Summary
Na/K-ATPase (NKA), besides its ion transporter function, is a signal transducer by regulating Src family kinases (SFK). The signaling NKA contributes to oxidized LDL-induced macrophage foam cell formation and interacts with TLR4. However, its role in lipopolysaccharides (LPS)-induced signaling and glycolytic switch in macrophages remains unclear. Using peritoneal macrophages from NKA α1 haploinsufficient mice (NKA α1+/−), we found that NKA α1 haploinsufficiency led to enhanced LPS-stimulated NF-κB pathway, ROS signaling, and pro-inflammatory cytokines. Intraperitoneal injection of LPS resulted in more severe lung inflammation and injury with lower survival rate in NKA α1+/− mice. Additionally, LPS induced a higher extent of the metabolic switch from oxidative phosphorylation to glycolysis. Mechanistically, NKA α1 interacted with TLR4 and Lyn. The presence of NKA α1 in this complex attenuated Lyn activation by LPS, which subsequently restricted the downstream ROS and NF-κB signaling. In conclusion, we demonstrated that NKA α1 suppresses LPS-induced macrophage pro-inflammatory signaling through Lyn.
Subject areas: Biological sciences, immunology, molecular biology
Graphical abstract

Highlights
-
•
NKA α1 interacts with TLR4 and SFKs
-
•
NKA α1 restricts LPS-stimulated pro-inflammatory responses in macrophages
-
•
NKA α1+/− mice injected with LPS show more lung injury and lower survival rate
Biological sciences; Immunology; Molecular biology.
Introduction
Pro-inflammatory response during infection is necessary for host protection, excessive production of pro-inflammatory cytokines and chemokines can promote immunopathology in diseases such as sepsis (Tsujimoto et al., 2008), cancer (So and Ouchi, 2010) and diabetes (Jialal et al., 2014). Therefore, down-regulation of inflammatory responses is an essential element of homeostasis and necessary to restore and maintain tissue and organ function in the setting of inflammatory stimuli. Macrophages play a critical role in innate immunity and LPS/TLR4 signaling is well-known to induce pro-inflammatory macrophages with cytokine secretion, ROS production, as well as a glycolytic metabolic switch to sustain the inflammatory status (Mills et al., 2016). Although mechanisms that block continuous LPS effects (e.g., endotoxin tolerance) have been characterized (Collins and Carmody, 2015), it is not clear how macrophages restrict initial LPS/TLR4 signaling.
The Na/K-ATPase (NKA) is an integral membrane protein formed by the assembly of α and β subunits in a 1:1 ratio (Lingrel and Kuntzweiler, 1994). It is an ion transporter critical for maintaining cellular homeostasis and also functions as a signal transducer (Xie and Cai, 2003). NKA’s α subunit contains large cytosolic domains that directly interact with many signaling proteins including Src (Tian et al., 2006), Lyn (Chen et al., 2015), caveolin-1 (Wang et al., 2004), PI3K (Yudowski et al., 2000), PLC-γ1 (Yuan et al., 2005), and cytoskeleton-associated proteins (Dash et al., 2018; Devarajan et al., 1994). The interaction of NKA with its binding partners is regulated by various extracellular ligands for distinct cellular functions and its role in macrophage inflammatory responses has been reported (Chen et al., 2015, 2017).
We previously showed that cardiotonic steroids, which are canonical ligands for NKA, stimulated a signaling complex consisting of NKA α1, TLR4, and Lyn in macrophages, leading to pro-inflammatory cytokine production (Chen et al., 2017). Therefore, we hypothesized that NKA α1 may also contribute to LPS/TLR4 signaling. Here, we present evidence that macrophages with reduced NKA α1 were hypersensitive to LPS as they displayed enhanced TLR4 signaling, NF-κB activation, pro-inflammatory cytokine production, ROS generation, and glycolytic switch. Bulk RNA sequencing analysis further revealed a profound and massive signaling and metabolism alteration introduced by the reduction in NKA α1. This phenomenon was explored in vivo by LPS intraperitoneal (IP) injection of mice partially deficient in NKA α1, which showed higher cytokine production in the blood as well as tissues, more severe lung injury, and significantly lower survival rate compared to their littermate WT control. Mechanistically, NKA α1 binds both TLR4 and Lyn and suppresses Lyn activation (Plociennikowska et al., 2015). The presence of NKA in the TLR4 signaling complex thus sequesters Lyn and limits the initial macrophage pro-inflammatory responses, facilitating tissue homeostasis.
Results
Na/K-ATPase haploinsufficiency in macrophages leads to enhanced inflammatory activation by LPS
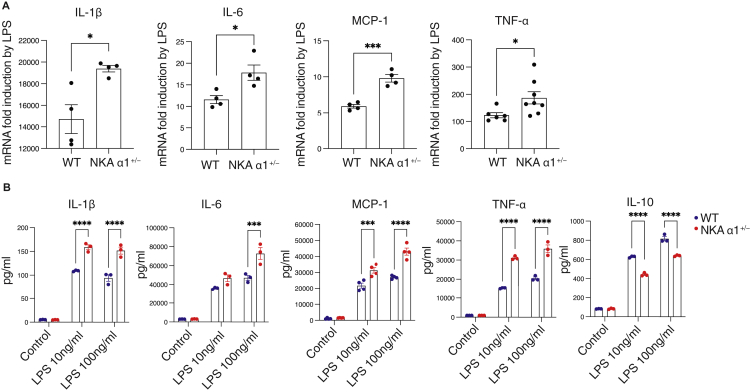
Peritoneal macrophages from atp1a1 haploinsufficient mice (NKA α1+/−) expressed approximately 45% of the NKA α1 protein levels compared to WT cells (Figure S1 and (Chen et al., 2015)). Compared to WT cells, LPS stimulation of NKA α1+/− macrophages induced significantly greater increases in transcripts of IL-1β, IL-6, MCP-1, and TNF-α (Figure 1A). Cytokine protein levels in culture media from LPS-treated NKA α1+/− macrophages were also 55-80% higher (Figure 1B). Conversely, secretion of the anti-inflammatory cytokine IL-10 was attenuated by ∼30% (Figure 1B). In an in vivo study, WT or NKA α1+/− mice were IP injected with LPS (15 mg/kg body weight); a concentration known to stimulate the production of cytokines, tissue damage, multiorgan dysfunction, and death (Alexander and Rietschel, 2001; Cohen, 2002; Engelberts et al., 1991; Liu et al., 2014). Although basal plasma cytokine levels were low with no obvious difference between the two genotypes, NKA α1+/− mice displayed 2.2- to 3.2-fold higher plasma levels of IL-1β, IL-6, and MCP-1 compared to WT littermates (Figure 2A). Furthermore, NKA α1+/− mice had significantly decreased survival rates compared to WT; 26% 7-day survival vs 61% (Figure 2B). To determine the role of monocytes/macrophages in this hyper-inflammatory phenotype, mice were injected with clodronate liposomes to selectively deplete resident macrophages (Biewenga et al., 1995; van Rooijen et al., 1996; van Rooijen and van Kesteren-Hendrikx, 2002) prior to LPS injection. As shown in Figure 2B, macrophage depletion improved 7 days survival rate of NKA α1+/− mice from 26% to 90%, suggesting that macrophages are responsible for the NKA haploinsufficiency-induced hyper-inflammatory response.
Figure 1.
NKA haploinsufficiency enhanced the production of pro-inflammatory cytokines induced by LPS
(A) WT and NKA α1+/− peritoneal macrophages were treated with 100 ng/mL LPS for 6 h. Total RNA was isolated and subjected to qRT-PCR.
(B) WT and NKA α1+/− peritoneal macrophages were treated with LPS for 24 h. Culture medium was collected and subjected to ELISA assay. The data are presented as mean ± S.E. n = 3–4 separate experiments. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 determined by Student’s t test or two-way ANOVA analysis.
Figure 2.
NKA haploinsufficiency in mice enhances systemic inflammation, lung injuries, and lethality induced by LPS
15 mg/kg body weight LPS or vehicle PBS (control) was IP injected into WT and NKA α1+/− mice.
(A) 24h after injection, plasma cytokines were measured by ELISA. n = 10–18 mice per group. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 determined by two-way ANOVA analysis.
(B) WT and NKA α1+/− mice were IP injected with LPS alone or together with clodronate liposomes (50μg/mouse) or NAC (150 mg/kg body weight). Mice survival rates were determined. The survival analysis was performed by the Kaplan-Meier method and statistical analysis was performed by the log-rank test. n = 10–18 mice for each group. ∗p < 0.05 compared with WT + LPS group; ##p < 0.01 compared with NKA α1+/− +LPS group.
(C) Lung injury was assessed by H&E staining and histopathological examination 16h after injection. Representative images of lung sections were shown under 20X magnification. Black arrows point to severe alveolar hemorrhage. Scale bar, 75 μm n = 3 mice for each group.
(D) Lung IL-1β, TNF-α, and MCP-1 mRNA expression were determined by qRT-PCR. n = 8–9 mice for each group. ∗∗p < 0.01 determined by Student’s t test.
(E) Lung tissue cells were examined by flow cytometry 16h after injection. The percentage of macrophages/monocytes and (F) neutrophils among CD45+ leukocytes were shown. The quantitative data are presented as mean ± S.E. n = 6 mice for each group. ∗p < 0.05, ∗∗p < 0.01 determined by two-way ANOVA analysis. See also Figure S2.
LPS/TLR4 signaling causes acute lung injury which contributes to mortality (Huang et al., 2018; Imai et al., 2008). Consistent with published studies, histopathological analysis of lung tissues revealed immune cell infiltration, alveolar hemorrhage, edema, and alveolar wall thickening in LPS-injected WT mice compared with PBS-injected WT mice (Figure 2C) (Hu et al., 2017). Lungs from LPS-injected NKA α1+/− mice showed more severe inflammation and structural damage compared with LPS-injected WT mice (Figure 2C). Consistent with previous reports that IL-1β contributed to acute lung injury (Kolb et al., 2001), significantly higher levels of IL-1β mRNA were seen in LPS-treated NKA α1+/− mice lung tissues compared with WT, although no significant difference was observed in TNF-α and MCP-1 (Figure 2D). As expected, LPS increased the percentage of macrophages (CD45+F4/80+Ly6C−Ly6Glo) and monocytes (CD45+F4/80loLy6C+Ly6Glo) in WT mice. A slightly higher macrophage/monocyte percentage was seen in NKA α1+/− mice compared with WT mice (Figure 2E). These results further support an inhibitory role for NKA in LPS-induced inflammation and lung injury.
No significant differences in circulating blood cell counts or immune cell subset distributions in spleen were seen after LPS challenge in NKA α1+/− mice compared to WT (Figures S2A and S2B) and no significant changes in lung neutrophil levels (CD45+F4/80-Ly6Ghigh) were observed (Figure 2F) possibly related to the time at which the studies were performed (16h after injection), as neutrophils are normally attracted early and quickly cleared by surrounding macrophages.
Na/K-ATPase haploinsufficiency enhances LPS-mediated TLR4 and NF-κB signaling
Bulk RNA sequencing analyses (RNA-seq) were performed on WT and NKA α1+/− macrophages treated with LPS to comprehensively examine transcriptomics alteration. Gene Set Enrichment Analysis (GSEA) indicated that gene transcripts of components of multiple signaling pathways, many of which are pro-inflammatory, were significantly upregulated by NKA haploinsufficiency (Table S1). As MAPKs and AKT are downstream of LPS/TLR4 signaling (McGuire et al., 2013), we assessed their activation status by immunoblot using antibodies specific for their active phosphorylated states. LPS stimulated time-dependent phosphorylation of AKT (Ser473), plateauing at 3h in WT macrophages. In NKA α1+/− macrophages, AKT phosphorylation increased by 2∼3-fold more between 2h and 6h in response to LPS (Figure S3A). However, no obvious difference was detected in the LPS-stimulated ERK1/2 pathway (Figure S3B).
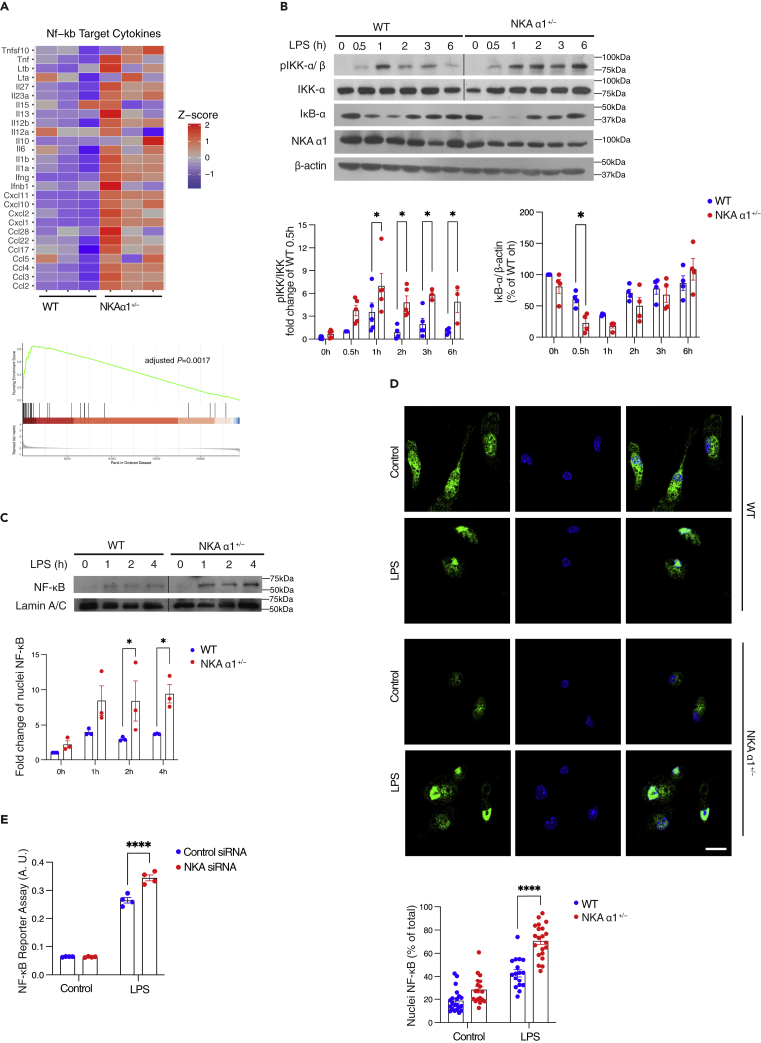
NF-κB is a critical downstream effector of LPS/TLR4 inflammatory responses (Bonizzi and Karin, 2004). GSEA of NF-κB target genes showed higher levels of pro-inflammatory cytokine gene expression in NKA α1+/− macrophages after LPS stimulation (Figure 3A), in agreement with the qRT-PCR and ELISA data shown in Figure 1. To examine NF-κB activation pathways, we assessed IκB kinase (IKK) activation as it is known to be downstream of LPS/TLR4. Immunoblots for phospho-IKKα/β (pIKK-α/β), the active form of IKK, showed that LPS induced up to 3-fold higher levels of phosphorylation in NKA α1+/− macrophages compared to WT. Furthermore, IKK phosphorylation was prolonged for at least 6h in the NKA α1+/− cells (Figure 3B). However, proteins involved in the LPS/TLR4 signaling pathway such as TLR4, MyD88, IKKα, IκB-α, and NF-κB (p65) showed no difference between WT and NKA α1+/− macrophages (Figure S1), indicating that the hypersensitivity to LPS in NKA α1+/− macrophages was not simply owing to elevated expression in LPS/TLR4 signaling proteins. Degradation of IκB-α, the substrate of IKK, was also enhanced between 0.5 and 2h in the NKA α1+/− cells (Figure 3B). Degradation of IκB-α initiates nuclear translocation of NF-κB (p65) (Baker et al., 2011; de Winther et al., 2005), so nuclear fractions from both WT and NKA α1+/− macrophages treated with LPS were isolated by centrifugation and immunoblotted for NF-κB. Figure 3C shows approximately 2-fold higher levels of nuclear NF-κB in NKA α1+/− macrophages. Immunofluorescence microscopy using anti-NF-κB (green) and DAPI (blue) showed a higher percentage of nuclei containing NF-κB (70.5 ± 3.2%) in NKA α1+/− macrophages compared to WT macrophages (42.9 ± 3.2%) after LPS exposure (Figure 3D), confirming the cell fractionation data. NF-κB transcriptional activity was then measured directly using a reporter system, THP-1 Dual cells which are derived from human THP-1 monocyte cell line by integration of NF-κB-inducible reporter construct (Chen et al., 2017; Dey et al., 2015). NKA α1 was specifically knocked down by 80% in these cells using siRNA-based techniques (Figure S4). Consistent with the increased LPS signaling seen in NKA α1+/− macrophages, we observed statistically significant higher NF-κB transcriptional activity in response to LPS compared to control siRNA transfected cells (Figure 3E). Taken together, these data suggest that NKA negatively regulates NF-κB activation, trafficking, and transcriptional activity induced by LPS.
Figure 3.
NKA haploinsufficiency leads to hyperactivation of NF-κB signaling induced by LPS
(A) Heatmap based on bulk RNA-seq Z score (upper panel) and Gene Set Enrichment Analysis (GSEA) (lower panel) on NF-κB target genes were shown from LPS-treated WT and NKA α1+/− peritoneal macrophages (100 ng/mL, 6h).
(B) WT and NKA α1+/− peritoneal macrophages were treated with 100 ng/mL LPS for the indicated time. Representative Western blot images of phosphorylated Ser176/180 IKK, IKK, IκB-α, and NKA α1 and β-actin were shown. Quantified data from 3 to 4 separate experiments were shown below.
(C) Representative Western blot images of NF-κB and Lamin A/C from nuclear fractions were shown.
(D) Representative fluorescent images for cellular distribution of NF-κB (green) are shown after 30min LPS treatment. Nuclei were stained with DAPI (blue). Scale bar = 10 μm. The ratio between nuclear and total NF-κB signals was calculated and shown in the dot plots below.
(E) THP-1 Dual cells transfected with control or NKA α1 siRNA for 48 h were treated with 100 ng/mL LPS for additional 24h. The medium was collected for NF-κB reporter assay. A.U. arbitrary unit. The quantitative data are presented as mean ± S.E. n = 3–5 separate experiments. ∗p < 0.05, ∗∗∗∗p < 0.0001 compared with WT + LPS group. Significance was determined by two-way ANOVA analysis. See also Table S1, Figures S1, S3, and S4.
LPS-induced ROS production contributes to macrophage pro-inflammatory responses and enhances NF-κB nuclear translocation (Gloire and Piette, 2009). NKA signaling also increases ROS generation in cardiac myocytes and renal proximal tubule cell lines (Pratt et al., 2018). We hypothesized that NKA regulates inflammatory signaling events through ROS production. Macrophage ROS levels were measured with a widely used fluorescent redox probe, carboxy-DCFDA (Crow, 1997). As shown in Figure 4A, LPS increased DCFDA fluorescence in both WT and NKA α1+/− macrophages, and NKA α1+/− cells showed ∼30% more fluorescence. To test whether mitochondria contribute to LPS-induced ROS generation, the mitochondria-targeted fluorescent superoxide probe, MitoNeoD (Shchepinova et al., 2017), was added to the cells before LPS stimulation. No stimulation was detected in any of the conditions (Figure 4A), suggesting that NKA-mediated enhanced ROS generation in response to LPS was not dependent on mitochondria. We, therefore, probed the role of the NADPH oxidase (NOX) pathway, another major source of cellular ROS (Bedard and Krause, 2007), using a cytoplasmic ROS inhibitor, N-acetyl cysteine (NAC) (Ezeriņa et al., 2018) and a NOX2 inhibitor, apocynin that blocks p47phox recruitment to membrane NOX2 complex (Touyz, 2008). Figure 4B shows that apocynin and NAC decreased the LPS-induced IL-1β, IL-6, and TNF-α production more in NKA α1+/− macrophages. As ROS are known to regulate NF-κB (Gloire and Piette, 2009), we isolated nuclear fractions and found that LPS-induced nuclear NF-κB translocation was attenuated by NAC in NKA α1+/− macrophages (Figure 4C). However, NAC did not affect nuclear NF-κB levels in WT macrophages, suggesting that pro-inflammatory signaling is more sensitive to ROS in NKA α1+/− macrophages. To further investigate the mechanism of NKA regulation on NOX2-derived ROS, we measured Rac GTPases (Rac1/⅔), which are activators of NOX2 (Heyworth et al., 1993; Hordijk, 2006). In TLR4 immunoprecipitates, significantly higher levels of Rac GTPases were recruited to TLR4 by LPS in NKA α1+/− macrophages compared to WT (Figure 4D). Consistent with the finding that LPS leads to enhanced oxidative stress in NKA α1+/− macrophages, we found through GSEA analysis that anti-oxidant gene transcripts were elevated in LPS-treated NKA α1+/− macrophages compared with WT (Figure S5). In vivo relevance of these data was demonstrated using the LPS injection model described in Figure 2 in WT and NKA+/− mice pre-treated with IP injection of NAC. Survival was significantly increased in NKA α1+/− groups (Figure 2B) and the enhanced plasma MCP-1 level seen in the LPS-treated NKA α1+/− mice was completely blocked by NAC pretreatment (Figure 4E). Taken together, these data suggest that NKA haploinsufficiency promoted LPS-induced inflammatory responses through enhanced NOX2-dependent ROS generation regulated by the upstream NKA/TLR4 complex.
Figure 4.
NKA haploinsufficiency enhanced LPS-induced inflammatory response through ROS signaling
(A) LPS-treated WT and NKA α1+/− peritoneal macrophages (100 ng/mL, 5 min) were incubated with carboxy-DCFDA (left) or MitoNeoD (Right). Mean fluorescence intensity (MFI) was quantified and shown in the bar graph.
(B) WT and NKA α1+/− peritoneal macrophages were treated with LPS (100 ng/mL) and apocynin (300 uM)/NAC (5mM) for 6 h. Culture media were collected and subjected to ELISA assay for IL-6, TNF-α, and IL-1β.
(C) Nuclear fractions were extracted and immunoblotted for NF-κB and Lamin A/C. Representative Western blot images were shown with quantified data in bar graphs below. The lanes were run on different gels.
(D) TLR4 precipitates or total cell lysates (Input) were immunoblotted for Rac1/2/3 and TLR4. Representative blots are shown with quantified data in bar graphs later below. The lanes were run on the same gel but were noncontiguous.
(E) WT and NKA α1+/− mice were IP injected with LPS alone or together with NAC (150 mg/kg body weight). After 24h, blood plasma MCP-1 was measured by ELISA assay. n = 6–18 mice for each group. The quantitative data are presented as mean ± S.E. n = 3–5 separate experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, determined by two-way ANOVA analysis. See also Figure S5.
Na/K-ATPase haploinsufficiency promotes glycolytic switch induced by LPS
Emerging evidence suggests that pro-inflammatory macrophage activation is accompanied by and dependent on a metabolic switch from oxidative phosphorylation (OXPHOS) to glycolysis (Palsson-McDermott et al., 2015). Analysis of the transcriptome from LPS-treated WT and NKA α1+/− macrophages showed that glycolysis pathway transcripts were significantly higher in NKA α1+/− macrophages compared to WT (Figure 5A). In contrast, most transcripts of genes in the TCA cycle were not significantly changed (Figure 5B). We, therefore, conducted metabolic assays using the Seahorse extracellular flux system in WT and NKA α1+/− macrophages. Consistent with the RNA-seq results, NKA α1+/− macrophages showed a 50% higher extracellular acidification rate (ECAR), which is indicative of increased glycolysis (Figure 5C) along with partial suppression of oxygen consumption rate (OCR) which is indicative of suppressed OXPHOS (Figure 5D), confirming a metabolic switch. These results suggest that NKA suppresses metabolic reprogramming induced by LPS.
Figure 5.
NKA haploinsufficiency promotes glycolytic switch induced by LPS
Heatmap based on bulk RNA-seq Z score (upper panel) and Gene Set Enrichment Analysis (GSEA) (lower panel) on (A) glycolysis and (B) TCA cycle genes were shown from LPS-treated WT and NKA α1+/− peritoneal macrophages (100 ng/mL, 6h).
(C) Representative glycolysis stress tests and (D) mito stress test curves from 24h LPS-treated WT and NKA α1+/− peritoneal macrophages were shown. Right, maximum ECAR and OCR were quantified and shown in bar graphs; n = 4 separate experiments. ∗∗p < 0.01, compared with WT + LPS group determined by Student’s t test.
Na/K-ATPase restricts LPS/TLR4 pro-inflammatory signaling through Lyn inhibition
Under basal conditions, the α1 chain of NKA interacts directly with Src-family kinases (SFK), including Lyn, holding them in an inactive state. Upon interaction with its ligands, a conformational change ensues, releasing the SFK which leads to autophosphorylation and activation (Chen et al., 2015; Haas et al., 2002; Lai et al., 2013; Yu et al., 2018; Zhang et al., 2020). SFK, especially Lyn, facilitates TLR4 signaling by being directly recruited to and phosphorylating TLR4 (Plociennikowska et al., 2015). We thus hypothesized that NKA-mediated downregulation of LPS signaling could be owing to suppressed Lyn activation within the NKA/TLR4 complex. WT peritoneal macrophages were, therefore, exposed to LPS at different times and TLR4 was immunoprecipitated (IP). Lyn activation (phosphorylation at Tyr396 or pY396-Lyn) was determined by immunoblot. The amount of NKA α1 co-precipitated with TLR4 was also quantified by immunoblot. Figure 6A shows that both NKA α1 and pY396-Lyn were precipitated by anti-TLR4 under basal conditions, suggesting tri-molecular complex formation. Although LPS treatment decreased the amount of NKA α1 in the complex by ∼70%, the amount of pY396-Lyn was increased to ∼2.5-fold. In total cell lysates, LPS stimulated pY396-Lyn for up to 30 min. To validate this finding, a reciprocal IP was performed with anti-NKA α1 followed by immunoblots for TLR4 and pY396-Lyn. Both TLR4 and pY396-Lyn were co-precipitated by anti-NKA α1 and LPS treatment led to a ∼30% reduction of TLR4 in the precipitates along with ∼1.7-fold increased pY396-Lyn (Figure 6B). We then compared pY396-Lyn in anti-TLR4 precipitates from LPS-treated WT and NKA α1+/− macrophages and found that NKA haploinsufficiency further increased the amount of pY396-Lyn associated with TLR4 and in the total cell lysates (Figure 6C). These data support our proposition that NKA restricts Lyn phosphorylation within the TLR4 signaling complex, perhaps by sequestering it in an inactive state. As a validation of the critical role of Lyn in LPS-induced inflammatory responses, inhibition of SFK activity with the pan-Src inhibitor PP2 suppressed LPS-induced production of IL-6, MCP-1, and TNF-α in both mouse peritoneal macrophages and human monocyte-derived macrophages (hMDM) (Figures 6D and 6E).
Figure 6.
NKA restricts LPS/TLR4 signaling by suppressing Lyn
WT and NKA α1+/− peritoneal macrophages were treated with 100 ng/mL LPS for 15min or other indicated times.
(A) TLR4 precipitates or total cell lysates (input) were immunoblotted for NKA α1, pTyr396-Lyn, Lyn, and TLR4. Representative blot images are shown. Quantified data were shown in bar graphs on the right.
(B) NKA α1 precipitates were immunoblotted for TLR4, pTyr396-Lyn, and NKA α1. Representative blot images are shown with quantified data in bar graphs.
(C) TLR4 precipitates or total cell lysates (input) from WT and NKA α1+/− peritoneal macrophages were immunoblotted for pTyr396-Lyn, Lyn, and TLR4. Representative blots are shown with quantified data in the bar graph. The lanes were run on the same gel but were noncontiguous. Culture media from (D) WT mouse peritoneal macrophages or (E) human monocyte-derived macrophages (hMDMs) pre-treated with 10 μM PP2 for 1h followed with 100 ng/mL LPS for 6h were subjected to ELISA assay for MCP-1, IL-6, and TNF-α. The quantitative data are presented as mean ± S.E. n = 3–4 separate experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 determined by one-way or two-way ANOVA analysis.
Discussion
In this study, we report that genetic knockdown of NKA α1 expression led to hyper-inflammatory responses to LPS in macrophages in vitro and in vivo, consistent with a role for NKA in suppressing LPS signaling. We further demonstrate that NKA α1 interacts with TLR4 and suppresses activation of the SFK Lyn, which is essential for LPS-induced pro-inflammatory signaling. As our previous studies as well as others have shown that SFK contribute to the activation of NF-κB pathway and NOX-dependent ROS production in response to various ligands including LPS (Byeon et al., 2012; Check et al., 2010; Kennedy et al., 2013; Lee et al., 2007; Yi et al., 2021), we propose here a novel role of NKA as a negative regulator of LPS/TLR4 signaling potentially by regulating SFK activity.
Although originally discovered as a cation transporter (Skou, 1957), NKA was shown to function as a signal transducer by directly regulating SFK and their downstream effectors including MAP kinases, PI3K pathways, caveolin-1, Ca2+ signaling, and ROS generation (Chen et al., 2015; Haas et al., 2002; Wang et al., 2004; Yuan et al., 2005; Zhang et al., 2020). More specifically, NKA α1 cytosolic domain directly interacts with SFK kinase domain and prevents SFK activation (Tian et al., 2006). Ligand binding (e.g. ouabain) to NKA α1 (Tian et al., 2006) or to its signaling partners (e.g. oxLDL binding to CD36 (Chen et al., 2015)) leads to a conformational change in the NKA α1 cytosolic domain that releases the SFK kinase domain for activation. Therefore, the amount of NKA α1 available to engage with the signaling complex titrates the amount of SFK that can be activated. Consistent with this model, we found that LPS reduced TLR4/NKA α1 complex formation and increased TLR4-associated pY396-Lyn (Figure 6). In NKA α1 haploinsufficient cells TLR4/NKA-α1 complex formation was reduced even more and TLR4-associated pY396-Lyn was further increased, leading to excess inflammation.
We have previously shown that NKA is involved in macrophage inflammatory responses independent of its ion transporter function (Chen et al., 2017). Binding of NKA to its specific ligand ouabain activates the NF-κB pathway through a signaling complex including NKA, TLR4, and CD36. Subsequently, macrophages respond by increasing the production and secretion of pro-inflammatory cytokines including IL-1β, MCP-1, IL-6, and TNF-α. The notion that NKA contributes to macrophage immune regulation is further supported by our finding that NKA/CD36 complex critically regulates pro-inflammatory signaling cross-talk between renal cells and macrophages, which facilitates renal injury and kidney diseases (Kennedy et al., 2013). Our follow-up study also indicates that NKA/CD36 complex promotes macrophage conversion to lipid-laden foam cells under atherosclerosis conditions (Chen et al., 2015). Thus, macrophage NKA appears to participate in the regulation of both immune signaling and metabolism in response to various extracellular stimulants. Nevertheless, while our previous studies showed a positive role of NKA in macrophage immune activation as induced by ouabain and oxidized LDL (Chen et al., 2015, 2017), this study shows that macrophage NKA can also play an inhibitory role in response to LPS. One major difference here is that LPS decreased interaction between NKA and TLR4 (Figure 6), while ouabain or oxidized LDL increased interaction between NKA and its partners. This may be one of the mechanisms through which macrophages are differentially activated in response to distinct extracellular ligands.
Besides the signaling events mentioned above, another interesting finding here is that NKA α1 level regulates the macrophage glycolytic switch (Figure 5). LPS induces a macrophage glycolytic switch, which not only provides ATP for energy consumption but also metabolic intermediates for de novo synthesis (O'Neill and Hardie, 2013). Moreover, some metabolites such as succinate accumulate intracellularly during the glycolytic switch and sustain pro-inflammatory responses as a second messenger (Tannahill et al., 2013). Therefore, evidence strongly indicates that macrophage metabolisms are coupled with and even determine their activation status. Here we show that reduced NKA α1 expression sensitizes macrophages to LPS-induced glycolytic switch, possibly through hyperactivation of NF-κB. However, it is not clear whether NKA-mediated mechanisms are involved, which warrants further investigation.
We have explored the physiological relevance by injecting LPS to WT and NKA α1+/− mice to mimic the condition of acute sepsis. Consistent with our in vitro findings, NKA α1+/− mice are hypersensitive to LPS showing significantly higher plasma pro-inflammatory cytokines with lower survival rate (Figure 2). When we eliminated macrophages in mice, we found both WT and NKA α1+/− mice are protected from LPS and there was no difference in survival rate, which supports the critical role of NKA in macrophages during acute inflammation. Moreover, reduced NKA expression levels were reported from human patient tissue samples of various diseases including sepsis (Maslove and Wong, 2014), chronic kidney disease (Nakagawa et al., 2015), Crohn’s disease (Haberman et al., 2014), Alzheimer’s disease (Liang et al., 2008), breast cancer (Best et al., 2015), and ovarian cancer (Yeung et al., 2013). It would be interesting to explore whether dysregulated immune responses owing to NKA reduction may play a role in those diseases.
Taken together, we have defined a novel mechanistic connection between NKA and LPS/TLR4 signaling. Because NKA antagonists, such as pNaKitde (Li et al., 2009) and rostafuroxin (Ferrari, 2010), have been developed. Our previous work has demonstrated that pNaKtide, by blocking NKA-mediated oxidative stress, effectively ameliorates various metabolic symptoms such as obesity (Sodhi et al., 2015), and uremic cardiomyopathy (Sodhi et al., 2020). It will be interesting to test whether they are effective against sepsis in vivo and therefore provide a novel therapeutic strategy.
Limitations of the study
In this study, we showed NKA suppressed pro-inflammatory response through a member of SFK, Lyn. However, we cannot exclude the potential contribution of other members of SFK as the phospho-Lyn antibody (pY396-Lyn) may cross-react with other activated SFK. In addition, the mechanism of how SFK promotes LPS-induced ROS and NF-κB pathway is still not well understood, which is beyond the scope of this work and should be explored by future studies. Although we provide aligned raw counts and normalized counts for the whole macrophage transcriptomics in Table S2, the pre-processed raw RNA-sequencing data are unfortunately not available owing to a technical mistake during data storage and maintenance.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IκB-α Rabbit Polyclonal Antibody | Cell Signaling Technology | Cat#9242; RRID: AB_331623 |
| NF-κB p65 (C22B4) Rabbit Monoclonal Antibody | Cell Signaling Technology | Cat#4764; RRID: AB_823578 |
| Phospho-p44/42 MAPK (pErk1/2) (Thr202/Tyr204) Rabbit Polyclonal Antibody | Cell Signaling Technology | Cat#9101; RRID: AB_331646 |
| Phospho-Akt (Ser473) Rabbit Monoclonal Antibody | Cell Signaling Technology | Cat#4060; RRID: AB_2315049 |
| p44/42 MAPK (Erk1/2) Rabbit Polyclonal Antibody | Cell Signaling Technology | Cat#9102; RRID: AB_330744 |
| Akt (pan) (40D4) Mouse Monoclonal Antibody | Cell Signaling Technology | Cat#2920; RRID: AB_1147620 |
| Lyn Rabbit Polyclonal Antibody | Cell Signaling Technology | Cat#2732; RRID: AB_10694080 |
| MyD88 (D80F5) Rabbit Monoclonal Antibody | Cell Signaling Technology | Cat#4283; RRID: AB_10547882 |
| Rac1/⅔ Antibody Rabbit Polyclonal Antibody | Cell Signaling Technology | Cat#2465; RRID: AB_2176152 |
| Phospho-IKKα/β (Ser176/180) Rabbit Monoclonal Antibody | Cell Signaling Technology | Cat#2697; RRID: AB_2079382 |
| IKKα (D3W6N) Rabbit Monoclonal Antibody | Cell Signaling Technology | Cat#61294; RRID: AB_2799606 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID: AB_2099233 |
| SRC Family (phospho Y418) Rabbit monoclonal antibody | Abcam | Cat#ab40660; RRID: AB_776106 |
| β-Actin Mouse Monoclonal Antibody | Sigma-Aldrich | Cat#A5316; RRID: AB_476743 |
| Lamin A/C (4C11) Mouse Monoclonal Antibody | Cell Signaling Technology | Cat#4777; RRID: AB_10545756 |
| NKA-α1 α6F Mouse Monoclonal Antibody | Developmental Studies Hybridoma Bank | Cat#α6F |
| NKA-α1 (C464.6) Mouse Monoclonal Antibody | Santa Cruz | Cat#sc-21712; RRID: AB_626713 |
| TLR4 Mouse Monoclonal Antibody | Santa Cruz | Cat#sc-293072; RRID: AB_10611320 |
| Goat anti-Mouse IgG-HRP | Santa Cruz | Cat#sc-2005; RRID: AB_631736 |
| Donkey anti-Rabbit IgG Secondary Antibody, Alexa Fluor™ 488 | Thermo Fisher Scientific | Cat#A21206; RRID: AB_2535792 |
| FITC anti-mouse CD45.2 Monoclonal Antibody | Biolegend | Cat#109805; RRID: AB_313442 |
| PE/Cyanine5 anti-mouse F4/80 Monoclonal Antibody | Biolegend | Cat#123111; RRID: AB_893494 |
| APC anti-mouse Ly-6C Monoclonal Antibody | Biolegend | Cat#128016; RRID: AB_1732076 |
| PE anti-mouse Ly-6G Monoclonal Antibody | Biolegend | Cat#127607; RRID: AB_1186104 |
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharides from Escherichia coli O 55:B5 | Sigma-Aldrich | Cat#L2880; EC: 297-473-0 |
| PP2 | Cayman chemical | Cat#13198; CAS: 172889-27-9 |
| N-acetylcysteine | Cayman chemical | Cat#20261; CAS: 616-91-1 |
| Apocynin | Cayman chemical | Cat#11976; CAS: 498-02-2 |
| Thioglycollate medium | Sigma | Cat#T9032 |
| RPMI media | Corning | REF#10-040-CV |
| X-VIVO™ 10 Hematopoietic Serum-Free Culture Media | Thermo Fisher Scientific | Cat#04-380Q |
| Human Serum | Sigma | Cat#H3667; MDL: MFCD00165829 |
| CM-H2DCFDA | Thermo Fisher Scientific | C6827 |
| MitoNeoD | Chen et al. (2019); Shchepinova et al. (2017) | N/A |
| Blotting-Grade Blocker | Bio-rad | Cat#1706404 |
| Viromer Blue transfection reagent | Origene | Cat#TT100300 |
| Critical commercial assays | ||
| Mouse CCL2 ELISA kit | Invitrogen | REF#88-7391-88 |
| Mouse IL6 ELISA kit | Invitrogen | REF#88-7064-88 |
| Mouse TNF alpha ELISA kit | Invitrogen | REF#88-7324-88 |
| Mouse IL10 ELISA kit | Invitrogen | REF#88-7105-88 |
| Mouse IL-1 beta ELISA kit | Invitrogen | REF#88-7013-88 |
| Standard Macrophage Depletion Kit (Clodrosome® + Encapsome®) | Encapsula NanoSciences | SKU#CLD-8901 |
| CelLytic M Cell Lysis Reagent | Sigma | Cat#C2978 |
| Pierce™ Protein A/G Agarose | Thermo Fisher Scientific | Cat#20421 |
| Chemiluminescent substrate | Thermo Fisher Scientific | REF#34580 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368813 |
| Deposited data | ||
| RNA sequencing data | This paper | https://data.mendeley.com/datasets/vxyjg9m7nm/1 |
| Experimental models: Cell lines | ||
| Murine Peritoneal macrophages | In-house | N/A |
| Human monocyte-derived macrophages | In-house | N/A |
| THP-1 Dual cell line | Gift from Dr. Kathryn J. Moore (New York University) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 mice | Charles River/NCI Research | STRAIN CODE: 556 |
| Mouse: Atp1a1 haplodefienct mice (NKA α1+/−) | James et al. (1999) | N/A |
| Oligonucleotides | ||
| NKA α1 siRNA (h) | Santa Cruz | Sc-36010 |
| Primer Tnf: 5′-TCTTTGAGATCCATGCC GTTG-3′; 5′-AGACCCTCACACTCAGATCA-3′ |
IDT DNA Technologies | N/A |
| Primer ccl2: 5′-AACTACAGCTTCTTT GGGACA-3′; 5′-CATCCACGTGTTGGCTCA-3′ |
IDT DNA Technologies | N/A |
| Primer IL1B: 5′-TTGAAGTTGACGGACC CCAAA-3′; 5′-TGTGCTGCTGCGAGATTTGAA-3′ |
IDT DNA Technologies | N/A |
| Primer ACTB: 5′-GGCTGTATTCCCCTC CATCG-3′; 5′-CCAGTTGGTAACAATGCCATGT-3′ |
IDT DNA Technologies | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software | Version 9.1.1 (223) |
| ImageJ | open-source image analysis software | 1.53 version |
| FlowJo | FlowJo Software | Version 10.8.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Yiliang Chen (yilchen@mcw.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines and cell culture
Peritoneal macrophages were harvested by lavage from mice of both sex injected with 2mL intraperitoneal (IP) administration of 4% thioglycolate (Sigma, Cat# T9032) 4 days prior to collection (Febbraio et al., 2000). Collected cells were counted, centrifuged at 250g for 5min, resuspended in RPMI media (Gibco by Life Technologies) supplemented with 10% FBS, 100U/mL penicillin and 100 μg/mL streptomycin (Sigma) and seeded into culture dishes or plates for further analysis. Cells were maintained at 37°C in a humidified incubator set at 5% CO2. Human monocyte-derived macrophages (HMDMs) were generated from monocytes isolated from human buffy coats by Ficoll density centrifugation and then differentiated over 7 days in vitro. (Chen et al., 2017; Johnson et al., 1977). HMDMs were maintained in X-Vivo 10 hematopoietic cell medium (Lonza, Cat# 04-380Q) containing 5% human serum (Sigma, Cat# H3667) and 1% penicillin/streptomycin. The cells were serum-starved overnight and then treated with LPS diluted in serum-free medium. LPS was purified from Escherichia coli Gram-negative bacteria consisting of a lipid A moiety linked to an antigenic O-polysaccharide (Smooth-form) (Sigma, Cat#L2880). The THP-1 Dual cell line was a kind gift from Dr. Kathryn J. Moore (New York University).
Experimental animals
Atp1a1 haploinsufficient mice (NKA α1+/−) were generated (James et al., 1999) and backcrossed with C57BL/6 mice (Jackson Laboratory) for more than 10 generations. NKA α1+/− mice are fertile with no obvious growth defect or health issues and are born with normal 1:1 female-to-male ratios (James et al., 1999). Mice of both sexes at 8 to 16 weeks of age were used to isolate peritoneal macrophages or other experiments. Littermate WT mice were used as control to compare with NKA α1+/− mice. All mice were kept in a 12-h dark/light cycle and fed standard chow ad libitum unless indicated otherwise. Animal handling followed AALAC and National Institutes of Health guidelines, and all procedures involving live animals were approved by the Institutional Animal Care and Use Committee at Medical College of Wisconsin.
Mouse in vivo LPS challenge
Mice of both sex were intraperitoneally (IP) injected with LPS (15 mg/kg body weight) or PBS (vehicle) and monitored every 12 h for 7 days for survival. Data was analyzed by the Kaplan Meier method and the log rank test was used to compare survival distributions and test differences between independent experimental groups. In some studies mice were IP injected with Clodronate liposomes (50μg per mouse) 2 days before LPS injection to deplete tissue resident macrophages (Godwin et al., 2013; van Rooijen and Hendrikx, 2010), or NAC (150 mg/kg body weight) one day before LPS injection following with repeated NAC injection for another 2 days.
Method details
Immunoprecipitation and immunoblot analysis
Cells were washed with PBS and lysed in ice-cold CelLytic M Cell Lysis Reagent (Sigma, Cat# C2978) with protease inhibitor cocktail (Roche) and phosphatase inhibitors (Sigma). For immunoprecipitation assays, the lysates were pre-cleared with 25 μL of A/G agarose beads (Thermo Scientific, Cat# 20,421) at 4°C for 1h and the supernatants containing ∼1 mg protein were incubated with 2 μg primary antibody overnight at 4°C; 25 μL of A/G agarose beads was then added and incubated overnight at 4°C. The beads were then washed with lysis buffer, boiled in SDS-PAGE buffer for 5 min, and then analyzed by immunoblot. The signals were detected with chemiluminescent substrate (Thermo Scientific) and quantified by ImageJ software. For immunoblotting, protein concentrations were determined by the NanoDrop One/OneC spectrophotometer (Thermo Scientific) and equal amounts of proteins were used in each lane.
Isolation of nuclei
Macrophages seeded in 6 cm dishes (3 × 106 cells/dish) were serum-starved for 24h and then treated with LPS (100 ng/mL) for indicated times. The cells were washed with ice-cold PBS, scraped and centrifuged for 10 s at 10,000 rpm. The cells were resuspended in 900 μL PBS containing 0.1% NP-40, triturated by pipetting, and centrifuged for 10 s at 10,000 rpm. The pellets were washed and resuspended in 30 μL 1X laemmli sample buffer, boiled, and subjected to immunoblot analysis.
Immunofluorescence and confocal microscopy
Macrophages grown on uncoated glass coverslips in 6-well plates were serum-starved for 24 h and then treated with LPS at 100 ng/mL for 30min. The cells were fixed in 4% paraformaldehyde for 15 min, permeabilized in 0.2% Triton X-100 for 10 min, blocked with 3% BSA (Biorad, Cat# 1706404) for 1h and then incubated with primary antibody overnight at 4°C. After three washes with PBS, cells were incubated with Alexa 488 conjugated secondary antibody for 3h at room temperature in the dark. Cells were then washed and counterstained with DAPI Vectashield mounting medium and imaged by laser confocal fluorescence microscopy (Olympus Laser Scanning Confocal and Multiphoton Microscope). The images were then analyzed and signal of nuclei NF-κB was quantified by MetaMorph software (Molecular Devices).
Transfection of siRNA and NF-κB reporter assay
The NF-κB reporter assay was performed following the manufacturer (InvivoGen) instructions. THP-1 Dual cells were first transfected with NKA α1 siRNA using Viromer Blue (Origene, Cat#TT100300) and then incubated with indicated doses of LPS at 37°C for 24h. Cell culture medium was then collected and cells removed by centrifugation at 1000g for 1 min. The QUANTI-Blue detection reagent was added to the supernatants for 2h at 37°C and NF-κB activation was then quantified by colorimetric assessment of SEAP activity.
Bulk RNA-Sequencing
WT or NKA α1+/− peritoneal macrophages were treated with control (cell culture media: RPMI1640 plus 1% Penicillin/Streptomycin), or LPS 100 ng/mL in cell culture media 6h (n = 3). Total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen) and quantified using a NanoDrop Onec spectrophotometer. Quality of extracted RNA was assessed with the Bioanalyzer RNA Nano Assay (Agilent) and all samples had observed RNA Integrity Number values > 7.4 with DV200 over 81%. RNA libraries were prepared (Illumina TruSeq Stranded mRNA, single indexed) and run on an Illumina High Seq-2500 for 125bp paired end reads at the MCW Genomic Sciences and Precision Medicine Center. Samples were sequenced to an average depth of 40 million reads. Sequence reads were aligned to the reference genome, mm10, and reference transcriptome, GRCm38, using the STAR aligner and MAPR-Seq workflow v3.0. Read count data were normalized using DESeq2. Hierarchical clustering and PCA analysis is performed for normalized counts for all the samples. Euclidean distance and the complete linkage clustering method is used for hierarchical clustering. Differential expression was calculated using EdgeR v3.8 and the gene-wise exact test on TMM-normalized levels. Differential expression was called between each unique pair of conditions. We defined genes as differentially expressed if they had an absolute fold-change ≥ 4 between two conditions and the corresponding FDR-adjusted p value < 0.05. All data was quality controlled using FastQC and RSeQC, followed by manual review and data visualization. Pathway analysis was conducted using “gseKEGG” method in “clusterProfiler” package to generate the differential pathway. GSEA plot was plotted by function “gseaplot2” in enrich plot library under bioconductor package.
Quantitative RT-PCRV
Total RNA from mouse peritoneal macrophages treated with LPS for 4h or from lung tissues removed from mice after LPS or PBS injection for 16h was extracted by RNeasy Mini kit (Qiagen), converted to cDNA by the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and then subjected to quantitative RT-PCR using the ABI 7500 Real-Time PCR System (Chen et al., 2019). Relative gene expression levels were calculated by the quantitative threshold (CT) method (ΔΔCT) using actb (β-actin gene) as the internal control.
Seahorse extracellular flux assay
Macrophages were plated in the specialized XF96 cell culture microplate (Seahorse Bioscience) at 8 × 104 cells/well. Cells were exposed to LPS (100 ng/mL, 24h) before glycolysis stress assay to measure extracellular acidification rate (ECAR) and mito-stress assay for oxygen consumption rate (OCR) by Seahorse Bioscience Extracellular Flux Analyzer (Agilent) as described (Chen et al., 2019). The cells were then lysed in RIPA buffer and subjected to Lowry protein assay (Bio-Rad). ECAR and OCR values were normalized to protein content.
Cytokine ELISA assays
The cell culture media or mouse plasma was subjected to ELISA assay using ELISA kits following the manufacturer (ThermoFisher Scientific) instructions.
Histopathology
Lungs were harvested from mice 16h after LPS challenge and fixed with 4% paraformaldehyde, dehydrated, paraffin-embedded and cut into 6 μm sections. The tissue sections were deparaffinized in xylene and stained with hematoxylin and eosin for microscopic analysis (20× magnification) (Sodhi et al., 2015).
Flow cytometry
Mouse lung tissue was washed with PBS, minced with scissors into pieces no larger than 2–3 mm, incubated at 37°C for 15 min in PBS and then filtered through a 40μm filter (Fisher). After centrifuging at 10,000 g for 1 min the pellets were resuspended and incubated for 15 min at room temperature with antibodies. Nonspecific binding was blocked using Fc-Block (Biolegend). Cells were then analyzed by flow cytometry with a BD LSR II instrument. FlowJo software 10.8.1 (BD) was used to analyze the data.
Measurement of cellular and mitochondrial ROS
Macrophages (1.5 × 106 cells/dish in 35mm culture dishes) were incubated with 1μM carboxy-DCFDA for 15 min at 37°C to detect total cellular ROS or with 5μM MitoNeoD for 15min to assess mitochondrial ROS (Chen et al., 2019; Shchepinova et al., 2017). Cells were then treated with LPS 100 ng/mL in culture medium for timed periods, then washed with PBS, scraped and immediately subjected to flow cytometry analysis.
Quantification and statistical analysis
Data are presented as means ± SE of at least 3 independent experiments. Statistical analysis was performed using the Student’s t test (two-tailed, unpaired, and equal variance) for comparisons between two groups or ANOVA followed by Bonferroni’s multiple comparison test for comparison among more than two groups. All statistical analyses were performed using GraphPad Prism 9 software. Statistical significance was accepted at p < 0.05. N = number of separate experiments for the in vitro studies, and n = number of animals for the in vivo studies.
Acknowledgments
We would like to thank Dr. Michael P. Murphy (University of Cambridge) and Richard C. Hartley (University of Glasgow) for providing MitoNeoD, Dr. Kathryn J. Moore (New York University) for providing the THP-1 Dual cell line. We thank Dr. Marie Schulte from the Versiti Blood Research Institute, Northwestern Mutual Microscopy Core for the technical support of FV1000 Olympus Laser Scanning Confocal and Multiphoton Microscope. In addition, we thank Liang Wu for helping with the high-resolution figure preparation. This work is supported by National Institutes of Health grants R01 HL153397 (to R.L. Silverstein, W. Cui, and Y. Chen), 1R15HL150721 (to K.Sodhi.), 5R01HL058012 (to D.Sahoo.), the BrickStreet Foundation (to J.I.Shapiro.), the Huntington Foundation (to J.I.Shapiro.) and American Heart Association Scientist Development Grant 17SDG661117 (to Y. Chen), Career Development Award 19CDA34660043 (to Z. Zheng) and MCW New Faculty Startup Fund (to Y. Chen and Z. Zheng). Graphical abstract was created with BioRender.com.
Author contributions
Y.C. and J.Z. conceived the study, designed and performed the experiments, analyzed the data, and wrote the article. J.C. performed the FACS experiments. M.A.B., W.D., and Y.Z. analyzed the data. W.H. conducted the qRT-PCR experiments on peritoneal macrophages. X.W. carried out spleen studies. S.S.P., H.V.L., and K.S. carried out histopathological experiments and qRT-PCR in lung tissues. W.C., Z.Z., J.S., D.S., and R.L.S. provided valuable ideas and suggestions and edited the article.
Declaration of interests
The authors have declared that no conflict of interest exists.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104963.
Supplemental information
Data and code availability
-
•
The aligned RNA-seq data including the raw counts and normalized counts for the whole macrophage transcriptomics have been deposited at Mendeley and are publicly available as of the date of publication. DOI link is listed in the Key resources table. Unfortunately, the pre-processed raw RNA-sequencing data is not available due to a technical mistake during data storage and maintenance.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alexander C., Rietschel E.T. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 2001;7:167–202. doi: 10.1177/09680519010070030101. [DOI] [PubMed] [Google Scholar]
- Baker R.G., Hayden M.S., Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- Best M.G., Sol N., Kooi I., Tannous J., Westerman B.A., Rustenburg F., Schellen P., Verschueren H., Post E., Koster J., et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biewenga J., van der Ende M.B., Krist L.F., Borst A., Ghufron M., van Rooijen N. Macrophage depletion in the rat after intraperitoneal administration of liposome-encapsulated clodronate: depletion kinetics and accelerated repopulation of peritoneal and omental macrophages by administration of Freund's adjuvant. Cell Tissue Res. 1995;280:189–196. doi: 10.1007/BF00304524. [DOI] [PubMed] [Google Scholar]
- Bonizzi G., Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Byeon S.E., Yi Y.S., Oh J., Yoo B.C., Hong S., Cho J.Y. The role of Src kinase in macrophage-mediated inflammatory responses. Mediators Inflamm. 2012;2012:512926. doi: 10.1155/2012/512926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Check J., Byrd C.L., Menio J., Rippe R.A., Hines I.N., Wheeler M.D. Src kinase participates in LPS-induced activation of NADPH oxidase. Mol. Immunol. 2010;47:756–762. doi: 10.1016/j.molimm.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Huang W., Yang M., Xin G., Cui W., Xie Z., Silverstein R.L. Cardiotonic steroids stimulate macrophage inflammatory responses through a pathway involving CD36, TLR4, and Na/K-ATPase. Arterioscler. Thromb. Vasc. Biol. 2017;37:1462–1469. doi: 10.1161/ATVBAHA.117.309444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Kennedy D.J., Ramakrishnan D.P., Yang M., Huang W., Li Z., Xie Z., Chadwick A.C., Sahoo D., Silverstein R.L. Oxidized LDL-bound CD36 recruits an Na(+)/K(+)-ATPase-Lyn complex in macrophages that promotes atherosclerosis. Sci. Signal. 2015;8:ra91. doi: 10.1126/scisignal.aaa9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang M., Huang W., Chen W., Zhao Y., Schulte M.L., Volberding P., Gerbec Z., Zimmermann M.T., Zeighami A., et al. Mitochondrial metabolic reprogramming by CD36 signaling drives macrophage inflammatory responses. Circ. Res. 2019;125:1087–1102. doi: 10.1161/CIRCRESAHA.119.315833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. The immunopathogenesis of sepsis. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- Collins P.E., Carmody R.J. The regulation of endotoxin tolerance and its impact on macrophage activation. Crit. Rev. Immunol. 2015;35:293–323. doi: 10.1615/critrevimmunol.2015015495. [DOI] [PubMed] [Google Scholar]
- Crow J.P. Dichlorodihydrofluorescein and dihydrorhodamine 123 are sensitive indicators of peroxynitrite in vitro: implications for intracellular measurement of reactive nitrogen and oxygen species. Nitric Oxide. 1997;1:145–157. doi: 10.1006/niox.1996.0113. [DOI] [PubMed] [Google Scholar]
- Dash B., Dib-Hajj S.D., Waxman S.G. Multiple myosin motors interact with sodium/potassium-ATPase alpha 1 subunits. Mol. Brain. 2018;11:45. doi: 10.1186/s13041-018-0388-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winther M.P.J., Kanters E., Kraal G., Hofker M.H. Nuclear factor kappaB signaling in atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2005;25:904–914. doi: 10.1161/01.ATV.0000160340.72641.87. [DOI] [PubMed] [Google Scholar]
- Devarajan P., Scaramuzzino D.A., Morrow J.S. Ankyrin binds to two distinct cytoplasmic domains of Na, K-ATPase alpha subunit. Proc. Natl. Acad. Sci. USA. 1994;91:2965–2969. doi: 10.1073/pnas.91.8.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B., Dey R.J., Cheung L.S., Pokkali S., Guo H., Lee J.H., Bishai W.R. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 2015;21:401–406. doi: 10.1038/nm.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberts I., von Asmuth E.J., van der Linden C.J., Buurman W.A. The interrelation between TNF, IL-6, and PAF secretion induced by LPS in an in vivo and in vitro murine model. Lymphokine Cytokine Res. 1991;10:127–131. [PubMed] [Google Scholar]
- Ezeriņa D., Takano Y., Hanaoka K., Urano Y., Dick T.P. N-acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H(2)S and sulfane sulfur production. Cell Chem. Biol. 2018;25:447–459.e4. doi: 10.1016/j.chembiol.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio M., Podrez E.A., Smith J.D., Hajjar D.P., Hazen S.L., Hoff H.F., Sharma K., Silverstein R.L. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari P. Rostafuroxin: an ouabain-inhibitor counteracting specific forms of hypertension. Biochim. Biophys. Acta. 2010;1802:1254–1258. doi: 10.1016/j.bbadis.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Gloire G., Piette J. Redox regulation of nuclear post-translational modifications during NF-kappaB activation. Antioxid. Redox Signal. 2009;11:2209–2222. doi: 10.1089/ars.2009.2463. [DOI] [PubMed] [Google Scholar]
- Godwin J.W., Pinto A.R., Rosenthal N.A. Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M., Wang H., Tian J., Xie Z. Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 2002;277:18694–18702. doi: 10.1074/jbc.M111357200. [DOI] [PubMed] [Google Scholar]
- Haberman Y., Tickle T.L., Dexheimer P.J., Kim M.O., Tang D., Karns R., Baldassano R.N., Noe J.D., Rosh J., Markowitz J., et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth P.G., Knaus U.G., Settleman J., Curnutte J.T., Bokoch G.M. Regulation of NADPH oxidase activity by Rac GTPase activating protein(s) Mol. Biol. Cell. 1993;4:1217–1223. doi: 10.1091/mbc.4.11.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordijk P.L. Regulation of NADPH oxidases: the role of Rac proteins. Circ. Res. 2006;98:453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- Hu X., Tian Y., Qu S., Cao Y., Li S., Zhang W., Zhang Z., Zhang N., Fu Y. Protective effect of TM6 on LPS-induced acute lung injury in mice. Sci. Rep. 2017;7:572. doi: 10.1038/s41598-017-00551-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Xiu H., Zhang S., Zhang G. The role of macrophages in the pathogenesis of ALI/ARDS. Mediators Inflamm. 2018;2018:1264913. doi: 10.1155/2018/1264913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H.C., Wang H., et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P.F., Grupp I.L., Grupp G., Woo A.L., Askew G.R., Croyle M.L., Walsh R.A., Lingrel J.B. Identification of a specific role for the Na, K-ATPase alpha 2 isoform as a regulator of calcium in the heart. Mol. Cell. 1999;3:555–563. doi: 10.1016/s1097-2765(00)80349-4. [DOI] [PubMed] [Google Scholar]
- Jialal I., Kaur H., Devaraj S. Toll-like receptor status in obesity and metabolic syndrome: a translational perspective. J. Clin. Endocrinol. Metab. 2014;99:39–48. doi: 10.1210/jc.2013-3092. [DOI] [PubMed] [Google Scholar]
- Johnson W.D., Jr., Mei B., Cohn Z.A. The separation, long-term cultivation, and maturation of the human monocyte. J. Exp. Med. 1977;146:1613–1626. doi: 10.1084/jem.146.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D.J., Chen Y., Huang W., Viterna J., Liu J., Westfall K., Tian J., Bartlett D.J., Tang W.H.W., Xie Z., et al. CD36 and Na/K-ATPase-alpha1 form a proinflammatory signaling loop in kidney. Hypertension. 2013;61:216–224. doi: 10.1161/HYPERTENSIONAHA.112.198770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb M., Margetts P.J., Anthony D.C., Pitossi F., Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J. Clin. Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F., Madan N., Ye Q., Duan Q., Li Z., Wang S., Si S., Xie Z. Identification of a mutant alpha1 Na/K-ATPase that pumps but is defective in signal transduction. J. Biol. Chem. 2013;288:13295–13304. doi: 10.1074/jbc.M113.467381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Moon C., Lee H.W., Park E.M., Cho M.S., Kang J.L. Src tyrosine kinases mediate activations of NF-kappaB and integrin signal during lipopolysaccharide-induced acute lung injury. J. Immunol. 2007;179:7001–7011. doi: 10.4049/jimmunol.179.10.7001. [DOI] [PubMed] [Google Scholar]
- Li Z., Cai T., Tian J., Xie J.X., Zhao X., Liu L., Shapiro J.I., Xie Z. NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells. J. Biol. Chem. 2009;284:21066–21076. doi: 10.1074/jbc.M109.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.S., Reiman E.M., Valla J., Dunckley T., Beach T.G., Grover A., Niedzielko T.L., Schneider L.E., Mastroeni D., Caselli R., et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingrel J.B., Kuntzweiler T. Na+, K(+)-ATPase. J. Biol. Chem. 1994;269:19659–19662. doi: 10.1016/S0021-9258(17)32067-7. [DOI] [PubMed] [Google Scholar]
- Liu A., Fang H., Wei W., Kan C., Xie C., Dahmen U., Dirsch O. G-CSF pretreatment aggravates LPS-associated microcirculatory dysfunction and acute liver injury after partial hepatectomy in rats. Histochem. Cell Biol. 2014;142:667–676. doi: 10.1007/s00418-014-1242-x. [DOI] [PubMed] [Google Scholar]
- Maslove D.M., Wong H.R. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol. Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire V.A., Gray A., Monk C.E., Santos S.G., Lee K., Aubareda A., Crowe J., Ronkina N., Schwermann J., Batty I.H., et al. Cross talk between the Akt and p38alpha pathways in macrophages downstream of Toll-like receptor signaling. Mol. Cell Biol. 2013;33:4152–4165. doi: 10.1128/MCB.01691-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I., et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Nishihara K., Miyata H., Shinke H., Tomita E., Kajiwara M., Matsubara T., Iehara N., Igarashi Y., Yamada H., et al. Molecular markers of tubulointerstitial fibrosis and tubular cell damage in patients with chronic kidney disease. PLoS One. 2015;10:e0136994. doi: 10.1371/journal.pone.0136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A.J., Hardie D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płóciennikowska A., Hromada-Judycka A., Borzęcka K., Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 2015;72:557–581. doi: 10.1007/s00018-014-1762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt R.D., Brickman C.R., Cottrill C.L., Shapiro J.I., Liu J. The Na/K-ATPase signaling: from specific ligands to general reactive oxygen species. Int. J. Mol. Sci. 2018;19:E2600. doi: 10.3390/ijms19092600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepinova M.M., Cairns A.G., Prime T.A., Logan A., James A.M., Hall A.R., Vidoni S., Arndt S., Caldwell S.T., Prag H.A., et al. MitoNeoD: a mitochondria-targeted superoxide probe. Cell Chem. Biol. 2017;24:1285–1298.e12. doi: 10.1016/j.chembiol.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skou J.C. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta. 1957;23:394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- So E.Y., Ouchi T. The application of Toll like receptors for cancer therapy. Int. J. Biol. Sci. 2010;6:675–681. doi: 10.7150/ijbs.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi K., Maxwell K., Yan Y., Liu J., Chaudhry M.A., Getty M., Xie Z., Abraham N.G., Shapiro J.I. pNaKtide inhibits Na/K-ATPase reactive oxygen species amplification and attenuates adipogenesis. Sci. Adv. 2015;1:e1500781. doi: 10.1126/sciadv.1500781. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sodhi K., Wang X., Chaudhry M.A., Lakhani H.V., Zehra M., Pratt R., Nawab A., Cottrill C.L., Snoad B., Bai F., et al. Central role for adipocyte Na, K-ATPase oxidant amplification loop in the pathogenesis of experimental uremic cardiomyopathy. J. Am. Soc. Nephrol. 2020;31:1746–1760. doi: 10.1681/ASN.2019101070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Cai T., Yuan Z., Wang H., Liu L., Haas M., Maksimova E., Huang X.Y., Xie Z.J. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol. Biol. Cell. 2006;17:317–326. doi: 10.1091/mbc.e05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touyz R.M. Apocynin, NADPH oxidase, and vascular cells: a complex matter. Hypertension. 2008;51:172–174. doi: 10.1161/HYPERTENSIONAHA.107.103200. [DOI] [PubMed] [Google Scholar]
- Tsujimoto H., Ono S., Efron P.A., Scumpia P.O., Moldawer L.L., Mochizuki H. Role of Toll-like receptors in the development of sepsis. Shock. 2008;29:315–321. doi: 10.1097/SHK.0b013e318157ee55. [DOI] [PubMed] [Google Scholar]
- van Rooijen N., Hendrikx E. Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol. Biol. 2010;605:189–203. doi: 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
- van Rooijen N., Sanders A., van den Berg T.K. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J. Immunol. Methods. 1996;193:93–99. doi: 10.1016/0022-1759(96)00056-7. [DOI] [PubMed] [Google Scholar]
- van Rooijen N., van Kesteren-Hendrikx E. Clodronate liposomes: perspectives in research and therapeutics. J. Liposome Res. 2002;12:81–94. doi: 10.1081/lpr-120004780. [DOI] [PubMed] [Google Scholar]
- Wang H., Haas M., Liang M., Cai T., Tian J., Li S., Xie Z. Ouabain assembles signaling cascades through the caveolar Na+/K+-ATPase. J. Biol. Chem. 2004;279:17250–17259. doi: 10.1074/jbc.M313239200. [DOI] [PubMed] [Google Scholar]
- Xie Z., Cai T. Na+-K+--ATPase-mediated signal transduction: from protein interaction to cellular function. Mol. Interv. 2003;3:157–168. doi: 10.1124/mi.3.3.157. [DOI] [PubMed] [Google Scholar]
- Yeung T.L., Leung C.S., Wong K.K., Samimi G., Thompson M.S., Liu J., Zaid T.M., Ghosh S., Birrer M.J., Mok S.C. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y.S., Kim H.G., Kim J.H., Yang W.S., Kim E., Jeong D., Park J.G., Aziz N., Kim S., Parameswaran N., Cho J.Y. Syk-MyD88 Axis is a critical determinant of inflammatory-response in activated macrophages. Front. Immunol. 2021;12:767366. doi: 10.3389/fimmu.2021.767366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Cui X., Zhang J., Xie J.X., Banerjee M., Pierre S.V., Xie Z. Heterogeneity of signal transduction by Na-K-ATPase alpha-isoforms: role of Src interaction. Am. J. Physiol. Cell Physiol. 2018;314:C202–C210. doi: 10.1152/ajpcell.00124.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Cai T., Tian J., Ivanov A.V., Giovannucci D.R., Xie Z. Na/K-ATPase tethers phospholipase C and IP3 receptor into a calcium-regulatory complex. Mol. Biol. Cell. 2005;16:4034–4045. doi: 10.1091/mbc.e05-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudowski G.A., Efendiev R., Pedemonte C.H., Katz A.I., Berggren P.O., Bertorello A.M. Phosphoinositide-3 kinase binds to a proline-rich motif in the Na+, K+-ATPase alpha subunit and regulates its trafficking. Proc. Natl. Acad. Sci. USA. 2000;97:6556–6561. doi: 10.1073/pnas.100128297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li X., Yu H., Larre I., Dube P.R., Kennedy D.J., Tang W.H.W., Westfall K., Pierre S.V., Xie Z., Chen Y. Regulation of Na/K-ATPase expression by cholesterol: isoform specificity and the molecular mechanism. Am. J. Physiol. Cell Physiol. 2020;319:C1107–C1119. doi: 10.1152/ajpcell.00083.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The aligned RNA-seq data including the raw counts and normalized counts for the whole macrophage transcriptomics have been deposited at Mendeley and are publicly available as of the date of publication. DOI link is listed in the Key resources table. Unfortunately, the pre-processed raw RNA-sequencing data is not available due to a technical mistake during data storage and maintenance.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.