Abstract
Chromatin regulation involves four subfamilies composed of ATP-dependent multifunctional protein complexes that remodel the way DNA is packaged. The SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex subfamily mediates nucleosome reorganization and hence activation/repression of critical genes. The SWI/SNF complex is composed of the BRG-/BRM-associated factor and Polybromo-associated BAF complexes, which in turn have multiple subunits. Significantly, ~20% of malignancies harbor alterations in >1 of these subunits, making the genes encoding SWI/SNF family members among the most vulnerable to genomic aberrations in cancer. ARID1A is the largest subunit of the SWI/SNF complex and is altered in ~40%–50% of ovarian clear cell cancers and ~15%–30% of cholangiocarcinomas, in addition to a variety of other malignancies. Importantly, outcome was improved after immune checkpoint blockade (ICB) in patients with ARID1A-altered versuss wild-type tumors, and this result was independent of microsatellite instability or tumor mutational burden. Another subunit—PBRM1—is mutated in ~40% of clear cell renal cell carcinomas and ~12% of cholangiocarcinomas; there are contradictory reports regarding ICB responsiveness. Two other SWI/SNF subunits of interest are SMARCA4 and SMARCB1. SMARCA4 loss is the hallmark of small cell carcinoma of the ovary hypercalcemic type (and is found in a variety of other malignancies); SMARCA4 germline alterations lead to rhabdoid tumor predisposition syndrome-2; SMARCB1 germline alterations, rhabdoid tumor predisposition syndrome-1. Remarkable, although anecdotal, responses to ICB have been reported in both SMARCA4-aberrant and SMARCB1-aberrant advanced cancers. This review focuses on the role that SWI/SNF chromatin remodeling subunits play in carcinogenesis, the immune microenvironment, and in immunotherapy responsiveness.
Keywords: Genetic Markers, Immunotherapy
Introduction
Chromatin remodeling elements refer to a group of proteins that remodel the way DNA architecture is packaged in order to permit access of condensed genomic DNA to the transcription machinery and thereby regulate gene expression. Chromatin itself is a complex of DNA and protein; its primary function is to package long DNA molecules into more compact structures.1 A primary protein component of chromatin is histones. Histones bind to DNA and function as ‘anchors’ around which the strands are wound. There are several levels of chromatin organization, and one of the important ones relate to nucleosomes. A nucleosome is the basic repeating unit of eukaryotic chromatin. A solitary nucleosome is made up of about 150 base pairs of DNA sequence blanketed around a core of histone proteins. DNA must be compacted into nucleosomes to fit into the nucleus and each human cell contains about 30 million nucleosomes.2
Although humans have four chromatin remodeler protein families—SWI/SNF (SWItch/Sucrose Non-Fermentable), CHD ((Chromodomain-Helicase-DNA binding), Imitation SWItch 1 and INO80 (inositol requiring 80)3–6—the best studied of these families of chromatin remodeler proteins are the SWI/SNF complexes. SWI/SNF is a subfamily of ATP-dependent chromatin remodeling complexes that associate to remodel the way DNA is parceled. This complex possesses a DNA-stimulated ATPase activity that can destabilize histone-DNA interactions in nucleosomes in an ATP-dependent manner. The SWI/SNF complex mediates nucleosome reorganization, allowing genes to be activated or repressed.3 6
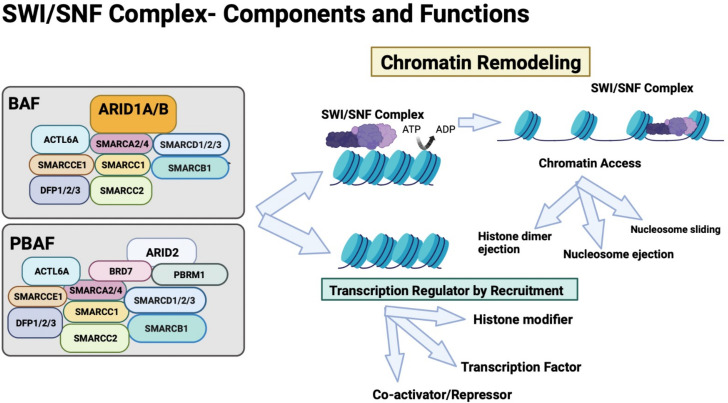
The SWI/SNF complex is composed of polymorphic BRG-associated/BRM-associated factor (BAF) and Polybromo-associated BAF (PBAF) complexes. BAF is made up of SMARCA2 or 4, ACTL6A, SMARCC1, SMARCC2, SMARCCE1, DFP1/2/3, ARID1A/B, SMARCD1/2/3 and SMARCB1 subunits; PBAF is made up of SMARCA2 or 4, ACTL6A, SMARCC1, SMARCC2, SMARCCE1, DFP1/2/3, SMARCD1/2/3 and SMARCB1, ARID2, BRD7, and PBRM1 subunits (figure 1). Importantly, ~20% of malignancies have an aberration in one of the genes encoding these subunits.7 Furthermore, abnormalities in SWI/SNF chromatin remodeling complex subunits have shown promise as markers for ICB responsiveness (table 1, figure 1).7–48
Figure 1.
SWI/SNF complex is an evolutionarily conserved ATP-dependent complex that contains multiple subunits. It has a key role in chromatin remodeling and regulation of transcription by recruitment of transcription factors, coactivators and repressors and histone modifiers. BRG1-associated or BRM1-associated factors (BAF) and Polybromo-associated BAF (PBAF) are two subclasses of this complex and differ in a few subunits as shown. These complexes consist of one of the two mutually exclusive catalytic ATPase subunits: SMARCA2 (Brahma or BRM) or SMARCA4 (BRG1), and other subunits such as SMARCB1, SMARCC1, and SMARCC2. PBAF complexes can be distinguished from BAF complexes because the former contains PBRM1 and ARID2 but lack ARID1A/B. SWI/SNF complexes regulate chromatin access by controlling the processes of histone dimer ejection, nucleosome ejection, and repositioning of nucleosomes by sliding. ARID1A binds DNA and may regulate the chromatin remodeling activity of the SWI/SNF complex through recruitment and binding of transcriptional factors. ARID subunits help with binding of the ATPase subcomplex. PBRM1 is essential for the stability of the SWI/SNF chromatin remodeling complex SWI/SNF-B (PBAF). ACTL6A, an actin domain, SMARCCE1, and DFP1/2/3 are accessory subunits common to both BAF and PBAF and are rarely mutated in cancers. SWI/SNF, SWItch/Sucrose Non-Fermentable.
Table 1.
Examples of genes controlling SWI/SNF chromatin remodeling and immune checkpoint blockade (ICB) responsiveness
| Name of gene | % Patients with cancer and alterations in the gene | Chromatin remodeling complex functions | Response to immunotherapy | Comment |
| ARID1A | ~3%–7% of all cancers1 7 Highest rates of mutation are seen in clear cell cancers of the ovary (~50% of patients), endometroid tumors (~40%), gastric cancers (~30%)7–9 |
|
Preclinical studies-treatment with anti-PD-L1 antibody show reduced tumor burden and increased survival in ARID1A loss mice ovarian tumors versus ARID1A wild type.11 ARID1A aberrations resulted in limited chromatin accessibility to interferon (IFN) responsive genes causing impaired IFN expression, poor T cell response and reduced tumor immune response.12 |
ARID1A is implicated in interactions with mismatch repair gene, MSH2, possibly compromising its function.18 |
| Pan-cancer patients with ARID1A alterations had significantly prolonged OS on ICB13 ARID1A alterations were a positive predictor for longer PFS after checkpoint blockade (HR (95% CI), 0.61 (0.39 to 0.94), p=0.02).14 PFS benefit after ICB was not dependent on MSI or TMB.14 Loss of ARID1A associated with: High PDL1 in gastric cancers15 and EBV16 (and both EBV and PDL1 predict for response in gastric cancer17) | ||||
| PBRM1 | ~3% of diverse cancers19 Highest rates of mutation seen in clear cell renal cell carcinoma tumor (seen in 40% of patients), cholangiocarcinomas (12%).19–21 |
|
Mixed data showing both that PBRM1 predicts response to ICB and does not predict response to ICB ICB response was associated with loss-of-function mutations in the PBRM1 gene (p=0.012) in metastatic ccRCC, a cancer not associated with MSI or high TMB.22 OS was significantly better with PBRM1 loss in 324 metastatic clear cell renal cell carcinomas on nivolumab (not reached vs 25 mos, p=0.05).23 However, in IMmotion in metastatic ccRCCs showed worse outcome for PBRM1-altered tumors in atezolizumab versus sunitinib group.24 A recent study with multivariate models of ccRCC patients treated with ICB (n=189), loss-of-function (LOF) mutations in PBRM1 were not associated with OS (HR=1.24, p=0.47) or time to treatment failure (HR=0.85, p=0.44), Pan cancer across 11 solid tumors (n=2936), LOF mutations not associated with improved OS (HR = 0.9, p = 0.7).25 In an NSCLC retrospective analysis, PBRM1 mutation predicted for worse prognosis on ICB regardless of high or low TMB (median OS of PBRM1-mutant versus wild-type patients was 6 vs 13 months)26 |
PBRM1 is thought to confer resistance to T cell–induced apoptosis, and a PBRM1 deletion in a B16F10 melanoma mouse model increases chances of response to anti–PD-1 and anti-CTLA4 agents’.12 Enhancement of immunostimulatory genes which play a role in hypoxia response and JAK–STAT signaling in PBRM1-mutant ccRCC cell lines.27 PBRM1 loss is consistent with a decreased immunogenic tumor microenvironment and instead upregulated angiogenesis.28 |
| SMARCA4 | About 5%–7% of all cancers29 Highest rates of mutation seen in SCCOHT (close to 100% patients)30 and NSCLC (10% of patients),31 but also seen in undifferentiated endometrial sacromas, SMARCA4-deficient thoracic sarcoma, gastric cancers, and a subset of multiple other tumor types32–34 |
|
SMARCA4 is hallmark of SCCOHT; 4/4 patients with objective response include three with durable CRs.36 SMARCA4 mutations were more prevalent in responders to PDL1 blockade in Keynote 012 trial in HNSCC.37 SMARCA4-mutated NSCLC was associated with improved outcomes with ICB in MSK-IMPACT study.38 Case reports show response to nivolumab or pembrolizumab in SMARCA4-mutated NSCLC.39 40 SMARCA4-deficient thoracic sarcoma (SMARCA4-DTS) case reports show response to nivolumab in third line therapy and another report shows rapid response to pembrozilumab in PDL1 positive SMARCA4-DTS.41 42 |
SMARCA4 alteration is sole mutation in rare SCCOHT which is characterized by high PDL-1 expression and T-cell infiltrate43; these tumors respond anecdotally to checkpoint blockade43 |
| SMARCB1 | Loss of SMARCB1 expression is seen in malignant rhabdoid tumors and epithelioid sarcomas. renal medullary carcinoma, cribriform neuroepithelial tumors, malignant mesotheliomas, synovial sarcomas, extracellular myxoid chondrosarcomas and familial schwannomatosis SMARCB1 loss is at the protein and DNA level in most tumors 44 45 |
|
Malignant rhabdoid tumors have low TMB across cancers, but case reports of 3 patients with SMARCB1-loss aggressive pediatric cancers demonstrate evidence of response to ICB.46 47% (14/30) of patients with SMARCB1-loss tumors had positive PD-L1 staining.45 46 Anecdotal reports show a pediatric renal medullary cancer with SMARCB1 loss responding to atezolizumab.47 An adult patient with recurrent SMARCB1-loss renal medullary carcinoma had a complete response to nivolumab lasting greater than 9 months despite low TMB.47 An advanced refractory, SMARCB1-deficient epitheloid sarcoma accomplished a complete remission to combined ipilimumab and nivolumab48 |
ARID1A, AT-rich Interactive Domain-containing protein; ccRCC, clear cell renal cell carcinoma; CR, complete remission; EBV, Epstein Barr virus; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous cell cancer; Jak-STAT, Janus kinase-signal transducer and activator of transcription; MSI, microsatellite instability; MSK, Memorial Sloan Kettering; NSCLC, non-small cell lung cancer; OS, overall survival; PBRM1, polybromo 1; PD-L1, programmed death ligand; PFS, progression-free survival; SCCOHT, small cell cancer ovarian hypercalcemic type; SMARCA4, SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin; SMARCA4-DTS, SMARCA4-deficient thoracic sarcomas; SWI/SNF, SWItching/Sucrose Non-Fermenting; TIM-3, T-cell immunoglobulin mucin-3; TMB, tumor mutational burden.
The development of ICB for cancer therapy, with several molecules now approved, signals a dynamic change in the field of immuno-oncology. Ipilimumab, which is anti-cytotoxic T-lymphocyte antigen-4 (CTLA-4), was the first approved immune checkpoint inhibitor for treating patients with advanced melanoma.49 Pembrolizumab, nivolumab, atezolizumab, avelumab, durvalumab, and cemiplimab, all of which are anti-programmed death 1 (anti-PD1) or anti-programmed death ligand 1 (anti PDL1) antibodies, have been subsequently approved.50 51 The overall response rates across tumors on checkpoint blockade therapy is, however, only in the order of 15%–20%, making it imperative to develop reliable and biologically based biomarkers to predict response.52 Some established biomarkers include deficient mismatch repair/microsatellite instability-high (dMMR/MSI-H)53 and high tumor mutational burden (TMB) (> 10 mutations/megabase),54 55 but there are also reports of new markers such as major histocompatibility presentation of neoantigens and immune response gene signature panels.56–59 Additionally, genomic markers such as PDL1 amplification,60 chromosome 9p21.3 loss,61 and novel markers such as chromatin remodeling gene aberrations—ARID1A, PBRM1, SMARCA4, and SMARCB114 21 37 46 —may also predict ICB responsiveness. Much of this data has just recently emerged and some of the clinical linkages between chromatin remodeling genes and immunotherapy remain anecdotal or a matter of debate. Even so, these linkages merit further understanding and exploration because of their high potential impact in the patient care setting. Here, we provide an overview of the emerging data on the valuable role of SWI/SNF chromatin remodeling genes in cancer and in immune responsiveness. We also balance the discussion with commentary on the pitfalls of using some of the chromatin remodeling gene aberrations such as PBRM1 loss as a marker for immunotherapy response, as the linkage may be with other aspects of tumor proliferation like angiogenesis.
Mutations in chromatin-remodeling SWI/SNF complex genes and tumor immunity
Alterations in various subunits of the SWI/SNF chromatin remodelers are found in about 20% of human cancers.7 The literature suggests that the most frequently altered subunit gene is ARID1A (BAF250A), which is aberrant in up to 7% of cancers, with other subunits such as PBRM1 and SMARCA4 altered in ~3%–7% of all malignant neoplasms.7 18 29
Importantly, tumor immunity may be affected by alterations in SWI/SNF chromatin remodeling genes in multiple ways: (1) loss of PBRM1(polybromo 1) and ARID2 (AT-rich Interactive Domain-containing protein), which leads to increased expression of genes that play a role in IFNγ (interferon-gamma) signaling and thus could increase responses to immunotherapy,12 62 as high IFNγ activates Janus kinase (JAK)–signal transducer and activator of transcription (STAT), which turns on PD-L1 expression63; (2) SMARCB1-mutant rhabdoid tumors show infiltration by subpopulations of clonally expanded T cells, suggesting a tumor-specific immune response 64; and (3) ARID1A may interact with MSH2, a MMR protein. ARID1A alterations are associated with improved outcome after checkpoint blockade; these responses appear to be independent of TMB and microsatellite stability status.11 14 65
ARID1A
ARID1A is the largest subunit of the SWI/SNF complex. It is located on chromosome 1p and is involved in chromatin remodeling activity via a conserved DNA binding domain, which enables binding transcription factors as well as transcriptional coactivator/corepressor complexes.1 10 Germline mutations in ARID1A are associated with truncating mutations that cause a severe form of Coffin-Siris syndrome that is characterized by feeding difficulties, bowel obstruction, and early severe respiratory problems as well as congenital heart disease(table 2).66
Table 2.
Examples of germline mutations in chromatin remodeling genes and their associated conditions
| Gene | Name of syndrome | Features of syndrome | Examples of cancers in patients with germline mutations | Comment |
| ARID1A | Coffin-Siris syndrome66 |
ARID1A alterations were found in ~7% of Coffin-Siris syndrome66 Features of Coffin-Siris syndrome include developmental delays, hypoplastic digits, hirsutism, and microcephaly66 |
Cancers not described | Coffin-Siris syndrome is caused by mutations in the ARID1A, ARID1B, SMARCA4, SMARCB1 or SMARCE1 genes.66 |
| ARID1B | Coffin-Siris syndrome | ARID1B mutations were the most common cause of Coffin- Siris syndrome (51%–75%) | Cancers not described | |
| PBRM1 | Familial renal cell carcinoma | A case report showed a family with renal cell carcinoma with germline mutations of PBRM175 | Renal cell carcinoma | Germline PBRM1 truncating mutation (p. Asp1333Glyfs) associated with renal cell carcinoma75 |
| SMARCB1 | Coffin Siris syndrome Rhabdoid tumor predisposition syndrome (RTPS1)83 |
Typically, infants and children present with cancers, while some affected patients present with benign schwannomas.96 | Rhabdoid tumors,83 96 | |
| SMARCA4 | Rhabdoid tumor predisposition syndrome (RTPS2)83 | Typically, infants and children present with cancers, and 11% of Coffin Siris82 syndrome have SMARCA4 germline mutations. | Rhabdoid tumors83 SCCOHT30 |
ARID1A, AT-rich Interactive Domain-containing protein; PBRM1, polybromo 1; SCCOHT, small cell carcinoma of the ovary hypercalcemic type; SMARCA4, SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin; SWI/SNF, mating-type SWItching (SWI) and sucrose fermentation (Sucrose Non-Fermenting - SNF).
ARID1A alterations occur in 3%–7% of cancers overall1 7; a variety of malignancies are affected, including but not limited to 46–50% of ovarian clear cell cancers,7–9 15%–27% of cholangiocarcinomas,67 endometrial carcinomas,68 colorectal cancers,69 10%–35% of gastric cancers,9 and many others.
From a functional viewpoint, as a subunit of SWI/SNF chromatin remodeler, ARID1A facilitates target-specific binding of SWI/SNF complexes to chromatin, thereby modifying the accessibility of chromatin to nuclear factors. In malignancies, ARID1A possesses the features of a gatekeeper in that it regulates cell cycle progression, and of a caretaker in that, it prevents genomic instability.70
Preclinical data in mice models suggest that ARID1A may be involved with MMR gene MMR and MSH2 protein.11 65 In ovarian cancer cell lines of a syngeneic mice model, there are increased tumor-infiltrating lymphocytes, and higher TMB and PDL1 expression.70 71 Additionally, ARID1A interacts on the carboxy-terminal with the Enhancer of Zeste 2 PRC2 subunit (EZH2) that functions as a catalytic subunit of the polycomb repressive complex 2 (PRC2). This interaction results in inhibition of the repressive function of EZH2 in IFN-responsive genes in human cancer cell lines.71 Furthermore, in gastric cancers, ARID1A aberrations are associated with Epstein-Barr positivity and high PDL1 expression which also strengthens the responsiveness to ICB.15–17 ARID1A is also vital for creating an open chromatin state on DNA damage and mediates the non-homologous end-joining (NHEJ) pathway.72 ARID1A deficient cells cannot mount NHEJ repair and combination treatment with low-dose radiation and Olaparib, a PARP inhibitor, elicited tumor responses in ovarian cancer mice model.72 These preclinical observations are the basis of the ATARI clinical trial with an ATR (Ataxia telangiectasia and Rad3 related) inhibitor in combination with a PARP inhibitor in gynecological cancers.73 Finally, alterations in ARID1A activate the phosphatidylinositol 3-kinase PI3K/serine-threonine kinase AKT/mammalian target of rapamycin mTOR pathways.74
In the clinical pan-cancer setting, MSI-H, as well as high TMB, were significantly more frequent in ARID1A-altered versus ARID1A wild-type tumors (20% vs 0.9%, p<0.001: and 26% vs 8.4%, p<0.001, respectively). Median PFS after checkpoint blockade immunotherapy was significantly longer in the patients with ARID1A-altered tumors than in those with ARID1A wild-type tumors (11 months vs 4 months, p=0.006). Multivariate analysis showed that ARID1A alterations predicted a better outcome after ICB and this result was not dependent on MSI or TMB.14 65
PBRM1
PBRM1 is a nucleosome-recognition subunit of the PBAF SWI/SNF chromatin-remodeling complex. Polybromo-1 (PBRM1), found on chromosome 3p21, functions as a tumor suppressor gene and is most commonly mutated in clear cell cancer of the kidney (~40% of patients) and cholangiocarcinomas (~12% of patients).19–21 Recently, a germline frameshift mutation in PBRM1 was identified as a predisposing factor for renal cell cancer (RCC).75 Preclinical studies show that PBRM1 itself may confer resistance to T cell-induced apoptosis and PBRM1-deficient murine melanomas had increased infiltration by cytotoxic T cells.12 Deletion of PBRM1 in a B16F10 melanoma mouse model increased susceptibility to anti-PD-1 and anti-CTLA4 agents.12
Some clinical studies have shown a response to ICB. For example, immunotherapy response was associated with loss-of-function mutations in the PBRM1 gene (p=0.012) in metastatic clear cell RCC (ccRCC),22 cancer not associated with MSI-H or high TMB, and overall survival (OS) was significantly better with PBRM1 loss in patients with metastatic ccRCC on nivolumab versus those not receiving nivolumab (not reached vs 25 months, p=0.05).23 Enhancement of immunostimulatory genes involved in hypoxia response and JAK-STAT signaling in PBRM1-mutant ccRCC lines may elucidate the immunotherapy response.27 28 However, contrary to the above, there are conflicting reports suggesting that PBRM1 mutations correlate with a decreased immunogenic tumor microenvironment in human RCC lines.24 25 PBRM1-deficient mouse subcutaneous renal tumors show resistance to ICB.28 Moreover, analysis of the IMmotion150 renal cell carcinoma study also suggests that PBRM1 mutations correlate with attenuated immunotherapy responsiveness.24 28 In multivariate models of ccRCC patients treated with ICB (n=189) at Memorial Sloan Kettering, loss-of-function mutations in PBRM1 were not associated with longer OS (HR=1.24, p=0.47) or time-to-treatment failure (HR=0.85, p=0.44).25
Similarly, in a non-small-cell lung cancer (NSCLC) retrospective analysis, PBRM1 mutations predicted a worse prognosis on ICB regardless of high or low TMB (median survival of PBRM1-mutant versus wild-type patients was 6 versus 13 months).26
There are notable limitations to using PBRM1 as a biomarker for immunotherapy response as the positive responses are in the setting of antiangiogenic therapy and not in the front-line setting. As seen in the IMMOTION 150 study, PBRM1 mutations are associated with high angiogenesis and renal cell carcinomas may benefit more from antiangiogenic therapy.76 77 Patients with PBRM1 mutated tumors had better progression-free survival with a multi- receptor tyrosine kinase inhibitor sunitinib than with ICB, suggesting that PBRM1 loss may be more useful as a marker for anti-angiogenesis therapy.78–80
A recent study of PBRM1 loss in diverse cancer types concluded that PBRM1 loss portended worse outcomes on ICB in ccRCC, adenocarcinomas of the lung, cutaneous melanoma, and bladder cancers even with high TMB.81 Thus, immunotherapy must be used with caution for these tumors and large-scale trials are needed to resolve this conflict.
SMARCA4 and SMARCB1
The SMARCA4 gene is situated on chromosome 19p and encodes the BRG1 protein. It belongs to the SWI/SNF chromatin remodeling complex and functions as an ATPase. There are two main categories of cancer-related SMARCA4 alterations: class 1 mutations—truncating mutations, fusions, and homozygous deletion (loss of function); and class 2 mutations—missense mutations (dominant-negative/gain of function through loss of function can also occur). Protein loss generally occurs with class 1 mutations.35
Overall, about 5%–7% of cancers have SMARCA4 alterations.29 Deficiency of SMARCA4 has been implicated in oncogenesis in small cell carcinoma of the ovary hypercalcemic type (SCCOHT),30 undifferentiated endometrial carcinomas, and uterine sarcomas,33 the aggressive SMARCA4-deficient thoracic sarcoma,32 and in some non-small-cell lung adenocarcinomas (NSCLCs).31
Germline mutations in SMARC4 have been detected in ~11% of those with Coffin-Siris syndrome (table 2). In particular, missense mutations with gain-of-function or dominant-negative effects are seen; some features of the syndrome are microphthalmia, intellectual disability, and lack of predisposition to cancers.82 Inactivating SMARCA4 mutations are seen in SCCOHT (almost all of which have SMARCA4 mutations, with about 40% having germline mutations).82 Germline mutations in SMARCA4 are also associated with the rhabdoid tumor predisposition syndrome-2 (RTPS2)(table 2).83 Most individuals diagnosed with SMARCA4-related RTPS inherited a pathogenic variant from an unaffected parent. The hallmark of RTPS2 is a notably increased predisposition to rhabdoid tumors which are rare but highly aggressive tumors arising in children under the age of four. Rhabdoid tumors can occur in almost any anatomic location but occur most frequently in the central nervous system, with >50% presenting in the cerebellum. Overall, ~35% of rhabdoid tumors are associated with germline SWI/SNF mutations.84
SCCOHT is a rare but aggressive monogenic tumor characterized by a low TMB, but still, exhibits a highly immunogenic tumor microenvironment with high PDL1 expression and infiltration by T cells.36 43Anecdotally, there are case reports of a sustained clinical response to anti-PDL1 therapy in patients with SCCOHT, suggesting that SMARCA4 mutations, even in the setting of low mutational burden can lead to an immunogenic tumor environment.36 85
Despite associations with aggressive disease, class 1 SMARCA4 mutations in NSCLC were associated with response to ICB.38 39 SMARCA4 genomic alterations are found in ~10% of all NSCLC.86 Recently a case report showed a more than 14-month sustained response when treated with nivolumab in a SMARCA4-deficient NSCLC with a high tumor mutation burden.39 Similarly, case reports of sustained tumor regression have been seen with nivolumab in SMARCA4-deficient thoracic sarcoma (SMARCA4-DTS) in third-line therapy after prior cytotoxic therapy.41 In another case report, there was a rapid response to pembrolizumab in a SMARCA4-deficient thoracic sarcoma overexpressing PDL1.40 Taken together these observations suggest that SMARCA4-deficient tumors merit prospective evaluation in clinical trials for their ICB responsiveness.
SMARCB1 (SWI/SNF-related matrixassociated actindependent regulator of chromatin subfamily B member 1) or INI1 (integrase interactor 1) is located at chromosomal position 22q11.2 and functions as a core subunit protein in the SWI/SNF complex.44 46 Loss of SMARCB1 expression/altered SMARCB1 is predominantly seen in pediatric renal and extrarenal malignant rhabdoid tumors (almost all of which show SMARCB1 alterations), epithelioid sarcomas, atypical central nervous system teratoid/rhabdoid tumors; renal medullary carcinoma, synovial sarcomas malignant mesotheliomas, sinonasal carcinomas, and cribriform neuroepithelial tumors.44 45 In particular, loss of SMARCB1 is present in a high number of epithelioid sarcomas.87 Interestingly, loss of SMARCB1 results in high expression of EZH2 leading to upregulation of several oncogenic pathways including Wnt/beta-catenin, Myc, and the Sonic Hedgehog pathways.88 89 Recent studies have shown promising results for EZH2 inhibitors in many cancer types including epithelioid sarcoma (which have SMARCB1 aberrations).90 Indeed, tazemetostat, an EZH2 inhibitor was granted accelerated approval in 2020 for use in metastatic and locally advanced epithelioid sarcomas.91
Additionally, recent studies point to other ways a SMARCB1 deficiency can modulate tumor immunogenicity, that is, by selecting for de-repression of endogenous retroviral elements (ERV). ERV de-repression can cause the accumulation of double-stranded RNA in the cytoplasm, which can stimulate a cell’s innate immune response through the IFN-α and IFN -λ pathways.92 DNA methyltransferase inhibitors have been shown to upregulate a cell’s immune signaling through a type 1 interferon response which is accompanied by ERV expression in many cancers, including ovarian cancer. Furthermore, ERV expression, especially expression of ERV3-2, is associated with ICB response in clear cell renal carcinomas.93 94 Additionally, association with DNA transposase, a piggyBac transposable element derived 5 (PGBD5) gene, can cause genetic rearrangements in lethal rhabdoid tumors. There is evidence to suggest that PGBD5 expression can cause SMARCB1 somatic inactivation in these tumors; this includes PGBD5-specific signals (PSS) sequences found in SMACRB1 deficient rhabdoid tumors.95
The archetype of a SMARCB1-deficient tumor is the malignant rhabdoid tumor, initially described in the kidney but also is seen in soft tissue, viscera, and the brain (where it is designated as an atypical teratoid rhabdoid tumor). These cancers are overwhelmingly diagnosed in the very young, and most have a fatal course. Pathologically, most but not all contain a population of ‘rhabdoid’ cells, typically cells with vast cytoplasm, eccentric vesicular nuclei, perinuclear spherical inclusions, and large inclusion-like nucleoli.96 Germline mutations occur as part of the RTPS and are of two subclasses. RTPS1 is caused by alterations in SMARCB1 and RTPS2 is caused by alterations in SMARCA4. Most cases are due to SMARCB1, but the features of RTPS1 and RTPS2 are clinically similar. In comparison to sporadic isolated rhabdoid tumors, the syndromic form is associated with an increased risk of developing multiple tumors at younger ages and schwannomas (benign nerve sheath tumors) that present primarily in adulthood.83 RTPS is inherited in an autosomal dominant manner. Many individuals have the disorder as the result of a de novo germline SMARCB1 pathogenic variant.83 96 Rhabdoid tumors are among the least mutated tumors with very low TMB but surprisingly have significant PDL1 expression and, in a small case series, patients showed responses to anti-PDL1 therapies.46 92
Renal medullary carcinoma is a rare but aggressive cancer characterized by balanced translocations that disrupt the tumor suppressor role of SMARCB1.97 There are anecdotal reports of responses to anti-PDL1 therapy in renal medullary cancers in both pediatric and adult patients but there are also reports of a lack of response.47 98 In a mouse model of rhabdoid tumors, there was complete tumor regression in 67% to 80% of treated mice on anti-PDL1 therapy.64 Additionally, there was clear evidence of tumor-infiltrating CD8+ T cells which had high expression of clinically targetable inhibitory immune-checkpoint receptors, including PD-1, LAG-3, and TIM-3.64 Finally, an advanced refractor, SMARCB1-deficient epithelioid sarcoma had a complete remission after taking a combined anti-PD1 immune checkpoint inhibitor therapy and anti-CTLA4 agent.48
Conclusions
The SWI/SNF chromatin-remodeling complex is vital for transcriptional activation of genes normally repressed by chromatin. The role that these complexes play in responses to commonly used immune checkpoint inhibitors is still being delineated, but recent data points to improved outcomes in patients with ARID1A-altered malignancies (across tumor types) even in the absence of traditionally used markers of immunotherapy response such as PDL1 and TMB.14 Some of the underlying mechanisms may include inhibition of the repressive function of EZH2 in IFN-responsive gene function,71 increased tumor-infiltrating lymphocytes,64 and interaction with MMR genes such as MSH2.11 While ARID1A loss has been posited as a reliable biomarker for ICB response,14 65 there are contradictory reports regarding PBRM1 loss, with some studies showing response and other pan-cancer trials showing a lack of response to ICB.25 26
SMARCA4 alterations are seen in rare but aggressive cancers such as SCCOHT, and SMARCA4-deficient thoracic and uterine sarcomas. SMARCA4 alterations are also seen in ~10% of NSCLC. Anecdotal studies suggest that some of these cancers may be responsive to ICB administration.36 37 The DART prospective clinical trial, which combines nivolumab and ipilimumab in rare tumors, also has a cohort that specifically addresses SCCOHT (NCT02834013). SMARCB1-deficient tumors have also been reported anecdotally to respond to ICB, including with compete for remissions in advanced diseases such as epithelioid sarcoma.48
Germline mutations in SMARCB1 and SMARCA4 are responsible for the rhabdoid tumor predisposition syndromes (RTPS1 and RTPS2, respectively) and there are reports of response to ICB in pediatric tumors that arise in patients with these cancers.83 96
A very recent study looked at nine patients with metastatic pancreatic cancer who harbored SWI/SNF alterations and found that 8/9 showed responsiveness to ICB even though only three were microsatellite unstable, and pancreatic cancers are usually resistant to ICB.99 These findings suggest a role for prospective clinical trials using SWI/SNF alterations as a predictive marker for ICB.
In conclusion, the SWI/SNF chromatin-remodeling complex is responsible for a multitude of functions that play an important role in gene transcription. The various subunits are mutated in a substantial subset of patients with malignancy (~20% of human cancers overall); germline mutations predisposing individuals to tumors also occur. The SWI/SNF chromatin remodeling complex has unique effects on the tumor immune microenvironment, translating to both preclinical and early clinical data suggesting responsiveness to commonly used immune checkpoint inhibitors across a variety of cancer types. Further prospective studies of immunotherapy in patients with SWI/SNF chromatin remodeling subunit gene aberrations and malignancies are urgently warranted.
Footnotes
Contributors: NK and RK wrote, reviewed, and revised the article. SK and SL reviewed, edited, and revised the article. All authors read and approved the final article.
Funding: Funded in part by National Cancer Institute grant P30 CA023100 and the Joan and Irwin Jacobs Fund philanthropic fund.
Competing interests: SK serves as a consultant for Foundation Medicine, NeoGenomics and CureMatch. He receives speaker’s fee from Roche and advisory board for Pfizer. He has research funding from ACT Genomics, Sysmex, Konica Minolta and OmniSeq. RK has the following disclosure information: Stock and Other Equity Interests (IDbyDNA, CureMatch, Inc., and Soluventis); Consulting or Advisory Role (Gaido, LOXO, X-Biotech, Actuate Therapeutics, Roche, NeoMed, Soluventis, Pfizer, and Merck); Speaker’s fee (Roche); Research Funding (Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Guardant Health, Grifols, Konica Minolta, DeBiopharm, Boerhringer Ingelheim, and OmniSeq [All institutional]); Board Member (CureMatch, Inc., and CureMetrix, Inc.). SL is the Co-founder of io9, advisor for SymptoHealth, and on the scientific advisory board for Biological Dynamics and Human Longevity Inc. NK has no disclosures to report.
Provenance and peer review: Commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009;78:273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 2.Annunziato A. DNA packaging: nucleosomes and chromatin. Nature Education 2008;1:26. [Google Scholar]
- 3.Clapier CR, Iwasa J, Cairns BR, et al. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 2017;18:407–22. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. In: Seminars in cell & developmental biology. 10. Academic Press, 1999: 189–95. 10.1006/scdb.1999.0300 [DOI] [PubMed] [Google Scholar]
- 5.Gursoy-Yuzugullu O, House N, Price BD. Patching broken DNA: nucleosome dynamics and the repair of DNA breaks. J Mol Biol 2016;428:1846–60. 10.1016/j.jmb.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A 2010;107:3458–62. 10.1073/pnas.1000398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med 2016;6:a026930. 10.1101/cshperspect.a026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones S, Wang T-L, Shih I-M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010;330:228–31. 10.1126/science.1196333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin C, Song W, Bi X, et al. Recent advances in the ARID family: focusing on roles in human cancer. Onco Targets Ther 2014;7:315. 10.2147/OTT.S57023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han L, Madan V, Mayakonda A, et al. Chromatin remodeling mediated by ARID1A is indispensable for normal hematopoiesis in mice. Leukemia 2019;33:2291–305. 10.1038/s41375-019-0438-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med 2018;24:556–62. 10.1038/s41591-018-0012-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan D, Kobayashi A, Jiang P, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770–5. 10.1126/science.aao1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang T, Chen X, Su C, et al. Pan-cancer analysis of ARID1A alterations as biomarkers for immunotherapy outcomes. J Cancer 2020;11:776. 10.7150/jca.41296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamura R, Kato S, Lee S, et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer 2020;8. 10.1136/jitc-2019-000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 2011;43:1219. 10.1038/ng.982 [DOI] [PubMed] [Google Scholar]
- 16.Kim Y-B, Ahn JM, Bae WJ, et al. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int J Cancer 2019;145:916–26. 10.1002/ijc.32140 [DOI] [PubMed] [Google Scholar]
- 17.Miliotis CN, Slack FJ. Multi-layered control of PD-L1 expression in Epstein-Barr virus-associated gastric cancer. J Cancer Metastasis Treat 2020;6. 10.20517/2394-4722.2020.12. [Epub ahead of print: 23 05 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allo G, Bernardini MQ, Wu R-C, et al. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol 2014;27:255–61. 10.1038/modpathol.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savas S, Skardasi G. The SWI/SNF complex subunit genes: their functions, variations, and links to risk and survival outcomes in human cancers. Crit Rev Oncol Hematol 2018;123:114–31. 10.1016/j.critrevonc.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 20.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011;469:539–42. 10.1038/nature09639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misumi K, Hayashi A, Shibahara J, et al. Intrahepatic cholangiocarcinoma frequently shows loss of BAP1 and PBRM1 expression, and demonstrates specific clinicopathological and genetic characteristics with BAP1 loss. Histopathology 2017;70:766–74. 10.1111/his.13127 [DOI] [PubMed] [Google Scholar]
- 22.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801–6. 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vano YA, Rioux-Leclercq N, Dalban C, et al. NIVOREN GETUG-AFU 26 translational study: association of PD-1, AXL, and PBRM-1 with outcomes in patients (PTS) with metastatic clear cell renal cell carcinoma (mccRCC) treated with nivolumab (N). J Clin Oncol 2020;38:618. [Google Scholar]
- 24.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749–57. 10.1038/s41591-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hakimi AA, Attalla K, DiNatale RG, et al. A pan-cancer analysis of PBAF complex mutations and their association with immunotherapy response. Nat Commun 2020;11:1. 10.1038/s41467-020-17965-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H, Liu J, Zhang Y, et al. PBRM1 mutation and preliminary response to immune checkpoint blockade treatment in non-small cell lung cancer. NPJ Precis Oncol 2020;4:1–4. 10.1038/s41698-020-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vuong L, Kotecha RR, Voss MH, et al. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov 2019;9:1349–57. 10.1158/2159-8290.CD-19-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X-D, Kong W, Peterson CB, et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nat Commun 2020;11:1–4. 10.1038/s41467-020-15959-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernando TM, Piskol R, Bainer R, et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun 2020;11:1–3. 10.1038/s41467-020-19402-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelinic P, Mueller JJ, Olvera N, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet 2014;46:424–6. 10.1038/ng.2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina PP, Romero OA, Kohno T, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat 2008;29:617–22. 10.1002/humu.20730 [DOI] [PubMed] [Google Scholar]
- 32.Perret R, Chalabreysse L, Watson S, et al. SMARCA4-deficient thoracic sarcomas. Am J Surg Pathol 2019;43:455–65. 10.1097/PAS.0000000000001188 [DOI] [PubMed] [Google Scholar]
- 33.Kolin DL, Quick CM, Dong F, et al. SMARCA4-deficient uterine sarcoma and undifferentiated endometrial carcinoma are distinct clinicopathologic entities. Am J Surg Pathol 2020;44:263–70. 10.1097/PAS.0000000000001375 [DOI] [PubMed] [Google Scholar]
- 34.Huang S-C, Ng K-F, Yeh T-S, et al. The clinicopathological and molecular analysis of gastric cancer with altered SMARCA4 expression. Histopathology 2020;77:250–61. 10.1111/his.14117 [DOI] [PubMed] [Google Scholar]
- 35.Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol 2010;102:122–8. 10.1016/j.pbiomolbio.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witkowski L, Goudie C, Ramos P, et al. The influence of clinical and genetic factors on patient outcome in small cell carcinoma of the ovary, hypercalcemic type. Gynecol Oncol 2016;141:454–60. 10.1016/j.ygyno.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanna GJ, Lizotte P, Cavanaugh M, et al. Frameshift events predict anti–PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoenfeld AJ, Bandlamudi C, Lavery JA, et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin Cancer Res 2020;26:5701-5708. 10.1158/1078-0432.CCR-20-1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naito T, Umemura S, Nakamura H, et al. Successful treatment with nivolumab for SMARCA4-deficient non-small cell lung carcinoma with a high tumor mutation burden: a case report. Thorac Cancer 2019;10:1285–8. 10.1111/1759-7714.13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henon C, Blay J-Y, Massard C, et al. Long lasting major response to pembrolizumab in a thoracic malignant rhabdoid-like SMARCA4-deficient tumor. Ann Oncol 2019;30:1401–3. 10.1093/annonc/mdz160 [DOI] [PubMed] [Google Scholar]
- 41.Iijima Y, Sakakibara R, Ishizuka M, et al. Notable response to nivolumab during the treatment of SMARCA4-deficient thoracic sarcoma: a case report. Immunotherapy 2020;12:563–9. 10.2217/imt-2019-0142 [DOI] [PubMed] [Google Scholar]
- 42.Takada K, Sugita S, Murase K, et al. Exceptionally rapid response to pembrolizumab in a SMARCA4-deficient thoracic sarcoma overexpressing PD-L1: a case report. Thorac Cancer 2019;10:2312–5. 10.1111/1759-7714.13215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jelinic P, Ricca J, Van Oudenhove E, et al. Immune-active microenvironment in small cell carcinoma of the ovary, hypercalcemic type: rationale for immune checkpoint blockade. J Natl Cancer Inst 2018;110:787–90. 10.1093/jnci/djx277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohashi K, Oda Y. Oncogenic roles of SMARCB1/INI1 and its deficient tumors. Cancer Sci 2017;108:547–52. 10.1111/cas.13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agaimy A, Hartmann A, Antonescu CR, et al. SMARCB1 (INI-1)-deficient Sinonasal Carcinoma: A Series of 39 Cases Expanding the Morphologic and Clinicopathologic Spectrum of a Recently Described Entity. Am J Surg Pathol 2017;41:458. 10.1097/PAS.0000000000000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forrest SJ, Al-Ibraheemi A, Doan D, et al. Genomic and immunologic characterization of INI1-deficient pediatric cancers. Clin Cancer Res 2020;26:2882-2890. 10.1158/1078-0432.CCR-19-3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beckermann KE, Jolly PC, Kim JY, et al. Clinical and immunologic correlates of response to PD-1 blockade in a patient with metastatic renal medullary carcinoma. J Immunother Cancer 2017;5:1–5. 10.1186/s40425-016-0206-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pecora A, Halpern S, Weber M, et al. Rapid and complete response to combination anti-CTLA-4 and anti-PD-1 checkpoint inhibitor therapy in a patient with stage IV refractory end-stage epithelioid sarcoma: a case report. J Immunother 2020;43:286–90. 10.1097/CJI.0000000000000332 [DOI] [PubMed] [Google Scholar]
- 49.Hargadon KM, Johnson CE, Williams CJ. Immune checkpoint blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors. Int Immunopharmacol 2018;62:29–39. 10.1016/j.intimp.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 50.Vaddepally RK, Kharel P, Pandey R, et al. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers 2020;12:738. 10.3390/cancers12030738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Migden MR, Rischin D, Schmults CD, et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med 2018;379:341–51. 10.1056/NEJMoa1805131 [DOI] [PubMed] [Google Scholar]
- 52.Meng X, Huang Z, Teng F, et al. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev 2015;41:868–76. 10.1016/j.ctrv.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 53.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodman AM, Sokol ES, Frampton GM, et al. Microsatellite-Stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res 2019;7:1570–3. 10.1158/2326-6066.CIR-19-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jardim DL, Goodman A, de Melo Gagliato D, et al. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 2021;39:154–73. 10.1016/j.ccell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodman AM, Castro A, Pyke RM, et al. MHC-I genotype and tumor mutational burden predict response to immunotherapy. Genome Med 2020;12:1–3. 10.1186/s13073-020-00743-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodman AM, Kato S, Chattopadhyay R, et al. Phenotypic and genomic determinants of immunotherapy response associated with squamousness. Cancer Immunol Res 2019;7:866–73. 10.1158/2326-6066.CIR-18-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boichard A, Pham TV, Yeerna H, et al. APOBEC-related mutagenesis and neo-peptide hydrophobicity: implications for response to immunotherapy. Oncoimmunology 2019;8:1550341. 10.1080/2162402X.2018.1550341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pham TV, Boichard A, Goodman A, et al. Role of ultraviolet mutational signature versus tumor mutation burden in predicting response to immunotherapy. Mol Oncol 2020;14:1680–94. 10.1002/1878-0261.12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018;4:1237–44. 10.1001/jamaoncol.2018.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han G, Yang G, Hao D, et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nature Communications 2021;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karachaliou N, Gonzalez-Cao M, Crespo G, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol 2018;10:1758834017749748. 10.1177/1758834017749748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villarino AV, Kanno Y, O'Shea JJ. Mechanisms and consequences of JAK-STAT signaling in the immune system. Nat Immunol 2017;18:374. 10.1038/ni.3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leruste A, Tosello J, Ramos RN, et al. Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell 2019;36:597–612. 10.1016/j.ccell.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 65.Mullen J, Kato S, Sicklick JK, et al. Targeting ARID1A mutations in cancer. Cancer Treat Rev 2021;100:102287. 10.1016/j.ctrv.2021.102287 [DOI] [PubMed] [Google Scholar]
- 66.Tsurusaki Y, Okamoto N, Ohashi H, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet 2012;44:376–8. 10.1038/ng.2219 [DOI] [PubMed] [Google Scholar]
- 67.Zou S, Li J, Zhou H, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 2014;5:1. 10.1038/ncomms6696 [DOI] [PubMed] [Google Scholar]
- 68.Wiegand KC, Lee AF, Al-Agha OM, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol 2011;224:328–33. 10.1002/path.2911 [DOI] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu R-C, Wang T-L, Shih I-M. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 2014;15:655–64. 10.4161/cbt.28411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Wang W, Zhang Y, et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest 2020;130:2712–26. 10.1172/JCI134402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park Y, Chui MH, Suryo Rahmanto Y, et al. Loss of ARID1A in tumor cells renders selective vulnerability to combined ionizing radiation and PARP inhibitor therapy. Clin Cancer Res 2019;25:5584–94. 10.1158/1078-0432.CCR-18-4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banerjee S, Stewart J, Porta N, et al. ATARI trial: ATR inhibitor in combination with olaparib in gynecological cancers with ARID1A loss or no loss (ENGOT/GYN1/NCRI). International Journal of Gynecologic Cancer 2021;31:1471–5. 10.1136/ijgc-2021-002973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samartzis EP, Gutsche K, Dedes KJ, et al. Loss of ARID1A expression sensitizes cancer cells to PI3K- and AKT-inhibition. Oncotarget 2014;5:5295. 10.18632/oncotarget.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benusiglio PR, Couvé S, Gilbert-Dussardier B, et al. A germline mutation in PBRM1 predisposes to renal cell carcinoma. J Med Genet 2015;52:426–30. 10.1136/jmedgenet-2014-102912 [DOI] [PubMed] [Google Scholar]
- 76.Briggs LG, Cone EB, Lee RJ, et al. Prognostic and predictive biomarkers for metastatic renal cell carcinoma. J Cancer Metastasis Treat 2021;7:46. 10.20517/2394-4722.2021.84 [DOI] [Google Scholar]
- 77.Moreira M, Pobel C, Epaillard N, et al. Resistance to cancer immunotherapy in metastatic renal cell carcinoma. Cancer Drug Resist 2020;3:454–71. 10.20517/cdr.2020.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braun DA, Ishii Y, Walsh AM, et al. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol 2019;5:1631–3. 10.1001/jamaoncol.2019.3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carril-Ajuria L, Santos M, Roldán-Romero JM, et al. Prognostic and predictive value of PBRM1 in clear cell renal cell carcinoma. Cancers 2019;12:16. 10.3390/cancers12010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Argentiero A, Solimando AG, Krebs M, et al. Anti-angiogenesis and immunotherapy: novel paradigms to envision tailored approaches in renal cell-carcinoma. J Clin Med 2020;9:1594. 10.3390/jcm9051594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Q, Shen R, Xu H, et al. Comprehensive analyses of PBRM1 in multiple cancer types and its association with clinical response to immunotherapy and immune infiltrates. Ann Transl Med 2021;9:465. 10.21037/atm-21-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Errichiello E, Mustafa N, Vetro A, et al. SMARCA4 inactivating mutations cause concomitant Coffin-Siris syndrome, microphthalmia and small-cell carcinoma of the ovary hypercalcaemic type. J Pathol 2017;243:9–15. 10.1002/path.4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nemes K, Bens S, Bourdeaut F. Rhabdoid Tumor Predisposition Syndrome. GeneReviews® [Internet. Seattle: University of Washington, 2017. [Google Scholar]
- 84.Bourdeaut F, Lequin D, Brugières L, et al. Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res 2011;17:31–8. 10.1158/1078-0432.CCR-10-1795 [DOI] [PubMed] [Google Scholar]
- 85.Mardinian K, Adashek JJ, Botta GP, et al. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol Cancer Ther 2021;20:2341–51. 10.1158/1535-7163.MCT-21-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dagogo-Jack I, Schrock AB, Kem M. Clinicopathologic characteristics of BRG1-Deficient non-small cell lung cancer. Journal of Thoracic Oncology 2020;15:766–76. [DOI] [PubMed] [Google Scholar]
- 87.Sullivan LM, Folpe AL, Pawel BR, et al. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod Pathol 2013;26:385–92. 10.1038/modpathol.2012.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet 2014;30:356–63. 10.1016/j.tig.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Italiano A. Targeting epigenetics in sarcomas through EZH2 inhibition. J Hematol Oncol 2020;13:33. 10.1186/s13045-020-00868-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang N, Eccleston M, Clermont P-L, et al. EZH2 inhibition: a promising strategy to prevent cancer immune editing. Epigenomics 2020;12:1457–76. 10.2217/epi-2020-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoy SM. Tazemetostat: first approval. Drugs 2020;80:513–21. 10.1007/s40265-020-01288-x [DOI] [PubMed] [Google Scholar]
- 92.Ngo C, Postel-Vinay S. Immunotherapy for SMARCB1-Deficient sarcomas: current evidence and future developments. Biomedicines 2022;10:650. 10.3390/biomedicines10030650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solovyov A, Vabret N, Arora KS, et al. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Rep 2018;23:512–21. 10.1016/j.celrep.2018.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henssen AG, Koche R, Zhuang J, et al. PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat Genet 2017;49:1005–14. 10.1038/ng.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pawel BR. SMARCB1-deficient tumors of childhood: a practical guide. Pediatr Dev Pathol 2018;21:6–28. 10.1177/1093526617749671 [DOI] [PubMed] [Google Scholar]
- 97.Calderaro J, Masliah-Planchon J, Richer W. Balanced translocations disrupting SMARCB1 are hallmark recurrent genetic alterations in renal medullary carcinomas. Eur Urol 2016;69. 10.1016/j.eururo.2015.09.027. [Epub ahead of print: 22 09 2021]. [DOI] [PubMed] [Google Scholar]
- 98.Sodji Q, Klein K, Sravan K, et al. Predictive role of PD-L1 expression in the response of renal medullary carcinoma to PD-1 inhibition. J Immunother Cancer 2017;5:62. 10.1186/s40425-017-0267-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Botta GP, Kato S, Patel H, et al. SWI/SNF complex alterations as a biomarker of immunotherapy efficacy in pancreatic cancer. JCI Insight 2021;6. 10.1172/jci.insight.150453. [Epub ahead of print: 22 09 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]