Abstract
The long noncoding RNA LINC00839 has been shown to be involved in the progression of some cancer types, such as bladder cancer, prostate cancer, breast cancer, and neuroblastoma. However, if LINC00839 has roles in colorectal cancer (CRC), it has not been elucidated so far. Here, we focus on the biological role and involved mechanisms of LINC00839 in CRC. We show that LINC00839 is selectively upregulated in CRC and locates to the nucleus. High expression of LINC00839 is associated with poor outcomes in CRC patients. Functional experiments show that LINC00839 promotes CRC proliferation, invasion, and metastasis in vitro and in vivo. Mechanistically, LINC00839 recruits Ruvb1 to the Tip60 complex and increases its acetylase activity. LINC00839 guides the complex to the NRF1 promoter and promotes acetylation of lysines 5 and 8 of histones H4, thereby upregulating the expression of NRF1. Subsequently, NRF1 activates mitochondrial metabolism and biogenesis, thereby promoting CRC progression. In summary, our study reports on a mechanism by which LINC00839 positively regulates NRF1, thus promoting mitochondrial metabolism and biogenesis, as well as CRC progression.
Keywords: histone acetylation, LINC00839, NRF1, OXPHOS, Tip60 complex
Subject Categories: Cancer, Metabolism, RNA Biology
Nuclear lncRNA LINC00839 serves as a scaffold to recruit and activate RUVBL1/TIP60 complexes. LINC00839 targets the NRF1 promoter, enhances its expression, promotes mitochondrial metabolism and biogenesis, and thus colorectal cancer progression.

Introduction
Colorectal cancer (CRC) is an aggressive malignancy, and it has the third highest incidence and second highest mortality rate of all cancers worldwide (Siegel & Miller, 2019). The initial events in CRC have been relatively well studied, and treatments for CRC have significantly improved over the past few decades; however, the mechanism underlying CRC tumorigenesis has not been fully elucidated, and effective diagnostic methods have not been fully identified until recently (Stein & Schlag, 2007). Therefore, the molecular mechanisms underlying CRC tumorigenesis and progression should be explored to facilitate the development of new diagnostics and the discovery of new therapeutic strategies for CRC, especially for patients with advanced disease.
Tumor cells require energy and material metabolism to provide a foundation for their survival and reproduction. Traditionally, enhanced aerobic glycolysis was considered an important hallmark of cancer (Hua & Ten Dijke, 2020). However, in recent years, metabonomic studies based on isotope labeling have revealed that the function and activity of mitochondria in tumors are complete (Porporato et al, 2018; Kreitz et al, 2019). The mitochondrial TCA cycle can also provide energy for tumor cell survival or proliferation. With the increasing evidences, the role of the TCA cycle in cancer metabolism needs to be re‐evaluated (Anderson et al, 2018; Huang et al, 2019; Cai et al, 2020). In addition, changes in mitochondrial function and mitochondrial biogenesis, which are regulated by many factors, also play important roles in cancer. Peroxisome proliferator‐activated receptor‐gamma (PPARγ) coactivator‐1alpha (PGC‐1α) activates nuclear respiratory factor 1 and 2 (NRF1 and NRF2) and subsequently mitochondrial transcription factor A (TFAM), thus regulating mitochondrial biogenesis (Collu‐Marchese et al, 2015). Abnormal mitochondrial biogenesis has been observed in many cancers (LeBleu et al, 2014; Torrens‐Mas et al, 2019; Si et al, 2020). Therefore, to treat cancer effectively, therapeutic interventions that target the mitochondria must be considered—future studies on combination therapies will be important for advancing cancer treatments.
LncRNAs are aberrantly expressed in a variety of tumors and are closely related to malignant transformation. The mechanisms underlying lncRNA functions are diverse. For example, lncRNA MALAT1 can suppress breast cancer metastasis by inhibiting transcriptional activation (Kim et al, 2018). The lncRNA UCA1, which sponges antitumor miRNAs, accelerates tumor progression in gastric cancer (Wang et al, 2019). LncRNAs can also serve as scaffolds to guide proteins to form ribonucleoprotein complexes (RNPs) or recruit chromatin‐modifying complexes to target genes in the nucleus (Huarte et al, 2010; Simon et al, 2013). The lncRNA LINC00839, located on human chromosome 10q11.21, is functional unknown, and reports of its role in cancer are scanty. To date, limited studies have documented that LINC00839 is involved in cancers, such as bladder cancer, prostate cancer, breast cancer, and neuroblastoma (Sahu et al, 2018; Xu et al, 2018; Vishnubalaji et al, 2019; Zhang et al, 2019, 2020; Chen et al, 2020). Evidence has shown that high glucose can induce the upregulation of LINC00839 in breast cancer cells (Hu et al, 2019). LINC00839 can interact with Lin28B to promote breast cancer chemoresistance (Chen et al, 2020), and it can also competitively bind to microRNA‐454‐3p and thus increase c‐Met expression (Zhang et al, 2020). Recent studies suggest that LINC00839 silencing represses neuroblastoma cell proliferation, invasion, and glycolysis and promotes apoptosis through the miR‐338‐3p/GLUT1 axis (Yang et al, 2021). However, the role of LINC00839 in CRC is not well characterized, and whether it exerts an effect on CRC has not yet been elucidated.
In this study, we report that LINC00839 is a critical oncogenic lncRNA that is capable of predicting adverse CRC prognosis. LINC00839 promotes cellular proliferation and metastasis. Mechanistically, LINC000839 exerts its oncogenic effects by recruiting Ruvb1 to the Tip60 complex and increasing its acetylase activity, resulting in the acetylation of histones at the NRF1 promoter and increased expression of NRF1; subsequently, NRF1 plays an important role in OXPHOS and mitochondrial biogenesis in CRC. This study systematically demonstrates the oncogenic role of LINC00839, as well as its underlying mechanism, and provides a potential therapeutic target for CRC.
Results
LINC00839 is highly expressed in CRC and positively associated with poor outcomes in patients
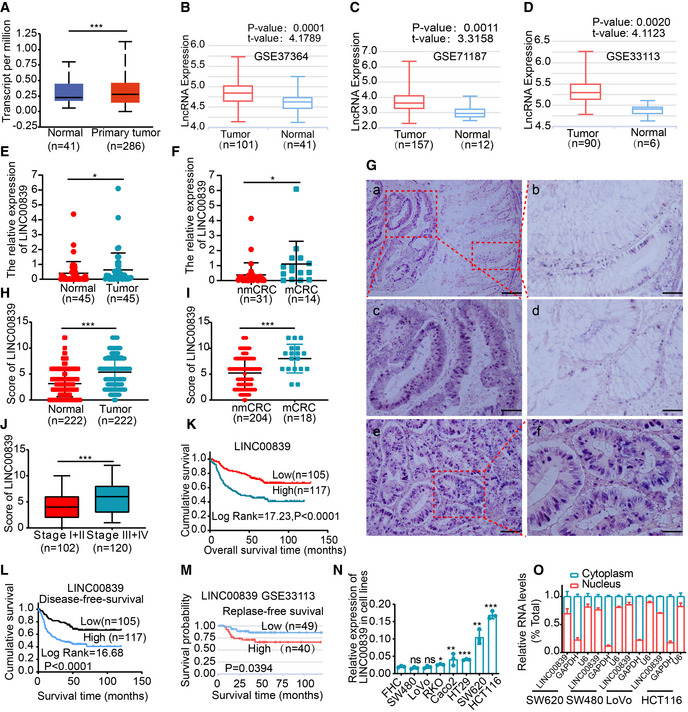
We analyzed the expression of LINC00839 using the UALCAN database (Chandrashekar et al, 2017) and lnCAR datasets, and the results suggested that LINC00839 was more highly expressed in CRC tissues than in matched normal adjacent tissues (NATs; Fig 1A–D). LINC00839 mRNA expression was increased in tumor specimens obtained from 45 CRC patients as determined by qPCR, and its expression was higher in patients with metastasis (Fig 1E and F). In situ hybridization (ISH) was used to verify the expression of LINC00839 in a cohort of 222 formalin‐fixed and paraffin‐embedded (FFPE) samples. The results also revealed that 52.7% (117/222) of patients had higher LINC00839 expression in tumor tissues than in NATs (Fig 1G and H). These data suggest that LINC00839 is highly expressed in CRC, and its high expression is correlated with metastasis.
Figure 1. LINC00839 is highly expressed in CRC and positively associated with poor outcomes in patients.
-
AThe expression of LINC00839 in CRC tissues and normal mucosa tissues in the UALCAN database.
- B–D
-
E, FThe expression levels of LINC00839 in tumor tissues and NATs from the CRC cohort (n = 45). nmCRC denotes CRC patients without metastases, and mCRC denotes CRC patients with metastases.
-
GRepresentative ISH staining of LINC00839 in paraffin‐embedded samples of CRC tumor tissues and NATs. (a) Expression of LINC00839 in tumor tissues and NATs. Scale bars, 100 μm. (b) Magnification of normal mucosal area and (c) magnification of the tumor area. Scale bars, 50 μm. (d) Weakly positive staining of LINC00839 in normal mucosa. Scale bars, 50 μm. (e) Strong positive staining of LINC00839 in tumors and (f) magnification of the local area. Scale bars, 100 μm.
-
HThe expression score of the tumors and NATs in the ISH cohort (n = 222).
-
IDifferential expression of LINC00839 in nonmetastatic CRC patients (nmCRC) and metastatic CRC patients (mCRC).
-
JExpression of LINC00839 in CRC patients with early‐stage (stage I and II) and advanced‐stage (stage III and IV) disease.
-
K, LKaplan–Meier analysis of overall survival and disease‐free survival rate of the ISH CRC cohort.
-
MKaplan–Meier analysis of the disease‐free survival rate of CRC patients in the LnCAR database (GSE33113).
-
NThe expression of LINC00839 in a colon mucosa epithelial cell line (FHC) and CRC cell lines.
-
OThe subcellular localization was determined by nucleocytoplasmic fractionation of several CRC cell lines.
Data information: In (A, B, C, D and J), the middle lines represent the median, and the upper and lower lines represent the upper and lower quartiles. In (E, F, N and O), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 technical replicates. In (H and I), the data are shown as the mean ± SD and were analyzed by Student's t‐test. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We further investigated whether LINC00839 expression is correlated with poor outcomes in CRC patients. In the 222 CRC ISH cohort, TNM stage, lymph node status, distant metastasis (Fig 1I), vascular invasion, and clinical stage (Fig 1J) were correlated with high expression of LINC00839 (Appendix Table S1). Survival analysis showed that patients with high LINC00839 expression had a lower overall survival rate and disease‐free survival rate (Fig 1K and L). Similar results were observed in the public lnCAR (Fig 1M). Taken together, these results indicate a correlation of increased LINC00839 expression with CRC metastasis and low survival probability.
Similar results were observed in cell lines. LINC00839 expression was higher in the RKO, HT‐29, SW620, HCT116, and CaCo2 CRC cell lines than in the FHC normal colon cell line (Fig 1N). However, the difference in the expression of LINC00839 between the SW480 and LoVo CRC cell lines was not significant. In addition, subcellular localization was assessed in the nucleocytoplasmic fractions from several CRC cell lines (Fig 1O). The results were consistent with those of the ISH analysis and showed that LINC00839 was predominantly expressed in the nucleus of colorectal epithelial cells and CRC cells.
Nuclear LINC00839 accelerates the development of CRC in vitro and in vivo
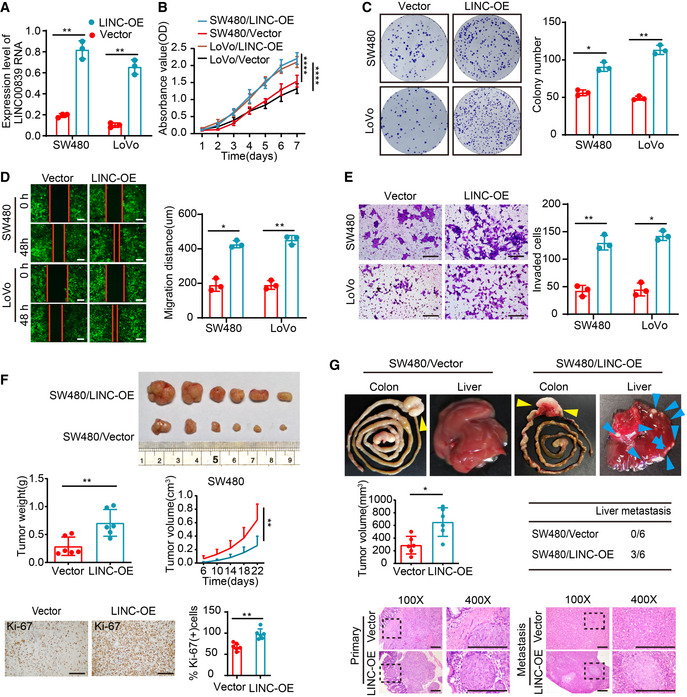
To elucidate the role of LINC00839 in CRC progression, LINC00839 was overexpressed and knocked down in CRC cells (Figs 2A and EV1A). The proliferation of cells was measured by CCK8 and colony formation assays, and the migration and invasion abilities were observed by wound‐healing and Transwell assays. These experiments showed that overexpression of LINC00839 significantly enhanced the proliferation, migration, and invasion of CRC cells compared with the control (Fig 2B–E). Knockdown of LINC00839 caused the opposite results (Fig EV1B–F). Furthermore, we established a cell‐derived xenograft (CDX) model, and the results demonstrated that overexpression of LINC00839 dramatically accelerated tumor growth (Fig 2F). Tumors derived from LINC00839‐overexpressing cells exhibited increased tumor volume and a high proliferation index as detected by Ki‐67. In contrast, knockdown of LINC00839 caused the opposite results (Fig EV1G).
Figure 2. Overexpression of LINC00839 promotes CRC cell growth and metastasis in vitro and in vivo .
-
AOverexpression of LINC00839 in SW480 and LoVo cells was confirmed by qPCR (mean ± SD, Student's t‐test, n = 3 technical replicates).
-
BThe proliferation of LINC00839‐overexpressing cells was determined by CCK8 assay (mean ± SD, ANOVA, n = 3 biological replicates).
-
CColony formation assay was performed with LINC00839‐overexpressing cells and vector control cells.
-
DWound‐healing assay was performed with LINC00839‐overexpressing cells and vector control cells. Scale bars, 100 μm.
-
EMatrigel invasion assay was performed with LINC00839‐overexpressing cells. Scale bars, 100 μm.
-
FThe CDX model showing the effect of LINC00839 on tumor growth. Representative image of tumors harvested from the mouse model (upper panel), the quantitative analysis of tumor weight and tumor volume (medium panel), and representative images and quantification of IHC staining of Ki‐67 in tumors (bottom panel; n = 6 for each cohort, and the results are presented as the mean ± SD, Student's t‐test (medium left and low right panel) and ANOVA (medium right). Scale bars, 100 μm (n = 6 biological replicates).
-
GThe CRC orthotopic mouse model revealed the effect of LINC00839 on tumor development. Representative images of primary tumors (yellow arrowheads) and liver metastasis (blue arrowheads) in the model (upper panel), analysis of tumor volume and the number of mice with liver metastasis (medium panel), and H&E staining of tumors and metastases (n = 6 for each cohort). Scale bars, 100 μm (n = 6 biological replicates).
Data information: In (C–E), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
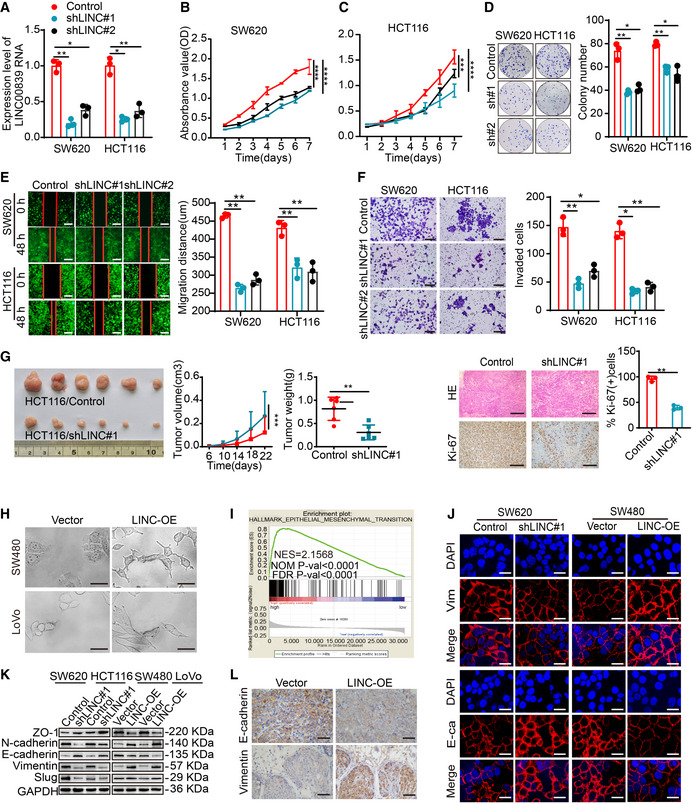
Figure EV1. LINC00839 promotes the proliferation, migration, invasion, and EMT of CRC cells.
-
AKnockdown of LINC00839 was confirmed in SW620 and HCT116 cells by qPCR (mean ± SD, Student's t‐test, n = 3 technical replicates).
-
B, CCell proliferation was measured by CCK8 assay in LINC00839‐knockdown SW620 and HCT116 cells (mean ± SD, ANOVA, n = 3 biological replicates).
-
DColony formation was assessed in LINC00839‐knockdown SW620 and HCT116 cells.
-
EWound‐healing assay was performed with LINC00839‐knockdown cells and control cells. Scale bars, 100 μm.
-
FMatrigel invasion assay was performed with LINC00839‐knockdown HCT116 and SW620 cells. Scale bars, 100 μm.
-
GThe CDX model revealed the effect of LINC00839 on tumor growth. Representative image of tumors harvested from the mouse model (left panel), quantitative analysis of tumor weight and tumor volume (medium panel), and representative images and quantification of IHC staining of Ki‐67 in tumor tissues (left panel; n = 6 for each cohort; the results are presented as the mean ± SD and were analyzed by Student's t‐test (medium and right panel) and ANOVA (left panel)). Scale bars, 100 μm (n = 6 biological replicates).
-
HCells overexpressing LINC00839 exhibited EMT‐like cell morphology: most cells lost their epithelial characteristics and exhibited a spindle‐shaped and fibroblast‐like morphology. Scale bars, 10 μm.
-
IGSEA showed the enrichment of epithelial–mesenchymal transition (EMT) signatures in the high HCT116 and SW620 cells expression group (TCGA cohort).
-
JIF staining of E‐cadherin and Vimentin in LINC00839‐overexpressing and LINC00839‐knockdown cells. Scale bars, 10 μm.
-
KEpithelial and mesenchymal marker expression in LINC00839‐overexpressing and LINC00839‐knockdown cells was measured by WB.
-
LRepresentative images of IHC staining of Vimentin and E‐cadherin in tumors from the orthotopic implantation model. Scale bars, 200 μm.
Data information: In (D–F), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
To investigate whether LINC00839 affects tumor metastasis, we established a CRC orthotopic mouse model by injecting the well‐described CRC cell line SW480 into the caecal termini of nude mice. Once one mouse became moribund, all the mice were simultaneously sacrificed to examine liver metastasis using HE staining. The results showed that LINC00839‐overexpressing cells exhibited enhanced liver colonization abilities compared with the control cells (Fig 2G). Together, these results reveal that LINC00839 can accelerate CRC cell proliferation and metastasis in vitro and in vivo.
LINC00839 induces EMT in CRC cells
Studies have shown that EMT is an important driver of the proliferation, invasion, and metastasis of CRC (Vu & Datta, 2017; Tang et al, 2020). We found that cells overexpressing LINC00839 exhibited EMT‐like cell morphology (Fig EV1H). Thus, we hypothesized that LINC00839 can accelerate the EMT of CRC cells. GSEA of TCGA datasets revealed that high LINC00839 expression was positively correlated with EMT (Fig EV1I). Furthermore, IF and WB results indicated that LINC00839 can downregulate the expression of E‐cadherin and ZO‐1 and promote the expression of N‐cadherin, Vimentin, and Slug (Fig EV1J and K). Importantly, by measuring the expression of E‐cadherin and Vimentin via IHC staining, we observed the same results in the tumors from the CRC orthotopic mouse model (Fig EV1L). These results demonstrate that LINC00839 can promote the progression of EMT in CRC.
LINC00839 serves as a scaffold to recruit Ruvb1 to the Tip60 complex and increases its acetylase activity
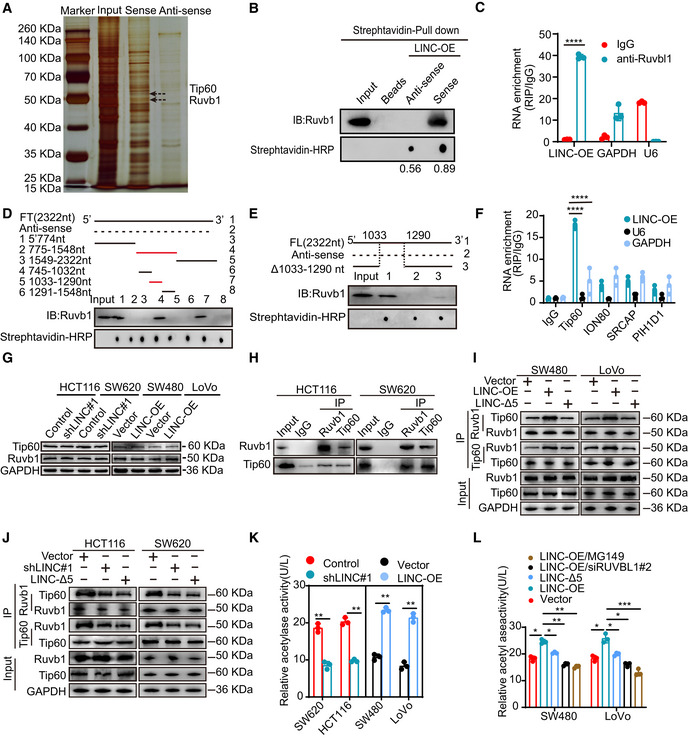
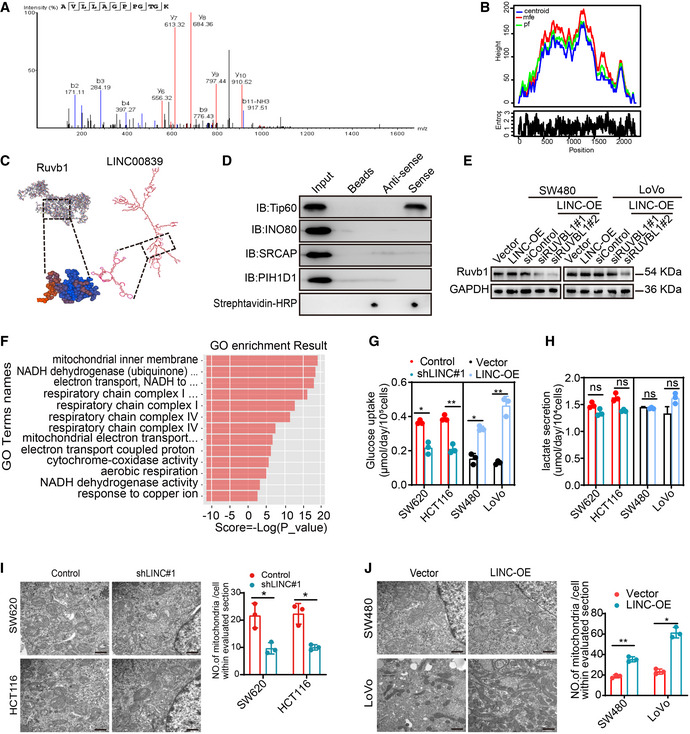
The subcellular localization of lncRNAs determines their function. As LINC00839 is mainly localized to the nucleus, we hypothesized that LINC00839 may play a role in epigenetic regulation, transcriptional regulation, or posttranscriptional regulation. Subsequently, we performed an RNA pull‐down assay and observed obvious enrichment of several proteins (Fig 3A). The enriched proteins were identified by liquid chromatography–tandem mass spectrometry (LC–MS/MS). A total of 131 potential binding proteins were identified (Dataset EV1). Among these proteins, we focused on Ruvb1, which is a vital component in some large multiprotein complexes (Fig EV2A). Next, RNA pull‐down assays and subsequent WB confirmed the binding of LINC00839 and Ruvb1 (Fig 3B). We also performed a RIP assay and found that LINC00839 was significantly enriched by the Ruvb1 antibody (Fig 3C). To identify their specific binding sites, RPISeq was used to predict the ability of different regions of LINC00839 to bind to Ruvb1 (Fig EV2B). According to the results, we generated different LINC00839 fragments and performed an RNA pull‐down experiment; the RNA hairpin structures were mostly preserved in each fragment we designed. The results showed that only nucleotides 775–1,548 of LINC00839 bound to Ruvb1 (Fig 3D). To further narrow down the specific binding sites, sequence containing nucleotides 775–1,548 was further divided into three fragments. The RNA pull‐down experiment results indicated that nucleotides 1,033–1,290 of LINC00839 bound to Ruvb1 (Fig 3D). Importantly, the LINC00839 fragment lacking nucleotides 1,033–1,290 showed little ability to bind to Ruvb1 (Fig 3E). These data indicate that nucleotides 1,033–1,290 of LINC00839 bind to Ruvb1. I‐TASSER software from Zhang's laboratory was used to predict the 3D structure of the Ruvb1 protein, and the C‐score of the predicted model was −0.59, which indicates model accuracy with a high degree of confidence. Then, we further used RNA fold analyses and structure software to predict the secondary structure of LINC00839. Fig EV2C presents the structure of the LINC00839‐Ruvb1 binding complex.
Figure 3. LINC00839 serves as a scaffold to recruit Ruvb1 to the Tip60 complex and increases its acetylase activity.
-
AAn RNA pull‐down assay was performed to identify proteins that bind to LINC00839 in HCT116 cells.
-
BRNA pull‐down assay and WB showed that biotinylated LINC00839 can bind to Ruvb1 in HCT116 cells. Dot blot of RNA–protein interactions indicating equal amounts of RNA used in the assay.
-
CRIP experiments were performed to confirm the binding of LINC00839 to Ruvb1. IgG was used as a negative control antibody. Primers specific for U6 and GAPDH were used as negative control primers.
-
DDifferent fragments of LINC00839 (solid line) and its antisense sequence (dotted line) were used for the RNA pull‐down assay and the subsequent WB.
-
EFull‐length LINC00839, antisense sequence of LINC00839, and LINC00839 sequence lacking nucleotides 1,033–1,290 were used for the RNA pull‐down assay and the subsequent WB.
-
FRIP experiments were performed by incubating Tip60, INO80, SRCAP, and PIH1D1 antibodies with HCT116 cell lysates.
-
GThe effect of LINC00839 on the expression of Tip60 and Ruvb1 was determined by WB.
-
HInteraction of endogenous Ruvb1 and Tip60 in CRC cells was confirmed by Co‐IP.
-
IThe interaction between Ruvb1 and Tip60 in cells overexpressing wild‐type LINC00839 and LINC‐Δ5 was confirmed by Co‐IP.
-
JThe interaction between Ruvb1 and Tip60 in LINC00839‐KD cells and cells expressing LINC‐Δ5 was confirmed by Co‐IP.
-
KAcetylase activity assays were performed with LINC00839‐overexpressing and LINC00839‐knockdown cells.
-
LAcetylase activity was measured in SW480 and LoVo cells subjected to different treatments.
Data information: In (C, F, K and L) the data are presented as the mean ± SD and were analyzed by unpaired t‐test, n = 3 technical replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
Figure EV2. LINC00839 interacts with the Ruvb1/Tip60 complex to promote OXHPOS and mitochondrial biogenesis.
-
ARuvb1 was identified by LC–MS.
-
BThe peak diagram of the centroid structure of LINC00839 was predicted by RNAfold.
-
CPattern of the structure of LINC00839 and Ruvb1 and their binding.
-
DRNA pull‐down and WB assays were performed to confirm the specific interaction between some large multiprotein complexes and LINC00839. Dot blot of RNA–protein binding samples indicating that equal amounts of RNA were used in the assay.
-
EThe Ruvb1 knockdown efficiency was confirmed by WB.
-
FGO enrichment analysis of the DEGs identified in the RNA‐seq.
-
GGlucose intake by LINC00839‐overexpressing cells and LINC00839‐KD cells was determined.
-
HLactate secretion by LINC00839‐overexpressing cells and LINC00839‐KD cells was measured.
-
I, JThe number of mitochondria in LINC00839‐knockdown cells and LINC00839‐overexpressing cells as confirmed by TEM. Scale bars, 500 nm.
Data information: In (G–J), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
As Ruvb1 plays an important role in some large multiprotein complexes, we wondered whether LINC00839 could interact with certain multiprotein complexes containing Ruvb1, such as the Tip60, INO80, and R2TP complexes. We performed RNA pull‐down and RIP using Tip60, ION80, SRCAP, and PIH1D1 antibodies and observed that LINC00839 was only enriched by the Tip60 antibody (Figs EV2D and 3F). Furthermore, Tip60 was identified at approximately 60 kDa by LC–MS (Dataset EV1). Surprisingly, WB analysis showed that the expression of Tip60 and Ruvb1 was not regulated by LINC00839 (Fig 3G). This prompted us to ask whether LINC00839 can serve as a scaffold to recruit Ruvb1 to the Tip60 complex. First, we confirmed the interaction of Ruvb1 and Tip60 in HCT116 and SW620 cells by coimmunoprecipitation (Co‐IP; Fig 3H). To further explore the role of LINC00839 in the interaction between Ruvb1 and the Tip60 complex, we performed Co‐IP experiments in LINC00839‐overexpressing cells, cells expressing a LINC00839 molecule lacking nucleotides 1,033–1,290 (LINC‐Δ5), and LINC00839‐knockdown cells. Our findings showed that LINC‐Δ5 expression and LINC00839 knockdown inhibited the binding of Ruvb1 and the Tip60 complex (Fig 3I and J), indicating that Ruvb1 interacted with Tip60 in a LINC00839‐dependent manner.
With this in mind, we sought to determine whether the interaction between LINC00839 and the Ruvb1/Tip60 complex affected the acetylase activity. We first measured the acetylase activity in cells overexpressing LINC00839 and LINC‐Δ5, as well as in LINC00839‐KD cells. Consequently, LINC00839 overexpression increased acetylase activity, and LINC‐Δ5 overexpression did not exert a similar effect; however, knockdown of LINC00839 decreased acetylase activity (Fig 3K and L). To further elucidate whether the increase in acetylase activity was due to the Ruvb1/Tip60 complex, we treated LINC00839‐overexpressing cells with 74 μM MG149, which is a histone acetyltransferase inhibitor that targets Tip60, and RUVBL1 siRNA. The knockdown efficiency of RUVBL1 siRNA was validated by WB (Fig EV2E). The results showed that both MG149 and RUVBL1 siRNA reversed the increase in acetylase activity caused by LINC00839 (Fig 3L). Collectively, these results indicated that LINC00839 can serve as a scaffold to recruit Ruvb1 to the Tip60 complex and increase its acetylase activity.
LINC00839 promotes NRF1‐mediated OXPHOS and mitochondrial biogenesis in CRC
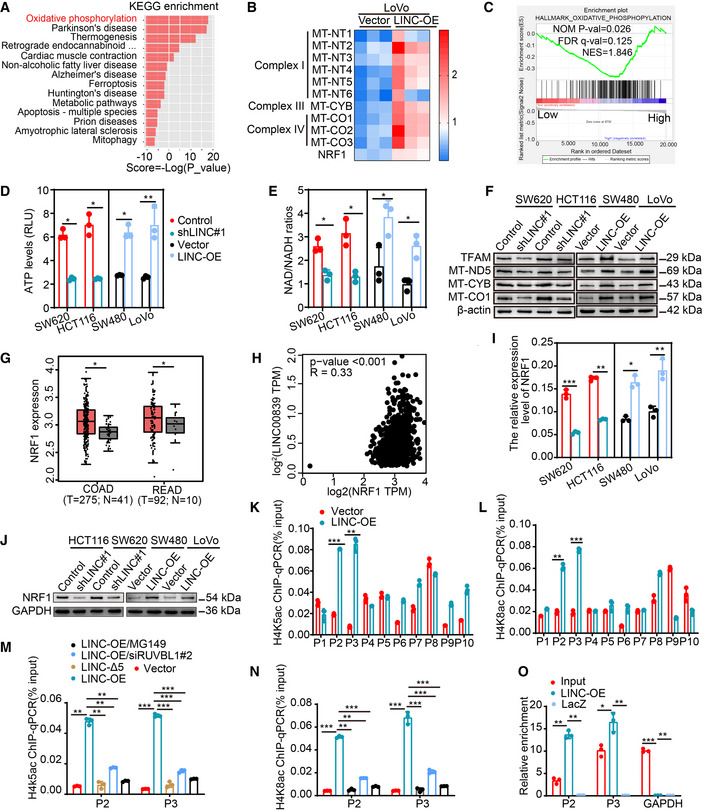
To elucidate the biological process in which LINC00839 may participate, we overexpressed LINC00839 in LoVo cells and identified the transcription profiles through RNA‐seq. Notably, differentially expressed gene (DEG) analysis revealed that 48 genes were upregulated and 51 genes were downregulated in LINC00839‐overexpressing cells (≥ 1.5‐fold change, P‐value < 0.05; Dataset EV2). KEGG pathway analysis showed that the DEGs were associated with oxidative phosphorylation (OXPHOS) and some diseases related to mitochondrial dysfunction (Fig 4A). GO enrichment analysis also identified some annotations correlated with mitochondrial components and functions (Fig EV2F). Most of the DEGs encode essential subunits of mitochondrial complexes I, III, and IV and are involved in mitochondrial biogenesis (Fig 4B). Similarly, GSEA of CRC datasets (GSE14333) also suggested a relationship between LINC00839 and OXPHOS (Fig 4C). Considering these results, we inferred that LINC00839 may facilitate OXPHOS and mitochondrial biogenesis in CRC. Glucose intake, lactate secretion, ATP levels, and NAD/NADH ratios were measured in LINC00839‐overexpressing and LINC00839‐knockdown CRC cell lines (Figs EV2G and H, and 4D and E). As the results showed, LINC00839 overexpression increased the intracellular ATP levels, NAD/NADH ratios, and glucose intake but did not affect the lactate secretion of CRC cells, suggesting that OXPHOS is enhanced and that glucose metabolism provides a substrate for OXPHOS but not for glycolysis. We also confirmed the expression of several genes involved in OXPHOS and mitochondrial biogenesis, including TFAM, MT‐ND5, MT‐CYB, and MT‐CO1, and found that the expression of these proteins was upregulated by LINC00839 overexpression (Fig 4F). Mitochondrial mass was also increased by LINC00839 overexpression, as measured by TEM (Fig EV2I and J). These results suggest that LINC00839 regulates OXPHOS and mitochondrial biogenesis in CRC.
Figure 4. LINC00839 promotes NRF1‐mediated OXPHOS and mitochondrial biogenesis in CRC.
-
AKEGG pathway analysis of the DEGs identified by RNA‐seq.
-
BHeatmap of DEGs related to OXPHOS and mitochondrial biogenesis. Expression levels are shown in log2 scale of normalized intensity values (see color gradient) ranging from low (blue) to high (red) expression.
-
CGSEA of LINC00839 in the OXPHOS pathway (GSE14333).
-
D, EThe ATP levels and the NAD/NADH ratio were measured in LINC00839‐overexpressing and LINC00839‐KD cells (mean ± SD, Student's t‐test, n = 3 biological replicates).
-
FExpression of MT‐ND5, MT‐CYB, MT‐CO1, and TFAM in LINC00839‐overexpressing and LINC00839‐KD cells as determined by WB.
-
GAnalysis of NRF1 expression in CRC tissues (T) and normal tissues (N) in the GEPIA database.
-
HCorrelation analysis of LINC00839 and NRF1 mRNA levels in the TCGA and GTEx databases (Pearson).
-
I, JExpression of NRF1 in LINC00839‐overexpressing and LINC00839‐KD cells was measured by qPCR and WB.
-
K, LThe enrichment of the NRF1 promoter (P1‐P10) by H4K5ac and H4K8ac antibodies in LINC00839‐overexpressing and control LoVo cells as determined by CHIP assay. Normal IgG was used as a negative control.
-
M, NThe enrichment of the P2 and P3 fragments by H4K5ac and H4K8ac antibodies in LoVo cells as determined by CHIP assay. Normal IgG was used as a negative control.
-
OThe enrichment of NRF1 with a probe targeting LINC00839 relative to a NC probe in HCT116 cells, as determined by ChIRP assay. LacZ served as a negative control (NC).
Data information: In (I, K, L, M, N, and O), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 technical replicates. In (G), the middle lines represent the median, and the upper and lower lines represent the upper and lower quartiles; the data were analyzed by ANOVA. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
As NRF1, which is highly expressed in cells overexpressing LINC000839 (Fig 4B), can regulate the expression of genes related to mitochondrial energy metabolism and biogenesis, we hypothesized that LINC00839 affects OXPHOS and mitochondrial biogenesis by regulating NRF1. To confirm this, we utilized the Gene Expression Profiling Interactive Analysis (GEPIA) database to determine the expression level of NRF1 and the correlation between the expression levels of LINC00839 and NRF1. The results showed that NRF1 expression was significantly upregulated in CRC patients (Fig 4G) and that there was a positive correlation between the expression of LINC00839 and NRF1 (Fig 4H). Additionally, NRF1 was upregulated by LINC00839 as determined by qPCR and WB (Fig 4I and J). Taken together, these results suggest that LINC00839 can promote NRF1‐mediated OXPHOS and mitochondrial biogenesis in CRC.
The Ruvb1/Tip60 complex acetylates histone H4 at the NRF1 promoter
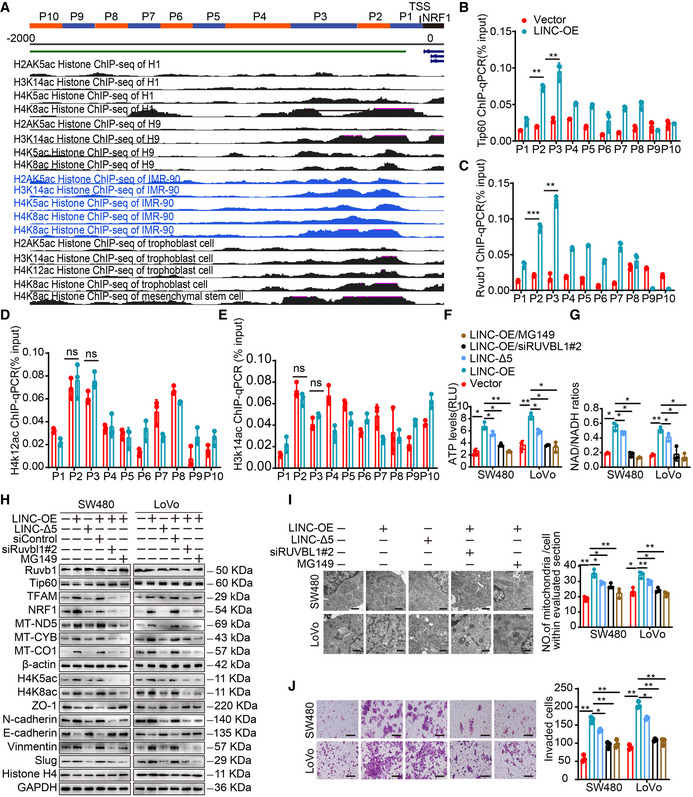
The result showing that LINC00839 can recruit Ruvb1 to the Tip60 complex and increase its acetylase activity prompted us to ask whether the Ruvb1/Tip60 complex can regulate the expression of NRF1 via posttranslational or chromatin modification. However, there are no Tip60‐specific modification sites in the NRF1 peptide sequence according to predictions with GPS‐PAIL. Thus, we hypothesized that Tip60 may acetylate histones at the NRF1 promoter. It has been reported that the Tip60 complex can acetylate H2A lysine 5 (H2AK5), H3 lysine 14 (H3K14), and H4 lysines 4, 8, and 12 (H4K5, H4K8, and H4K12; Yamagata & Kitabayashi, 2009; Nammo et al, 2018) and function as a transcriptional activator. ENCODE data in the UCSC Genome Browser revealed that H4 lysines 5, 8, and 12 (H4K5, H4K8, and H4K12) and H3 lysine 14 (H3K14) were acetylated at the NRF1 promoter (Fig EV3A). Based on these results, we wondered whether H4K5, H4K8, H4K12, and H3K14 at the NRF1 promoter were acetylated by the Tip60 complex. We divided the promoter sequence (2,000 bp before the transcriptional initiation site of NRF1) into 10 fragments (Fig EV3A) and carried out a series of ChIP–qPCR analyses. Ruvb1 and Tip60 were enriched in the NRF1 promoter, especially in the P2 (−324 ~ −174 bp) and P3 (−474 ~ −325 bp) fragments, in LINC00839‐overexpressing cells compared with control cells (Fig EV3B and C). Furthermore, the nucleosomes H4K5ac and H4K8ac, but not H4K12ac and H3K14ac, were acetylated at the NRF1 gene promoter (Figs 4K and L, and EV3D and E). Together, these results demonstrate that the Ruvb1/Tip60 complex can bind to the NRF1 promoter in the P2 and P3 sequences, and H4K5 and H4K8 are acetylated at these sites, while H4K12 and H3K14 are not markedly acetylated. Importantly, we also found that overexpression of LINC00839 can enhance the binding of the complex and the acetylation of histones at the P2 and P3 sites.
Figure EV3. LINC00839 mediates the acetylation of the NRF1 promoter by the Ruvb1/Tip60 complex to regulate OXPHOS, mitochondrial biogenesis, and CRC progression.
-
AThe pattern of 10 NRF1 promoter fragments (upper panel) and ChIP‐seq uniform peaks in the NRF1 promoter region was analyzed by ENCODE projects and visualized by UCSC Genome Browser (bottom panel).
-
B–EThe enrichment of the NRF1 promoter (P1‐P10) by Tip60 (B), Ruvb1 (C), H4K12ac (D), and H3K14ac (E) antibodies in LINC00839‐overexpressing and control LoVo cells as determined by CHIP assay. Normal IgG was used as a negative control.
-
F, GATP levels and NAD/NADH ratios in SW480 and LoVo cells were measured.
-
HExpression of genes related to mitochondrial biogenesis and EMT in SW480 and LoVo cells as determined by WB.
-
IThe number of mitochondria in SW480 and LoVo cells was confirmed by TEM. Scale bars, 500 nm.
-
JInvasion of SW480 and LoVo cells was assessed by Matrigel invasion assay. Scale bars, 100 μm.
Data information: In (B–G), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 technical replicates. In (I, J), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
To understand the effect of LINC00839‐mediated complex recruitment on histone modification and to elucidate whether the Ruvb1/Tip60 complex was essential for the acetylation of the NRF1 promoter, we performed ChIP–qPCR in cells expressing LINC‐Δ5, cells treated with MG149, and cells in which RUVBL1 was knocked down. The results showed that overexpression of LINC‐Δ5 had a negligible impact on the acetylation of the NRF1 promoter compared with the negative control (Fig 4M and N). Furthermore, treatment with MG149 and knockdown of RUVBL1 largely reversed the LINC00839‐induced increase in histone acetylation (Fig 4M and N). Taken together, these data indicated that LINC00839 increased the acetylase activity of the Ruvb1/Tip60 complex and acetylated histones H4K5 and H4K8 at the P2 and P3 sites of the NRF1 promoter. These results also motivated us to ask whether LINC00839 is associated with the NRF1 promoter. Thus, CHIRP‐qPCR was performed, and the results showed significant enrichment of LINC00839 in the NRF1 promoter (Fig 4O). This finding indicates that LINC00839 binds to the NRF1 promoter. Collectively, these data support a model in which LINC00839, as a molecular scaffold of the Ruvb1/Tip60 complex, promotes histone acetylation at the NRF1 promoter and facilitates NRF1 expression.
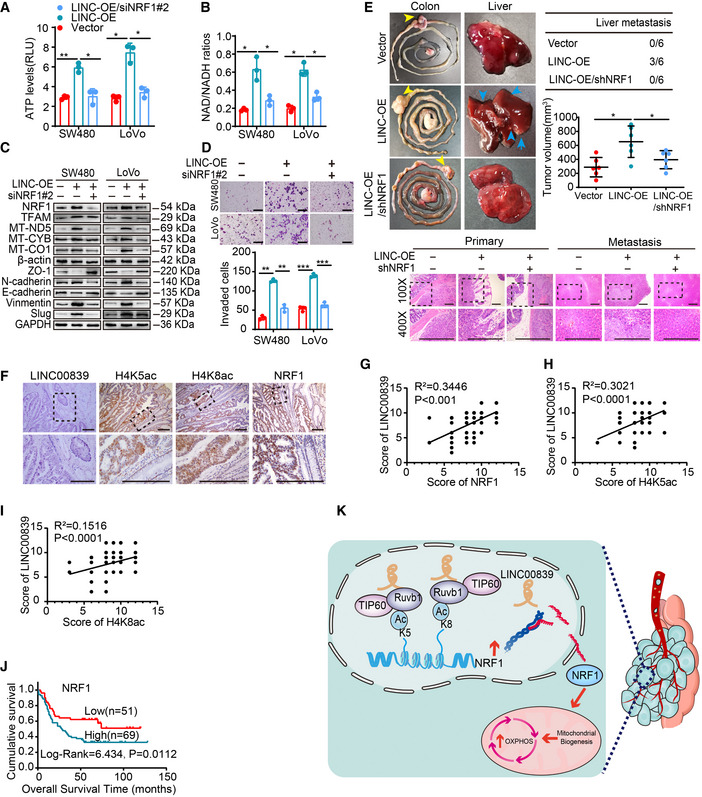
A LINC00839‐mediated Ruvb1/Tip60‐NRF1 axis regulates OXPHOS, mitochondrial biogenesis, and CRC progression
We next explored whether the effect of LINC00839 on OXPHOS and mitochondrial biogenesis depends on Ruvb1/Tip60 and NRF1. First, we evaluated the OXPHOS levels of cells by measuring the ATP levels, the NAD/NADH ratio, and the expression of genes related to mitochondrial biogenesis. The results showed that LINC00839 overexpression promoted OXPHOS, while the effect of LINC‐Δ5 overexpression was significantly lower (Fig EV3F–H). Treatment with MG149 and knockdown of RUVBL1 in LINC00839‐overexpressing cells reversed the LINC00839‐induced increase in OXPHOS (Fig EV3F–H). Mitochondrial biogenesis was also observed in the TME, and the result was similar to that described above (Fig EV3I). Next, we determined whether the effect of the Ruvb1/Tip60 complex on acetylation is essential for CRC progression. A Transwell invasion assay and WB were performed. As expected, the results showed that LINC‐Δ5 impaired the invasion and EMT of CRC cells compared with wild‐type LINC00839, and treatment with MG149 and knockdown of RUVBL1 reversed the LINC00839‐induced invasion and EMT (Fig EV3J). Together, these results indicated that Ruvbl1/Tip60 is pivotal for LINC00839‐induced OXPHOS, mitochondrial biogenesis, and CRC progression.
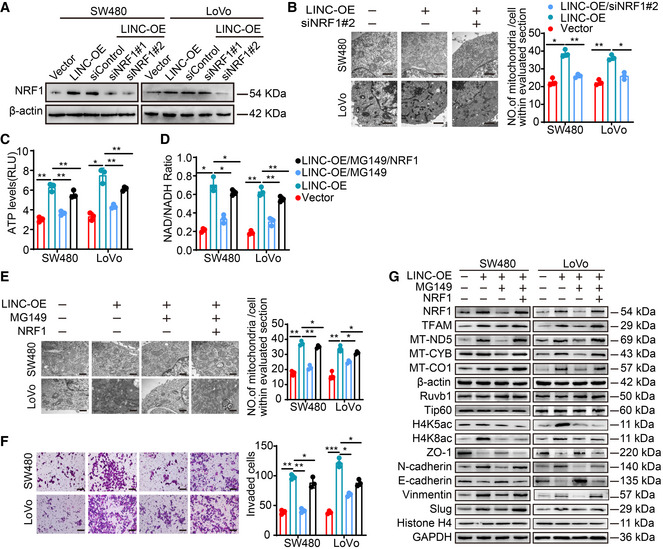
Similarly, we also knocked down NRF1 in LINC00839‐overexpressing cells (Fig EV4A) and measure OXPHOS, mitochondrial biogenesis, and CRC progression. As the results showed, NRF1 knockdown reversed the LINC00839‐induced promotion of OXPHOS, mitochondrial biogenesis, and CRC progression (Figs 5A–D and EV4B). The key role of NRF1 in metastasis was also confirmed in a CRC orthotopic mouse model; the model was established by injecting LINC00839‐overexpressing and NRF1‐knockdown SW480 cells into the caecal termini of nude mice. After all the mice were sacrificed, the tumor volume was calculated, and metastasis was confirmed by HE staining. The results showed that knocking down NRF1 attenuated the LINC00839 overexpression‐induced tumor growth and metastasis (Fig 5E) as well as the LINC00839 overexpression‐induced OXPHOS, mitochondrial biogenesis, invasion, and EMT in vitro.
Figure EV4. LINC00839 promotes OXPHOS, mitochondrial biogenesis, and CRC invasion through Ruvb1/Tip60‐NRF1 signaling.
-
AThe NRF1 knockdown efficiency was confirmed by WB.
-
BThe number of mitochondria in SW480 and LoVo cells was confirmed by TEM. Scale bars, 500 nm.
-
C, DATP levels and NAD/NADH ratios in SW480 and LoVo cells were determined.
-
EThe number of mitochondria in SW480 and LoVo cells was confirmed by TEM. Scale bars, 500 nm.
-
FInvasion of SW480 and LoVo cells was determined by Matrigel invasion assay. Scale bars, 100 μm.
-
GExpression of genes related to mitochondrial biogenesis and EMT markers in SW480 and LoVo cells as determined by WB.
Data information: In (B, E, and F), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. In (C and D), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 technical replicates. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
Figure 5. LINC00839 promotes NRF1‐mediated OXHPOS and CRC progression.
-
A, BATP levels and NAD/NADH ratios were measured in SW480 and LoVo cells.
-
CExpression of genes related to mitochondrial biogenesis and EMT in SW480 and LoVo cells as determined by WB.
-
DInvasion of SW480 and LoVo cells was assessed by Matrigel invasion assay. Scale bars, 100 μm.
-
ERepresentative images of primary tumors (yellow arrowhead) and metastases in the orthotopic implantation models (n = 6 for each cohort, left panel). The number of mice with liver metastasis (blue arrowhead) was determined. The primary tumor volume in each group was calculated (right panel). H&E staining of tumors and metastases (bottom panel). Scale bars, 200 μm (n = 6 biological replicates).
-
FISH staining of LINC00839 and IHC staining of NRF1, H4K5ac, and H4K8ac in paraffin‐embedded samples of CRC tumor tissues and NATs. Scale bars, 200 μm.
-
G–ICorrelation analysis of the expression of LINC00839 and NRF1 (G), H4K5ac (H), and H4K8ac (I) in CRC patients (n = 120).
-
JCorrelation of NRF1 expression with the overall survival rate in patients with CRC (n = 120). NRF1 expression was stratified as high and low according to the median expression level.
-
KA schematic illustration of the mechanism by which LINC00839 recruits the Ruvb1/Tip60 complex to acetylate histones at the NRF1 promoter and regulates CRC tumor progression via OXPHOS.
Data information: In (A, B, and D), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. In (G–I) show the Pearson correlation coefficient results. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
To further elucidate whether the expression of NRF1 is dependent on the acetylation activities of the Ruvb1/Tip60 complex, we treated LINC00839‐overexpressing cells with MG149. The expression of NRF1 was decreased, and OXPHOS, mitochondrial biogenesis, and CRC progression were also significantly inhibited (Fig EV4C–G). We next overexpressed NRF1 on the basis of MG149 treatment, and the inhibition of these phenomena was reversed (Fig EV4C–G).
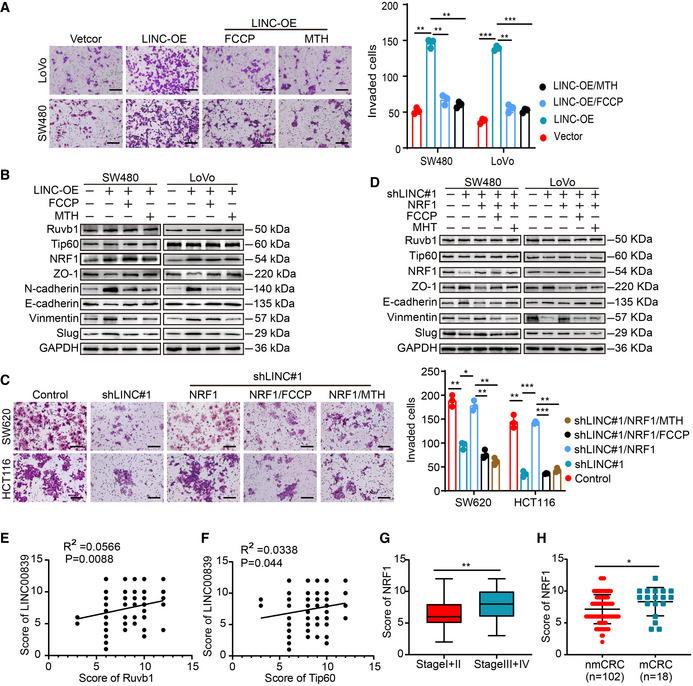
FCCP, which is an effective uncoupler of oxidative phosphorylation, disrupts ATP synthesis by transporting protons. Metformin (MTH) can target complex I and decrease ATP production (Ashton et al, 2018). Transwell assays showed that treatment with FCCP and MTH inhibited the increase in cell invasion, as well as EMT, induced by LINC00839 overexpression (Fig EV5A and B). Moreover, we overexpressed NRF1 in LINC00839‐KD cells. The decreased invasion and EMT of cells were reversed, while treatment with FCCP and MTH inhibited the malignant phenotype again (Fig EV5C and D). These results suggested that NRF1‐mediated OXPHOS is essential for the EMT and tumor progression induced by LINC00839.
Figure EV5. LINC00839 promotes CRC progression through Ruvb1/Tip60‐NRF1‐mediated OXPHOS.
-
AInvasion of LINC00839‐overexpressing cells treated with FCCP and MTH was determined by Matrigel invasion assay. Scale bar, 100 μm.
-
BEMT marker expression levels in LINC00839‐overexpressing cells treated with FCCP and MTH were measured by WB.
-
CInvasion of LINC00839‐knockdown cells in which NRF1 was overexpressed and to which FCCP and MTH were added was measured by Matrigel invasion assay. Scale bar, 100 μm.
-
DEMT marker expression levels in LINC00839‐knockdown cells in which NRF1 was overexpressed and to which FCCP and MTH were added were measured by WB.
-
EThe correlation between the expression of LINC00839 and Ruvb1 in the CRC cohort (n = 120, Pearson).
-
FThe correlation between the expression of LINC00839 and Tip60 in the CRC cohort (n = 120, Pearson).
-
GThe expression levels of NRF1 in patients with early‐stage (stage I and II) and advanced‐stage (stage III and IV) disease.
-
HExpression of NRF1 in nonmetastatic CRC patients (nmCRC) and metastatic CRC patients (mCRC).
Data information: In (A and C) the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 biological replicates. In (G), the middle lines represent the median, and the upper and lower lines represent the upper and lower quartiles. In (H), the data are presented as the mean ± SD and were analyzed by Student's t‐test. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
High LINC00839 expression is associated with high histone acetylation levels and high NRF1 expression, indicating poor prognosis in CRC patients
To determine the clinical relevance of our findings, the protein expression of LINC00839, H4K5ac, H4K8ac, and NRF1 was evaluated in 120 CRC patients. The expression of H4K5ac, H4K8ac, and NRF1 was upregulated in CRC tissues compared with NATs (Fig 5F). The expression of LINC00839 was correlated with that of H4K5ac, H4K8ac, and NRF1, which is consistent with previous predictions about NRF1 expression and its correlates in CRC (Fig 5G–I). There was no correlation among the expression levels of Ruvb1, Tip60, and LINC00839 (Fig EV5E and F). Moreover, the high expression of NRF1 was positively correlated with advanced stage and metastasis of CRC (Fig EV5G and H) and indicated poor outcomes in CRC patients (Fig 5J). Taken together, these results indicated that the LINC00839‐Ruvb1/Tip60‐NRF1 regulatory axis can promote CRC progression and that LINC00839 might be a potential marker for the outcome of CRC. In summary, our conclusion shows the regulatory process by which LINC00839 guides the Ruvb1/Tip60 complex to the NRF1 promoter and regulates mitochondrial function, thereby promoting EMT and CRC progression (Fig 5K).
Discussion
LINC00839 is a novel oncogenic lncRNA in CRC. Our study shows that LINC00839 is highly expressed in CRC and that high LINC00839 expression indicates advanced stage of disease and poor outcomes in CRC patients. In vitro and in vivo assays confirmed that LINC00839 promotes the metastasis and progression of CRC, which was consistent with our clinical data. Consistent with the reported function of LINC00839 in EMT (Hu et al, 2019), our study demonstrates that LINC00839 induces the EMT of CRC. Furthermore, we reveal that LINC00839, as a scaffold, recruits Ruvb1 to the Tip60 complex and increases its acetylase activity. The complex promotes NRF1 expression by acetylating histones in its promoter region and regulates OXPHOS and mitochondrial biogenesis in CRC. Previous studies have shown that LINC00839 is involved in the occurrence and progression of many cancers, but most of these studies identified LINC00839 as a miRNA sponge (Zhang et al, 2020). Our study reveals a new functional mechanism by which LINC00839 regulates NRF1 expression via chromatin epigenetic modification.
RUVBL1 belongs to the ATPases associated with various cellular activities (AAA+) family and participates in a wide range of cellular processes, including chromatin remodeling. As a part of multisubunit complexes that play key roles in transcription, Ruvb1 can transcriptionally regulate gene expression and is associated with multiple cancers. Previous studies have shown that Ruvb1 is a coactivator of TIP60 and increases the transcriptional activity of TCF4 via histone H4 acetylation (Feng et al, 2003). Ruvb1 is required for the lysine acetyltransferase activity of the TIP60 complex (Jha et al, 2013). Our study showed that LINC00839 can promote the binding of Ruvb1 and the Tip60 complex but does not influence the expression of Ruvb1 and Tip60. The binding of Ruvb1 and the Tip60 complex enhances the acetyltransferase activity of the complex, suggesting the pivotal role of LINC00839 in the acetylation of histones by the Ruvb1/Tip60 complex.
To explore the cellular process in which LINC00839 may participate, the RNA profile was analyzed, and our experiments revealed that LINC00839 can promote OXPHOS and mitochondrial biogenesis. It is widely reported that NRF1 can regulate the expression of genes related to mitochondrial energy metabolism and biogenesis, and LINC00839 can also promote the expression of NRF1. Thus, we hypothesize that NRF1 may be a hub gene in LINC00839‐induced OXPHOS and mitochondrial biogenesis. We also validated the essential role of NRF1 in this process by knocking down NRF1. To illustrate the crucial function of histone acetylation, we treated the cells with MG149. As a result, OXPHOS and mitochondrial biogenesis were inhibited. We also overexpressed NRF1 in the cells treated with MG149, and the inhibited phenotype was reversed again. All these data demonstrated the importance of LINC00839‐Ruvb1/Tip60‐NRF1 signaling in OXPHOS and mitochondrial biogenesis in CRC.
As we revealed, LINC00839 can target the Ruvb1/Tip60 complex and direct the complex to the NRF1 promoter. LINC00839 guides the expression of NRF1 in trans, as they are distantly located genes. However, whether LINC00839 directs the Ruvb1/Tip60 complex only to NRF1 is unclear. As the function of the Ruvb1/Tip60 complex has been widely reported in cancer, we hypothesize that the function of LINC00839 as a guide is not unique. Importantly, considering the hub role of NRF1 in the effects of LINC0839 on OXPHOS and CRC progression, the role of LINC0839 as a guide that directs the Ruvb1/Tip60 complex to the NRF1 promoter is essential and pivotal.
NRF1‐mediated OXPHOS promotes the progression of CRC, but whether the increase in mitochondrial biogenesis promotes EMT and CRC progression has not been verified. In summary, our study demonstrated the vital role of LINC00839 in the progression of CRC. LINC00839 binds to the Ruvb1/Tip60 complex and promotes the acetylation of histones H4K5 and H4K8 at the NRF1 promoter, increasing NRF1 transcription and expression. Upregulated NRF1 expression promotes OXPHOS and mitochondrial biogenesis and then promotes EMT and tumor progression in CRC. Future research should focus on the expression and biological function of LINC00839 in multiple cancers and determine the specificity and heterogeneity of its expression patterns and mechanism of action in order to provide a basis for precision treatment of cancer.
Materials and Methods
Ethics approval and consent to participate
All the experiments performed in this study followed the ethical standards of Southern Medical University (Guangzhou, China) and the Declaration of Helsinki. Informed consent does not apply to all the data that were analyzed anonymously. All the animal experiments were approved by the Animal Care and Use Committee of Southern Medical University and complied with the Guidelines of the Institutional Animal Care and Use Committee of Southern Medical University during all animal studies.
Cell culture and tissue samples
The human CRC cell lines HT‐29, LoVo, SW480, SW620, HCT116, RKO, and CaCo2 and the normal colon cell line FHC were obtained from ATCC (Manassas, VA, USA). The identities of all the cell lines were confirmed by STR profiling. The cells were cultured in incubators in 5% CO2 at 37°C and were maintained in RPMI 1640 medium (Gibco, CA, USA) supplemented with 10% FBS (Gibco). CRC tissues and adjacent mucosa tissues were collected from Nanfang Hospital. The study was approved by the Human Research Ethics Committees of Southern Medical University.
Vector construction
Human LINC00839 was amplified by PCR and cloned into pcDNA3.1 by MssBio (Guangzhou, China). Different mutants of LINC00839 (∆1,023–1,290) and Ruvb1 were cloned into pcDNA3.1 by IGE Biotechnology Ltd. (Guangzhou, China). The lentiviral vectors used in this study were constructed by Genechem (Shanghai, China). The shRNA sequences used are listed in Appendix Table S2.
RNA isolation, reverse transcription, and quantitative real‐time PCR (qPCR)
TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA from cells and fresh tissues. A PrimeScript RT reagent Kit (TaKaRa, Dalian, China) was used to synthesize cDNA. qPCR was performed with an ABI7500 Real‐time PCR system (Applied Biosystems, CA, USA) using a SYBR Green Real‐Time RT–PCR kit (TaKaRa, Dalian, China). The expression of related genes was calculated with the comparative 2−ΔΔCT method. For tissue samples, the minimum CT value of the normal group was set to 1. For cell samples, the average CT value of the control group was set to 1. For all the data, the expression of GAPDH served as the internal control. The primer pairs used for qPCR are listed in Appendix Table S3.
Western blotting
Proteins from cell lysates were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and electrotransferred onto PVDF membranes (Pall Corp, D.C., NY). Then, the membranes were blocked with 5% skim milk and incubated with the following primary antibodies at 4°C: anti‐GAPDH (Proteintech, #60004‐1‐Ig, 1:5,000), anti‐ACTB (Proteintech, #20536‐1‐AP, 1:5,000), anti‐Ruvb1 (Abcam, #ab226001, 1:2,000), anti‐Tip60 (CST, #12058, 1:500), anti‐NRF1 (Abcam, #ab175932, 1:500), anti‐H4K8ac (Abcam, #ab45166, 1:5,000), anti‐H4K5ac (Abcam, #8647, 1:5,000), anti‐H4K12ac (Abcam, #ab46983, 1:2,000), anti‐H3K27ac (CST, #8173, 1:1,000), anti‐H4 (CST, #13919, 1:1,000), anti‐H3 (CST, #4499, 1:5,000), anti‐E‐cadherin (CST, #3195T, 1:500), anti‐Vimentin (CST, #5741, 1:500), anti‐N‐cadherin (CST, #13116, 1:500), anti‐ZO‐1 (CST, #8193T, 1:500), anti‐Slug (CST, #9585T, 1:500), anti‐INO80 (Proteintech, #18810‐1‐AP, 1:500), anti‐TFAM (Proteintech, #22586‐1‐AP, 1:5,000), anti‐MT‐CYB (Proteintech, #55090‐1‐AP, 1:500), anti‐MT‐CO1 (Proteintech, #55071‐1‐AP, 1:500), anti‐MT‐NT5 (Proteintech, #66613‐1‐Ig, 1:500), and anti‐SRCAP (Abcam, #ab99408, 1:500). Then, the membranes were incubated with the appropriate secondary antibodies (1:5,000, Proteintech). Signals were detected using FDbio‐Femto ECL (Fudebio, Hangzhou, China) and a chemiluminescence system (Tanon 5200, Shanghai, China).
Cell transfection
Cells were cultured in 6‐well plates and then transfected using Lipofectamine 3000 Transfection Reagent (Invitrogen) according to the manufacturer's instructions. The cells were transfected with lentivirus (Genechem, Shanghai, China) according to the manufacturer's instructions. To obtain stably transfected cell lines, these cells were treated with the appropriate concentration of puromycin (2–8 μg/ml).
Cell number counting, CCK8, colony formation, wound‐healing, and Transwell assays
For cell number counting, cultured cells were digested with 0.25% trypsin and counted using a counter. For the CCK8 assay, CCK8 reagent was added to cells cultured in 96‐well plates at a 1:9 ratio. After incubation at 37°C for 2 h, the absorbance was measured at a wavelength of 490 nm with a microplate spectrophotometer. For the colony formation assay, cells (500 per well) were plated in 6‐well plates and cultured in medium for 2 weeks. Then, the cells were fixed with 4% paraformaldehyde and stained with Giemsa to count the colony numbers. For the wound‐healing assay, GFP‐labeled cells were cultured in 6‐well plates, and the migration of cells was observed under a fluorescence microscope at 0 and 48 h after lines were drawn in the cell culture monolayer. Cells were cultured with serum‐free media during the assays. For the Transwell assay, cells (10,000 per well) were suspended in serum‐free media and seeded in the upper chamber of 8‐μm‐pore Transwell plates, and media supplemented with 10% FBS was used as a chemoattractant in the lower compartment. After being cultured at 37°C for 2 days, the cells that successfully passed through the pore were fixed and stained with Giemsa for 15 min, five visual fields were randomly selected under the microscope, and the cells were counted and quantified by ImageJ (NIH). Matrigel (Corning, NY, USA) was also added to the upper chamber to assess cell invasion. Two‐tailed Student's t‐test was used to analyze the data from three independent experiments.
In situ hybridization (ISH)
ISH staining was performed according to the instructions of the ISH Kit (Roche, Basel, Switzerland). The sequence of the probe used in the research was ATGTTGCTTGCTGGGACTGATGTCTTCCTTAT (MssBio, Guangzhou, China). The staining scores were evaluated by two pathologists who were blinded to the clinical materials. A quick scoring system from 0 to 12 that combined the intensity and percentage of the positive signal was used. Briefly, a signal of 0 indicates no staining, 1 indicates weak staining, 2 indicates intermediate staining, and 3 indicates strong staining. According to the percentage of the area with positive staining, the degree of staining was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). The product of the intensity and range scores was considered the final score of LINC00839 (0–12).
Immunohistochemistry (IHC)
IHC was performed to investigate the expression of proteins in tissues, and the experiments followed a previously described protocol (33042270). The slides were incubated overnight with the following primary antibodies: anti‐NRF1 (Abcam, #ab175932, 1:1,000), anti‐H4K8ac (Abcam, #ab45166, 1:500), anti‐H4k5ac (Abcam, #8647, 1:2,000), anti‐E‐cadherin (CST, #3195T, 1:400), anti‐Vimentin (CST, #5741, 1:200), and anti‐Ki‐67 (CST, #5741, 1:200). An HRP‐conjugated secondary antibody and DAB staining kit (ZSGB‐BIO, Beijing, China) were used in the experiment. The scoring method was similar to that of ISH.
RNA pull‐down assay
Biotin‐labeled RNAs (LINC00839 and its antisense RNA) were transcribed in vitro with the Biotin RNA labeling Mix (Thermo Fisher Scientific, Waltham, USA) and T7 RNA polymerase (Life Technologies, Gaithersburg, MD). Biotinylated RNAs were mixed with streptavidin agarose beads (Thermo Fisher Scientific) at 4°C overnight. Total cell lysates were freshly prepared and added to each binding reaction with a protease/phosphatase inhibitor cocktail and RNase inhibitor, and then, the mixture was incubated for 1 h at 4°C. After washing thoroughly three times, the RNA–protein binding mixture was boiled in SDS buffer, and the eluted proteins were assessed by western blotting and mass spectrometry.
Chromatin immunoprecipitation (ChIP)
The ChIP experiment was performed using a SimpleChIP® Enzymatic Chromatin IP Kit (Cell Signaling Technology, MA, USA) following the manufacturer's instructions. Briefly, cells are fixed with formaldehyde to cross‐link the histone and nonhistone proteins to DNA. Then, chromatin was digested with micrococcal nuclease into 150–900 bp DNA/protein fragments. Antibodies specific for the RUVBL1, Tip60, H4K5ac, and h4k8ac proteins were added, and the complex coprecipitates were captured by Protein G magnetic beads. Finally, the cross‐linking was reversed, and the target DNA sequence was purified; then, the DNA sequences were ready for qPCR analysis. One‐tenth of the input chromatin was also treated in the same way and purified. The percentage of enriched DNA fragments in the input indicated the degree of enrichment.
RNA immunoprecipitation (RIP)
RIP was performed according to the instructions of the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, MA, USA). Briefly, lysates were prepared in lysis buffer supplemented with a protease inhibitor cocktail and RNase inhibitor. Then, protein A/G magnetic beads were prepared for incubation with 5 g of purified antibodies per immunoprecipitation reaction for 30 min at room temperature. Furthermore, to precipitate protein–RNA complexes, the mixture was incubated at 4°C overnight. Finally, RNA was purified, evaluated, and analyzed by qPCR.
Animal model
The animal experiments were approved by the Animal Care and Use Committee of Southern Medical University. The animals were randomly divided into the control and treatment groups. Four‐week‐old BALB/C‐nu/nu nude mice were purchased from the Animal Center of Southern Medical University (Guangzhou, China). The mice were housed under specific pathogen‐free conditions in temperature‐ and humidity‐controlled cages. To establish the xenograft model, cells were subcutaneously injected into the left and right groins of the mice. The tumor volume (volume = (length*width2)/2) was measured every 4 days. To establish the CRC orthotopic mouse model, cells (2 × 106 per mouse) were injected into the caecum termini of the nude mice. Once one mouse became moribund, all the mice were simultaneously sacrificed, and the intestinal and liver tissues were harvested for further examination. Representative images of the tumors were taken before surgical resection. Then, the samples were dehydrated, fixed, embedded in paraffin, and sectioned. The slides were stained with hematoxylin‐eosin or IHC and observed under a microscope.
Chromatin isolation by RNA purification (CHIRP)
CHIRP assays were performed using the chromatin isolation by RNA purification (CHIRP) Kit (gzscbio, China) following the manufacturer's instructions. First, 1 × 107 cells were collected for each reaction. The cells were lysed using 1 ml of complete lysis buffer, and the DNA fragments were then sheared into small pieces by ultrasonication. Second, the lysates were incubated with biotin‐labeled probes that could hybridize with LINC00839. Third, the probes combined with DNA were mixed with streptavidin magnetic beads and washed with wash buffer. Finally, RNA and DNA were isolated and purified for qRT–PCR. The probes are listed in the Appendix Table S4.
Coimmunoprecipitation (Co‐IP)
Cells were harvested by centrifugation at 4°C 1,400 g for 5 min and washed with cold PBS three times. Then, the cell pellets were resuspended in precooled RIPA buffer on ice for 20 min and centrifuged at 4°C 1,400 g for 15 min. The supernatants were collected in new tubes. Antibodies (1–2 g) were added to 1 ml of each cell lysate and incubated at 4°C for 12 h. After the addition of protein A/G agarose beads (Santa Cruz Biotechnology), the incubation was continued for 8–12 h at 4°C. The immunoprecipitants were washed with washing buffer five times and eluted with 2× SDS loading buffer by boiling for 5 min. Finally, the samples were separated by SDS–PAGE and analyzed by WB.
Transmission electron microscopy (TEM)
Cells were fixed in 2.5% glutaraldehyde at 4°C overnight. Then, the cells were fixed in 1% osmium tetroxide for 1 h, embedded in resin, sectioned, doubly stained with uranyl acetate and lead citrate, and observed using a transmission electron microscope (JEOL, Japan).
Nucleoplasm separation for RNA extraction
Subcellular isolation and RNA extraction were performed according to the manual of the PARIS™ Kit (Life Technologies, Gaithersburg, MD).
Detection of human histone acetyltransferase (HAT) activity
The HAT activity of the cells was measured following the protocol of an ELISA kit (BOSHEN, Jiangsu, China).
Measurement of NAD/NADH, ATP, glucose intake, and lactate secretion
The NAD/NADH levels in cells were measured using the Amplite™ Colorimetric NAD/NADH Ratio Assay Kit (AAT Bioquest, Sunnyvale, CA). ATP levels were measured with an ATP detection kit (Beyotime, Shanghai, China). The glucose levels in the cell culture supernatants were measured with the GOD‐PAP method. Levels of lactate secretion were measured with a Glycolysis Cell‐Based Assay Kit (Cayman, MI, USA).
Quantification and statistical analysis
All the statistical analyses were carried out using the SPSS 19.0 statistical software package (SPSS, Chicago, USA). All the statistical graphs were generated with GraphPad Prism version 7 (GraphPad Software, CA). Student's t‐tests and one‐way ANOVA were carried out to compare differences between two or more groups. Pearsonʼs chi‐squared (χ2) test was used to analyze the correlation between the expression levels and clinicopathological characteristics. Survival analyses were performed using the Kaplan–Meier method. P < 0.05 was considered statistically significant, and the levels of significance are presented as follows: * indicates P < 0.05, ** indicates P < 0.01, and *** indicates P < 0.001.
Author contributions
Xiaoting Liu: Formal analysis; visualization; methodology; writing – original draft. Jianxiong Chen: Methodology; writing – review and editing. Sijing Zhang: Data curation; formal analysis; validation. Xunhua Liu: Data curation; formal analysis. Xiaoli Long: Investigation; methodology. Jiawen Lan: Software; validation. Miao Zhou: Data curation; formal analysis. Lin Zheng: Data curation. Jun Zhou: Conceptualization; formal analysis; funding acquisition.
In addition to the CRediT author contributions listed above, the contributions in detail are:
JZ led the study design and prepared the manuscript. XTL and JXC contributed to the experiments. LZ designed the research and analyzed the data. XHL performed the statistical analysis. XLL assisted in tissue sample collection. JWL prepared the figures. MZ and SJZ performed data analysis and interpretation. All the authors read and approved the final manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant numbers 81272763, 81672466, 81972334, and 82173297), the National Key R&D Program of China (2017YFC1309002), and the Natural Science Foundation of Guangdong Province (grant nos. 2017A030313550 and 2019A1515011205).
EMBO reports (2022) 23: e54128
Data availability
The datasets produced in this study are available in the following databases: RNA‐Seq data: Gene Expression Omnibus GSE197706 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197706).
References
- Anderson NM, Mucka P, Kern JG, Feng H (2018) The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 9: 216–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton TM, McKenna WG, Kunz‐Schughart LA, Higgins GS (2018) Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res 24: 2482–2490 [DOI] [PubMed] [Google Scholar]
- Cai Z, Li CF, Han F, Liu C, Zhang A, Hsu CC, Peng D, Zhang X, Jin G, Rezaeian AH et al (2020) Phosphorylation of PDHA by AMPK drives TCA cycle to promote cancer metastasis. Mol Cell 80: 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce‐Rodriguez I, Chakravarthi B, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Shen H, Zhu X, Liu Y, Yang H, Chen H, Xiong S, Chi H, Xu W (2020) A nuclear lncRNA Linc00839 as a Myc target to promote breast cancer chemoresistance via PI3K/AKT signaling pathway. Cancer Sci 111: 3279–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu‐Marchese M, Shuen M, Pauly M, Saleem A, Hood DA (2015) The regulation of mitochondrial transcription factor a (Tfam) expression during skeletal muscle cell differentiation. Biosci Rep 35: e00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Lee N, Fearon ER (2003) TIP49 regulates beta‐catenin‐mediated neoplastic transformation and T‐cell factor target gene induction via effects on chromatin remodeling. Cancer Res 63: 8726–8734 [PubMed] [Google Scholar]
- Hu Q, Li C, Wang S, Li Y, Wen B (2019) LncRNAs‐directed PTEN enzymatic switch governs epithelial–mesenchymal transition. Cell Res 29: 286–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W, Ten Dijke P (2020) TGFβ‐induced metabolic reprogramming during epithelial‐to‐mesenchymal transition in cancer. Cell Mol Life Sci 77: 2103–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Chen Z, Cheng P, Jiang Z, Wang Z, Huang Y, Yang C, Pan J, Qiu F, Huang J (2019) LYRM2 directly regulates complex I activity to support tumor growth in colorectal cancer by oxidative phosphorylation. Cancer Lett 455: 36–47 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann‐Broz D, Khalil AM, Zuk O, Amit I, Rabani M et al (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Gupta A, Dar A, Dutta A (2013) RVBs are required for assembling a functional TIP60 complex. Mol Cell Biol 33: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN et al (2018) Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 50: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz J, Schönfeld C, Seibert M, Stolp V, Alshamleh I, Oellerich T, Steffen B, Schwalbe H, Schnütgen F, Kurrle N et al (2019) Metabolic plasticity of acute myeloid leukemia. Cell 8: 805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM et al (2014) PGC‐1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16: 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nammo T, Udagawa H, Funahashi N, Kawaguchi M, Uebanso T (2018) Genome‐wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet‐induced obesity in mice. Diabetologia 61: 2608–2620 [DOI] [PubMed] [Google Scholar]
- Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L (2018) Mitochondrial metabolism and cancer. Cell Res 28: 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu D, Ho SY, Juan HF, Huang HC (2018) High‐risk, expression‐based prognostic long noncoding RNA signature in neuroblastoma. JNCI Cancer Spectr 2: pky015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Fu J, Liu W, Hayashi T, Nie Y, Mizuno K, Hattori S, Fujisaki H, Onodera S, Ikejima T (2020) Silibinin inhibits migration and invasion of breast cancer MDA‐MB‐231 cells through induction of mitochondrial fusion. Mol Cell Biochem 463: 189–201 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD (2019) Cancer statistics, 2019. CA Cancer J Clin 69: 7–34 [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg‐Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT (2013) High‐resolution Xist binding maps reveal two‐step spreading during X‐chromosome inactivation. Nature 504: 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein U, Schlag PM (2007) Clinical, biological, and molecular aspects of metastasis in colorectal cancer. Recent Results Cancer Res 176: 61–80 [DOI] [PubMed] [Google Scholar]
- Tang Q, Chen J, Di Z, Yuan W, Zhou Z, Liu Z, Han S, Liu Y, Ying G, Shu X et al (2020) TM4SF1 promotes EMT and cancer stemness via the Wnt/β‐catenin/SOX2 pathway in colorectal cancer. J Exp Clin Cancer Res 39: 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens‐Mas M, Hernández‐López R, Pons DG, Roca P, Oliver J, Sastre‐Serra J (2019) Sirtuin 3 silencing impairs mitochondrial biogenesis and metabolism in colon cancer cells. Am J Physiol Cell Physiol 317: C398–C404 [DOI] [PubMed] [Google Scholar]
- Vishnubalaji R, Shaath H, Elkord E, Alajez NM (2019) Long non‐coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non‐favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov 5: 109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T, Datta PK (2017) Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancer 9: 171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, Zhang ZZ (2019) The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti‐tumor miRNAs. Mol Cancer 18: 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Wu YP, Yin HB, Xue XY, Gou X (2018) Molecular network‐based identification of competing endogenous RNAs and mRNA signatures that predict survival in prostate cancer. J Transl Med 16: 274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kitabayashi I (2009) Sirt1 physically interacts with Tip60 and negatively regulates Tip60‐mediated acetylation of H2AX. Biochem Biophys Res Commun 390: 1355–1360 [DOI] [PubMed] [Google Scholar]
- Yang L, Pei L, Yi J (2021) LINC00839 regulates proliferation, migration, invasion, apoptosis and glycolysis in neuroblastoma cells through miR‐338‐3p/GLUT1 Axis. Neuropsychiatr Dis Treat 17: 2027–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cao H, Ye L, Wen X, Wang S, Zheng W, Zhang Y, Huang D, Gao Y, Liu H et al (2019) Cancer‐associated methylated lncRNAs in patients with bladder cancer. Am J Transl Res 11: 3790–3800 [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo H, Ma L, Chen X, Chen G (2020) Long noncoding RNA LINC00839 promotes the malignant progression of osteosarcoma by competitively binding to MicroRNA‐454‐3p and consequently increasing c‐met expression. Cancer Manag Res 12: 8975–8987 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


