Abstract
The B cell translocation gene 1 (BTG1) and BTG2 play a key role in a wide range of cellular activities including proliferation, apoptosis, and cell growth via modulating a variety of central biological steps such as transcription, post-transcriptional, and translation. BTG1 and BTG2 have been identified by genomic profiling of B-cell leukemia and diverse lymphoma types where both genes are commonly mutated, implying that they serve as tumor suppressors. Furthermore, a low expression level of BTG1 or BTG2 in solid tumors is frequently associated with malignant progression and poor treatment outcomes. As physiological aspects, BTG1 and BTG2 have been discovered to play a critical function in regulating quiescence in hematopoietic lineage such as Hematopoietic stem cells (HSCs) and naïve and memory T cells, highlighting their novel role in maintaining the quiescent state. Taken together, emerging evidence from the recent studies suggests that BTG1 and BTG2 play a central antiproliferative role in various tissues and cells, indicating their potential as targets for innovative therapeutics.
Keywords: Antiproliferation, B cell malignancy, BTG1/2, T cell quiescence, Tumor suppressor
INTRODUCTION
The regulation of cell growth and proliferation is controlled by interactions of proteins that take part in antiproliferative functions such as tumor suppressors. Dysregulation of cell growth and proliferation transforms non-neoplastic cells into a tumorigenic state resulting from multiple genetic alterations (1). Cell cycle progression, differentiation, and apoptosis are all regulated by the B cell translocation gene (BTG)/TOB family of antiproliferation proteins. The mammalian BTG/Tob family consists of six proteins that control various cellular activities in a number of cell types (2). In particular, BTG1 and BTG2, have been identified as antiproliferative mediators, with the ability to promote programmed cell death or survival. Furthermore, BTG1 and BTG2 are being studied as tumor suppressors against lymphoma and solid tumors. The BTG1 and BTG2’s ability to protect cells from neoplastic transformation is associated with their function to control gene expression through interaction with transcriptional cofactors, as well as restrict messenger RNA (mRNA) abundance at the posttranscriptional level. In this context, BTG1 and BTG2 are involved in major mechanisms driving T cell quiescence as well as growth control in tumor cells, implying that BTG1 and BTG2 enhance deadenylation and degradation of mRNA. This review will provide updates on the recent information on BTG1 and BTG2 in various physiological and cellular contexts.
DISCOVERY OF BTG/TOB FAMILY: THE STRUCTURE AND FUNCTION OF ITS TRANSCRIPTIONAL REGULATION
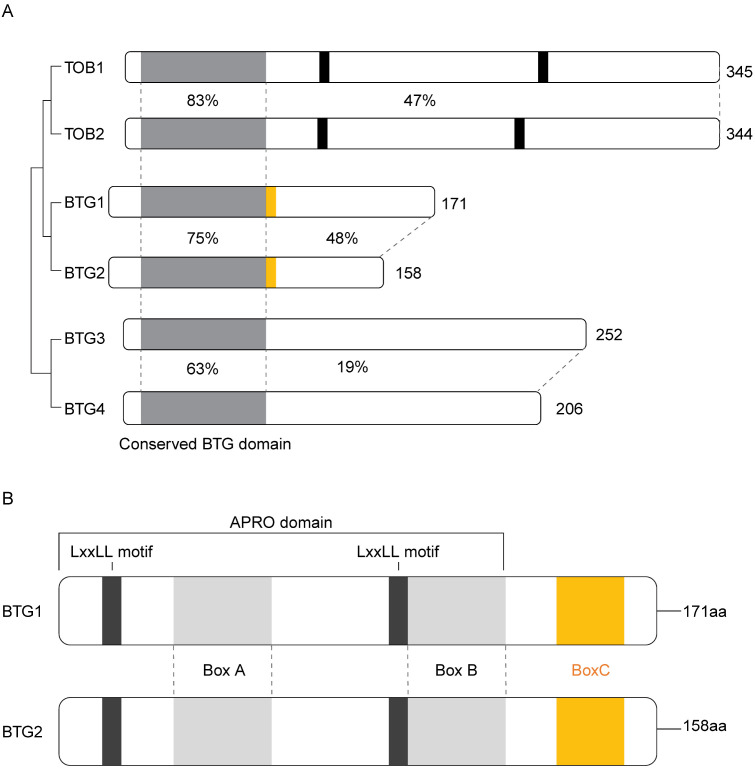
In human cells, the BTG/Tob family consists of six protein members; BTG1, BTG2/Tis21/PC3, BTG3/ANA, BTG4/PC3B, TOB1/TOB, and TOB2. BTG2 was the first protein identified in the BTG/Tob protein family. It was identified as an early-immediate response gene in rat PC12 cells and in mouse 3T3 fibroblasts in response to tetradecanoylphorbol acetate (TPA) treatment (3). The BTG1 gene, which is closely related to a gene that is induced by mitogens, was identified around the same time (4). A few years later, with functional and structural similarities to BTG1, TOB1 was discovered. The other three members of the BTG/Tob family were discovered by homology of the N-terminal domain which is highly conserved in this protein family. Amino acid sequence similarity suggests that BTG1 and BTG2, and Tob1 and Tob2 are substantially comparable, whereas BTG3 and BTG4 are relatively distant family members (Fig. 1A). Anti-proliferative (APRO) domain is a key conserved domain of BTG1 and BTG2. The conserved APRO domain in the BTG/Tob protein family includes two motifs, box A and box B. A third motif, box C, only exists in BTG1 and BTG2, which are the focus of this review, sharing 66% amino acid identity. The only difference between BTG1 and BTG2 is that the C-terminal region of BTG1 is slightly longer (5). The core regions of BTG1 and BTG2 include two LxxLL motifs, which are known to enhance protein-protein interactions (Fig. 1B).
Fig. 1.
Summary of human BTG/Tob protein family. (A) The schematic dia-gram shows amino acid sequence-based similarities between the BTG/Tob family members. Indicated are the total length of the proteins and simi-larity rate (percentage) of amino acids in the BTG domain (light gray) and the C-terminal region. Also, the conserved PAM2 (black) and box c domain are shown in the schematic representations. (B) Domains of BTG1 and BTG2. The APRO domain, which is conserved in BTG1 and BTG2, contains three motifs; box A, box B, and box C. These boxes make it easier for proteins to interact with one another. Box C (yellow) is found only in BTG1 and BTG2. Box A is known to interact with CNOT7/8 and nuclear receptors. Box B is known for its association with CNOT7/8. Box C is required for interacting with PRMT1 and PABPC1. The core regions of BTG1 and BTG2 in-clude two LxxLL motifs (black), which are known to enhance nuclear receptor interaction.
Chromosomes 12q22 and 1q32 contain the human BTG1 and BTG2 genes, respectively. Transcripts and proteins encoded from these genes are extremely unstable. The proteasome involves ubiquitination by the SCF-bTrCP1 complex (6). Further, the SCF-Skp2 complex regulates BTG2 protein stability (7). The 3’ untranslated regions of BTG2 contain up to 17 miRNA binding sites, implying that these molecules play a significant role in regulating BTG2 transcript levels (8).
BTG1 and BTG2 are found in both the nucleus and cytoplasm, and their functions are thought to be influenced by their intracellular location (9). Generally, the BTG1 and BTG2 N-terminal regions are required for cytoplasmic maintenance, and the C-terminal regions are required for regulating nuclear localization. Although both genes are generally expressed regardless of their location, BTG1 expression is particularly abundant in the pancreas, heart, and hematopoietic tissues, while BTG2 is highly expressed in the pancreas, thymus, CNS, kidneys, prostate, lungs, and skeletal muscles. Considering the antiproliferative properties of BTG1 and BTG2, they are downregulated during the G1-S transition phase in the cell cycle progression, and show a very high expression level in quiescent cells (10).
BTG2 was first identified to induce p53, which is a tumor suppress gene. When DNA damage occurs in cells, BTG2 expression increases, which is a result of elevated expression of tumor suppressor p53 since a loss-of-function mutant of p53 cannot induce BTG2 accumulation in DNA damaged cells (5). BTG2 has also been demonstrated to be responsive to the activation of nuclear factor-kB (NF-kB). Various genotoxic chemicals (UV, ionizing radiation), growth factors, interleukin 6, and cyclic adenosine monophosphate (cAMP) can be stimulating factors of BTG2 expression. BTG1 can also be induced by DNA damage, but it is different from BTG2 in that it is independent of p53 (11). Glucocorticoids, 4-Hydroxytamoxifen, transforming growth factor β (TGF β), and angiogenic growth factors all increase BTG1 transcript levels. As a result, both BTG1 and BTG2 are influenced by a number of intracellular signaling pathways.
C-TERMINAL DOMAIN OF BTG1 AND BTG2: PROVIDE KEY PROTEIN INTERACTIONS
Both BTG1 and BTG2 are transcriptional coactivators and physiologically interact with various intracellular transcription factors. Homeobox B9, which is a member of the Abd-B homeobox family, is one of the transcription factors that BTG1 and BTG2 bind to. During embryonic development, BTG1 and BTG2 regulate the Homeobox B9 transcription to influence pattern formation (12). Furthermore, the LxxLL motif of BTG1 and BTG2 allows binding to a wide range of nuclear receptors, including androgen receptor, estrogen receptor alpha, and T3 receptor (13, 14). The Box C domain, present only in BTG1 and BTG2, facilitates interaction with protein arginine methyltransferase 1 (PRMT1) (15); PRMT1 is one of the enzyme families involved in posttranslational modification by inducing methylation at arginine residues on several protein substrates. PRMT1 regulates several biological processes including hepatic gluconeogenesis, T cell activation, and even cancer cells (16). The interaction between PRMT1 and BTG2 is particularly well known for retinoic acid receptor-mediated gene regulation (17). BTG2 increases the activity of PRMT1 as a transcriptional coactivator, and eventually induces methylation at histone H4 arginine 3 residue. In the absence of retinoic acid in the nucleus, PRMT1 and BTG2 exist as RAR-containing complexes. When retinoic acid enters the nucleus, PRMT1 and BTG2 are recruited at the RARβ promoter and induce epigenetic alteration, resulting in elevated histone H4 acetylation levels (17). The interaction between PRMT1 and BTG2 is a necessary step for BTG2 antiproliferative function (18). The BTG1 and BTG2 are different from the other BTG/Tob family members owing to the presence of the box c domain near the C-terminal (Fig. 1B). Among the B cell lymphomas, lymphoblastic lymphoma (LBL) is an uncommon neoplasm of immature B cells, and when this cell line is treated with anti-IgM, BTG/Tob family proteins other than the BTG1 and BTG2 are upregulated. On the other hand, when BTG1 and BTG2 were overly expressed, lymphoma proliferation was inhibited, and when a deleted Box C domain mutant was expressed, the proliferation inhibitory effect was reduced, suggesting that PRMT1 is involved. Moreover, treatment with adenosine dialdehyde (AdOx), which can indirectly block methyltransferase activity, or knockdown of PRMT1 largely alleviates the progression of the cell cycle induced by BTG1/2 mediated antiproliferation (18).
Smad1 and Smad8, which is a transcriptional modulators activated by BMP receptor kinase, are also linked with BTG2. When BTG2 was overexpressed, the activity of a BMP-dependent synthetic reporter was dramatically increased (19).
Meanwhile, Ser-147 and Ser-149 residues in the C-terminal region of BTG2 can be phosphorylated by Erk1 and Erk2. Phosphorylation at the Ser-147 residue can induce the binding of BTG2 by PIN-1 (20). In a previous study, yeast two-hybrid screening was performed to find a protein that interacts with BTG2. As a result, it was found that PICK1, which functions by binding to the protein kinase PKCα, also interacts with BTG2. However, the physiological role of the PICK1 and BTG2 interaction is not yet identified (21).
Generally, BTG1 and BTG2 proteins are regulated by the ubiquitin/proteasome pathway (22). Meanwhile, domains located at the C-terminal of the BTG protein do not correlate with each other in controlling the total amount of protein. It has been re-ported that residues located in the C-terminal are not involved in the protein abundance because stability is reduced when fused with GFP protein (22).
BTG1 AND BTG2: GLOBAL REGULATORS OF MRNA ABUNDANCE
Members of the BTG family regulate gene expression in quiescent cells by limiting mRNA abundance. BTG1 and BTG2 bind to the CCR4-NOT complex, which is a multisubunit complex that plays an essential role in RNA metabolism in eukaryotes. Specifically, BTG1 and BTG2 bind to Ccr4-associated factor 1 (CAF1), a protein constituting the CCR4-NOT complex. CAF1 degrades the poly A tail of mRNA and greatly reduces mRNA stability, which results in an increased threshold for mRNA expression level in the cells (23, 24). BTG2 forms a complex for mRNA deadenylation with CAF1 and CCR4, thereby accelerating mRNA decay (24). In particular, as the CAF1 deadenylase activity increases, mRNA decay is promoted. At this time, BTG2 binds to poly A tail binding protein, PABPC1 so that BTG2 regulates the poly A tail length of mRNA, thereby lowering the number of transcripts in the cells (25). It has been reported that the CNOT7 and CNOT8 deadenylase subunits, which are components of the CCR4-NOT complex, interact with BTG protein to decrease the transcription of several genes, but the basic mechanism of this process is still unknown (26). The fact that BTG1 and BTG2 keep the cell transcription level low suggests an increase in the threshold for reaching the mRNA level required for cell activation. In particular, for the immune cell to perform effector functions, it needs to be sufficiently activated. Notably, the BTG1/2 expression levels in the lymph nodes and white blood cells were high among the BTG/Tob family, suggesting that they have specific functions in the immune system (23). Specifically, BTG1 and BTG2 are mainly expressed in the naïve and memory T cells among several T cell subsets, and this phenomenon is consistent with the expression pattern of Klf2, Il7r, and Foxo1, known as quiescence markers. As with other non-dividing cells, both BTG1 and BTG2 interact with PABP and CCR4-NOT7 deadenylase complex in quiescence T cells to regulate global mRNA abundance. When naïve T cells are activated, BTG1 and BTG2 rapidly decrease to increase mRNA abundance, creating conditions for escape from the quiescent state. Therefore, BTG1/2-mediated deadenylation is a mechanism that directly restricts mRNA abundance consequently maintaining the quiescent state in the T cells while inhibiting spontaneous activation (23).
PHYSIOLOGICAL ROLE OF BTG1 AND BTG2
A variety of physiological roles of BTG1 and/or BTG2 have been reported as their universal knock-out (KO) and conditional KO mice were generated. BTG1 and BTG2 are highly expressed in hematopoietic lineages, suggesting that they have an impact on hematopoietic stem cells (HSC), B cells, and T cells. BTG1 is known to be essential for resetting the quiescent state of HSC once they are activated (27). In addition, BTG1 or BTG2 single deficiency impaired B cell development in bone marrow and spleen. Interestingly, BTG1 and BTG2 double KO mice showed a more significant loss of B cell progenitor cells, suggesting that they are involved in early B cell commitment (28). Another group found that BTG2 regulates thymocyte development by modulating the proliferation in the double negative (DN) stage; DN1 and DN3 (29). Recently, our group showed that BTG1 and BTG2 have a pivotal role in regulating the maintenance of quiescent naïve and memory T cells in the periphery. Although single conditional KO mice did not affect T cells, BTG1, and BTG2 double conditional KO T cells showed a significantly increased population of effector T cells when compared to WT T cells, indicating their significant involvement in maintaining the quiescent state (23).
The role of BTG1 and BTG2 in non-hematopoietic cells is also observed. It has been demonstrated that BTG1 is overexpressed in adult neurogenic niches like the dentate gyrus and subventricular zone (SVZ). In mice, deletion of BTG1 lowers the ability of the adult stem cells and progenitor cells to proliferate in the dentate gyrus and SVZ, as well as causing apoptosis. This means that loss of the BTG1 disrupted cell cycle regulation during the transition from G1 to S phase (30). In breast cancer, overexpression of BTG1 resulted in the reduction of cell cycle-related proteins, inducing inhibition of cell proliferation (31). BTG2 expression is increased in vivo during neurogenesis while deletion of this gene promoted programmed cell death in vitro. These results indicate that BTG2 is involved in the process of neural differentiation, and it is essential for terminally differentiated cells to survive (4). During adult hippocampus neurogenesis, BTG2 is necessary for the regulation of newborn neuron proliferation and terminal differentiation (32). One of the most important stages in the development of neuronal circuits is neurite outgrowth. By controlling arginine methylation in the nucleus, BTG2, along with the arginine methyltransferase PRMT1, regulates neurite outgrowth (33). BTG2 expression increases and cyclin D1 levels decrease during the myogenic differentiation of myoblast cells (34). Also, it is known that BTG2 is downregulated in the JAK2-Stat3 signaling pathway. One study observed that Btg2 knockdown increased lipid accumulation and the expression of adipogenic marker genes. This means that the proadipogenic activity of the Stat3 signaling pathway inhibits the negative effect of BTG2 on adipogenesis (35). The upregulation of BTG2 genes after 12-O-tetra-decanoylphorbol-13-acetate (TPA) or retinoic acid (RA) stimulation may be involved in the differentiation of HL-60 cells (36). We have summarized various physiological effects of BTG1 and BTG2 in Table 1.
Table 1.
The physiological role of BTG1 and BTG2 in various tissues and cell types
| Type of cell or tissue | Observation | Mechanism | Reference |
|---|---|---|---|
| T lymphocyte | BTG1/2 deficiency allows naïve T cell easily to exit from the quiescence state | Lowering mRNA abundance by inducing constant deadenylation via interacting with PABP and CNOT7/8 | (23) |
| Ectopic expression of BTG2 inhibit DN1 and DN3 thymocyte expansion | (38) | ||
| HSC | High level of BTG1 requires to reset the quiescent state | (36) | |
| BTG2-deficient bone marrow shows an elevated hematopoietic progenitor cells expansion | BTG2 depresses AKT phosphorylation and inhibits mTOR signaling | (65) | |
| B lymphocyte | BTG1/2 deficiency reduces B cell progenitors in bone marrow and spleen | (37) | |
| Liver | Knockdown of BTG1 induces liver steatosis | BTG1 decreases SCD1 via suppressing ATF4 | (66) |
| Axial skeleton | BTG1/2 deficiency results in abnormal patterning of the axial skeleton | (67) | |
| Dentate gyrus and SVZ* | Deletion of BTG1 reduces the number of dividing adult stem and progenitor cells | (39) | |
| Adipocyte | Knockdown of BTG2 increased lipid accumulation and differentiation | STAT3 signaling pathway inhibits the negative effect of Btg2 on adipogenesis | (44) |
| Primary fibroblast | Ectopic expression of BTG2 induces senescence independently of p53 | BTG2 antagonizes the cell cycle regulator PIN1 | (33) |
| MEF* | Ablation of BTG1 gives a survival advantage under stress conditions | BTG1 enhances the activity of ATF4 via facilitating PRMT1 binding | (49) |
*SVZ: subventricular zone, MEF: Mouse embryonic fibroblast.
BTG1 AND BTG2: REGULATION OF CELL CYCLE AND APOPTOSIS
BTG1 expression is highest during the G0/G1 phases of the cell cycle and decreases as the cell progresses through G1. Furthermore, transfection experiments show that BTG1 inhibits cell proliferation (5). BTG2, which has a structure similar to BTG1, is also known to affect the cell cycle. BTG2 overexpression causes a partial suppression of cell growth in a variety of cell lines and has been shown to enhance apoptosis in pancreatic cancer cells (24). Additionally, overexpression of BTG2 reduces the expression of cyclin D1, MMP-1, and MMP-2, as well as lung cancer cell growth (37). It is known that inhibition of cyclin D1 levels depends on the binding of BTG2 to histone deacetylases, HDAC1, HDAC4, and HDAC9 (38). And its downregulation leads to the suppression of retinoblastoma (Rb) phosphorylation and G1 arrest (39). BTG2 suppresses the transition from G1 to the S phase by lowering the level of cyclin E and cyclin‐dependent kinase (cdk4) in the absence of a functional Rb (40). The Cdk4 is a direct target of the BTG2-PRMT1 complex in B cells, and cdk4 methylation causes protein degradation (41). BTG2 can induce G2/M arrest in a p53-independent manner. Overexpression of BTG1 and BTG2, which is not dependent on p53 expression, is one trait of drug-induced cellular senescence in human cancer cell lines (42). In normal fibroblasts, BTG2 expression causes cellular senescence via disrupting the cell cycle regulator pin1 (43). Later, it was discovered that BTG1, BTG2, BTG3, and TOB1 were regulated by the tumor suppressor p19 (Arf) ir-respective of p53, resulting in cell cycle arrest (44). Expression of BTG1 and BTG2 may be involved in regulating apoptosis and inducing cell cycle arrest. Forced BTG1 expression has been shown to promote cell death in various cell types like human breast cancer cells, murine fibroblasts, and microglia. BTG1 overexpression in the brain makes microglial cells more susceptible to inflammatory death.
Moreover, recent work showed that BTG1 and BTG2 maintain T cell quiescence by regulating proliferation and activation (23). In the majority of breast cancers, an antiapoptotic protein Bcl-2 is upregulated and linked to a reduced apoptotic response. BTG1 is a Bcl-2-regulated apoptotic mediator (45). However, in atherosclerotic lesions, BTG1 expression is limited to apoptotic cells in specific macrophage-rich regions.
BTG1 AND BTG2 AS TUMOR SUPPRESSORS
BTG1 and BTG2: DNA damage repair and stress response
The most famous tumor suppressor gene, p53, can upregulate BTG2 and downregulate cyclins D1 and E during hepatocarcinogenesis (46). Once the DNA is damaged, p53 is activated, causing DNA repair and a G1/S-phase cell cycle arrest. If the damage can’t be repaired, p53 promotes apoptosis or programmed cell death. BTG2 has been shown to have an antitumor impact via the Ras signal transduction pathway, which is dependent on p53 (47). As a result, BTG2 gene expression is significantly increased when the DNA is damaged. Various DNA-damaging factors can stimulate BTG2 expression through p53 and cause cell cycle arrest in the G1 phase by suppressing cyclin D1, therefore facilitating DNA damage repair. Inhibition of BTG2 in primary fibroblasts mimics the loss of p53 function in cooperation with oncogenic Ras (HRasv12), allowing cells to avoid replicative senescence while triggering transformation and immortalization. In this oncogenic context, inhibiting BTG2 increases the levels of cyclins D1 and E1 as well as Rb phosphorylation, which is similar to earlier findings (39). Furthermore, BTG2 was discovered to influence p53 activity through posttranslational modification. In bladder cancer cells expressing oncogenic Ras and mutant p53, BTG2-mediated p53 regulation causes a switch from senescence to apoptosis, which lowers tumorigenicity (48). One study suggested that BTG1 interacts with Activating Transcription Factor 4 (ATF4) and regulates its activity by binding the protein arginine methyl transferase PRMT1. The loss of BTG1 gives a survival benefit to primary mouse embryonic fibroblasts (MEFs) under stress conditions in BTG1 knockout mice. Regulation of ATF4, a major modulator of cellular stress responses, is involved in this pro-survival impact. Therefore, BTG1/PRMT1 complex is a regulator of ATF4-mediated stress responses (49).
BTG1 and BTG2 in solid tumors
Downregulated BTG1/2 expression and function have been observed in various solid tumors. BTG2 has been recognized as a possible biomarker for cancer patients’ prognosis in several research studies (50). Autophagy supports the survival of tumor cells under metabolic stress. miR-22 suppressed autophagy and increased apoptosis, increasing the sensitivity of colorectal cancer (CRC) cells to 5-fluorouracil (5-FU) treatment. Notably, BTG1 is a target of miR-22. Accordingly, by post-transcriptional suppression of BTG1, miR-22 may serve as a key switch between autophagy and apoptosis to modulate 5-FU sensitivity (51). Overexpression of miR-511, involved in cancer development, induced the proliferation of hepatoma cells by targeting BTG1 (52). In addition, in colon cancer, increased levels of miR-301A reduce the expression of BTG1 and increase tumorigenesis (53). It is known that BTG2 is related to tumor suppressor genes like RB, p53, and p73 (46, 47). In breast cancer, decreased BTG2 expression is found to be substantially associated with increasing tumor size and cyclin D1 protein overexpression (9). BTG2 affects cell cycle distribution, increases radiation-induced apoptosis, and suppresses DNA repair-related protein expression, all of which can improve the radiosensitivity of breast cancer cells (54). In liver cancer, BTG2 expression is often downregulated and, consequently, the level of cyclin-D1/cyclin E is increased (46). Furthermore, BTG2 inhibits cancer cell proliferation through downregulation of STAT3 activity and IL-6 expression in human dermal fibroblasts (55). In this context, in most solid tumors, BTG1/2 serves as tumor suppressors. However, more studies are needed to reveal the underlying detailed mechanisms. We described the known observation and mechanism of BTG1 and BTG2 in various cancers in Table 2.
Table 2.
BTG1 and BTG2 function as a tumor suppressor in various cancers
| Type of cancer | Observation | Mechanism | Reference |
|---|---|---|---|
| BL, FL, DLBCL, HBL* | BTG1 or BTG2 are frequently mutated or deleted | (57, 58, 61, 68-71) | |
| ALL | Deletion of BTG1 can be a cancer driver gene in leukemogenesis | Excessive proliferation due to deletion of BTG1 where RAG1/RAG2 mediated recombination occurs | (62) |
| AML | BTG1 is downregulated | Ectopic expression of BTG1inhibits proliferation | (72) |
| Liver cancer | BTG2 expression is downregulated | BTG1 is downregulated by miR-511 overexpression, promoting proliferation of hepatoma cells | (52) |
| BTG1 expression is downregulated | (73) | ||
| Breast cancer | BTG1 expression is downregulated | (40) | |
| BTG2 can affect radiation-induced apoptosis | Lack of BTG2 induces overexpression of cyclin D1 | (9) | |
| Ovarian cancer | Downregulation of BTG2 is associated with poor prognosis | BTG2 inhibits proliferation and cell-cycle via AKT and ERK signaling | (74) |
| Laryngeal carcinoma | BTG2 expression is downregulated | BTG2 is suppressed via miR-21 | (75) |
| Gastric cancer | BTG2 expression is downregulated | BTG2 is suppressed via miR-27a-3p | (76) |
| Colorectal cancer | BTG1 expression is downregulated | Post-transcriptional suppression of BTG1 by miR-22 might balance between autophagy and apoptosis | (51) |
| Lung epithelial cancer | Ectopic expression of BTG2 inhibits the growth, proliferation | By reducing the expression of cyclin D1, MMP-1 and MMP-2 | (27) |
| Bladder cancer | Ectopic expression of BTG2 induces a switch from senescence to apoptosis | By translocating of p53 protein | (48) |
*BL: Burkitt lymphoma, FL: Follicular lymphoma, HBL: High-grade B cell lymphoma, DLBCL: diffuse large B-cell lymphoma, ALL: Acute lymphocytic leukemia, AML: Acute myeloid leukemia.
BTG1 and BTG2 in B-cell malignancies
BTG1 and BTG2 have been shown to be frequently mutated in various B-cell malignancies in several studies (56, 57). For example, in the Burkitt lymphoma (BL) subtype, the mutation of RBL2/p130 which regulates the expression of BTG1 affects the development of BL (58). Follicular lymphoma (FL), represented by DLBCL (Diffuse Large B Cell Lymphoma), was also found to have point mutations in BTG1 and BTG2 (59). And BTG1 mutations were linked to a lower chance of survival in ABC DLBCL patients (60). One study showed that BTG1 may play a critical role in the development of DLBCL, according to bioinformatics analysis. 401 genes were identified as BTG1-associated DLBCL genes through an overlapping study of 407 BTG1-associated genes and 22,187 DLBCL-associated genes. BTG1-associated DLBCL genes were linked to tumor progression and DLBCL signaling pathways, from a pathway analysis. Thus, BTG1 could be used as a prognostic biomarker for DLBCL (61). In Acute Lymphoblastic leukemia (ALL), BTG1 deletions may drive the leukemogenesis at sites that are likely to have abnormal RAG1/RAG2 mediated recombination (62). In B‐cell precursor, acute lymphoblastic leukemia (BCP‐ALL), the most frequent type of cancer in children, deletions and mutations affecting the lymphoid transcription factor IKZF1 (IKAROS) are linked to an increased risk of recurrence and a poor prognosis. One study observed that single-copy deletions of BTG1 were highly enriched in IKZF1-deficient BCP‐ALL. The deficiency of BTG1 alone did not influence leukemia development, but both BTG1 and IKZF1 deficiency did (63). Resistance to glucocorticoids, which is a crucial component of ALL therapy, has been associated with the loss of NR3C1 and BTG1. These findings support previous research associating NR3C1 deficiency with poor outcomes in ALL recurrence (64). Further research is required to determine whether these negative results are directly linked to glucocorticoid resistance.
PERSPECTIVE
BTG1 and BTG2 play a wide range of essential cellular functions via governing proliferation, apoptosis, activation, cell growth, quiescence, and differentiation. Even though their functional heterogeneity is likely dependent on the different cellular contexts, a bulk of studies suggest that BTG1 and BTG2 mainly serve as antiproliferative proteins. It is likely that these proteins have different functions depending on their cellular location. For example, BTG1 and BTG2 may play a prominent role in transcription by serving as transcriptional cofactors via interacting with nuclear receptors and transcriptional factors, thereby modulating gene-specific programs. BTG1 and BTG2 however likely govern the post-transcriptional programs by recruiting and inducing the deadenylation machinery, thereby controlling global mRNA stability. According to previous observations (23, 28), BTG1 and BTG2 are likely functionally redundant in many tissues based on their structural similarities. Therefore, it would be meaningful to investigate s a variety of functional aspects of BTG1 and BTG2 using a double deficient conditional KO system in many cell types such as tissue-specific stem cells and cancer cells. In addition, the regulatory mechanism for controlling the activation or inactivation of BTG1 and BTG2 needs to be ap-propriately addressed in the near future as they quickly respond to extrinsic signals to rapidly adapt to environmental changes. Recent technological advances such as single cell RNA-sequencing (scRNA-seq), mRNA TAIL-sequencing (mTAIL-seq), and thiol(SH)-linked alkylation for the metabolic sequencing of RNA (SLAM-seq) would be valuable in addressing the precise molecular mechanism of the antiproliferative role of BTG1 and BTG2 in various pathophysiological contexts.
ACKNOWLEDGEMENTS
We thank all members of the Department of Biochemistry and Molecular Biology of Yonsei University College of Medicine for administrative support. This work was supported by the Brain Korea 21 FOUR Project for Medical Science, Yonsei University College of Medicine. This work of SSH has supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2021R1A2C2093640 and 2018R1A5A2025079) and a faculty research grant (6-2021-0156) and a new faculty research seed money grant (2021-32-0055) of Yonsei University College of Medicine.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Coleman WB. Genomic catastrophe and neoplastic transformation. Am J Pathol. 2015;185:1846–1849. doi: 10.1016/j.ajpath.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Wang C, Wu J, Li L. BTG/Tob family members Tob1 and Tob2 inhibit proliferation of mouse embryonic stem cells via Id3 mRNA degradation. Biochem Biophys Res Commun. 2015;462:208–214. doi: 10.1016/j.bbrc.2015.04.117. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida Y, Matsuda S, Ikematsu N, et al. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–2693. doi: 10.1038/sj.onc.1201805. [DOI] [PubMed] [Google Scholar]
- 4.el-Ghissassi F, Valsesia-Wittmann S, Falette N, Duriez C, Walden PD, Puisieux A. BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene. 2002;21:6772–6778. doi: 10.1038/sj.onc.1205888. [DOI] [PubMed] [Google Scholar]
- 5.Rouault JP, Rimokh R, Tessa C, et al. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasajima H, Nakagawa K, Kashiwayanagi M, Yokosawa H. Polyubiquitination of the B-cell translocation gene 1 and 2 proteins is promoted by the SCF ubiquitin ligase complex containing betaTrCP. Biol Pharm Bull. 2012;35:1539–1545. doi: 10.1248/bpb.b12-00330. [DOI] [PubMed] [Google Scholar]
- 7.Park TJ, Kim JY, Park SH, Kim HS, Lim IK. Skp2 enhances polyubiquitination and degradation of TIS21/BTG2/PC3, tumor suppressor protein, at the downstream of FoxM1. Exp Cell Res. 2009;315:3152–3162. doi: 10.1016/j.yexcr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Fei JF, Haffner C, Huttner WB. 3' UTR-dependent, miR-92-mediated restriction of Tis21 expression maintains asymmetric neural stem cell division to ensure proper neocortex size. Cell Rep. 2014;7:398–411. doi: 10.1016/j.celrep.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 9.Kawakubo H, Brachtel E, Hayashida T, et al. Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res. 2006;66:7075–7082. doi: 10.1158/0008-5472.CAN-06-0379. [DOI] [PubMed] [Google Scholar]
- 10.Rouault JP, Falette N, Guehenneux F, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 11.Cortes U, Moyret-Lalle C, Falette N, et al. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog. 2000;27:57–64. doi: 10.1002/(SICI)1098-2744(200002)27:2<57::AID-MC1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Prevot D, Voeltzel T, Birot AM, et al. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 13.Busson M, Carazo A, Seyer P, et al. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene. 2005;24:1698–1710. doi: 10.1038/sj.onc.1208373. [DOI] [PubMed] [Google Scholar]
- 14.Hu XD, Meng QH, Xu JY, et al. BTG2 is an LXXLL-dependent co-repressor for androgen receptor transcriptional activity. Biochem Biophys Res Commun. 2011;404:903–909. doi: 10.1016/j.bbrc.2010.12.064. [DOI] [PubMed] [Google Scholar]
- 15.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 16.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passeri D, Marcucci A, Rizzo G, et al. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26:5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hata K, Nishijima K, Mizuguchi J. Role for Btg1 and Btg2 in growth arrest of WEHI-231 cells through arginine methylation following membrane immunoglobulin engagement. Exp Cell Res. 2007;313:2356–2366. doi: 10.1016/j.yexcr.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Cho JW, Kim JJ, Park SG, et al. Identification of B-cell translocation gene 1 as a biomarker for monitoring the remission of acute myeloid leukemia. Proteomics. 2004;4:3456–3463. doi: 10.1002/pmic.200400968. [DOI] [PubMed] [Google Scholar]
- 20.Hong JW, Ryu MS, Lim IK. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J Biol Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- 21.Lin WJ, Chang YF, Wang WL, Huang CY. Mitogen-stimulated TIS21 protein interacts with a protein-kinase-Calpha-binding protein rPICK1. Biochem J. 2001;354:635–643. doi: 10.1042/bj3540635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasajima H, Nakagawa K, Yokosawa H. Anti-proliferative proteins of the BTG/Tob family are degraded by the ubiquitin-proteasome system. Eur J Biochem. 2002;269:3596–3604. doi: 10.1046/j.1432-1033.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 23.Hwang SS, Lim J, Yu Z, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020;367:1255–1260. doi: 10.1126/science.aax0194. [DOI] [PubMed] [Google Scholar]
- 24.Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupfler B, Birck C, Seraphin B, Mauxion F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat Commun. 2016;7:10811. doi: 10.1038/ncomms10811.d3496cb77c9e4475a194e5471d760c2a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS. The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell. 2009;20:3840–3850. doi: 10.1091/mbc.e09-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venezia TA, Merchant AA, Ramos CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301.5e6624498529416cab81c0d6816cd11e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tijchon E, van Emst L, Yuniati L, et al. Tumor suppressors BTG1 and BTG2 regulate early mouse B-cell development. Haematologica. 2016;101:e272–276. doi: 10.3324/haematol.2015.139675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konrad MA, Zuniga-Pflucker JC. The BTG/TOB family protein TIS21 regulates stage-specific proliferation of developing thymocytes. Eur J Immunol. 2005;35:3030–3042. doi: 10.1002/eji.200526345. [DOI] [PubMed] [Google Scholar]
- 30.Farioli-Vecchioli S, Micheli L, Saraulli D, et al. Btg1 is required to maintain the pool of stem and progenitor cells of the Dentate Gyrus and Subventricular Zone. Front Neurosci. 2012;6:124. doi: 10.3389/fnins.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu R, Zou ST, Wan JM, Li W, Li XL, Zhu W. BTG1 inhibits breast cancer cell growth through induction of cell cycle arrest and apoptosis. Oncol Rep. 2013;30:2137–2144. doi: 10.3892/or.2013.2697. [DOI] [PubMed] [Google Scholar]
- 32.Farioli-Vecchioli S, Saraulli D, Costanzi M, et al. Impaired terminal differentiation of hippocampal granule neurons and defective contextual memory in PC3/Tis21 knockout mice. PLoS One. 2009;4:e8339. doi: 10.1371/journal.pone.0008339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyata S, Mori Y, Tohyama M. PRMT1 and Btg2 regulates neurite outgrowth of Neuro2a cells. Neurosci Lett. 2008;445:162–165. doi: 10.1016/j.neulet.2008.08.065. [DOI] [PubMed] [Google Scholar]
- 34.Evangelisti C, Astolfi A, Gaboardi GC, et al. TIS21/BTG2/PC3 and cyclin D1 are key determinants of nuclear diacylglycerol kinase-zeta-dependent cell cycle arrest. Cell Signal. 2009;21:801–809. doi: 10.1016/j.cellsig.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Hong JW, Park KW. B cell translocation gene 2 (Btg2) is regulated by Stat3 signaling and inhibits adipocyte differentiation. Mol Cell Biochem. 2016;413:145–153. doi: 10.1007/s11010-015-2648-z. [DOI] [PubMed] [Google Scholar]
- 36.Cho BO, Jeong YW, Kim SH, et al. Up-regulation of the BTG2 gene in TPA- or RA-treated HL-60 cell lines. Oncol Rep. 2008;19:633–637. doi: 10.3892/or.19.3.633. [DOI] [PubMed] [Google Scholar]
- 37.Wei S, Hao C, Li X, Zhao H, Chen J, Zhou Q. Effects of BTG2 on proliferation inhibition and anti-invasion in human lung cancer cells. Tumour Biol. 2012;33:1223–1230. doi: 10.1007/s13277-012-0370-y. [DOI] [PubMed] [Google Scholar]
- 38.Micheli L, D'Andrea G, Leonardi L, Tirone F. HDAC1, HDAC4, and HDAC9 bind to PC3/Tis21/Btg2 and are required for its inhibition of cell cycle progression and cyclin D1 expression. J Cell Physiol. 2017;232:1696–1707. doi: 10.1002/jcp.25467. [DOI] [PubMed] [Google Scholar]
- 39.Guardavaccaro D, Corrente G, Covone F, et al. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol. 2000;20:1797–1815. doi: 10.1128/MCB.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim IK, Lee MS, Ryu MS, et al. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol Carcinog. 1998;23:25–35. doi: 10.1002/(SICI)1098-2744(199809)23:1<25::AID-MC4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 41.Dolezal E, Infantino S, Drepper F, et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat Immunol. 2017;18:911–920. doi: 10.1038/ni.3774. [DOI] [PubMed] [Google Scholar]
- 42.Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB. Molecular determinants of terminal growth arrest induced in tumor cells by a chemothera-peutic agent. Proc Natl Acad Sci U S A. 2002;99:389–394. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheaton K, Muir J, Ma W, Benchimol S. BTG2 antagonizes Pin1 in response to mitogens and telomere disruption during replicative senescence. Aging Cell. 2010;9:747–760. doi: 10.1111/j.1474-9726.2010.00601.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuo ML, Duncavage EJ, Mathew R, et al. Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res. 2003;63:1046–1053. [PubMed] [Google Scholar]
- 45.Nahta R, Yuan LX, Fiterman DJ, et al. B cell translocation gene 1 contributes to antisense Bcl-2-mediated apoptosis in breast cancer cells. Mol Cancer Ther. 2006;5:1593–1601. doi: 10.1158/1535-7163.MCT-06-0133. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Chen C, Wang G, et al. Aberrant expression of the p53-inducible antiproliferative gene BTG2 in hepatocellular carcinoma is associated with overexpression of the cell cycle-related proteins. Cell Biochem Biophys. 2011;61:83–91. doi: 10.1007/s12013-011-9164-x. [DOI] [PubMed] [Google Scholar]
- 47.Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi OR, Ryu MS, Lim IK. Shifting p53-induced senescence to cell death by TIS21(/BTG2/Pc3) gene through posttranslational modification of p53 protein. Cell Signal. 2016;28:1172–1185. doi: 10.1016/j.cellsig.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Yuniati L, van der Meer LT, Tijchon E, et al. Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress. Oncotarget. 2016;7:3128–3143. doi: 10.18632/oncotarget.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagener N, Bulkescher J, Macher-Goeppinger S, et al. Endogenous BTG2 expression stimulates migration of bladder cancer cells and correlates with poor clinical prognosis for bladder cancer patients. Br J Cancer. 2013;108:973–982. doi: 10.1038/bjc.2012.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanda M, Oya H, Nomoto S, et al. Diversity of clinical implication of B-cell translocation gene 1 expression by histopathologic and anatomic subtypes of gastric cancer. Dig Dis Sci. 2015;60:1256–1264. doi: 10.1007/s10620-014-3477-8. [DOI] [PubMed] [Google Scholar]
- 52.Zhang SQ, Yang Z, Cai XL, et al. miR-511 promotes the proliferation of human hepatoma cells by targeting the 3'UTR of B cell translocation gene 1 (BTG1) mRNA. Acta Pharmacol Sin. 2017;38:1161–1170. doi: 10.1038/aps.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He C, Yu T, Shi Y, et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhibiting BTG1. Gastroenterology. 2017;152:1434–1448. doi: 10.1053/j.gastro.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 54.Hu X, Xing L, Jiao Y, et al. BTG2 overexpression increases the radiosensitivity of breast cancer cells in vitro and in vivo. Oncol Res. 2013;20:457–465. doi: 10.3727/096504013X13685487925211. [DOI] [PubMed] [Google Scholar]
- 55.Quy LN, Choi YW, Kim YH, Chwae YJ, Park TJ, Lim IK. TIS21(/BTG2/PC3) inhibits interleukin-6 expression via downregulation of STAT3 pathway. Cell Signal. 2013;25:2391–2399. doi: 10.1016/j.cellsig.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 56.Hunter ZR, Xu L, Yang G, et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood. 2014;123:1637–1646. doi: 10.1182/blood-2013-09-525808. [DOI] [PubMed] [Google Scholar]
- 57.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Falco G, Leucci E, Lenze D, et al. Gene-expression analysis identifies novel RBL2/p130 target genes in endemic Burkitt lymphoma cell lines and primary tumors. Blood. 2007;110:1301–1307. doi: 10.1182/blood-2006-12-064865. [DOI] [PubMed] [Google Scholar]
- 59.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6:130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan W, Li SX, Gao H, Yang W. Identification of B-cell translocation gene 1-controlled gene networks in diffuse large B-cell lymphoma: a study based on bio-informatics analysis. Oncol Lett. 2019;17:2825–2835. doi: 10.3892/ol.2019.9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waanders E, Scheijen B, van der Meer LT, et al. The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution. PLoS Genet. 2012;8:e1002533. doi: 10.1371/journal.pgen.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheijen B, Boer JM, Marke R, et al. Tumor suppressors BTG1 and IKZF1 cooperate during mouse leukemia development and increase relapse risk in B-cell precursor acute lymphoblastic leukemia patients. Haematologica. 2017;102:541–551. doi: 10.3324/haematol.2016.153023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irving JA, Enshaei A, Parker CA, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2016;128:911–922. doi: 10.1182/blood-2016-03-704973. [DOI] [PMC free article] [PubMed] [Google Scholar]