Abstract
LINE‐1 (L1) retroelements have retained their ability to mobilize. Mechanisms regulating L1 mobility include DNA methylation in somatic cells and the piRNA pathway in the germline. During preimplantation stages of mouse embryonic development, however, both pathways are inactivated leading to a window necessitating alternate means of L1 regulation. We previously reported an increase in L1 levels in Dicer_KO mouse embryonic stem cells (mESCs), which was accompanied by only a marginal increase in retrotransposition, suggesting additional mechanisms suppressing L1 mobility. Here, we demonstrate that L1 ribonucleoprotein complexes (L1 RNP) accumulate as aggregates in the cytoplasm of Dicer_KO mESCs along with the RNA helicase MOV10. The combined overexpression of L1 ORF1p and MOV10 is sufficient to create L1 RNP aggregates. In Dicer_KO mESCs, MOV10 is upregulated due to the loss of its direct regulation by miRNAs. The newly discovered posttranscriptional regulation of Mov10, and its role in preventing L1 retrotransposition by driving cytosolic aggregation, provides routes to explore for therapy in disease conditions where L1s are upregulated.
Keywords: LINE‐1, miRNA, retrotransposition, MOV10, cytosolic aggregates
Subject Categories: Chromatin, Transcription & Genomics
L1 high expression but low mobility in miRNA_KO mESCs is due to upregulation of the RNA helicase MOV10, triggered by the lack of MOV10‐targeting miRNAs. Increased MOV10 expression induces L1 RNP aggregation in the cytoplasm, preventing L1 mobility.
Introduction
Approximately 17–20% of human and mouse genomes are composed of long interspersed elements 1 (LINE‐1 or L1; Lander et al, 2001; Waterston et al, 2002). These elements, ranging from 6 to 7 kb in length, encode enzymatic activities necessary for retrotransposition. In mouse, L1s are composed of a 5′ untranslated region (UTR) harboring an RNA polymerase II (Pol II) promoter encoding a bicistronic transcript. The two open reading frames (ORF) encode for L1 ORF1 protein (ORF1p) that is speculated to function as an RNA chaperone and L1 ORF2 protein (ORF2p) that has endonuclease and reverse transcriptase activities. The transcript harbors a 3'UTR and a poly adenylation (poly(A)) signal. Only a full‐length poly(A) transcript is capable of transposing. Upon export from the nucleus, L1 RNA is translated in the cytoplasm. L1 RNA, ORF1p, and ORF2p associate to form ribonucleoprotein particles (L1 RNPs), which are imported back together into the nucleus. Once in the nucleus, the L1 RNA is reverse‐transcribed and integrated into a new genomic location by a coupled reverse transcription. During this mobilization mechanism, the retrotransposon sequence is prone to truncations and inversions, resulting in the insertion of mutated copies unable to jump a second time (Beck et al, 2011; Jachowicz & Torres‐Padilla, 2016). Nevertheless, 100 (Brouha et al, 2003) and 3,000 (Goodier et al, 2001) full‐length L1 elements in human and mouse genomes, respectively, retain the ability to encode the machinery necessary for production of the RNA intermediate, its reverse transcription, and consequent integration into a new genomic location. In mouse, active L1s are divided into three subfamilies: Tf, Gf, and A, which are defined by the variable sequence and numbers of monomers (tandem repeat units of 200 bp) contained in their 5'UTR (Naas et al, 1998; Deberardinis & Kazazian, 1999; Goodier et al, 2001).
While transposable elements are indispensable for genome variation and evolution, rogue and/or rampant transposition leads to disease (Beck et al, 2011). Elucidating mechanisms that regulate L1 transcription and mobility have been an active area of research since their discovery. DNA methylation in somatic cells and Piwi‐interacting RNA (piRNA) pathway in the germline are well‐established regulators of L1 retrotransposition (DiGiacomo et al, 2013; Goodier, 2016; Newkirk et al, 2017). At the blastocyst stage of embryonic development, however, both the above‐mentioned pathways are inactivated leading to a window necessitating alternate mechanisms of L1 regulation. The microRNA (miRNA) effector protein DICER has been implicated in modulating expression of L1 during this stage of development (Bodak et al, 2017). MiRNAs are 21–24 nucleotide (nt) long Pol II transcripts that play a major role in fine‐tuning gene expression posttranscriptionally (Ha & Kim, 2014; Bartel, 2018). Briefly, miRNAs are transcribed as primary (pri) miRNAs and processed into precursor (pre) miRNAs by DGCR8/DROSHA microprocessor complex in the nucleus. Upon export into the cytoplasm, DICER cleaves pre‐miRNAs to give rise to mature miRNAs. The mature miRNA duplex is loaded onto ARGONAUTE (AGO) proteins, upon unwinding of the duplex, one of the two strands is degraded. Along with accessory proteins, AGO loaded with the guide miRNA strand forms the RNA‐induced silencing complex (RISC) and acts as the effector. Base pairing of miRNA at its seed sequence with complementary miRNA response elements (MREs) typically found in the 3'UTR sequence of mRNAs induces translational repression or mRNA degradation. Preimplantation mouse embryos deleted for Dicer present an upregulation of L1 elements (Svoboda et al, 2004; Kanellopoulou et al, 2005). In human cancer cells, miR‐128 was shown to regulate L1 transposition via two mechanisms. First, miR‐128 repressed L1 expression directly by binding to a noncanonical binding site in L1 ORF2 RNA (Hamdorf et al, 2015), and second, miR‐128 bound to a canonical binding site in the 3'UTR sequence of TNPO1 an import factor that regulates entry of L1 RNP complex into the nucleus posttranslation (Idica et al, 2017). This mode of regulation via miR‐128, however, does not appear to be conserved in mESCs (Bodak et al, 2018). Recently, the direct binding of miRNA let‐7 to L1 mRNA was shown to impair L1 ORF2 translation and consequently retrotransposition (Tristán‐Ramos et al, 2020). As processing of pri‐let7 miRNA to mature let‐7 miRNA is blocked in mESCs (Viswanathan et al, 2008), this mechanism of fine‐tuning L1 expression is also not conserved in mESCs. To delve deeper into the role of Dicer in regulating L1 during embryonic development, our laboratory utilized mouse embryonic stem cells (mESCs) as a model. In Dicer_Knockout (KO) mESCs, while a 6–8‐fold increase in L1 transcription was observed, a concomitant increase in the rate of retrotransposition was not uncovered (Bodak et al, 2017). In this study, we demonstrate that miRNAs are involved in the regulation of L1 retrotransposition in mESCs through the direct regulation of the RNA helicase Mov10. Upon loss of miRNAs, MOV10 is strongly upregulated and accumulates in the cytoplasm of mESCs, driving sequestration of L1 RNPs into aggregates, thereby preventing L1 mobility.
Results
L1 RNA and protein accumulate in cytoplasmic foci of Dicer_KO mESCs
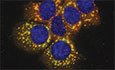
To better understand why the strong upregulation of L1 RNAs does not lead to a subsequent retrotransposition in Dicer_KO mESCs (Bodak et al, 2017), we looked at the localization of L1 RNA and protein in wild‐type (WT) and mutant cells. We probed for L1 RNA derived from the Tf L1 family by RNA fluorescent in situ hybridization (RNA FISH) along with L1 ORF1p by indirect immunofluorescence (IF). While in WT mESCs, we observed diffused signal for both L1 Tf RNA and ORF1p, they co‐localized as L1 ribonucleoprotein (L1 RNP) foci in cytoplasm of the two independent Dicer_KO clones (Fig 1A). The median number of L1 RNP foci in the cytoplasm per cell in Dicer_KO1 and Dicer_KO2 mESCs was 9 and 7, respectively, as compared to 0 in WT cells. In addition, in 30–35% of Dicer_KO cells, L1 RNP were observed to co‐localize in larger foci (Fig 1A). These observations led us to hypothesize that sequestration of L1 RNP in the cytoplasm of Dicer_KO mESCs is preventing L1 retrotransposition.
Figure 1. L1 RNPs accumulate as cytoplasmic foci in Dicer_KO mESCs.
-
AMaximum intensity projections across Z stacks of example images from indicated mESCs stained for L1 Tf RNA (red) combined with immunostaining for L1 ORF1p (green) and nuclei stained with DAPI (blue). The gray square marks position of the inset. White arrow heads point to cytoplasmic foci where L1 RNA and ORF1p co‐localize. Scatter plot shows the number of co‐localized L1 Tf‐ORF1 foci in the cytoplasm per cell (n = 275 WT, 304 Dicer_KO1, 311 Dicer_KO2 cells). Red arrow heads point to relatively larger sized L1 RNP foci. Bar graphs are mean values of percentage of cells with large L1 Tf‐ORF1 foci co‐localizing in the cytoplasm (n = 3 technical replicates).
-
BMaximum intensity projections across Z stacks of example images from indicated mESCs immunostained for L1 ORF1p (red), MOV10 (green) and nuclei stained with DAPI (blue). The gray square marks position of inset in the zoomed image. White arrow heads point to cytoplasmic foci where L1 ORF1p and MOV10 co‐localize. Scatter plot shows the number of co‐localized ORF1‐MOV10 foci in the cytoplasm per cell (n = 293 WT, 295 Dicer_KO1, 295 Dicer_KO2 cells). Red arrow heads point to relatively larger sized L1 ORF1‐MOV10 foci. Bar graphs are mean values of percentage of cells with large ORF1‐MOV10 foci co‐localizing in the cytoplasm (n = 3 technical replicates).
Data information: Data are depicted as scatter plots where circles are single data points representing number of co‐localized L1 Tf‐ORF1 foci in the cytoplasm per cell (A), or co‐localized L1 ORF1‐MOV10 foci in the cytoplasm per cell (B) red bar is median. Statistical analysis was performed using Mann–Whitney U test and ****P‐value < 0.0001. Bar graphs represent mean ± SD. Scale bar 5 μm.
Source data are available online for this figure.
L1 RNPs co‐localize with MOV10 helicase in cytoplasmic foci of Dicer_KO mESCs
To characterize L1 RNP foci, we aimed to identify other cellular components that might share their location with them. We therefore tested whether known interactors of human L1 proteins might co‐localize with L1 RNP cytoplasmic foci in Dicer_KO mESCs (Doucet et al, 2010; Goodier et al, 2012; Taylor et al, 2013, 2018). Amongst the list of candidates interacting with both L1 ORF1p and L1 ORF2p (Taylor et al, 2018), we looked at RNA helicases UPF1 and MOV10 by IF. While UPF1 was observed to have diffused cytoplasmic staining (Appendix Fig S1), MOV10 co‐localized with L1 RNPs in the cytoplasm of Dicer_KO mESCs (Fig 1B). Further analysis revealed MOV10 to co‐localize with L1 ORF1p in Dicer_KO cells with a median of 3 foci in WT cells and 12 and 15, respectively, in Dicer_KO1 and Dicer_KO2 mESCs. Percentage of cells with large ORF1‐MOV10 foci was 26–47% in the two Dicer_KO lines (Fig 1B). The higher frequency of ORF1‐MOV10 foci as compared with Tf‐ORF1p foci in Dicer_KO cells is most likely due to the lower sensitivity for detecting Tf RNA by RNA FISH. As MOV10 co‐localization with L1 ORF1 foci in Dicer_KO mESCs was high and due to the absence of good antibodies available for L1 proteins raised in hosts other than rabbit for co‐staining IF experiments, we further used MOV10 as a proxy for L1 RNP localization.
L1 RNP foci are aggregates of RNA and proteins
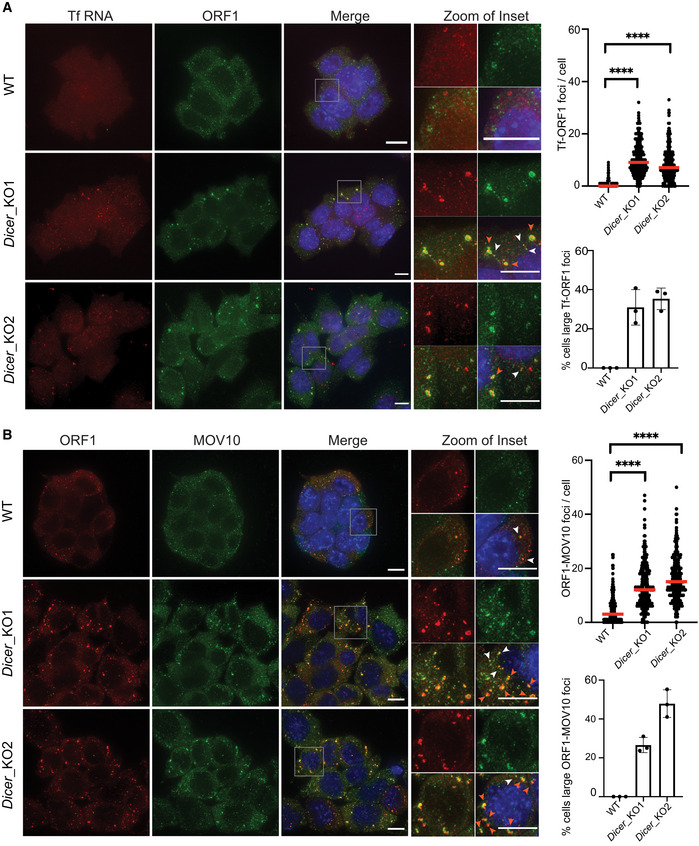
Localization of L1 RNPs as cytoplasmic foci was previously reported for human L1 proteins upon their ectopic overexpression in HEK293T cells (Goodier et al, 2007). L1 ORF1 foci were furthermore shown to co‐localize with stress granules and RNA‐binding proteins including components of the RISC complex (Goodier et al, 2007). To assess the nature of the observed mouse L1 RNP foci, we co‐stained WT and Dicer_KO mESCs for G3BP1, a marker for stress granules (Kedersha et al, 2016), along with MOV10. The signal for G3BP1 was mainly diffused cytoplasmic in both WT and Dicer_KO mESCs, indicating that unlike human cancer cells, mouse L1 ORF1‐MOV10 foci are not stress granules (Fig 2A). However, treatment with 0.5 mM sodium arsenite for 20 min to induce stress caused MOV10 to co‐localize with G3BP1 as cytoplasmic bodies in Dicer_KO cells (Fig 2A). These data led us to hypothesize that L1 RNP foci in Dicer_KO mESCs might be poised but are not as yet mature stress granules.
Figure 2. Cytosolic L1 RNP foci are poised to be stress granules that co‐localize with multiple small DDX6 foci.
-
AWT and Dicer_KO mESCs were treated with 0.5 mM sodium arsenite (NaAsO2) for 20 min or left untreated prior to fixation with formaldehyde. Maximum intensity projections across Z stacks of example images from indicated mESCs immunostained for G3BP1 (red) and MOV10 (green) with nuclei stained with DAPI (blue).
-
BRepresentative Western blots showing low AGO2 protein levels in Dicer_KO as compared to WT mESCs (right side). LAMINB1 served as loading control. On the left, maximum intensity projections across Z stacks of example images from indicated mESCs stained for L1 Tf RNA FISH (red) combined with immunostaining for a resident protein of P‐bodies, DDX6 (green) and nuclei stained with DAPI (blue). The gray square marks position of the inset. Yellow arrow heads point to cytoplasmic foci where L1 RNA and DDX6 protein co‐localize.
Data information: (A and B) are representative images of three independent experiments. Bar graphs represent mean ± SD. 94–150 cells per cell line were analyzed for each experiment. Scale bar 5 μm.
Source data are available online for this figure.
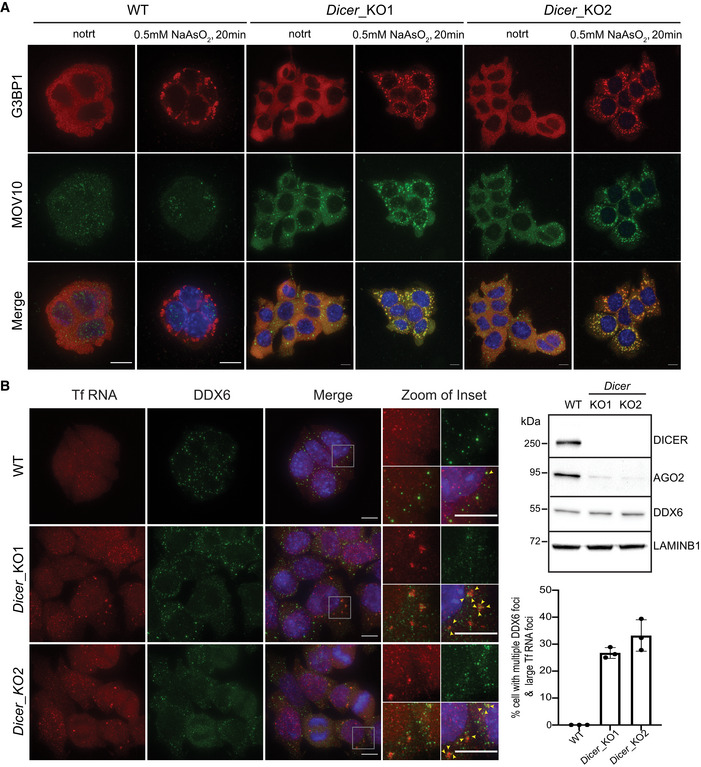
Partitioning of stress granule proteins as liquid–liquid phase separation (LLPS) is emerging as a main driver for shifting dynamics from being near soluble to condensate formation thereby impacting their biological function (Wheeler et al, 2016). RNA and RNA‐binding proteins (RBPs) are key components of these cytoplasmic condensates (Roden & Gladfelter, 2021). Recently, by microscopy and NMR spectroscopy, human L1 ORF1p was shown to form liquid droplets in vitro in a salt‐dependent manner (Newton et al, 2021). To test whether L1 ORF1 foci in mESCs undergo similar LLPS, we treated Dicer_KO mESCs with 3% 1,6 hexanediol for 15 min, a concentration at which proteins undergoing LLPS have been previously observed to change solubility from being in foci to becoming diffused in mESCs (Valsecchi et al, 2021). No overt change in L1 ORF1‐MOV10 foci was observed in cells treated with 1,6 hexanediol (Fig EV1A), suggesting that L1 ORF1‐MOV10 foci are not LLPS condensates.
Figure EV1. L1 RNP cytosolic aggregates are not sensitive to treatment with 3% 1,6 Hexanediol.
-
AMaximum intensity projections across Z stacks of example images from indicated mESCs immunostained for L1 ORF1p (red) and MOV10 (green) with nuclei stained with DAPI (blue). Gray box mark position of the insets (n = 3 biological replicates).
-
BMaximum intensity projections across Z stacks of example images from indicated mESCs immunostained for LC3B (red) and MOV10 (green) with nuclei stained with DAPI (blue). The gray square depicts position of inset (n = 3 technical replicates).
Data information: Scale bar 5 μm.
Human L1 ORF1p are also known to associate with processing body (P‐body) enriched mRNAs (Briggs et al, 2021). While elucidation of the functional relevance of P‐bodies is an active area of research, it is well established that these cytoplasmic granules also undergo LLPS (Luo et al, 2018). As the L1 RNP foci are not sensitive to 1,6 hexanediol treatment and most likely not undergoing LLPS, our data argue against L1 RNP foci being components of P‐body in mutant mESCs. In addition, the protein ARGONAUTE2 (AGO2), a known component of P‐bodies (Sen & Blau, 2005) and an effector of the miRNA biogenesis pathway, is required for P‐body formation (Pauley et al, 2006; Eulalio et al, 2007). In Dicer_KO mESCs, due to the absence of miRNAs, AGO2 protein levels are reduced and the protein destabilized (Bodak et al, 2017; Figs 2B and EV3C). However, protein levels of DDX6, another known constituent of P‐bodies (Ayache et al, 2015), were unchanged as compared with WT cells (Fig 2B). We therefore looked at the cellular localization of DDX6 to assess P‐body integrity and association with L1 RNP foci. Unlike WT cells where DDX6 formed droplet‐like foci characteristic of P‐bodies in the cytoplasm, in Dicer_KO cells, DDX6 was more diffusely localized in the cytoplasm. In 26–32% of Dicer_KO mESCs, multiple small DDX6 foci were observed co‐localizing with larger L1 Tf RNA foci (Fig 2B). The partial co‐localization with DDX6 in cells with low AGO2 levels suggest that L1 RNP foci are not canonical P‐bodies, corroborating earlier studies enumerating the requirement of intact miRNA biogenesis in P‐body fidelity (Pauley et al, 2006; Eulalio et al, 2007).
Figure EV3. Posttranscriptional regulation of Mov10 by miRNAs in mESCs.
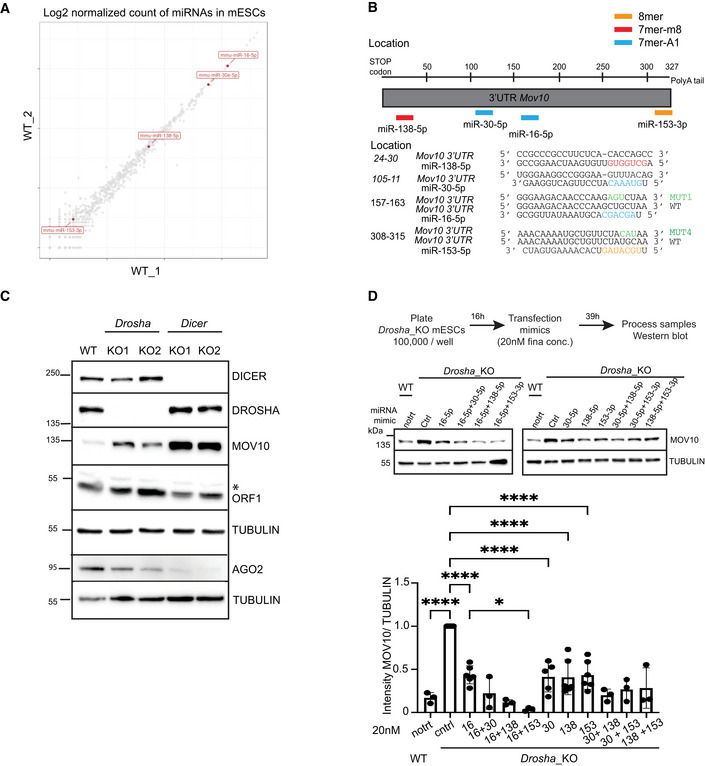
- A
-
BSchematic of 3'UTR sequence of Mov10 RNA helicase. Location and sequence of WT and mutant (MUT) miRNA response elements (MREs) for mouse miR‐138‐5p (red), miR‐30‐5p (blue) miR‐16‐5p (blue) and miR‐153‐3p (orange) predicted to target Mov10 (ENSMUST00000168015.8) are color coded based on seed type matching for respective miRNAs.
-
CWestern Blot analysis to assess the expression of L1 ORF1p, MOV10, AGO2 in the indicated cell lines, immunoblot with TUBULIN served to control for loading. KO status was confirmed by probing for DROSHA and DICER (n = 3 technical replicates). Asterisk marks nonspecific band.
-
DSchematic depicting design of experiment for processing samples along with representative gels for WB analysis and quantitation of MOV10 and TUBULIN signals in Drosha_KO cells transfected with ctrl or mimic miRNA either singly or in pairs (n = 3–6 biological replicates). Untreated WT cells are shown for comparison of MOV10 expression.
Data information: Bar graphs are mean intensity ± SD of MOV10 normalized by TUBULIN. Values are relative to transfection for the Ctrl mimic that was set to 1. P‐values were computed using ordinary one‐way ANOVA test ****P‐value < 0.0001 and *P‐value < 0.05.
Source data are available online for this figure.
Finally, we ascertained that L1 RNP foci were not autophagosomes (Guo et al, 2014) as LC3B a marker for autophagosomes did not co‐localize with MOV10 in mESCs by IF (Fig EV1B). We therefore called L1 RNPs present in cytoplasmic foci of Dicer_KO mESCs, aggregates as they contain an assembly of RNA and proteins without undergoing phase separation.
Generation of mESCs to upregulate L1 expression using CRISPRa
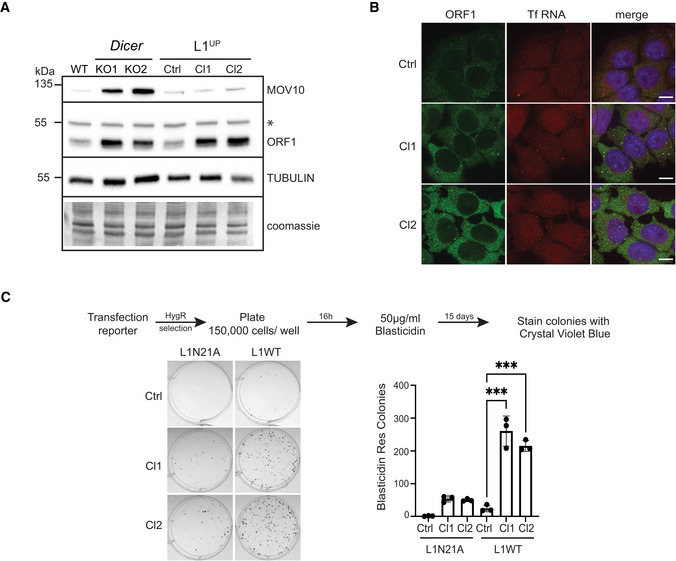
L1 upregulation is amongst the many changes in gene expression observed upon deleting Dicer in mESCs (Bodak et al, 2017; Cirera‐Salinas et al, 2017). To find out whether as observed in human cultured cells overexpression of L1s was sufficient for cytoplasmic sequestration (Goodier et al, 2007, 2012), we engineered WT mESCs to endogenously upregulate L1 using CRISPRa (L1UP; Figs 3A and EV2A). We designed single‐guide RNAs (sgRNAs) to target dCas9 fused with VP160 to the 5'UTR sequence of the L1 Tf family (Fig EV2B). For the generation of independent clones (Cl), L1UP Cl1 cells were transfected with one sgRNA, while two sgRNA pairs were used to upregulate L1 in L1UP Cl2. A 2.5‐fold increase in L1 Tf transcript levels as compared with the control cell line (Ctrl) transfected with an empty sgRNA vector was observed (Fig EV2C) in L1UP clones. Given the sequence homology of the three L1 families, we also observed a threefold increase in transcript levels of L1 A family. An increase in expression of L1 Gf family was also observed; however, the variability between experiments rendered the increase to be statistically significant only for Cl1 (Fig EV2C). While L1 transcript levels in L1UP cells was lower than in Dicer_KO (Fig EV2C), the expression of L1 ORF1p in L1UP was similar to that observed in Dicer_KO cells (Fig 3A).
Figure 3. Endogenous L1 upregulation leads to increased L1 retrotransposition.
-
ARepresentative Western blots showing L1 ORF1p and MOV10 levels in the indicated cell lines. Immunoblot with antibody recognizing TUBULIN and coomassie stained membranes depict the loading, asterisk marks position of nonspecific band in the ORF1 immunoblot (n = 3 technical replicates).
-
BMaximum intensity projections across Z stacks of example images from indicated mESCs immunostained for L1 ORF1p (green) combined with RNA FISH for L1 Tf RNA (red) and nuclei stained with DAPI (blue) (n = 3 technical replicates). Scale bar 5 μm.
-
CRepresentative images of BlastR colonies stained with crystal violet blue of indicated cell lines is shown on the left. Cells were transfected with either mutant reporter plasmid (L1N21A) or retrotransposition competent reporter (L1WT) as shown in the scheme with timeline for the experiment on the top. Bar graphs on the right depict the average number of BlastR colonies (n = 3 biological replicates).
Data information: In (C) bar graphs represent mean ± SD, P‐value was determined using ordinary one‐way ANOVA test. ***Represent P‐value < 0.001.
Source data are available online for this figure.
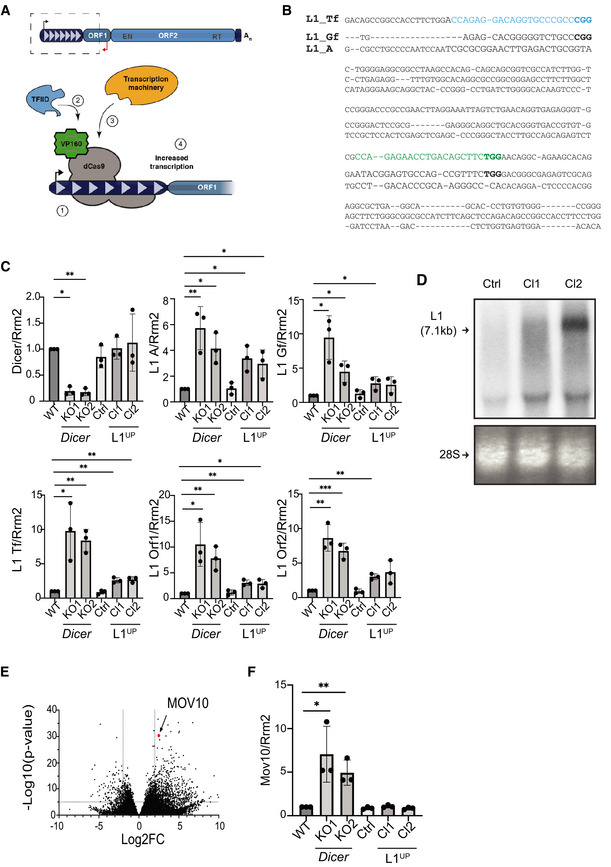
Figure EV2. Generation of mESCs upregulating L1 expression using CRISPRa.
-
ASchematic depicting full‐length L1 element and summary of CRISPRa. To generate L1UP cells, mESCs were co‐transfected with plasmid encoding catalytically dead Cas9 protein (dCas9) fused to VP160 and sgRNAs that (1) targeted the fusion protein to the 5'UTR sequence of Tf L1 family allowing (2) recruitment of transcription factors and (3) transcription machinery to (4) upregulate L1 transcription.
-
BSequence alignment of 5'UTR sequences of murine L1 Tf, Gf and A subfamilies. The two sgRNA sequences used to upregulate L1 expression are indicated in blue and green, with protospacer adjacent motifs (PAM) in bold.
-
CRT–qPCR analysis to quantitate Dicer, and L1 RNA expression levels in the depicted cell lines. Rrm2 was utilized for normalization, and graphs depict fold change in transcript levels compared to WT which was set to one (n = 3 technical replicates).
-
DRepresentative northern blot analysis probed for L1 RNA to assess L1 transcript length and expression levels in the engineered L1UP Cl1, Cl2 as compared with Ctrl cells. Arrow points to full‐length L1 transcript. Ethidium bromide staining of 28S RNA was used to confirm equal loading (n = 3 technical replicates).
-
EDifferential gene expression from RNA‐seq analysis of Dicer_KO vs. WT mESCs plotted using previously published data (Bodak et al, 2017).
-
FRT–qPCR analysis to confirm upregulation of Mov10 in Dicer_KO cells. Rrm2 was utilized for normalization, and graphs depict fold change in transcript levels in the indicated cell lines as compared to WT which was set to one (n = 3 technical replicates).
Data information: In (C and F) bar graphs are mean ± SD. P‐value computed using unpaired t‐test comparing results from individual cells to WT mESCs. ***P‐value < 0.0005, **P‐value < 0.001, *P‐value < 0.05. In (E) each dot represents a single gene. Position for Mov10 in the graph is marked. Values for Log2 fold change (Log2FC) were plotted on the x‐axis and Log10 of the P‐value on the y‐axis.
Source data are available online for this figure.
L1UP mESCs are prone to retrotransposition
To assess whether L1 elements upregulated with CRISPRa were competent for retrotransposition, we primarily performed northern blot analysis. While for Cl2, a discrete band similar to full‐length transcript expressed in Dicer_KO mESCs was observed, the signal for L1 in Cl1 was more of a smear (Bodak et al, 2017; Fig EV2D). The difference may arise from having used two guides for generation of Cl2 as opposed to Cl1 L1UP cells. Importantly, this level of upregulation of L1 RNA was not sufficient to cause L1 RNP accumulation in cytoplasmic aggregates in L1UP mESCs (Fig 3B). Using a plasmid‐based retrotransposition assay (Kopera et al, 2016), we tested whether in the absence of L1 RNP cytosolic sequestration, there was an enhanced rate of L1 retrotransposition in the engineered L1UP mESCs. We transfected Ctrl, L1UP Cl1, and L1UP Cl2 with either wild‐type JJ‐L1SM (L1WT) or a plasmid with a missense mutation in the endonuclease domain of ORF2 rendering it incompetent for jumping (L1N21A) that carried Blasticidin resistance (BlastR) as a reporter gene and Hygromycin (HygR) as a selection marker (Maclennan et al, 2017). Unlike in Dicer_KO cells (Bodak et al, 2017), L1 upregulation was accompanied by an increase in the rate of mobility depicted by the higher number of BlastR colonies observed in L1UP Cl1 and Cl2 as compared with Ctrl mESCs (Fig 3C). BlastR colonies observed in the two L1UP cell lines transfected with L1N21A reporter confirm previous observation of mobilization of mutant L1s aided by endogenous full‐length L1s in the cell, but at relatively low frequencies (Wei et al, 2001). To conclude, forced endogenous upregulation of L1 active elements in WT mESCs is not sufficient to create L1 RNP cytoplasmic aggregates and leads to an increase in retrotransposition.
MOV10 expression is upregulated in Dicer_KO mESCs
Given that upregulation of L1 in mESCs was not sufficient to induce L1 RNP aggregation in the cytoplasm (Fig 3B), and our finding that MOV10 co‐localized with L1 RNP in Dicer_KO cells (Fig 1B), we speculated that cytosolic aggregation of L1 RNPs might be driven by the upregulation of MOV10 observed in Dicer_KO mESCs at RNA and protein levels (Figs 3A and EV2E). MOV10 upregulation in Dicer_KO mESCs was confirmed by RT–qPCR analysis (Fig EV2F). In addition, no changes in MOV10 expression were observed either at RNA (Fig EV2F) or at protein levels in L1UP mESCs (Fig 3A). We therefore ruled out L1 overexpression as the driver for MOV10 upregulation and investigated the role of miRNAs in posttranscriptional regulation of Mov10 as miRNA biogenesis is impaired in Dicer_KO mESCs.
MOV10 expression is regulated by several miRNAs in mESCs
Using TargetScan software (Agarwal et al, 2015), we identified multiple miRNAs (miR‐138‐5p, miR‐30‐5p, miR‐16‐5p, and miR‐153‐5p) as predicted to target the 3'UTR sequence of Mov10 (Fig EV3B). The relative expression of each miRNA in WT cells was determined using previously published small RNA sequencing data from our laboratory (Ngondo et al, 2018; Fig EV3A). MiR‐16‐5p and miR‐30‐5p are highly expressed in WT mESCs compared with the intermediate expression of miR‐138‐5p, and the low expression of miR‐153‐3p (Fig EV3A). We tested whether the predicted miRNAs might directly regulate Mov10 expression by performing a luciferase reporter assay (Jin et al, 2013). We subcloned the 3'UTR sequence of Mov10 downstream of the Renilla luciferase reporter gene in a plasmid that also encoded Firefly luciferase as a normalizer. Transient transfection of this plasmid along with the respective miRNAs into HEK293T followed by measurement of the respective luminescence showed that for the tested mimics, RENILLA expression was significantly sensitive to transfection with miR‐16‐5p and miR‐153‐3p (Fig 4A). To confirm that downregulation of the luciferase reporter was specific and due to the interaction of miRNA mimics with the corresponding miRNA response element (MRE) in the 3'UTR sequence of Mov10, we generated three additional luciferase reporter plasmids. We mutated the MRE in the 3'UTR sequence of Mov10 that is complementary to the seed sequence of miR‐16‐5p (CTG > ATG: MUT1), miR‐153‐3p (TGC > CAT: MUT4), and a third plasmid where both the MREs are mutated (MUT1 + MUT4; Fig EV3B). Indeed, mutating the MREs rendered expression of RENILLA luciferase to be insensitive to transfection with the corresponding miRNA mimic, providing further proof for the capability of miRNA‐mediated posttranscriptional regulation of Mov10 (Fig 4B). To corroborate that the upregulation of MOV10 in Dicer_ KO cells is indeed mediated by miRNAs and is not a consequence of noncanonical function of Dicer, we tested whether a similar upregulation of MOV10 is present in Drosha_KO cells where the canonical miRNA biogenesis pathway is also impaired (Cirera‐Salinas et al, 2017). Western blot (WB) analysis on Drosha_KO cells revealed that MOV10 is indeed upregulated in these cells (Fig EV3C). To validate the miRNA‐mediated regulation of Mov10 expression, we transiently transfected Drosha_KO mESCs with the respective miRNA mimics either singly or in pairs and measured MOV10 expression. Unlike as previously observed with the luciferase assay, the expression of MOV10 was downregulated to a similar extent upon transfection with each of the four tested miRNA mimics (Fig EV3D). Interestingly, only paired transfection of miR‐16‐5p with miR‐138‐5p or miR‐153‐3p acted synergistically to reduce MOV10 protein levels down to WT levels (Fig EV3D). Recent studies highlight cautious interpretation of results when using high concentrations of miRNA mimics for transfections (Jin et al, 2015; preprint: Mockly et al, 2022). Therefore, to further rule out nonspecificity of miRNA‐mediated regulation of Mov10 expression, we did a titration of miR‐16‐5p and miR‐153‐3p mimic concentrations from 1 to 20 nM for dual transfections into Drosha_KO cells. A decrease in MOV10 protein levels was observed for all concentrations tested (Fig 4C). The knockdown (KD) of MOV10 expression was statistically significant for concentrations 2 nM and above, and the best efficiency of KD was achieved with 4 nM final concentration, which was slightly better than what we obtained with 20 nM concentration for transfections (Fig 4C). We were limited to perform the above transfections with miRNA mimics to modulate MOV10 expression only in Drosha_KO cells and not in Dicer_KO as AGO2, where miRNA mimics need to be loaded to exhibit an effect, was destabilized to a much larger extent in Dicer_KO mESCs (Fig EV3C). Collectively, these data reveal a role for miRNAs in fine‐tuning MOV10 expression in mESCs, explaining the observed MOV10 upregulation in Dicer_KO and Drosha_KO mESCs (Figs 3A and EV3C).
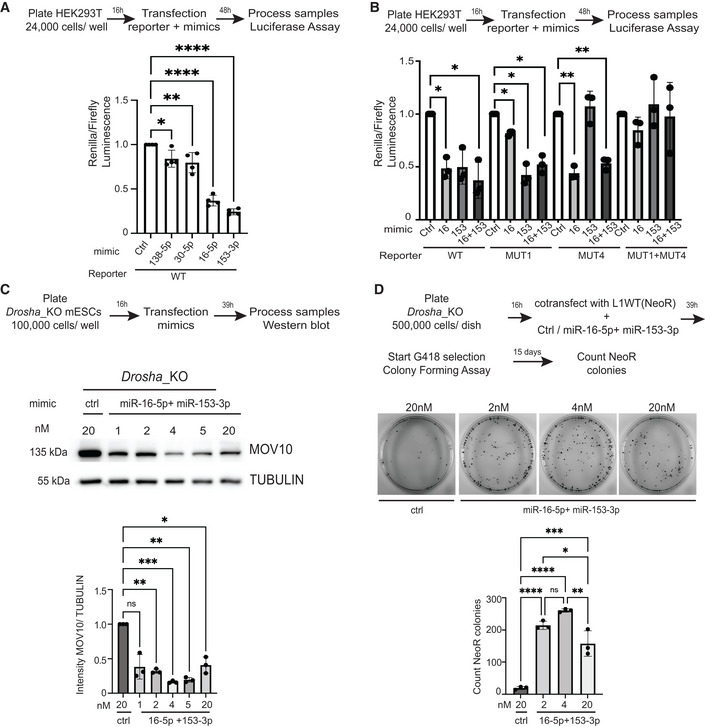
Figure 4. Multiple miRNAs regulate MOV10 expression and L1 retrotransposition in mESCs.
-
AScheme and quantification of relative luminescence from luciferase assays validating posttranscriptional regulation of MOV10 by miRNAs in HEK293T cells (n = 4 biological replicates).
-
BScheme and quantification of relative luminescence from luciferase assays in Hek293T cells to confirm specificity of posttranscriptional regulation of MOV10 by miRNAs with either wild‐type (WT) or mutant (MUT) reporters where the MicroRNA response element (MRE) for the depicted miRNAs in the 3'UTR of MOV10 were mutated (n = 3 biological replicates).
-
CScheme and representative images for western blot analysis and corresponding quantification to assess miRNA‐mediated regulation on MOV10 expression in mESCs (n = 3 biological replicates).
-
DScheme and representative images from colony‐forming assays for plasmid‐based retrotransposition analysis where miRNA‐mediated downregulation of MOV10 in Drosha_KO mESCs is accompanied by an increase in L1 retrotransposition (n = 3 biological replicates).
Data information: In (A–D), bar graphs represent mean ± SD, P‐value determined using ordinary one‐way ANOVA test ****P‐value < 0.0001, ***P‐value < 0.001, **P‐value < 0.005, *P‐value < 0.05.
Source data are available online for this figure.
Cytosolic aggregation of L1 RNPs also occurs in Drosha_KO cells
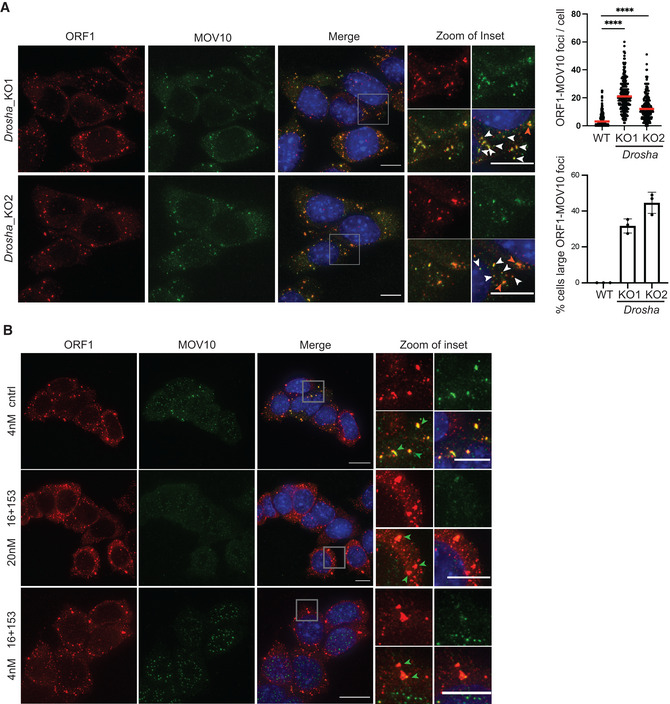
Given the upregulation of MOV10 and L1 ORF1p in Drosha_KO cells as compared with WT (Fig EV3C), we next assessed whether L1 RNP correspondingly also aggregate in the cytoplasm of these miRNA mutants. We performed IF with L1 ORF1p and MOV10 antibodies in two independent Drosha_KO clones and observed MOV10 co‐localizing with L1 RNPs in the cytoplasm of Drosha_KO mESCs (Fig EV4A). The median ORF1‐MOV10 aggregates per cell were 21 and 12 in Drosha_KO1 and Drosha_KO2 mESCs, respectively (Fig EV4A). Percentage of cells with large ORF1‐MOV10 foci was 31–44% in the two Drosha_KO lines (Fig EV4A), similar to that observed in Dicer_KO cells (Fig 1B).
Figure EV4. Cytosolic L1 RNP foci in Drosha_KO cells.
-
AMaximum intensity projections across Z stacks of example images from indicated Drosha_KO mESCs immunostained for L1 ORF1p (red) combined with MOV10 (green) and nuclei stained with DAPI (blue). The gray square marks position of the inset. White arrow heads point to cytoplasmic foci where ORF1p and MOV10 co‐localize. Scatter plot shows the number of co‐localized L1 Tf‐ORF1 foci in the cytoplasm per cell (288 Drosha_KO1, 299 Drosha_KO2 cells, 293 WT (same as Fig 1)). Red arrow heads point to relatively larger sized L1 RNP foci. Bar graphs are mean values of percentage of cells with large ORF1‐MOV10 foci co‐localizing in the cytoplasm (n = 3 technical replicates).
-
BMaximum intensity projections across Z stacks of example images from Drosha_KO2 cells transfected with either control or indicated concentration of miR‐16‐5p and miR‐153‐3p miRNA mimics (n = 3). The gray square marks position of the inset. Green arrow heads point to relatively large cytoplasmic foci where ORF1p and MOV10 co‐localize.
Data information: In (A) data are depicted as scatter plots where circles are single data points representing number of co‐localized ORF1p‐MOV10 foci in the cytoplasm per cell, red bar is median. Statistical analysis was performed using Mann–Whitney U test and ****P‐value < 0.0001. Bar graphs represent mean ± SD. Scale bar 5 μm.
Source data are available online for this figure.
Restoring MOV10 expression in Drosha_KO mESCs leads to L1 retrotransposition
To confirm our hypothesis that aggregation of L1 RNPs driven by MOV10 overexpression was preventing L1 retrotransposition, we examined whether reducing MOV10 expression in Drosha_KO cells would allow L1 mobilization. We used a plasmid‐based retrotransposition assay (Kopera et al, 2016) and transiently co‐transfected Drosha_KO cells with pCEP‐L1WT reporter plasmid that carried Neomycin resistance (NeoR; Maclennan et al, 2017) as a reporter along with either 20 nM Ctrl mimic or 2, 4 and 20 nM final concentration of miR‐16‐5p and miR‐153‐3p mimic together to downregulate MOV10 expression. A total of 500,000 cells were plated for each condition for the colony‐forming assay (CFA), and media was supplemented with G418 39 h post transfection. The mean NeoR colonies obtained 15‐day postselection was 20 in the Drosha_KO clones transfected with Ctrl mimics from three independent experiments. A statistically significant increase in NeoR colonies in cells transfected with miRNA mimics was observed with the mean increasing to 214, 260, and 157 for transfections with 2, 4, and 20 nM mimic concentration, respectively (Fig 4D). We next assessed whether transfections with 4 or 20 nM of miR‐16‐5p and miR‐153‐3p mimic impacted L1 ORF1‐MOV10 aggregate formation in Drosha_KO cells by IF. We did not observe any overt change in aggregate formation in any of the tested conditions (Fig EV4B). From the CFA experiment, we learnt that on an average retrotransposition is only observable in approximately 200 of the 500,000 cells that were initially plated. We conclude that IF analysis does not afford the same sensitivity to observe the breakdown of the aggregates. Finally, our results are in line with data from human cancer cells supporting the role for MOV10 as a negative regulator of retrotransposition (Arjan‐Odedra et al, 2012; Goodier et al, 2012; Li et al, 2013; Choi et al, 2018; Warkocki et al, 2018), and to our knowledge, the first to report a role for miRNAs in fine‐tuning Mov10 expression.
Ectopic expression of MOV10 in L1UP cells can induce L1 RNP aggregate formation and decreases L1 retrotransposition
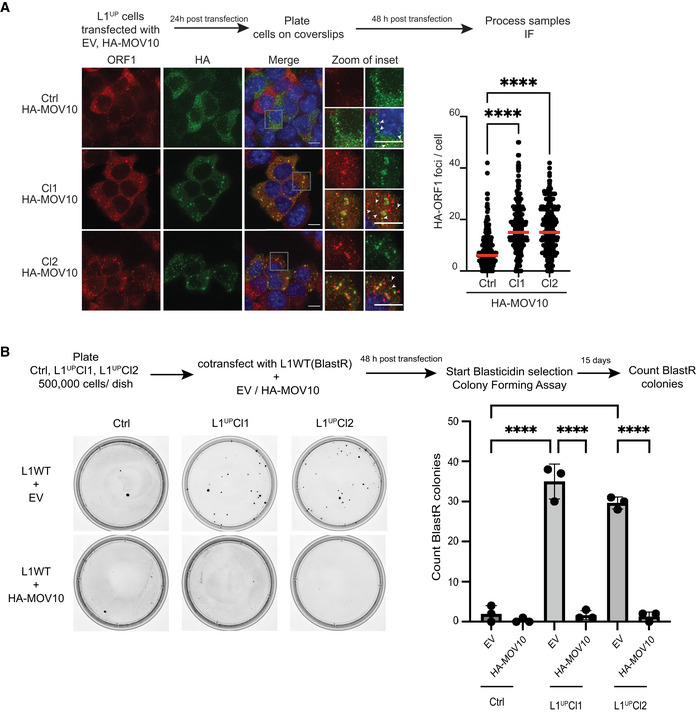
Mature miRNAs might regulate multiple mRNAs and an mRNA can be targeted by several miRNAs (Peter, 2010). While we show that transfection with miR‐16‐5p and miR‐153‐3p mimics downregulates MOV10 expression leading to increased L1 mobility, we cannot unequivocally rule out that changes in expression of another gene targeted by these miRNAs might be responsible for the observed increase in transposition. To assess whether MOV10 expression is sufficient to induce L1 RNP aggregation in the cytosol, we transiently transfected Ctrl, L1UP Cl1, and L1UP Cl2 mESCs with a plasmid encoding HA tagged human MOV10 (HA‐MOV10). In IF experiments with an antibody against HA to detect exogenously expressed HA‐MOV10 along with anti‐L1 ORF1p antibody, we detected HA‐MOV10‐ORF1 aggregates in the cytoplasm of L1UP Cl1 and L1UP Cl2 significantly more than in Ctrl cell line (Pval < 0.001). The median number of foci observed in Ctrl was 6 per cell while in the two L1UP clones, this was 15 (Fig 5A). In addition, the morphology of the larger HA‐MOV10‐ORF1 aggregates observed in L1UP clones was reminiscent of those observed in Dicer_KO mESCs (Figs 5A and 1B).
Figure 5. MOV10 upregulation is sufficient to create L1 RNP aggregates in L1UP mESCs abrogating L1 retrotransposition.
-
AScheme of experiment and representative images along with quantitation showing induction of L1 RNP aggregate formation upon ectopic MOV10 expression. Images are maximum intensity projections across Z stacks from indicated mESCs stained for L1 ORF1p (red) combined with immunostaining for HA (green) to detect ectopically expressed MOV10 tagged with HA, and nuclei stained with DAPI (blue). White arrow heads point to cytoplasmic foci where L1 ORF1p and HA‐MOV10 co‐localize. Data collected from 289 Ctrl, 275 L1UP Cl1, and 296 L1UP Cl2 mESCs (n = 3 biological replicates).
-
BScheme and representative images from colony‐forming assays for plasmid‐based retrotransposition analysis in Ctrl and L1UP mESCs comparing rate of retrotransposition upon transfection with Empty Vector (EV) or ectopic expression of HA‐MOV10 (n = 3 biological replicates).
Data information: In (A) scatter plots where circles are single data points representing number of co‐localized HA‐ORF1 foci in the cytoplasm per cell. Red bar marks median for the distribution. P‐value was determined using Mann–Whitney U test and **** represent P‐value < 0.0001. In (B) bar graphs represent mean ± SD, P‐value determined using ordinary one‐way ANOVA test ****P‐value < 0.0001.
Source data are available online for this figure.
To prove that MOV10‐induced L1 RNP aggregation restricts L1 mobility, we then transiently co‐transfected Ctrl, L1UPCl1, and L1UPCl2 mESCs with JJ‐L1WT reporter plasmid that carries BlastR reporter (Maclennan et al, 2017) along with either empty vector (EV) or HA‐MOV10 plasmids. The mean BlastR colonies was 35 and 29 for the two L1UP clones and 2 in Ctrl cells, corroborating our earlier observation of increased L1 mobility in L1UP cells as compared with Ctrl (Figs 5B and 3C). Importantly, a statistically significant decrease in BlastR colonies was observed in L1UP clones transfected with HA‐MOV10 when compared to EV with a mean of 1 BlastR colony obtained from the transfection in both the clones (Fig 5B).
Ectopic expression of L1 ORF1p along with MOV10 in WT mESCs is sufficient to induce L1 RNP aggregate formation
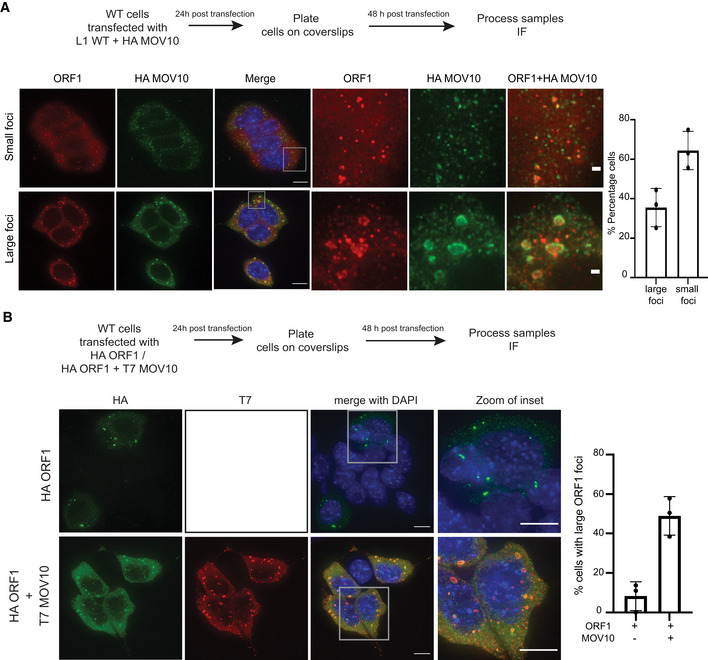
Guide RNAs can have off‐target effects (O'Geen et al, 2015). To rule this out as influencing our observations regarding induction of L1 RNP aggregate formation in L1UP cells, we transiently transfected the JJ‐L1WT reporter as a mean to upregulate L1 expression in WT mESCs in conjunction with HA‐MOV10. IF analysis was performed with an antibody specific for L1 ORF1p and HA to visualize the ectopic expression of MOV10 protein. We observed the induction of ORF1‐MOV10 aggregate formation in 35% of the transfected cells that were positive for HA (Fig 6A). These results are in line with our observations in L1UP cells where HAMOV10 was overexpressed similarly in a transient manner (Fig 5A), and in Dicer_KO and Drosha KO cells where endogenous L1 and MOV10 are upregulated (Figs 1B and EV4A).
Figure 6. Overexpression of L1 ORF1p and MOV10 is sufficient to induce cytosolic aggregation in WT mESCs.
-
AScheme of experiment and representative images showing RNP aggregate formation upon ectopic expression of full‐length L1 and HA‐MOV10 in WT mESCs. Images are maximum intensity projections across Z stacks stained for L1 ORF1p (red) combined with immunostaining for HA (green) to detect ectopically expressed MOV10, and nuclei stained with DAPI (blue). The bottom panel shows cells where accumulation of ORF1‐MOV10 was observed in relatively large foci compared with cells where co‐localization of the two proteins was mostly in relatively small foci (top panel), 68–119 transfected cells were analyzed per experiment (n = 3 biological replicates).
-
BScheme of experiment and representative images showing RNP aggregate formation upon ectopic expression of HA‐ORF1 or HA‐ORF1 in conjunction with T7‐MOV10 in WT mESCs. Images are maximum intensity projections across Z stacks stained for HA (green) and T7 (red) to detect ectopically expressed ORF1p and MOV10 respectively, and nuclei stained with DAPI (blue). A total of 100–147 transfected cells were analyzed per experiment (n = 3 biological replicates).
Data information: In (A and B) bar graph is mean ± SD. Scalebar 5 μm.
Source data are available online for this figure.
Interestingly, we were also able to induce accumulation of L1 ORF1p in large cytoplasmic foci when we transiently transfected WT mESCs with plasmids expressing only HA‐ORF1 along with T7‐MOV10 (Fig 6B). The percentage of cells with large ORF1 foci increased from 8% in the control to 48% when co‐transfected with the T7‐MOV10 plasmid (Fig 6B).
Together, our data implicate that MOV10 is playing a direct role in cytosolic sequestration of L1 RNP, thereby restricting retrotransposition and maintaining genome integrity in mESCs (Fig 7).
Figure 7. miRNAs‐mediated control of MOV10 expression regulates L1 retrotransposition in mESCs.
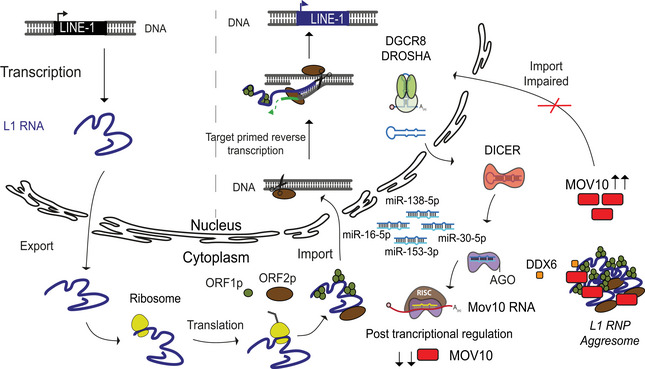
The life cycle of L1 retrotransposition is depicted as a model. Only full‐length L1 elements get transcribed driven by the promoter residing in its 5'UTR sequence. The bicistronic L1 RNA is exported from the nucleus into the cytosol and translated to give rise to L1 ORF1 (ORF1p) and L1 ORF2 (ORF2p) proteins. The L1 RNA and proteins form a complex (L1 RNP) and are imported back into the nucleus. Endonuclease activity of ORF2 nicks the target DNA and using a mechanism referred to as target primed reverse transcription a new copy of L1 element is inserted into the genome via a copy past mechanism of mobilization (Beck et al, 2011; Goodier, 2016). A key regulatory step for retrotransposition is the import of L1RNP back into the nucleus. The canonical miRNA biogenesis pathway illustrates the miRNAs discovered in this study that regulate expression of RNA helicase Mov10 a known modulator of L1 mobility. In the absence of miRNAs when either DICER or DROSHA proteins are deleted in mESCs, both L1 and MOV10 expression are upregulated. Our data suggest that in miRNA, mutant mESCs MOV10 induces L1 RNP aggregate formation in the cytoplasm, the impaired import consequently prevents L1 retrotransposition despite high L1 expression. While DDX6 was also found to co‐localize with the larger L1 RNP particles, the identification of molecular partners and biochemical activities intrinsic to the L1 RNP aggregates should unveil the bottle neck afforded to prevent import.
Discussion
The role of MOV10 in inhibiting retrotransposition in human tissue culture was discovered almost 10 years ago (Goodier et al, 2012). Since then, multiple reports have corroborated this seminal function, where it participates either directly or along with protein partners in curbing retrotransposition (Arjan‐Odedra et al, 2012; Li et al, 2013; Skariah et al, 2017; Choi et al, 2018; Warkocki et al, 2018; Nawaz et al, 2022). Here, we discover cytosolic‐body formation induced by MOV10 as a line of defense for sequestration of L1 RNP particles to prevent deleterious L1 retrotransposition in mESCs. While sequestration as a mean to limit retrotransposition has been previously observed (Goodier et al, 2007, 2012; Guo et al, 2014; Goodier, 2016), it appears that L1 RNP aggregates in miRNA mutant mESCs are different from those observed upon ectopic overexpression of MOV10 and L1 in human cancer cells as the latter unlike in our study were found to be stress granules.
MOV10 is a known interactor of proteins that are a part of the miRNA‐induced silencing complex (RISC) and plays an important role in mRNA decay (Kenny et al, 2014). It also localizes with AGO and TNRC6 proteins in P‐bodies (Meister et al, 2005). L1 ORF1p has been previously reported to interact with P‐body enriched proteins and RNA (Goodier et al, 2007; Briggs et al, 2021). We hypothesize that the absence of AGO2 and mature miRNAs in the miRNA mutant mESCs prevent P‐body formation and hinders similarly L1 ORF1 partitioning and LLPS. We think that the observed aggregates in mESCs have evolved as a specialized compartment where diverse activities for L1 RNP metabolism are brought together, which will require further dissection. MOV10 is a 5′ to 3' RNA helicase (Gregersen et al, 2014) and its catalytic activity is essential for inhibiting human L1 retrotransposition (Goodier et al, 2012). Whether this activity is essential for inducing L1 RNP aggregate formation could provide further mechanistic insight.
Given the plethora of functions MOV10 has been implicated in, it is not surprising that mechanisms have evolved to regulate its expression and activity (Nawaz et al, 2022). Posttranslational modification of MOV10 occurs via ubiquitination in neuron cultures derived from rat hippocampus resulting in its degradation (Banerjee et al, 2009). Moreover, phosphorylation and acetylation of MOV10 have been observed to occur in human cancer cell lines and speculated to regulate its activity and levels (Nawaz et al, 2022). Data presented here, to the best of our knowledge, are a first to unveil miRNA‐mediated posttranscriptional regulation of Mov10 expression. As MOV10 expression levels observed in Dicer_KO were higher than those in Drosha_KO mESCs (Fig EV3C), it is possible that the expression of MOV10 might also be modulated by microprocessor‐independent miRNAs. While transient transfection with all four tested miRNAs resulted in downregulation of MOV10, the absence of synergistic effect for miR‐16‐5p and miR‐30‐5p may rise from the inherent closeness of the two MREs in the 3'UTR of Mov10 causing steric hindrance and preventing the large RISC complex from binding the two simultaneously. MREs in Mov10 for all four tested miRNAs miR‐138‐5p, miR‐30‐5p, miR‐16‐5p, and miR‐153‐3p in mESCs are conserved in the 3'UTR sequence of hMOV10, raising the possibility that this mechanism regulating MOV10 expression may also be conserved in humans. Of note, miR‐138‐5p and miR‐153‐3p are highly expressed in the human brain (Ludwig et al, 2016), and both miRNAs are downregulated in brain pathologies from Alzheimer's disease patients (Long et al, 2012; preprint: Dobricic et al, 2021). The activation of expression and mobility of transposable elements has been reported in a majority of neurological disorders (Terry & Devine, 2020) and certain cancers (Xiao‐Jie et al, 2016). In case the mode of L1 regulation uncovered here in mESCs is conserved, fine‐tuning MOV10 expression in disease conditions using miRNA mimics to downregulate or conversely antagomirs to upregulate MOV10 expression can afford novel means of therapy.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| E14TG2a mESC (Mus musculus) | ATCC | CRL‐1821 |
| HEK293T (Homo sapiens) | ATCC | CRL‐3216 |
| Recombinant DNA | ||
| psiCHECK2‐mMov10‐3'UTR (WT) (mMov10 3'UTR) | Addgene this study | Cat # 178905 |
| psiCHECK2‐mMOV103'UTR‐MRE16‐mut1 | Addgene this study | Cat # 178906 |
| psiCHECK2‐mMOV103'UTR‐MRE153‐mut4 | Addgene this study | Cat # 178909 |
| psiCHECK2‐mMOV103'UTR‐MRE16+MRE153‐mut | Addgene this study | Cat # 178910 |
| pCDNA3‐T11HA‐hMOV10‐WT (human Mov10 cDNA) | Addgene this study | Cat # 178907 |
| T10‐T7‐hMOV10 (human Mov10 cDNA) | Addgene this study | Cat # 185052 |
| T11‐HA‐hORF1 (human L1 ORF1p cDNA) | Addgene this study | Cat # 185053 |
| pCEP‐L1SM‐WT (hygro) (mouse L1 neoR cassette) | Maclennan et al (2017) | |
| JJ‐L1SM WT (mouse L1 BlastR cassette) | Maclennan et al (2017) | |
| JJ‐L1SM N21A (mouse nutant L1 BlastR cassette) | Maclennan et al (2017) | |
| pKLV‐U6gRNA(BbsI)‐PGKpuro2ABFP‐L1mono3 | Addgene this study | Cat # 73542 |
| pKLV‐U6gRNA(BbsI)‐PGKpuro2ABFP‐L1mono1 | Addgene this study | Cat # 73543 |
| Antibodies | ||
| rabbit polyclonal anti‐ORF1p (IF 1:1,000, WB 1:4,000) | kind gift from Prof. O'Carroll | NA |
| mouse monoclonal 15C1BB anti‐MOV10 (IF 1:500) | Bethyl Laboratories Inc | A500‐009A‐T |
| rabbit polyclonal anti‐G3BP1 (IF 1:500) | Bethyl Laboratories Inc | A302‐033A |
| rabbit polyclonal anti‐LC3B antibody (IF 1:250) | Cell Signaling Technology | 2775 |
| rabbit polyclonal anti‐DDX6 (IF 1:500) | GeneTex | GTX102795 |
| rat monoclonal anti‐HA (IF 1:500) | Roche | 3F10 |
| Rabbit monoclonal T7‐Tag (D9E1X) XP (IF 1:250) | Cell Signaling Technology | 13246 |
| Goat anti‐RENT1 antibody (IF 1:250) | Bethyl laboratories | A300‐038A |
| Alexa fluor 488 goat anti‐rat IgG (IF 1:4,000) | Life Technologies | 11006 |
| Alexa fluor 488 donkey anti‐mouse IgG (IF 1:4,000) | Life Technologies | A21202 |
| Alexa fluor 546 donkey anti‐rabbit IgG (IF 1:4,000) | Life Technologies | A10040 |
| Alexa fluor 647 donkey anti‐mouse IgG (IF 1:4,000) | Life Technologies | A31571 |
| Alexa fluor 488 donkey anti‐goat IgG (IF 1:4,000) | Life Technologies | A11055 |
| rabbit anti MOV10 antibody (WB 1:2,000) | Proteintech | 10370‐1‐AP |
| rabbit polyclonal anti‐Dicer (WB 1:2,000) | Sigma | SAB42000087 |
| rabbit polyclonal anti‐Argonaute2 (WB 1:2,000) | Cell Signaling Technologies | C34C6 |
| rabbit anti‐Drosha (WB 1:2,000) | Cell Signaling Technologies | D28B1 |
| mouse anti‐Tubulin antibody (WB 1:10,000) | GenScript | A01410 |
| rabbit anti‐LaminB1 (WB 1:5,000) | Abcam | ab16048 |
| anti‐rabbit IgG HRP‐linked (WB 1:10,000) | Cell Signaling Technologies | 7076 |
| anti‐rat IgG HRP‐linked antibody (WB 1:10,000) | Cell Signaling Technologies | 7077 |
| Oligonucleotides and sequence‐based reagents | ||
| Experiment | Fwd Primer | Rev Primer |
| mMOV10 3'UTR PCR amplification | taggcgatcgctcgaggccacagccgcccgcctt | ttgcggccagcggccttttgcatagaacagcattttgt |
| miR16‐5p (CTG>AGT) mut1 mutageneis | acccaagagtctaaaactcggaggaaggggg | tttagactcttgggttgtcttccctagc |
| miR153‐3p (TGC>CAT) mut4/mut1+mut4 mutagenesis | tgttctacataaaaggccgctggccgca | cttttatgtagaacagcattttgtttttctt |
| hMOV10 cDNA PCR amplification | ggtcggaggcggatccatgcccagtaagttcagctgc | gatatctgcagaattctcagagctcattcctccactc |
| hORF1 cDNA PCR amplification | ggtcggaggcggatccatggggaaaaaacagaac | gatatctgcagaattctcattacattttggcatgattttgc |
| Guide RNA L1UP mono3 | caccgccagagaacctgacagcttc | |
| Guide RNA L1UP mono1 | caccgccagaggacaggtgcccgcc | |
| mmu‐miR‐16‐5p mimic | uagcagcacguaaauauuggcg | |
| mmu‐miR‐30e‐5p mimic | uguaaacauccuugacuggaag | |
| mmu‐miR‐138‐5p mimic | agcugguguugugaaucaggccg | |
| mmu‐miR‐153‐3p mimic | uugcauuagucacaaaagugauc | |
| miRIDIAN microRNA negative control 1 | catalog no CN‐001000‐01‐05 | |
| qPCR Rrm2 | ccgagctggaaagtaaagcg | atgggaaagacaacgaagcg |
| qPCR Mov10 | gacgatttacaaccacgacttca | gccagatttgcgatcttcattcc |
| qPCR Dicer | ccgatgatgcagcctctaatag | tccatctcgagcaattctctca |
| qPCR L1‐Tf | cagcggtcgccatcttg | caccctctcacctgttcagactaa |
| qPCR L1‐A | ggattccacacgtgatcctaa | tcctctatgagcagacctgga |
| qPCR L1‐Gf | ctccttggctccgggact | caggaaggtggccggttgt |
| qPCR L1‐ORF1 | actcaaagcgaggcaacact | ctttgattgttgtgccgatg |
| qPCR L1‐ORF2 | ggagggacatttcattctcatca | gctgctcttgtatttggagcataga |
| NB L1specifc | gagtttttgagtctgtatcc | ctctccttagtttcagtgg |
| Chemicals, enzymes and other reagents | ||
| Fibronectin | Sigma | FC010 |
| Puromycin | Sigma | P8833 |
| Hygromycin | Invitrogen | 10687010 |
| G418 | Invitrogen | 10131035 |
| Blasticidin | Invitrogen | R21001 |
| DMEM Media | Sigma | D6429‐500ML |
| Penicillin/Streptomycin | Sigma | P0781‐100ML |
| 0.05% Trypsin‐EDTA | Life Technologies | 25300054 |
| PBS1X | Life Technologies | 10010015 |
| 2‐ß‐mercaptoethanol | Life Technologies | 31350010 |
| FBS | Life Technologies | 10270‐106 |
| Lipofectamine™ RNAiMAX Transfection Reagent | Life Technologies | 13778030 |
| Lipofectamine™ 2000 Transfection Reagent | Life Technologies | 11668019 |
| Lipofectamine™ 3000 Transfection Reagent | Life Technologies | L3000015 |
| 1,6‐Hexanediol | Sigma | H11807 |
| Sodium Arsenite | Sigma | S7400 |
| Tris(carboxyethyl)phosphine | Sigma | 68957 |
| Tris‐HCl | AppliChem | A2937 |
| NaCl | Merck | 1.06404.1000 |
| MgCl2 | Sigma | M8266 |
| IGEPAL CA‐630 | Sigma | I3021‐50ML |
| Sodium dodecyl sulfate | Sigma | L6026 |
| Protease inhibitor cocktails | Roche | 5892791001 |
| Tween20 | Sigma | P1379 |
| Coomassie | VWR | 443283M |
| Triton‐X | Sigma | 93443 |
| PIPES | Sigma | 80635 |
| NotI‐HF | NEB | R3189S |
| BamH1‐HF | NEB | R3136 |
| XhoI‐HF | NEB | R0146 |
| EcoRI‐HF | NEB | R0101 |
| Bbs1 | NEB | R0539 |
| In‐Fusion® Snap Assembly Master Mix | Takara | 638944 |
| CloneAmp HiFi PCR Premix | Takara | 639298 |
| Vectashield | Vector Laboratories | H‐1000 |
| DAPI | Sigma | D9542 |
| Trizol | Life Technologies | 15596018 |
| Nick translation kit | Abbot Molecular | 06J40‐020 |
| Red‐dUTP | Enzo Life sciences | ENZ 42854 |
| Ribonucleoside Vanadyl Complex | NEB | S1420S |
| Trisodium citrate | Sigma | S4641 |
| DNA Clean & Concentrator‐25 | Zymo Research | D4033 |
| Clarify™ Western ECL substrate | BioRad | 1705060 |
| SuperSignal™ West Femto Maximum Sensitivity Substrate | Therno Scientific | 34094 |
| RQ1 Rnase‐Free DNase kit | Promega | M6101 |
| GoScript™ Reverse Transcriptase Kit | Promega | A5001 |
| KAPA SYBR® FAST qPCR Kit Optimized for Light Cycler® 480 | KAPA Biosystems, Sigma | KK4610 |
| PerfectHybTM Plus | Sigma | H7033 |
| Dual‐Glow Luciferase Assay kit | Promega | E2920 |
| Crystal Violet | Sigma | 1014080025 |
| Software | ||
| Prism 9.2.0 | GraphPad | https://www.graphpad.com |
| Fiji | Fiji ImageJ | Nature Methods, 9(7), 676–682 |
| Microsoft Excel | Microsoft | |
| ImageLab v6.1.0 | Bio‐Rad Laboratories | https://www.bio‐rad.com |
| Other | ||
| GloMax® Discover Multimode Microplate Reader | Promega | |
| Roche Light Cycler 480 | Roche | |
Methods and Protocols
Cell culture
E14TG2a mESC (ATCC CRL‐1821) were used as wild‐type cells. Dicer_KO (Bodak et al, 2017) and Drosha_KO (Cirera‐Salinas et al, 2017) were previously generated from E14TG2a in our laboratory using a paired CRISPR‐Cas9 approach (Wettstein et al, 2016). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen) supplemented with 15% preselected batch of FBS (GIBCO) tested for optimal mESCs growth, 1,000 U/ml of LIF (Millipore), 0.1 mM of 2‐β‐mercapto‐ethanol (Life Technologies), 0.05 mg/ml of streptomycin, and 50 U/ml of penicillin (Sigma). For routine culturing, cells were grown on 0.2% gelatin‐coated cell culture grade plastic vessels in the absence of feeder cells. For microscopy, coverslips were coated with 10 μg/ml fibronectin (Sigma, FC010) for at least 2 h at 37°C, coverslips were washed three times with 1× PBS, and cells were seeded 16–18 h before processing them for microscopy. HEK293T cells were grown in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen) supplemented with 10% FBS (GIBCO), 0.05 mg/ml of streptomycin, and 50 U/ml of penicillin (Sigma). All cell lines were routinely tested for mycoplasma contamination. Concentration of various antibiotics used was as follows 1 μg/ml puromycin (Sigma), 100 μg/ml hygromycin (Invitrogen), 250 μg/ml G418 (Sigma), 50 μg/ml blasticidin (Invitrogen).
Plasmids
3'UTR sequence of mouse MOV10 transcript ENSMUST00000168015.8 was PCR amplified using Fwd 5′‐taggcgatcgctcgaggccacagccgcccgcctt‐3′ and Rev 5′‐ttgcggccagcggccttttgcatagaacagcattttgt‐3′ primers and cDNA generated with random primers from mESCs as template. The PCR product was subcloned into plasmid psiCHECK2 (Promega) previously digested with NotI using the In‐Fusion cloning kit (Takara Bio) giving rise to plasmid psiCHECK2‐mMov10‐3'UTR (WT; addgene 178905). Using psiCHECK2‐mMov10‐3'UTR (WT) and Primers 5′‐acccaagagtctaaaactcggaggaaggggg‐3′ and 5′‐tttagactcttgggttgtcttccctagc‐3′, the MRE for miR16‐5p was mutated (CTG > AGT) using the infusion cloning kit to generate plasmid psiCHECK2‐mMOV103'UTR‐MRE16‐mut1 (addgene 178906). Using psiCHECK2‐mMov10‐3'UTR (WT) plasmid as template and Primers 5′‐tgttctacataaaaggccgctggccgca‐3′ and 5′‐cttttatgtagaacagcattttgtttttctt‐5′, the MRE for miR153‐3p was mutated (TGC > CAT) using the infusion cloning kit to generate plasmid psiCHECK2‐mMOV103'UTR‐MRE153‐mut4 (addgene 178909). Plasmid psiCHECK2‐mMOV103'UTR‐MRE16 + MRE153‐mut (addgene 178910) was generated using plasmid psiCHECK2‐mMOV103'UTR‐MRE16‐mut1 as template and primers Primers 5′‐tgttctacataaaaggccgctggccgca‐3′ and 5′‐cttttatgtagaacagcattttgtttttctt‐5′ with the infusion cloning kit.
Human MOV10 was PCR amplified with primers Fwd 5′‐ggtcggaggcggatccatgcccagtaagttcagctgc‐3′ and Rev 5′‐gatatctgcagaattctcagagctcattcctccactc‐3′ using plasmid pFLAG/HA‐MOV10 (addgene 10976; Meister et al, 2005) as template and subcloned into BamH1 and Xho1 digested pCDNA3‐T11‐HA plasmid (Foglieni et al, 2017) a kind gift from Prof. Polymenidou using In‐Fusion cloning kit (Takara) to yield plasmid pCDNA3‐T11HA‐hMOV10‐WT (addgene 178907). Plasmid pCDNA3‐T11HA‐hMOV10‐WT was digested with BamH1 and Xho1 to generate the hMOV10 insert that was subsequently subcloned into pCDNA3‐T10‐T7 a kind gift from Prof. Polymenidou to generate plasmid T10‐T7‐hMOV10 (addgene 185052). These were used in transient transfections to overexpress hMOV10 in L1Up Ctrl and WT cells. Plasmid T11‐HA‐hORF1 (addgene 185053) was generated using the infusion cloning kit. Primers 5′‐ggtcggaggcggatccatggggaaaaaacagaac‐3′ and 5′‐gatatctgcagaattctcattacattttggcatgattttgc‐3′ were used to PCR amplify L1‐hORF1 using plasmid L1‐RP as template (Xie et al, 2011) and subcloned into BamH1 and EcoR1 digested pCDNA3‐T11‐HA plasmid. T11‐HA‐ORF1 was used for ectopic expression of L1 ORF1p in WT mESCs. Plasmids used for the retrotransposition assay with mneo1 cassette as reporter was pCEP‐L1SM (hygro) and with mblast1 cassette was JJ‐L1SM WT and JJ‐L1SM N21A (hygro), all gifts from Prof. Garcia‐Perez.
Generation of L1UP mESCs using CRISPRa
L1UP mESCs were generated from E14TG2a mESCs using the CRISPRa approach (Cheng et al, 2013). Single gRNAs (sgRNAs) were designed using the L1 Tf consensus sequences (Fig EV2B; Naas et al, 1998). Sequence alignments (Schichman et al, 1993; Naas et al, 1998; Goodier et al, 2001) were performed using T‐Coffee (Notredame et al, 2000). SgRNAs to upregulate L1 Tf were individually subcloned into the plasmid pKLV‐U6gRNA(BbsI)‐PGKpuro2ABFP a gift from Prof. Yusa (addgene 50946, Koike‐Yusa et al, 2014), using the BbsI restriction site. Guide sequence used for generating Cl1 was 5′‐caccgccagagaacctgacagcttc‐3′ (addgene 73542). For Cl2, two guide pairs were used 5′‐caccgccagagaacctgacagcttc‐3′ (addgene 73542, same as for Cl1) and 5′‐caccgccagaggacaggtgcccgcc‐3′ (addgene 73543). AC95‐pmax‐dCas9VP160‐2A‐neo was a gift from Prof. Jaenish (addgene 48227; Cheng et al, 2013). Cells were transfected with 1 μg of each plasmid, and 24 h post transfection, they were cultured in the presence of puromycin (1 μg/ml) and G418 (250 μg/ml). Single clones were picked 1 week post transfection. The first screening for the selection of L1UP candidates was performed at the protein level for ORF1 expression by immunoblot analysis.
Ectopic protein expression
L1UP Ctrl mESC lines were transiently transfected with 2 μg T11HA‐hMOV10‐WT plasmid (addgene 178907) for ectopic expression of hMOV10 or T11HA‐EV (Foglieni et al, 2017) as empty vector control using Lipofectamine 3000 (Invitrogen). WT mESCs were transiently transfected with 2 μg pCEP‐L1SM along with 2 μg T11HA‐hMOV10‐WT plasmid (addgene 178907) for ectopic expression of L1 and hMOV10. WT mESCs were transiently transfected with 2 μg T11‐HA‐ORF1 (addgene 185053) and T10‐T7‐MOV10 (addgene 185052) or T10T7‐EV for ectopic expression of L1 hORF1p and hMOV10. Transfection complex was removed 6 h post transfection. Cells were trypsinized 32 h post transfection and plated on fibronectin‐coated cover slips. Samples were processed 48 h post transfection for indirect immunofluorescence (IF).
MiRNA mimic transfections in mESCs
A total of 100,000 Drosha_KO mESCs were seeded per well in a six‐well plates in duplicate for respective miRNA mimic transfections. Cells were grown in antibiotic‐free media and transfected with 20 nM mimic when transfected singly or 20, 5, 4, 2, or 1 nM respective mimic as described for dual transfections using RNAimax reagent (Invitrogen). Cells were harvested 39 h post transfection, and duplicate samples were pooled for protein extraction and subsequent Western blot analysis. The following miRNA mimics (Dharmacon, A horizon discovery Group company) were used:
mmu‐miR‐16‐5p 5'‐UAGCAGCACGUAAAUAUUGGCG‐3′ (C‐310511‐05‐05).
mmu‐miR‐30e‐5p 5'‐UGUAAACAUCCUUGACUGGAAG‐3′ (C‐310466‐07‐0002).
mmu‐miR‐138‐5p 5'‐AGCUGGUGUUGUGAAUCAGGCCG‐3′ (C310414‐07‐0002).
mmu‐miR‐153‐3p 5'‐UUGCAUUAGUCACAAAAGUGAUC‐3' (C310428‐05‐0002).
miRIDIAN microRNA negative control 1 (CN‐001000‐01‐05).
Indirect immunofluorescence (IF)
Cells grown on coverslips were washed with 1× PBS, fixed with 3.7% formaldehyde (Sigma) in 1× PBS for 10 min at room temperature. Post fixation cells were washed three times in 1× PBS and permeabilized with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES pH 6.8, and 0.5% Triton‐X) for 4 min on ice. After three further washes with 1× PBS, blocking was initiated in 1× PBS supplemented with 1% BSA and 0.1% Tween‐20 for 30 min at room temperature. Samples were incubated with primary antibody diluted in blocking buffer for 1 h at room temperature, there after washed three times with 1× PBS‐0.1% Tween‐20, incubated with secondary antibody diluted in blocking solution for 1 h and counterstained with 100 ng/ml DAPI (Sigma) in 1× PBS for 4 min before mounting on slides in Vectashield (Vector Labs). The primary antibodies diluted in blocking buffer used were as follows: rabbit polyclonal anti‐ORF1p (1:1,000, kind gift from Prof. O'Carroll), mouse monoclonal 15C1BB anti‐MOV10 (1:500, A500‐009A‐T Bethyl Laboratories Inc), rabbit polyclonal anti‐G3BP1 (1:500 A302‐033A, Bethyl Laboratories Inc), rabbit polyclonal anti‐LC3B antibody (1:250, 2775, Cell Signaling Technology), rabbit polyclonal anti‐DDX6 (1:500, GTX102795, GeneTex), rat monoclonal anti‐HA (1:500, 3F10, Roche), Rabbit monoclonal T7‐Tag (D9E1X) XP (1:250, 13246, Cell Signaling Technology), and Goat anti‐RENT1 antibody (1:250, A300‐038A, Bethyl laboratories). Secondary antibody used were Alexa fluor 488 goat anti‐rat IgG (1:4,000, 11006, Life Technologies), Alexa fluor 488 donkey anti‐mouse IgG (1:4,000, A21202, Life technologies), Alexa fluor 546 donkey anti‐rabbit IgG (1:4,000, A10040, Life technologies), Alexa fluor 647 donkey anti‐mouse IgG (1:4,000, A31571, Life technologies), and Alexa fluor 488 donkey anti‐goat IgG (1:4,000, A11055, Life technologies). Images were acquired using the Deltavision multiplex system equipped with an Olympus 1X71 (inverse) microscope, pco.edge 5.5 camera and 60× 1.4NA DIC Oil PlanApoN objective. Z stacks were taken 0.2 μm apart, images de‐convolved using Softworx software. Further image analysis and processing were performed using ImageJ. Excel (Microsoft) and Prism 9 (Graphpad) were used for data analysis and statistical testing.
Combined RNA FISH and IF
Cells grown on coverslips were first processed for IF following the protocol described above except all buffers and solution other than the fixative were also supplemented with 10 mM ribonucleoside vanadyl complex (NEB). After incubation with the secondary antibody, cells were fixed with 3.7% formaldehyde in 1× PBS for 10 min at room temperature and blocked in 1× PBS supplemented with 1% BSA, 0.1% Tween‐20, 2 mM glycine, and 10 mM RVC for 15 min. Cells were next washed and incubated in 2× SSC (0.03 M sodium citrate in 0.3 M sodium chloride) for 5 min. Probe specific for Tf L1 family was labeled with Red‐dUTP (Enzo Life sciences) using a nick translation kit (Abbot). Two microgram TFkan plasmid kind gift from Prof. Heard (Deberardinis & Kazazian, 1999; Chow et al, 2010) was incubated with 0.2 mM labeled dUTP, 0.1 mM dTTP, 0.1 mM dNTP mix and 2.5 μl nick translation enzyme in a 50 μl final volume as per guidelines from the kit. The reaction was incubated at 15°C for 15 h. A PCR purification column (zymogen) was used to clean the probe which was eluted in 50 μl water. The volume of the probe was decreased down to 5 μl using a speed vac, and the probe was diluted in 100 μl hybridization solution (1‐part 20× SSC, 2‐parts 10 mg/ml BSA, 2 parts 50% dextran sulfate, and 5 parts deionized formamide). The probe solution was denatured at 78°C for 5 min, placed on ice for 5 min, and 7 μl probe was spotted on a prebaked slide for each sample. During the overnight hybridization at 37°C in a humid chamber, the overturned coverslips were sealed using rubber cement. Post hybridization washes were performed with 50% formamide in 2× SSC thrice for 5 min followed by three washes with 2× SSC. DNA was counterstained with 100 ng/ml DAPI in 2× SSC and mounted on slides with Vectashield. Image acquisition and analysis were as for IF.
Western blot analysis
Total cellular protein was extracted from mESC pellets using a NP40‐based lysis buffer (1% NP40, 137 mM NaCl, 20 mM Tris–HCl, 1 mM EDTA) complemented with EDTA‐free protease inhibitor cocktail (Roche). Protein concentrations were determined by Bradford Assay (Bio‐Rad). 10–20 μg of total cellular protein was separated in 8 or 10% SDS‐PAGE gels and transferred on PVDF membranes. The antibodies used were as follows: rabbit polyclonal anti‐L1 ORF1p (1:5,000, gift from Prof. O'Carroll), rabbit polyclonal anti‐Dicer (1:2,000, SAB42000087, Sigma), rabbit polyclonal anti‐MOV10 antibody (1:2,000, 10,370‐1‐AP, Proteintech), rabbit polyclonal anti‐Argonaute2 (1:2,000 C34C6 Cell Signaling Technologies), rabbit anti‐Drosha (1:2,000, D28B1 Cell Signaling Technology), rat monoclonal anti‐HA (1:500, 3F10, Roche), mouse anti‐Tubulin antibody (1:10,000, A01410, GenScript), rabbit anti‐LaminB1 (1:5,000, ab16048, Abcam), rabbit anti‐DDX6 (1:2,000, GTX102795 GeneTex), anti‐rabbit IgG HRP‐linked (1:10,000, 7074, Cell Signaling Technologies), anti‐mouse IgG HRP‐linked (1:10,000, 7076, Cell Signaling Technologies), and anti‐rat IgG HRP‐linked antibody (1:10,000, 7077, Cell Signaling Technologies). Immunoblot blot were developed using the Clarify™ Western ECL substrate (Bio‐Rad) kit or SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Scientific) and detected using ChemiDoc™ MP imaging system (Bio‐Rad). All membranes were stained with coomassie to ensure equal loading.
RT–qPCR analysis
Total cellular RNA was extracted from cell pellets using TRizol® Reagent (Life Technologies). Extract quality was verified by loading 1 μg of total cellular RNA on a 1% Agarose gel. One microgram cellular RNA was treated with DNase (RQ1 Rnase‐Free DNase kit Promega) and reverse‐transcribed following the GoScript™ Reverse Transcriptase Kit (Promega) manufacturer's instructions. The produced cDNAs were diluted fivefold in distilled water. For each extract, PCR on the Rrm2 gene were performed, with and without reverse transcriptase treatment, to insure the absence of genomic DNA contamination. The quality‐controlled cDNAs were diluted two times in distilled water. Amplifications were performed on the Light Cycler® 480 (Roche) using 2 μl of the diluted cDNAs and the KAPA SYBR® FAST qPCR Kit Optimized for Light Cycler® 480 (KAPA biosystems). Differences between samples and controls were calculated based on the 2−ΔCT method. RT–qPCR assays were performed in biological triplicate. Primers utilized for the RT–qPCR assays are as follows: Rrm2fwd 5′‐ccgagctggaaagtaaagcg‐3′, Rrm2rev 5′‐atgggaaagacaacgaagcg‐3′, Mov10fwd 5′‐gacgatttacaaccacgacttca‐3′, Mov10rev 5′‐gccagatttgcgatcttcattcc‐3′, Dicerfwd 5′‐ccgatgatgcagcctctaatag‐3' Dicerrev 5′‐tccatctcgagcaattctctca‐3′, L1‐Tffwd 5′‐cagcggtcgccatcttg‐3′, L1‐Tfrev 5′‐caccctctcacctgttcagactaa‐3′,
L1‐Afwd 5′‐ggattccacacgtgatcctaa‐3′, L1‐Arev 5′‐tcctctatgagcagacctgga‐3′, L1‐Gffwd 5′‐ctccttggctccgggact‐3′, L1‐Gfrev 5′‐caggaaggtggccggttgt‐3′, L1‐ORF1fwd 5′‐actcaaagcgaggcaacact‐3′ L1‐ORF1rev 5′‐ctttgattgttgtgccgatg‐3′, L1‐ORF2fwd 5′‐ggagggacatttcattctcatca‐3′, L1‐ORF2rev 5′‐gctgctcttgtatttggagcataga‐3′.
Northern blot analysis
Northern blot analysis was performed as previously described (Kuramochi‐Miyagawa et al, 2008; Bodak et al, 2017). Thirty microgram of total RNAs extracted using Trizol was run on a denaturing 1% agarose gel with 1% formaldehyde. Following capillary transfer to nylon membranes overnight, the membrane was cross‐linked by UV radiation. PerfectHybTM Plus was used for prehybridization blocking and hybridization at 42°C. Post hybridization washes were performed in 2× SSC + 0.1% SDS. For detection of full‐length L1 transcripts, random primer extension labeling was carried out. DNA used for the reaction was PCR amplified using E14TG2a mESCs genomic DNA as template and L1‐specifc primers Fwd 5′‐gagtttttgagtctgtatcc‐3′ and Rev 5′‐ctctccttagtttcagtgg‐3′.
Dual luciferase reporter assay
A total of 70,000 HEK293T cells were plated per well in a 24‐well plates 16 h prior to transfection with Lipofectamine 2000 (Invitrogen). 0.5 μg of plasmid psiCHECK2‐3'UTR‐Mov10'UTR (WT or MUT) was co‐transfected with 50 nM indicated miRNA mimics or control mimic. Transfection complexes were removed 6 h post transfection. Luciferase activity was measured on a GloMax® Discover Multimode Microplate Reader (Promega, USA) after processing cells using the Dual‐Glow Luciferase Assay kit (E2920 Promega) 48 h post transfection. Results are means and error bars are standard deviation (SD) from three to four independent experiments.
Retrotransposition reporter and colony‐forming assays
1 × 106 L1UP and Ctrl mESCs were seeded in 10 cm dish 16 h prior to transfection with 6 μg of JJ‐L1SM (WT and L1N21A) plasmid using Lipofectamine 3000 (Invitrogen). Media exchange was initiated 6 h post transfection, and hygromycin supplemented media was added 48 h post transfection to select for stably transfected cells. Once the mock transfected cells were dead, 150,000 hygromycin resistant cells were seeded per well in a six‐well plates in triplicate and grown in media sans hygromycin for 16 h after which the media was supplemented with Blasticidin. Media exchange with fresh antibiotics was performed every 48 h for approximately 15 days, when individual Blasticidin‐resistant colonies were visible with the naked eye. Cells were washed with 1× PBS and stained with 1% crystal violet blue, 1% formaldehyde, 1% methanol for 20 min at room temperature, followed by washes with tap water. Plates were air‐dried and imaged using the ChemiDoc™ MP system (Bio‐Rad). Individual colonies were counted using ImageJ. Results are means and error bars are SD from three independent transfections.
Transient transfections of reporter plasmids were carried out using Lipofectamine 3000 (Invitrogen) when co‐transfections with miRNA mimics or plasmids for ectopic expression of hMOV10 were assayed for retrotransposition. A total of 500,000 cells were seeded for transient transfection with 6 μg of reporter plasmid and either 10 nM mimic for mmu‐miR‐16‐5p + 10 nM mmu‐miR‐153‐3p mimic or 6 μg of plasmid T11HA‐EV or T11HA‐hMOV10. Media exchange was initiated 6 h post transfection. 39 h post transfection, cells were grown in media supplemented with antibiotic resistance encoded by the respective cassette. Subsequent media exchanges, staining and counting of colonies, were the same as stated for stably transfected cells. Results are means and error bars are SD from three independent transfections.
Author contributions
Rajika Arora: Conceptualization; formal analysis; supervision; funding acquisition; investigation; visualization; methodology; writing – original draft. Maxime Bodak: Conceptualization; formal analysis; validation; visualization; methodology. Laura Penouty: Validation; methodology. Cindy Hackman: Validation; methodology. Constance Ciaudo: Conceptualization; resources; formal analysis; supervision; funding acquisition; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
Conceptualization: RA, MB, and CC; laboratory experiments: RA, MB, LP, and CH; writing original draft preparation: RA and CC; writing, review, and editing: CC; visualization: RA, MB, and CC; supervision: CC; funding acquisition: CC and RA. All authors have read and agreed to the published version of the manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We would like to thank the members of the Ciaudo laboratory and Dr Tobias Beyer for fruitful discussions and the critical reading of this manuscript. This work was supported by the Swiss National Science Foundation (grants 31003A_173120 and 310030_196861) and Novartis Foundation for Medical‐Biological Research (grant 19A018) to CC. In addition, RA and CC were supported by the NCCR RNA and Disease (Swiss National Science Foundation grant 182880) and the laboratory of Prof. Frédéric Allain at ETH. We also want to thank the ScopeM facility at ETH Zürich for their support with microscopy and image analysis. Open access funding provided by Eidgenossische Technische Hochschule Zurich.
EMBO reports (2022) 23: e54458.
Data availability
No primary datasets have been generated and deposited in public databases.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan‐Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH (2012) Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 9: 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayache J, Bénard M, Ernoult‐Lange M, Minshall N, Standart N, Kress M, Weil D (2015) P‐body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol Biol Cell 26: 2579–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS (2009) A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64: 871–884 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2018) Metazoan microRNAs. Cell 173: 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia‐Perez JL, Badge RM, Moran JV (2011) LINE‐1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12: 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodak M, Cirera‐Salinas D, Yu J, Ngondo RP, Ciaudo C (2017) Dicer, a new regulator of pluripotency exit and LINE‐1 elements in mouse embryonic stem cells. FEBS Open Bio 7: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodak M, Yu J, Ciaudo C (2018) Regulation of LINE‐1 elements by miR‐128 is not conserved in mouse embryonic stem cells. Front Genet 9: 683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs EM, McKerrow W, Mita P, Boeke JD, Logan SK, Fenyö D (2021) RIP‐seq reveals LINE‐1 ORF1p association with p‐body enriched mRNAs. Mob DNA 12: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz‐Prigge S, Farley AH, Morant JV, Kazazian HH (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 100: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R (2013) Multiplexed activation of endogenous genes by CRISPR‐on, an RNA‐guided transcriptional activator system. Cell Res 23: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hwang SY, Ahn K (2018) Interplay between RNASEH2 and MOV10 controls LINE‐1 retrotransposition. Nucleic Acids Res 46: 1912–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E et al (2010) LINE‐1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969 [DOI] [PubMed] [Google Scholar]
- Cirera‐Salinas D, Yu J, Bodak M, Ngondo RP, Herbert KM, Ciaudo C (2017) Noncanonical function of DGCR8 controls mESC exit from pluripotency. J Cell Biol 216: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Kazazian HH (1999) Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics 56: 317–323 [DOI] [PubMed] [Google Scholar]
- DiGiacomo M, Comazzetto S, Saini H, DeFazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D (2013) Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608 [DOI] [PubMed] [Google Scholar]
- Dobricic V, Schilling M, Schulz J, Zhu L‐S, Zhou C‐W, Fuß J, Franzenburg S, Zhu L‐Q, Parkkinen L, Lill CM, et al (2021) Differential microRNA expression analyses across two brain regions in Alzheimer's disease. bioRxiv 10.1101/2021.05.31.446406 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB (2010) Characterization of LINE‐1 ribonucleoprotein particles. PLoS Genet 6: e1001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm‐Ansmant I, Schweizer D, Izaurralde E (2007) P‐body formation is a consequence, not the cause, of RNA‐mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglieni C, Papin S, Salvadè A, Afroz T, Pinton S, Pedrioli G, Ulrich G, Polymenidou M, Paganetti P (2017) Split GFP technologies to structurally characterize and quantify functional biomolecular interactions of FTD‐related proteins. Sci Rep 7: 14013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL (2016) Restricting retrotransposons: a review. Mob DNA 7: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH (2012) MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8: e1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian J (2001) A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Zhang L, Vetter MR, Kazazian HH (2007) LINE‐1 ORF1 protein localizes in stress granules with other RNA‐binding proteins, including components of RNA interference RNA‐induced silencing complex. Mol Cell Biol 27: 6469–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M (2014) MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Molecular Cell 54: 573–585. 10.1016/j.molcel.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Gagnon D, Meng L, Perez‐Iratxeta C, Lagace D, Gibbings D (2014) Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 5: 1–11 [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524 [DOI] [PubMed] [Google Scholar]
- Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, Pedersen IM (2015) miR‐128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol 22: 824–831 [DOI] [PubMed] [Google Scholar]
- Idica A, Sevrioukov EA, Zisoulis DG, Hamdorf M, Daugaard I, Kadandale P, Pedersen IM (2017) MicroRNA miR‐128 represses LINE‐1 (L1) retrotransposition by down‐regulating the nuclear import factor TNPO1. J Biol Chem 292: 20494–20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Torres‐Padilla ME (2016) LINEs in mice: features, families, and potential roles in early development. Chromosoma 125: 29–39 [DOI] [PubMed] [Google Scholar]
- Jin HY, Gonzalez‐Martin A, Miletic AV, Lai M, Knight S, Sabouri‐Ghomi M, Head SR, Macauley MS, Rickert RC, Xiao C (2015) Transfection of microRNA mimics should be used with caution. Front Genet 6: 340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Chen Z, Liu X, Zhou X (2013) Evaluating the MicroRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol 936: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer‐deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P et al (2016) G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. Journal of Cell Biology 212: 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S (2014) MOV10 and FMRP regulate AGO2 association with MicroRNA recognition elements. Cell Rep 9: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike‐Yusa H, Li Y, Tan EP, Velasco‐Herrera MDC, Yusa K (2014) Genome‐wide recessive genetic screening in mammalian cells with a lentiviral CRISPR‐guide RNA library. Nat Biotechnol 32: 267–273 [DOI] [PubMed] [Google Scholar]
- Kopera HC, Larson PA, Moldovan JB, Richardson SR, Liu Y, Moran JV (2016) LINE‐1 cultured cell Retrotransposition assay. Methods Mol Biol 1400: 139–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi‐Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW et al (2008) DNA methylation of retrotransposon genes is regulated by piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, Guo F, Liang C, Cen S (2013) The MOV10 helicase inhibits LINE‐1 mobility. J Biol Chem 288: 21148–21160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Ray B, Lahiri DK (2012) MicroRNA‐153 physiologically inhibits expression of amyloid‐β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem 287: 31298–31310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E et al (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res 44: 3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Na Z, Slavoff SA (2018) P‐bodies: composition, properties, and functions. Biochemistry 57: 2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclennan M, García‐Cañadas M, Reichmann J, Khazina E, Wagner G, Playfoot CJ, Salvador‐Palomeque C, Mann AR, Peressini P, Sanchez L et al (2017) Mobilization of LINE‐1 retrotransposons is restricted by Tex19.1 in mouse embryonic stem cells. eLife 6: e26152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T (2005) Identification of novel argonaute‐associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Mockly S, Houbron É & Seitz H (2022) A rationalized definition of general tumor suppressor microRNAs excludes miR‐34a. bioRxiv 10.1101/2021.02.11.430795 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Fäh T, Schaefer M, Hermes V, Luitz J, Stalder P, Arora R, Ngondo RP, Ciaudo C (2022) AGO1 regulates pericentromeric regions in mouse embryonic stem cells. Life Sci Alliance 5: e202101277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas TP, Deberardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH (1998) An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz A, Shilikbay T, Skariah G, Ceman S (2022) Unwinding the roles of RNA helicase MOV10. WIREs RNA 13. 10.1002/wrna.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden BN, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD et al (2017) Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci USA 114: E5635–E5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JC, Naik MT, Li GY, Murphy EL, Fawzi NL, Sedivy JM, Jogl G (2021) Phase separation of the LINE‐1 ORF1 protein is mediated by the N‐terminus and coiled‐coil domain. Biophys J 120: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngondo RP, Cirera‐Salinas D, Yu J, Wischnewski H, Bodak M, Vandormael‐Pournin S, Geiselmann A, Wettstein R, Luitz J, Cohen‐Tannoudji M et al (2018) Argonaute 2 is required for extra‐embryonic endoderm differentiation of mouse embryonic stem cells. Stem Cell Reports 10: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T‐coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- O'Geen H, Yu AS, Segal DJ (2015) How specific is CRISPR/Cas9 really? Curr Opin Chem Biol 29: 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EKL (2006) Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep 7: 904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME (2010) Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29: 2161–2164 [DOI] [PubMed] [Google Scholar]
- Roden C, Gladfelter AS (2021) RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schichman SA, Adey NB, Edgell MH, Hutchison CA (1993) L1 A‐monomer tandem arrays have expanded during the course of mouse L1 evolution. Mol Biol Evol 10: 552–570 [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM (2005) Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Skariah G, Seimetz J, Norsworthy M, Lannom MC, Kenny PJ, Elrakhawy M, Forsthoefel C, Drnevich J, Kalsotra A, Ceman S et al (2017) Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biol 15: 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM (2004) RNAi and expression of retrotransposons MuERV‐L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285 [DOI] [PubMed] [Google Scholar]
- Taylor MS, Altukhov I, Molloy KR, Mita P, Jiang H, Adney EM, Wudzinska A, Badri S, Ischenko D, Eng G et al (2018) Dissection of affinity captured LINE‐1 macromolecular complexes. eLife 7: e30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, LaCava J, Mita P, Molloy KR, Huang CRL, Li D, Adney EM, Jiang H, Burns KH, Chait BT et al (2013) Affinity proteomics reveals human host factors implicated in discrete stages of LINE‐1 retrotransposition. Cell 155: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry DM, Devine SE (2020) Aberrantly high levels of somatic LINE‐1 expression and retrotransposition in human neurological disorders. Front Genet 10: 1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristán‐Ramos P, Rubio‐Roldan A, Peris G, Sánchez L, Amador‐Cubero S, Viollet S, Cristofari G, Heras SR (2020) The tumor suppressor microRNA let‐7 inhibits human LINE‐1 retrotransposition. Nat Commun 11: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi CIK, Basilicata MF, Georgiev P, Gaub A, Seyfferth J, Kulkarni T, Panhale A, Semplicio G, Manjunath V, Holz H et al (2021) RNA nucleation by MSL2 induces selective X chromosome compartmentalization. Nature 589: 137–142 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of MicroRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Krawczyk PS, Adamska D, Bijata K, Garcia‐Perez JL, Dziembowski A (2018) Uridylation by TUT4/7 restricts Retrotransposition of human LINE‐1s. Cell 174: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad‐Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV (2001) Human L1 Retrotransposition: Cis preference versus trans complementation. Mol Cell Biol 21: 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein R, Bodak M, Ciaudo C (2016) Generation of a knockout mouse embryonic stem cell line using a paired CRISPR/Cas9 genome engineering tool. Methods Mol Biol 1341: 321–343 [DOI] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R (2016) Distinct stages in stress granule assembly and disassembly. eLife 5: e18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao‐Jie L, Hui‐Ying X, Qi X, Jiang X, Shi‐Jie M (2016) LINE‐1 in cancer: Multifaceted functions and potential clinical implications. Genet Med 18: 431–439 [DOI] [PubMed] [Google Scholar]
- Xie Y, Rosser JM, Thompson TL, Boeke JD, An W (2011) Characterization of L1 retrotransposition with high‐throughput dual‐luciferase assays. Nucleic Acids Res 39: e16 [DOI] [PMC free article] [PubMed] [Google Scholar]


