will illuminate underlying mechanisms of neurological disorders and likely help to increase our understanding of oncogenesis.
Author contributions
Bondita Dehingia: Writing – original draft; writing – review and editing; visualization. Małgorzata Milewska: Writing – original draft; writing – review and editing; visualization. Marcin Janowski: Writing – original draft. Aleksandra Pękowska: Conceptualization; resources; supervision; funding acquisition; visualization; writing – original draft; project administration; writing – review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
AP – conceptualization. All the authors contributed to manuscript writing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgements
Work in the Pękowska lab is funded by the Dioscuri Grant (Dioscui is a programme initiated by the Max Planck Society (MPG), jointly managed with the National Science Centre in Poland (NCN), and mutually funded by the Polish Ministry of Education and Science and the German Federal Ministry of Education and Research (BMBF)), by the OPUS17 (UMO‐2019/33/B/NZ2/02437) and Sonata Bis 11 (UMO‐2021/42/E/NZ2/00392) grants from the NCN, and by the EMBO Installation Grant. The authors acknowledge Ewa Demianiuk for help with figure rendering and the Pękowska lab members for critical reading of the manuscript.
See the Glossary for abbreviations used in this article.
References
- Aitken SJ, Ibarra‐Soria X, Kentepozidou E, Flicek P, Feig C, Marioni JC, Odom DT (2018) CTCF maintains regulatory homeostasis of cancer pathways. Genome Biol 19: 106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B (2013) Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927 [DOI] [PubMed] [Google Scholar]
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD (2019) Live‐cell imaging reveals enhancer‐dependent Sox2 transcription in the absence of enhancer proximity. elife 8: e41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour E, Marko JF (2012) Self‐organization of domain structures by DNA‐loop‐extruding enzymes. Nucleic Acids Res 40: 11202–11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljahani A, Hua P, Karpinska MA, Quililan K, Davies JOJ, Oudelaar AM (2022) Analysis of sub‐kilobase chromatin topology reveals nano‐scale regulatory interactions with variable dependence on cohesin and CTCF. Nat Commun 13: 2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G, Schopflin R, Jerkovic I, Heinrich V, Ibrahim DM, Paliou C, Hochradel M, Timmermann B, Haas S, Vingron M et al (2017) Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res 27: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E, Daub CO, Vitting‐Seerup K, Andersson R, Lilje B, Drabløs F, Lennartsson A, Rönnerblad M, Hrydziuszko O, Vitezic M et al (2015) Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Zabidi MA, Pagani M, Rath M, Schernhuber K, Kazmar T, Stark A (2017) Genome‐wide assessment of sequence‐intrinsic enhancer responsiveness at single‐base‐pair resolution. Nat Biotechnol 35: 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Burcin M, Kaiser B, Muller M, Renkawitz R (1996) DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic Acids Res 24: 2640–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Mäueler W, Bassili G, Lutz M, Burke L, Epplen TJ, Renkawitz R (2000) The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA‐DRB1(*)0401 gene. Gene 253: 209–214 [DOI] [PubMed] [Google Scholar]
- Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, Lutz M, Neiman PE, Collins SJ, Renkawitz R, Lobanenkov VV et al (1999) Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem 274: 27092–27098 [DOI] [PubMed] [Google Scholar]
- Bailey CG, Gupta S, Metierre C, Amarasekera PM, O'young P, Kyaw W, Laletin T, Francis H, Semaan C, Singh KP et al (2021) Somatic mutations in CTCF zinc fingers produce cellular phenotypes explained by structure‐function relationships. bioRxiv 10.1101/2021.01.08.425848 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad A, Steiner C, Köhne AC, Renkawitz R (1990) Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 [DOI] [PubMed] [Google Scholar]
- Barisic D, Stadler MB, Iurlaro M, Schübeler D (2019) Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 569: 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington C, Georgopoulou D, Pezic D, Varsally W, Herrero J, Hadjur S (2019) Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat Commun 10: 2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth R, Bystricky K, Shaban HA (2020) Coupling chromatin structure and dynamics by live super‐resolution imaging. Sci Adv 6: eaaz2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Blencowe BJ, Rinn JL (2019) Differential contribution of steady‐state RNA and active transcription in chromatin organization. EMBO Rep 20: e48068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Maass PG, Lewandowski JP, Weiner CL, Rinn JL (2018) A TAD boundary is preserved upon deletion of the CTCF‐rich Firre locus. Nat Commun 9: 1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DCA, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR et al (2017) Complex multi‐enhancer contacts captured by genome architecture mapping. Nature 543: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF‐dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA (2019) Decreased enhancer‐promoter proximity accompanying enhancer activation. Mol Cell 76: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su J‐H, Sinnott‐Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X (2018) Super‐resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J‐P, Tanay A et al (2017) Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Gabriele M, Hansen AS (2021) Tracking and interpreting long‐range chromatin interactions with super‐resolution live‐cell imaging. Curr Opin Cell Biol 70: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R (1997) Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol 17: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM (2017) Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon L, Weiss FD, Beagan JA, Oliveira MS, Georgieva R, Wang Y‐F, Carroll TS, Dharmalingam G, Gong W, Tossell K et al (2022) Cohesin‐dependence of neuronal gene expression relates to chromatin loop length. Elife 11: e76539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou M t, Carone DM, Weng Z, Fazzio TG, Rando OJ (2014) High‐resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T (2018) Dynamic interplay between enhancer–promoter topology and gene activity. Nat Genet 50: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian Y, Shu W, Bo X, Wang S (2012) Comprehensive identification and annotation of cell type‐specific and ubiquitous CTCF‐binding sites in the human genome. PLoS ONE 7: e41374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergström R, Kwon Y‐W, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D et al (2007) CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome‐wide. Mol Cell Biol 27: 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ (2005) Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20: 483–489 [DOI] [PubMed] [Google Scholar]
- Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, Yost KE, Kim J, He J, Nevins SA et al (2018a) Promoter of lncRNA gene PVT1 is a tumor‐suppressor DNA boundary element. Cell 173: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W‐K, Spille J‐H, Hecht M, Lee C, Li C, Grube V, Cisse II (2018b) Mediator and RNA polymerase II clusters associate in transcription‐dependent condensates. Science 361: 412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5' element of the chicken beta‐globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila . Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing‐Simmons E, Masella S, Robles‐Rebollo I, Xiao X, Wang Y‐F, Barozzi I et al (2018) Control of inducible gene expression links cohesin to hematopoietic progenitor self‐renewal and differentiation. Nat Immunol 19: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh T‐Y, Cui K, Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Niemitz EL, Ravenel JD, Onyango P, Brandenburg SA, Lobanenkov VV, Feinberg AP (2001) Loss of imprinting of insulin‐like growth factor‐II in Wilms' tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res 61: 4947–4950 [PubMed] [Google Scholar]
- Damaschke NA, Gawdzik J, Avilla M, Yang B, Svaren J, Roopra A, Luo JH, Yu YP, Keles S, Jarrard DF et al (2020) CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin Epigenetics 12: 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao LTM, Galindo‐Albarrán AO, Castro‐Mondragon JA, Andrieu‐Soler C, Medina‐Rivera A, Souaid C, Charbonnier G, Griffon A, Vanhille L, Stephen T et al (2017) Genome‐wide characterization of mammalian promoters with distal enhancer functions. Nat Genet 49: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters J‐M (2019) DNA loop extrusion by human cohesin. Science 366: 1338–1345 [DOI] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ (2013) XCumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaugny RE, Skok JA (2020) CTCF and CTCFL in cancer. Curr Opin Genet Dev 61: 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Verma‐Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D et al (2011) CCCTC‐binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro‐B cells. Proc Natl Acad Sci USA 108: 9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L (2016) The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M et al (2019) Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet 51: 1263–1271 [DOI] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ et al (2017) A tiling‐deletion‐based genetic screen for cis‐regulatory element identification in mammalian cells. Nat Methods 14: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz‐bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W et al (2015) Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K et al (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159: 374–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schübeler D (2013) Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet 9: e1003994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c‐myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett NA, Lobanenkov VV (1998) A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer 22: 26–36 [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ (2001) CTCF‐binding sites flank CTG/CAG repeats and form a methylation‐sensitive insulator at the DM1 locus. Nat Genet 28: 335–343 [DOI] [PubMed] [Google Scholar]
- Filippova GN, Ulmer JE, Moore JM, Ward MD, Hu YJ, Neiman PE, Collins SJ, Qi CF, Loukinov DI, Pugacheva EM et al (2002) Tumor‐associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA‐binding specificity. Cancer Res 62: 48–52 [PubMed] [Google Scholar]
- Fitzpatrick GV, Pugacheva EM, Shin J‐Y, Abdullaev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ (2007) Allele‐specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol Cell Biol 27: 2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, Shareef SJ, Javed NM, Raut CP, Eschle BK et al (2019) Altered chromosomal topology drives oncogenic programs in SDH‐deficient GISTs. Nature 575: 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer‐Rachamimov AO, Suvà ML, Bernstein BE (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana‐Konwalski K (2017) Single‐nucleus Hi‐C reveals unique chromatin reorganization at oocyte‐to‐zygote transition. Nature 544: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schopflin R, Kraft K, Kempfer R, Jerkovic I, Chan W‐L et al (2016) Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269 [DOI] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z (2008) The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA (2015) Formation of chromosomal domains by loop extrusion. Cell Rep 15: 024620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, Levine M (2018) Developmental enhancers and chromosome topology. Science 361: 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele M, Brandão HB, Grosse‐Holz S, Jha A, Dailey GM, Cattoglio C, Hsieh T‐HS, Mirny L, Zechner C, Hansen AS (2022) Dynamics of CTCF‐ and cohesin‐mediated chromatin looping revealed by live‐cell imaging. Science 376: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, Chesi A, Manduchi E, Wells AD, Grant SFA et al (2017) A dementia‐associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet 101: 643–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters J, Mirny LA, Tachibana K (2017) A mechanism of cohesin‐dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi‐Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM (2019) Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 51: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi‐Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM (2014) Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96–100 [DOI] [PubMed] [Google Scholar]
- Golfier S, Quail T, Kimura H, Brugués J (2020) Cohesin and condensin extrude DNA loops in a cell‐cycle dependent manner. Elife 9: e53885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Lazaris C, Sakellaropoulos T, Lozano A, Kambadur P, Ntziachristos P, Aifantis I, Tsirigos A (2018) Stratification of TAD boundaries reveals preferential insulation of super‐enhancers by strong boundaries. Nat Commun 9: 542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Perez A, Perez‐Llamas C, Deu‐Pons J, Tamborero D, Schroeder MP, Jene‐Sanz A, Santos A, Lopez‐Bigas N (2013) IntOGen‐mutations identifies cancer drivers across tumor types. Nat Methods 10: 1081–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogné M, Pinaud E, Alt FW (2009) Long‐range oncogenic activation of Igh‐c‐myc translocations by the Igh 3′ regulatory region. Nature 462: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S et al (2013) De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet 93: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, Wysocka J (2020) Opposing effects of cohesin and transcription on CTCF organization revealed by super‐resolution imaging. Mol Cell 80: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J (2018) Transcription‐coupled changes in nuclear mobility of mammalian cis‐regulatory elements. Science 359: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y et al (2015) CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YA, Chang MM, Huang W, Ooi WF, Xing M, Tan P, Skanderup AJ (2018) Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat Commun 9: 1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yanez‐Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B et al (2017) The Cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P (1996) Fab‐7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10: 3202–3215 [DOI] [PubMed] [Google Scholar]
- Hansen AS, Amitai A, Cattoglio C, Tjian R, Darzacq X (2020) Guided nuclear exploration increases CTCF target search efficiency. Nat Chem Biol 16: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hsieh THS, Cattoglio C, Pustova I, Saldaña‐Meyer R, Reinberg D, Darzacq X, Tjian R (2019) Distinct classes of chromatin loops revealed by deletion of an RNA‐binding region in CTCF. Mol Cell 76: 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6: e25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen LLP, Kassouf MT, Oudelaar AM, Biggs D, Preece C, Downes DJ, Gosden M, Sharpe JA, Sloane‐Stanley JA, Hughes JR et al (2017) Tissue‐specific CTCF‐cohesin‐mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol 19: 952–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Shearwin KE, Dodd IB (2019) Positive and negative control of enhancer‐promoter interactions by other DNA loops generates specificity and tunability. Cell Rep 26: 2419–2433 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation‐sensitive enhancer‐blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X (2017) Structural basis for the versatile and methylation‐dependent binding of CTCF to DNA. Mol Cell 66: 711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikichi T, Kohda T, Kaneko‐Ishino T, Ishino F (2003) Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain‐specific promoters and mouse‐specific CTCF‐binding sites. Nucleic Acids Res 31: 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintrau AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA et al (2016) Activation of proto‐oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren C, Kanduri C, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R (2001) CpG methylation regulates the Igf2/H19 insulator. Curr Biol 11: 1128–1130 [DOI] [PubMed] [Google Scholar]
- Hsieh T‐HS, Cattoglio C, Slobodyanyuk E, Hansen AS, Darzacq X & Tjian R (2021) Enhancer‐promoter interactions and transcription are maintained upon acute loss of CTCF, cohesin, WAPL, and YY1. bioRxiv 10.1101/2021.07.14.452365 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X (2020) Resolving the 3D landscape of transcription‐linked mammalian chromatin folding. Mol Cell 78: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua P, Badat M, Hanssen LLP, Hentges LD, Crump N, Downes DJ, Jeziorska DM, Oudelaar AM, Schwessinger R, Taylor S et al (2021) Defining genome architecture at base‐pair resolution. Nature 595: 125–129 [DOI] [PubMed] [Google Scholar]
- Huang H, Zhu Q, Jussila A, Han Y, Bintu B, Kern C, Conte M, Zhang Y, Bianco S, Chiariello AM et al (2021) CTCF mediates dosage‐ and sequence‐context‐dependent transcriptional insulation by forming local chromatin domains. Nat Genet 53: 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM (2017) Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228 [DOI] [PubMed] [Google Scholar]
- Hyle J, Zhang Y, Wright S, Xu B, Shao Y, Easton J, Tian L, Feng R, Xu P, Li C (2019) Acute depletion of CTCF directly affects MYC regulation through loss of enhancer‐promoter looping. Nucleic Acids Res 47: 6699–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski M, Milewska M, Zare P, Pękowska A (2021) Chromatin alterations in neurological disorders and strategies of (Epi)genome rescue. Pharmaceuticals 14: 765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang J, Lun K, Li B, Zheng H, Li Y, Zhou R, Duan W, Wang C, Feng Y et al (2020) Genome‐wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol 21: 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS et al (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Taylor MS, Semple CA (2016) Mutational biases drive elevated rates of substitution at regulatory sites across cancer types. PLoS Genet 12: e1006207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L (2011) Genome architectures revealed by tethered chromosome conformation capture and population‐based modeling. Nat Biotechnol 30: 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin‐specific and methylation‐sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, Gylfe AE, Ristolainen H, Hänninen UA, Cajuso T et al (2015) CTCF/cohesin‐binding sites are frequently mutated in cancer. Nat Genet 47: 818–821 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1991) A position‐effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950 [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, Rabaia NA, Gurley KE, Guinney J, Busch SE et al (2014) CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep 7: 1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, van Blerkom P, Kieffer‐Kwon P, Kieffer‐Kwon K‐R, Park S, Wang X et al (2019) A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer‐Kwon K‐R, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P et al (2017) Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol Cell 67: 566–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer‐Kwon K‐R, Tang Z, Mathe E, Qian J, Sung M‐H, Li G, Resch W, Baek S, Pruett N, Grøntved L et al (2013) Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H (2019) Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c‐myc gene, is an 11‐Zn‐finger protein differentially expressed in multiple forms. Mol Cell Biol 13: 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad EDH, Nardini N, Caliebe A, Nagel I, Young D, Horvath G, Santoro SL, Shuss C, Ziegler A, Bonneau D et al (2019) CTCF variants in 39individuals with a variable neurodevelopmental disorder broaden the mutational andclinical spectrum. Genet Med 21: 2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Ishii H, Xiong X, Bianco S, Meitinger F, Hu R, Hocker JD, Conte M, Gorkin D, Yu M et al (2021) Promoter‐proximal CTCF binding promotes distal enhancer‐dependent gene activation. Nat Struct Mol Biol 28: 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Kesner B, An JY, Ahn JY, Cifuentes‐Rojas C, Colognori D, Jeon Y, Szanto A, del Rosario BC, Pinter SF et al (2015) Locus‐specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 57: 361–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506 [DOI] [PubMed] [Google Scholar]
- Laugsch M, Bartusel M, Rehimi R, Alirzayeva H, Karaolidou A, Crispatzu G, Zentis P, Nikolic M, Bleckwehl T, Kolovos P et al (2019) Modeling the pathological long‐range regulatory effects of human structural variation with patient‐specific hiPSCs. Cell Stem Cell 24: 736–752 [DOI] [PubMed] [Google Scholar]
- Lavagnolli T, Gupta P, Hörmanseder E, Mira‐Bontenbal H, Dharmalingam G, Hörmanseder E, Gurdon JB, Fisher AG, Merkenschlager M (2015) Initiation and maintenance of pluripotency gene expression in the absence of cohesin. Genes Dev 29: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Kang M‐K, Kim Y‐J, Yang B, Shim H, Kim S, Kim K, Yang CM, Min B‐G, Jung W‐J et al (2022) CTCF‐mediated chromatin looping provides a topological framework for the formation of phase‐separated transcriptional condensates. Nucleic Acids Res 50: 207–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C (2008) The LPS‐induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Haarhuis JHI, Sedeño Cacciatore Á, Oldenkamp R, van Ruiten MS, Willems L, Teunissen H, Muir KW, de Wit E, Rowland BD et al (2020) The structural basis for cohesin–CTCF‐anchored loops. Nature 578: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rivera CM, Ishii H, Jin F, Selvaraj S, Lee AY, Dixon JR, Ren B (2014) CRISPR reveals a distal super‐enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS ONE 9: e114485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO et al (2009) Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Ferguson‐Smith AC, Schultz RM, Bartolomei MS (2011) Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol Cell Biol 31: 3094–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EM, Martinez‐Fundichely A, Diaz BJ, Aronson B, Cuykendall T, MacKay M, Dhingra P, Wong EWP, Chi P, Apostolou E et al (2019) Identification of cancer drivers at CTCF insulators in 1,962 whole genomes. Cell Systems 8: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NQ, Maresca M, van den Brand T, Braccioli L, Schijns MMGA, Teunissen H, Bruneau BG, Nora EP, de Wit E (2021) WAPL maintains a cohesin loading cycle to preserve cell‐type‐specific distal gene regulation. Nat Genet 53: 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R (2016) Editing DNA methylation in the mammalian genome. Cell 167: 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R (2011) Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov V, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH (1990) A novel sequence‐specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC‐motif in the 5'‐flanking sequence of the chicken c‐myc gene. Oncogene 5: 1742–1753 [PubMed] [Google Scholar]
- Lower KM, Hughes JR, de Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane‐Stanley J, Vernimmen D et al (2009) Adventitious changes in long‐range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci USA 106: 21771–21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J, Xiang G, Gómez‐García PA, Tome JM, Zhang Z, Vermunt MW, Zhang H, Huang A, Keller CA, Giardine BM et al (2021) Distinct properties and functions of CTCF revealed by a rapidly inducible degron system. Cell Rep 34: 108783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R et al (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene‐enhancer interactions. Cell 161: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, Schultheiss H, Brehm A, Kouzarides T, Lobanenkov V et al (2000) Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res 28: 1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Chadwick BP, Boss JM (2008) The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long‐distance chromatin interactions. J Exp Med 205: 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino MM, Rega C, Russo R, Valletta M, Gentile MT, Esposito S, Baglivo I, de Feis I, Angelini C, Xiao T et al (2019) Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF. J Biol Chem 294: 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshal AD, Bailey CG, Champ K, Vellozzi M, O'Young P, Metierre C, Feng Y, Thoeng A, Richards AM, Schmitz U et al (2017) CTCF genetic alterations in endometrial carcinoma are pro‐tumorigenic. Oncogene 36: 4100–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TD, Patel RS, Cook DR, Choi MY, Patil A, Liang AC, Li MZ, Haigis KM, Elledge SJ (2021) The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 373: 1327–1335 [DOI] [PubMed] [Google Scholar]
- Mifsud B, Tavares‐Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA et al (2015) Mapping long‐range promoter contacts in human cells with high‐resolution capture Hi‐C. Nat Genet 47: 598–606 [DOI] [PubMed] [Google Scholar]
- Miron E, Oldenkamp R, Brown JM, Pinto DMS, Xu CS, Faria AR, Shaban HA, Rhodes JDP, Innocent C, de Ornellas S et al (2020) Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci Adv 6: eaba8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T (2020) The self‐organizing genome: principles of genome architecture and function. Cell 183: 28–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Imai Y, Hori S, Nishio M, Loo TM, Okada R, Yang L, Nakadai T, Maruyama R, Fujii R et al (2021) Pericentromeric noncoding RNA changes DNA binding of CTCF and inflammatory gene expression in senescence and cancer. Proc Natl Acad Sci USA 118: e2025647118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D (2011) A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145 [DOI] [PubMed] [Google Scholar]
- Nakahashi H, Kwon KRK, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A et al (2013) A genome‐wide map of CTCF multivalency redefines the CTCF code. Cell Rep 3: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Bulajié M, Dekker J, Mazzoni EO, Reinberg D (2016) CTCF‐mediated topological boundaries during development foster appropriate gene regulation. Genes Dev 30: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D (2015) CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347: 1017–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745 [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT (2016) Rapid protein depletion in human cells by auxin‐inducible degron tagging with short homology donors. Cell Rep 15: 210–218 [DOI] [PubMed] [Google Scholar]
- Nora EP, Caccianini L, Fudenberg G, So K, Kameswaran V, Nagle A, Uebersohn A, Hajj B, Saux A le, Coulon A, et al (2020) Molecular basis of CTCF binding polarity in genome folding Nat Commun 11, 5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A‐L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG (2017) Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J et al (2012) Spatial partitioning of the regulatory landscape of the X‐inactivation centre. Nature 485: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Aguilar R, Kesner B, Lee HG, Kriz AJ, Chu HP, Lee JT (2021a) Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 184: 6157–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Shao J, Mitra J, Xiong F, D'Antonio M, Wang R, Garcia‐Bassets I, Ma Q, Zhu X, Lee JH et al (2021b) Enhancer release and retargeting activates disease‐susceptibility genes. Nature 595: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E (2010) Does CTCF mediate between nuclear organization and gene expression? BioEssays 32: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich T, Vega‐Sendino M, Tillo D, Wu W, Zolnerowich N, Pavani R, Tran AD, Domingo CN, Franco M, Markiewicz‐Potoczny M et al (2021) CTCF is a barrier for 2C‐like reprogramming. Nature Communications 12: 4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreskovic E, Wheeler EC, Mengwasser KE, Fujimura E, Martin TD, Tothova Z, Elledge SJ (2022) Genetic analysis of cancer drivers reveals cohesin and CTCF as suppressors of PD‐L1. Proc Natl Acad Sci USA 119: e2120540119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortabozkoyun H, Huang P‐Y, Cho H, Narendra V, LeRoy G, Gonzalez‐Buendia E, Skok JA, Tsirigos A, Mazzoni EO, Reinberg D (2022) CRISPR and biochemical screens identify MAZ as a cofactor in CTCF‐mediated insulation at Hox clusters. Nat Genet 54: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar AM, Harrold CL, Hanssen LLP, Telenius JM, Higgs DR, Hughes JR (2019) A revised model for promoter competition based on multi‐way chromatin interactions at the α‐globin locus. Nat Commun 10: 5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachano T, Sánchez‐Gaya V, Ealo T, Mariner‐Faulí M, Bleckwehl T, Asenjo HG, Respuela P, Cruz‐Molina S, Muñoz‐San Martín M, Haro E et al (2021) Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness. Nat Genet 53: 1036–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua D, Massagué J (2009) Roles of TGFbeta in metastasis. Cell Res 19: 89–102 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T et al (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Pekowska A, Klaus B, Xiang W, Severino J, Daigle N, Klein FA, Oles M, Casellas R, Ellenberg J, Steinmetz LM et al (2018) Gain of CTCF‐anchored chromatin loops marks the exit from naive pluripotency. Cell Syst 7: 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, van Bortle K, Spacek D, Hess GT, Shamim MS, Machol I, Love MI, Aiden EL, Bassik MC, Snyder MP (2017) Static and dynamic DNA loops form AP‐1‐bound activation hubs during macrophage development. Mol Cell 67: 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos RC, Thoms JAI, Guan YF, Unnikrishnan A, Pimanda JE, Wong JWH (2016) Functional mutations form at CTCF‐cohesin binding sites in melanoma due to uneven nucleotide excision repair across the motif. Cell Rep 17: 2865–2872 [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, Strunnikov AV, Zentner GE, Ren B, Lobanenkov VV (2020) CTCF mediates chromatin looping via N‐terminal domain‐dependent cohesin retention. Proc Natl Acad Sci USA 117: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke WW, Taheny MJ, Fochtmann LJ, Vostrov AA (2000) Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res 28: 3370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, Powell S, Yashaswini C, LaMarca EA, Kassim B et al (2018) Neuron‐specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362: eaat4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang S‐C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer‐Kwon K‐R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID et al (2017) Cohesin loss eliminates all loop domains. Cell 171: 305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko JEJ, Klenova EM, Leon J, Filippova GN, Loukinov DI, Vatolin S, Robinson AF, Jia HY, Ulmer J, Ward MD et al (2001) Cell growth inhibition by the multifunctional multivalent zinc‐finger factor CTCF. Cancer Res 61: 6002–6007 [PubMed] [Google Scholar]
- Recillas‐Targa F, Bell AC, Felsenfeld G (1999) Positional enhancer‐blocking activity of the chicken beta‐globin insulator in transiently transfected cells. Proc Natl Acad Sci USA 96: 14354–14359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas‐Targa F, Pikaart MJ, Burgess‐Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G (2002) Position‐effect protection and enhancer blocking by the chicken  ‐globin insulator are separable activities. Proc Natl Acad Sci USA 99: 6883–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redolfi J, Zhan Y, Valdes‐Quezada C, Kryzhanovska M, Guerreiro I, Iesmantavicius V, Pollex T, Grand RS, Mulugeta E, Kind J et al (2019) DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nat Struct Mol Biol 26: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, Larson DR, Zhao K (2017) CTCF‐mediated enhancer‐promoter interaction is a critical regulator of cell‐to‐cell variation of gene expression. Mol Cell 67: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda M, Baglivo I, Burgess‐Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV (2007) Critical DNA binding interactions of the insulator protein CTCF: A small number of zinc fingers mediate strong binding, and a single finger‐DNA interaction controls binding at imprinted loci. J Biol Chem 282: 33336–33345 [DOI] [PubMed] [Google Scholar]
- Ribeiro‐Dos‐Santos AM, Hogan MS, Luther RD, Brosh R, Maurano MT (2022) Genomic context sensitivity of insulator function. Genome Res 32: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Carballo E, Lopez‐Delisle L, Zhan Y, Fabre PJ, Beccari L, El‐Idrissi I, Nguyen Huynh TH, Ozadam H, Dekker J, Duboule D (2017) The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev 31: 2264–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando‐Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG (2017) Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio‐Perez C, Tamborero D, Schroeder MP, Antolín AA, Deu‐Pons J, Perez‐Llamas C, Mestres J, Gonzalez‐Perez A, Lopez‐Bigas N (2015) In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27: 382–396 [DOI] [PubMed] [Google Scholar]
- Saldaña‐Meyer R, González‐Buendía E, Guerrero G, Narendra V, Bonasio R, Recillas‐Targa F, Reinberg D (2014) CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 28: 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña‐Meyer R, Rodriguez‐Hernaez J, Escobar T, Nishana M, Jácome‐López K, Nora EP, Bruneau BG, Tsirigos A, Furlan‐Magaril M, Skok J et al (2019) RNA interactions are essential for CTCF‐mediated genome organization. Mol Cell 76: 412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S‐C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J et al (2015) Chromatin extrusion explains key features of loop and domain formation in wild‐type and engineered genomes. Proc Natl Acad Sci USA 112: E6456–E6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long‐range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Manteiga JC, Weintraub AS, Day DS, Zamudio AV, Hnisz D, Lee TI, Young RA (2018) Transcriptional dysregulation of MYC reveals common enhancer‐docking mechanism. Cell Rep 23: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe‐Mie Y, Fonseca NA, Huber W, Haering H, Mirny L et al (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G (2012) Three‐dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472 [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV et al (2012) A map of the cis‐regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan W‐C, Georgopoulou D, Vietri Rudan M, Mira‐Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S (2013) Cohesin‐mediated interactions organize chromosomal domain architecture. EMBO J 32: 3119–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soochit W, Sleutels F, Stik G, Bartkuhn M, Basu S, Hernandez SC, Merzouk S, Vidal E, Boers R, Boers J et al (2021) CTCF chromatin residence time controls three‐dimensional genome organization, gene expression and DNA methylation in pluripotent cells. Nat Cell Biol 23: 881–893 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra R‐J, Klous P, Grosveld F, Galjart N, de Laat W (2006) CTCF mediates long‐range chromatin looping and local histone modification in the beta‐globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D et al (2011) DNA‐binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c‐myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stik G, Vidal E, Barrero M, Cuartero S, Vila‐Casadesús M, Mendieta‐Esteban J, Tian T, Choi J, Berenguer C, Abad A et al (2020) CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat Genet 52: 655–661 [DOI] [PubMed] [Google Scholar]
- Sun S, del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT (2013) Jpx RNA activates xist by evicting CTCF. Cell 153: 1537–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, Spitz F (2016) The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev Cell 39: 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F (2014) Functional and topological characteristics of mammalian regulatory domains. Genome Res 24: 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó PE, Tang SHE, Rentsendorj A, Pfeifer GP, Mann JR (2000) Maternal‐specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10: 607–610 [DOI] [PubMed] [Google Scholar]
- Szabo Q, Donjon A, Jerković I, Papadopoulos GL, Cheutin T, Bonev B, Nora EP, Bruneau BG, Bantignies F, Cavalli G (2020) Regulation of single‐cell genome organization into TADs and chromatin nanodomains. Nat Genet 52: 1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B et al (2015) CTCF‐mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron‐Herger M, Mallm JP, Marth C, Höfer T, Rippe K (2012) Genome‐wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19: 1185–1192 [DOI] [PubMed] [Google Scholar]
- Thiecke MJ, Wutz G, Muhar M, Tang W, Bevan S, Malysheva V, Stocsits R, Neumann T, Zuber J, Fraser P et al (2020) Cohesin‐dependent and ‐independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep 32: 107929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F et al (2017) A pathology atlas of the human cancer transcriptome. Science 357: eaan2507 [DOI] [PubMed] [Google Scholar]
- Ushiki A, Zhang Y, Xiong C, Zhao J, Georgakopoulos‐Soares I, Kane L, Jamieson K, Bamshad MJ, Nickerson DA, Shen Y et al (2021) Deletion of CTCF sites in the SHH locus alters enhancer–promoter interactions and leads to acheiropodia. Nat Commun 12: 2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uuskula‐Reimand L, Hou H, Samavarchi‐Tehrani P, Rudan MV, Liang M, Medina‐Rivera A, Mohammed H, Schmidt D, Schwalie P, Young EJ et al (2016) Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol 17: 182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta M, Russo R, Baglivo I, Russo V, Ragucci S, Sandomenico A, Iaccarino E, Ruvo M, de Feis I, Angelini C et al (2020) Exploring the interaction between the swi/snf chromatin remodeling complex and the zinc finger factor ctcf. Int J Mol Sci 21: 8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer‐Kwon KR, Jung S, Baranello L, Huang SC, de Khattabi L, Dose M, Pruett N et al (2018) The energetics and physiological impact of cohesin extrusion. Cell 173: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S (2015) Comparative Hi‐C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10: 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos ESM, Valdes‐Quezada C, Huang Y, Allahyar A, Verstegen MJAM, Felder A‐K, van der Vegt F, Uijttewaal ECH, Krijger PHL, de Laat W (2021) Interplay between CTCF boundaries and a super enhancer controls cohesin extrusion trajectories and gene expression. Mol Cell 81: 3082–3095 [DOI] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R et al (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22: 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su J‐H, Beliveau BJ, Bintu B, Moffitt JR, Wu C, Zhuang X (2016) Spatial organization of chromatin domains and compartments in single chromosomes. Science 353: 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ren G, Hong N, Jin W (2019) Exploring the changing landscape of cell‐to‐cell variation after CTCF knockdown via single cell RNA‐seq. BMC Genomics 20: 1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhang L, Ruan H, Li G (2020) Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells. Nucleic Acids Res 48: 5939–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T et al (2008) Cohesin mediates transcriptional insulation by CCCTC‐binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Weth O, Paprotka C, Günther K, Schulte A, Baierl M, Leers J, Galjart N, Renkawitz R (2014) CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res 42: 11941–11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechens N, Singh V, Gkikopoulos T, Schofield P, Rocha S, Owen‐Hughes T (2016) The chromatin remodelling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet 12: e1005940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehle L, Thorn GJ, Raddatz G, Clarkson CT, Rippe K, Lyko F, Breiling A, Teif VB (2019) DNA (de)methylation in embryonic stem cells controls CTCF‐dependent chromatin boundaries. Genome Res 29: 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemin A, Lopez‐Delisle L, Bolt CC, Gadolini ML, Duboule D, Rodriguez‐Carballo E (2021) Induction of a chromatin boundary in vivo upon insertion of a tad border. PLoS Genet 17: e1009691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Lettice LA, Hill RE, Bickmore WA (2016) Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143: 2994–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes‐Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W (2015) CTCF binding polarity determines chromatin looping. Mol Cell 60: 676–684 [DOI] [PubMed] [Google Scholar]
- Wutz G, Ladurner R, St Hilaire BG, Stocsits RR, Nagasaka K, Pignard B, Sanborn A, Tang W, Varnai C, Ivanov MP et al (2020) ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinstag1 from WAPL. elife 9: e52091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ et al (2017) Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: e201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL (2000) Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res 10: 1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Li X, Felsenfeld G (2021) The Myc‐associated zinc finger protein (MAZ) works together with CTCF to control cohesin positioning and genome organization. Proc Natl Acad Sci USA 118: e52091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Dong P, Chen X, Hsieh T‐HS, Banala S, de Marzio M, English BP, Qi Y, Jung SK, Kieffer‐Kwon K‐R et al (2020) 3D ATAC‐PALM: super‐resolution imaging of the accessible genome. Nat Methods 17: 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Wang H, Wright S, Hyle J, Zhang Y, Shao Y, Niu M, Fan Y, Rosikiewicz W, Djekidel MN et al (2021) Acute depletion of CTCF rewires genome‐wide chromatin accessibility. Genome Biol 22: 244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini‐Otero RD, Felsenfeld G (2010) Mediation of CTCF transcriptional insulation by DEAD‐box RNA‐binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24: 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, Wang Y (2017) Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res 27: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, Piel JC et al (2005) Rasgrf1 imprinting is regulated by a CTCF‐dependent methylation‐sensitive enhancer blocker. Mol Cell Biol 25: 11184–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C et al (2004) Poly(ADP‐ribosyl)ation regulates CTCF‐dependent chromatin insulation. Nat Genet 36: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A (2015) Enhancer–core‐promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Huang P, Sharma M, Keller CA, Giardine B, Zhang H, Gilgenast TG, Phillips‐Cremins JE, Hardison RC, Blobel GA (2020) Alteration of genome folding via contact domain boundary insertion. Nat Genet 52: 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lam J, Zhang D, Lan Y, Vermunt MW, Keller CA, Giardine B, Hardison RC, Blobel GA (2021) CTCF and transcription influence chromatin structure re‐configuration after mitosis. Nat Commun 12: 5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millán‐Ariño L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS et al (2015) PARP1‐ and CTCF‐mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell 59: 984–997 [DOI] [PubMed] [Google Scholar]
- Zhong X‐P, Krangel MS (1997) An enhancer‐blocking element between and gene segments within the human T cell receptor/locus. Proc Natl Acad Sci USA 94: 5219–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M (1996) The Fab‐7 element of the bithorax complex attenuates enhancer‐promoter interactions in the Drosophila embryo. Genes Dev 10: 3195–3201 [DOI] [PubMed] [Google Scholar]
- Zhou J, Levine M (1999) A novel cis‐regulatory element, the PTS, mediates an anti‐insulator activity in the Drosophila embryo. Cell 99: 567–575 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yu J, Gu J, Xue C, Zhang L, Chen J, Shen L (2021) Relaxed 3D genome conformation facilitates the pluripotent to totipotent‐like state transition in embryonic stem cells. Nucleic Acids Res 49: 12167–12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ et al (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 111: 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Roth G, Zhan Y, Cramard J, Redolfi J, Piskadlo E, Mach P, Kryzhanovska M, Tihanyi G, Kohler H et al (2022) Nonlinear control of transcription through enhancer‐promoter interactions. Nature 604: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]


