Abstract
With the emergence of CRISPR-mediated genome editing, there is an increasing desire for easy-to-use tools that can process and overview the spectra of outcomes. Here, we present Sequence Interrogation and Quantification (SIQ), a simple-to-use software tool that enables researchers to retrieve, data-mine and visualize complex sets of targeted sequencing data. SIQ can analyse Sanger sequences but specifically benefit the processing of short- and long-read next-generation sequencing data (e.g. Illumina and PacBio). SIQ facilitates their interpretation by establishing mutational profiles, with a focus on event classification such as deletions, single-nucleotide variations, (templated) insertions and tandem duplications. SIQ results can be directly analysed and visualized via SIQPlotteR, an interactive web tool that we made freely available. Using insightful tornado plot visualizations as outputs, we illustrate that SIQ readily identifies sequence- and repair pathway-specific mutational signatures in a variety of model systems, such as nematodes, plants and mammalian cell culture.
INTRODUCTION
The broad implementation of CRISPR technology in biological and biomedical research has led to an expansion of approaches that rely on robust and correct interpretation of sequence changes that result from repair of single-strand or double-strand DNA breaks (DSBs). Upon experimental perturbations, deep sequencing of PCR amplicons using next-generation sequencing (NGS) techniques has also become a valuable tool to obtain detailed information of the underlying mutagenic mechanisms: all outcomes combined represent the repair profile of a particular DNA damage in a particular cellular or organismal context. Unfortunately, the vast majority of the tools developed to analyse such data require training in informatics. To meet the increasingly growing demand for data-mining complex sets of NGS data by non-bioinformatically trained researchers, we designed and created SIQ, for Sequence Interrogation and Quantification. SIQ can be run on any computer system and uses the raw sequencing files as input to classify and quantify the identified sequence variants. It can run multiple files simultaneously and the resulting Excel file can be data-mined but also uploaded in SIQPlotteR, an interactive web tool we designed that allows for extensive data visualization and exploration (https://siq.researchlumc.nl/SIQPlotteR/). To be able to explore the utility of SIQ, we also uploaded all data used in this manuscript to be directly tried in SIQPlotteR.
MATERIALS AND METHODS
Cell culture and transfection
129/Ola-derived IB10 mouse embryonic stem (mES) cells were cultured on gelatin-coated plates in Buffalo rat liver (BRL)-conditioned mES cell medium [Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM GlutaMAX, 1 mM sodium pyruvate, 1× non-essential amino acids, 100 μM β-mercaptoethanol (all from Gibco), 10% foetal calf serum and leukaemia inhibitory factor]. HPRT-eGFP wild-type, Polq−/− and Ku80−/− mES cells were generated as previously described (1). Cells were transfected in suspension using a Lipofectamine 2000 (Invitrogen):DNA ratio of 2.4:1. Briefly, 1.5 × 106 cells were transfected using 3 μg of total DNA and incubated for 30 min at 37°C and 5% CO2 in round-bottom tubes; subsequently, cells were seeded on gelatin-coated plates containing BRL-conditioned medium.
HPRT-targeting assay
spSpCas9(BB)-2A-GFP (a gift from Feng Zhang, Addgene plasmid #48138), pU6-(BbsI)_CBh-Cas9-T2A-mCherry (a gift from Ralf Kuehn, Addgene plasmid #64324) and CBh-Cas9-Nickase-T2A-mCherry constructs containing single-guide RNAs (sgRNAs) were used to transfect mES cells (1). One day after transfection, the medium was refreshed. Cells were subcultured and HPRT-mutant cells were selected 7 days post-transfection either by sorting ≥100 000 GFP-negative cells on a BD FACSAria III (using BD FACSDiva software version 9.0.1, BD Biosciences) or by seeding 500 000 cells in 6-thioguanine (6-TG)-containing medium; subsequently, cells were allowed to grow for 5–7 days. See Supplementary Table S2 for sgRNA sequences.
Targeted sequencing of Cas9-induced repair outcomes
Samples for short-read (Illumina) sequencing were prepared essentially as described before (1). Briefly, genomic DNA was isolated and primers specific for the targeted regions were selected (Supplementary Table S1) that yield a ∼150–200 bp product on wild-type alleles and that contain adaptors for the p5 and p7 index primers (5′- GATGTGTATAAGAGACAG-3′ and 5′-CGTGTGCTCTTCCGATCT-3′, respectively). These primers were used to amplify the targeted region, PCR products were subsequently purified using AMPure XP beads (Beckman Coulter) according to the manufacturer’s protocol and DNA was eluted in 20 μl MQ. Flow-cell adaptor sequences were added by performing PCRs with 5 μl purified PCR product and 0.3 μM of p5 and p7 index primers. The PCR products were purified with AMPure XP beads and eluted in 20 μl MQ. PCR samples were pooled at equimolar concentrations per target-specific PCR. The quality and quantity of these pools were analysed using a high-sensitivity DNA chip on a Bioanalyzer (Agilent), which was used to generate an equimolar library that was sequenced on a NovaSeq6000 or HiSeq4000 (Illumina) by 150-bp paired-end sequencing.
For PacBio sequencing, 5′ Amino Modifier C6 (5AmMC6)-modified primers (IDT) were designed (Supplementary Table S1) to yield a ∼3500 bp product on wild-type alleles and that are tailed with universal sequences (5′-5AmMC6/GCAGTCGAACATGTAGCTGACTCAGGTCAC/Forward_sequence-3′ and 5′-5AmMC6/TGGATC-ACTTGTGCAAGCATCACATCGTAG/Reverse_sequence-3′). These primers were used to amplify the targeted region in 25 μl reactions using the PrimeSTAR GXL kit (Takara) and the following conditions: 98°C for 30 s, 20 cycles of 95°C for 15 s, 60°C for 15 s and 68°C for 4 min, and the final extension of 68°C for 7 min. Next, 2.5–3.5 ng round-one PCR product and barcoded universal primers were used in a second-round PCR with PrimeSTAR GXL and the following conditions: 98°C for 30 s, 20 cycles of 95°C for 15 s, 64°C for 15 s and 68°C for 4 min, and the final extension of 68°C for 7 min. DNA concentrations were measured using the Quant-iT dsDNA assay kit and the Qubit Fluorometer (both Thermo Fisher Scientific) according to the manufacturer’s protocol and PCR samples were pooled at equimolar concentrations to contain 1000–2000 ng of DNA in total; the quality of these pools was analysed on the Femto Pulse system (Agilent). SMRTbell library preparation was performed on 1000 ng purified PCR pool following the Procedure & Checklist—Amplicon Template Preparation and Sequencing (PN 100-815-000 version 04, Pacific Biosciences) and using SMRTbell Express Template Prep Kit 2.0. The library was sequenced on SequelII using Sequencing Primer V4, Sequencing Kit 2.0 and Binding Kit 2.0 on an 8M SMRT cell with a movie time of 30 h. Circular consensus sequences were generated with ccs version 6.0.0 (commit v6.0.0-2-gf165cc26) and barcodes were demultiplexed using lima 2.0.0 (commit v2.0.0).
SIQ implementation
SIQ is implemented in Java to be run on any operating system and requires at least Java 8 to run. As an initial check, SIQ checks whether all files can be located. In addition, the user can (strongly recommended) define flanks, which define the expected target site (e.g. a CRISPR cut site). The middle between the left and right flanks defines the expected target site and that location is set to 0. The provided flanks should be ≥15 bp and are required to be present in the reference sequence. For target where two cuts are made (e.g. if two sgRNAs are used), the flanks can be separated: the end of the left flank defines one target site and the start of the right flank defines the second target site. The primer used to perform the experiment can also be supplied (recommended) and need to be present in the reference DNA sequence as well. The primer sequences are used to ensure that reads start within the defined primers. If both R1 and R2 NGS files are supplied, SIQ attempts to merge the paired-end data using Flash (v2.2.00). SIQ then uses the merged file (or only the file in R1 if that was supplied) to map. For short-read data, it will initially check the orientation of the reads and assume that the same orientation is used throughout. For PacBio data, the reads are used in both forward and reverse complement orientations, depending on the read. Bases below a base quality threshold are cut off, leaving a high-quality read. That read is then mapped to the supplied reference using a k-mer mapping approach. The read is first mapped to the left side of the expected cut site and then extended as far as possible. The remaining part of the read (if any) is then mapped to the reference. For short-read sequencing, the read should start within the primer binding sites and the detected event should start at least five bases (optional and configurable) away from the primer binding sites. This ensures that mutagenic events are only detected if the primers annealed at the intended location in the DNA. For Sanger sequence and PacBio reads, the mapping is allowed to jump over locations with single-nucleotide variations (SNVs) and small indels if they are located >100 bp from within the target site. This ensures that only events in close vicinity to the expected target site are included in the output. SIQ classifies the reads based on the difference with the provided reference sequence and outputs an Excel table. Importantly, SIQ only works on Sanger sequences containing a single mutation (e.g. after colony PCR or PCR on individual clones). SIQ does not decompose Sanger sequence reads, such as TIDE, ICE and DECODR (2–4).
Templated insertions are insertions that are copied from a nearby stretch of DNA. To determine whether a delins (deletion with insertion) is a templated insertion, the inserted sequence is searched around the deletion junction. This is only performed for delins with an insertion of ≥6 bp as it is not possible for smaller insertions to determine the origin of the insertion (random chance of finding that sequence is too high). The search space is predefined to 100 bp (configurable) up- and downstream of the left junction (start point of the deletion) and the right junction (endpoint of the deletion) in both forward and reverse complement orientations and selects the largest overlapping sequence with the insert. A test is then performed to ensure that the probability of finding such a match is <10% when the junctions are shuffled. So only if an insertion with a large enough match in the flank is found it is classified as a templated insertion (tins).
Tandem duplications are insertions that exactly match the left or right junction and are ≥6 nucleotides long. Tandem duplication compounds (TD+) are insertions where part of the insertion exactly matches the left or right junction.
We have included a comprehensive user guide as a supplementary file to this manuscript to aid in running SIQ.
SIQPlotteR implementation
SIQPlotteR code was written using R (https://www.r-project.org) and RStudio (https://www.rstudio.com). To run the app, several freely available packages are required: shiny, ggplot2, dplyr, lobstr, colourpicker, grid, gridExtra, readxl, shinyWidgets, tidyr, RColorBrewer, sortable, ggpubr, ggrepel, DT, gplots, FactoMineR, factoextra and umap. Up-to-date code and new releases will be made available on GitHub, together with information on running the shiny app locally (https://github.com/RobinVanSchendel/SIQ).
The GitHub page of SIQ is the preferred way to communicate issues and request features (https://github.com/RobinVanSchendel/SIQ/issues). Alternatively, users can contact the developers by e-mail or Twitter. Contact information is found on the GitHub page. SIQPlotteR can be installed locally or you can use the available website to analyse SIQ output (https://siq.researchlumc.nl/SIQPlotteR/).
We have included a comprehensive user guide as a supplementary file to this manuscript to aid in creating visualizations using SIQPlotteR on SIQ-analysed data.
Generation of in silico datasets
To generate in silico datasets, we generated 200 target sites based on the human genome. For each dataset, set we created 11 subsets with a variable mutation frequency ranging from 0 to 1 with a step size of 0.1. For each subset, we created 10 000 reads that were either wild type or contained deletions and insertions ranging from −25 to + 25 bp. To introduce sequencing errors into our sets, we ran ART (5) to obtain a set with sequencing errors (options used: -na -ss HSXn -qs 10 -qs2 10). These sets were subsequently analysed by SIQ, CRISPResso2 (6) and AmpliCan (7).
Caenorhabditis elegans G4 experiment
A single animal of the strain XF1520 with genotype dog-1(gk10) was put on a 6 cm dish containing NGM and OP50. One week after plating, the plate was full and animals were rinsed off in MQ, washed five times with MQ and DNA was isolated using a DNA Blood & Tissue Culture Kit (Qiagen) following the manufacturer’s protocol. One microlitre of DNA was PCRed using primers at the G4 site qua2478 (see Supplementary Table S1) and processed as described above to generate an NGS library.
Arabidopsis thaliana Cas9 ADH-targeting experiment
The construct used to target the ADH locus is based on pDE-Cas9 (8) containing a T-DNA sequence with the phosphinothricin resistance gene bar, and CAS9 and sgRNA expression cassettes. pDE-CasADH, targeting the alcohol dehydrogenase 1 (At1G77120) gene, was constructed by inserting the sgRNA sequence ATCTTCGGCCATGAAGCTGG into pDE-Cas9. pDE-CasADH was introduced in the disarmed hypervirulent Agrobacterium tumefaciens strain AGL1 (9) using electroporation (10).
Root transformation in Columbia-0 ecotype was performed as described by Vergunst et al. (11). After cocultivation, the root explants were transferred to shoot induction medium containing 100 mg/l vancomycin and 100 mg/l timentin. Selective medium was additionally supplemented with 30 mg/l phosphinothricin. After an incubation period of 21 days, pools of 20 calli were frozen in liquid N2 and subsequently disrupted in a TissueLyser (Retch). DNA was isolated from disrupted frozen callus material using phenol/chloroform extraction as described by de Pater et al. (12) and PCR was performed using specific primers (see Supplementary Table S1) and processed as described above to generate an NGS library.
RESULTS
SIQ method
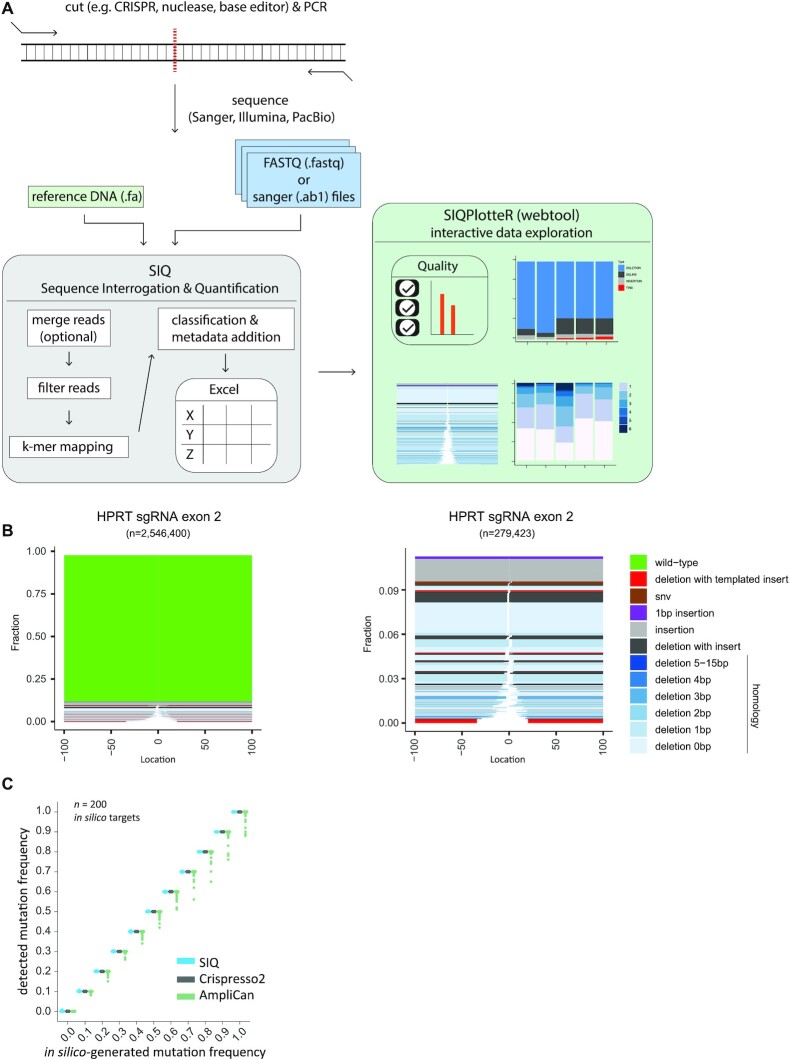
SIQ utilizes data obtained by sequencing PCR products covering a target site of interest that, for instance, has been targeted in vivo by CRISPR (or any other nuclease) and subsequently has been repaired by cellular repair pathways (Figure 1A). It can process collections of capillary (Sanger) sequences, each containing only a single mutation, but the true strength of SIQ is that it can identify mutational profiles in pooled DNA containing an extensive mix of mutational outcomes and deep sequenced by NGS methods. For SIQ analysis of experiments where short-read sequencing will be applied (i.e. Illumina paired-end sequencing), we recommend a PCR amplicon of <290 bp (for 2 × 150 bp paired-end reads) or <580 bp (for 2 × 300 bp paired-end reads) to ensure that the reads contain some overlap. For long-read sequencing, these criteria do not apply and larger amplicons can be used (e.g. >3 kb).
Figure 1.
Implementation of SIQ. (A) Schematic illustration of SIQ. NGS or Sanger sequencing on a PCR amplicon is required as input for SIQ, together with a reference DNA fasta file. Reads are optionally merged and SIQ produces an Excel output table, which can also directly be analysed by SIQPlotteR. (B) Examples of SIQPlotteR tornado plot visualizations for a target site in the mouse HPRT gene exon 2. Each colour represents an event type and the height of each coloured bar represents the contribution to the total fraction. The white space represents the deletion size for each event. For deletions, additional colouring is added, based on the extent of microhomology found at the junctions or the presence of insertions. The left panel shows all events, and the right panel excludes the wild-type events. (C) Two hundred in silico-generated target sites with varying mutation frequency, ranging from 0 to 1, were analysed by SIQ, CRISPResso2 and AmpliCan. For each site, 10 000 reads per mutation rate were generated and analysed. The results are displayed as a boxplot for each mutation frequency.
NGS data can directly be analysed by SIQ, which includes a graphical user interface, designed to run on any operating system (Windows, Linux or MacOS; Supplementary Figure S3). SIQ can also run from the command line, if desired. Apart from sequence data, SIQ requires a reference DNA sequence as input. What sets SIQ apart from other mapping approaches is that it is specifically designed to detect sequence changes at any target site (e.g. a CRISPR/Cas9, Cas12, I-SceI, TALEN, base editor or AsiSi site) and to focus on identifying variants at or in close proximity to the expected target site. This is achieved by performing a k-mer mapping strategy that detects matching sequences flanking the target site. If paired-end reads are used as input, they are first merged into a single read (via FLASH2 (13)). Reads are then passed through various filters, which includes removing low-quality and non-informative bases. Reads that pass the filters are mapped to the reference DNA file (Figure 1A).
Subsequently, event classification is carried out (Supplementary Figure S1) using categories that reflect common genetic variations, such as deletion, insertion, SNVs and wild type. Additional classification concerns deletions that also contain unmatched bases in between the deletion junctions, hence reflecting insertions. For cases in which (part of) the insertion can be reliably matched (surviving statistical scrutiny) to DNA sequences surrounding the mutation, the event is classified as a templated insertion (Supplementary Figure S1). Such templated insertions have been previously shown to be a unique hallmark of the DSB repair pathway theta-mediated end joining (TMEJ) (14). Another classification that SIQ reports includes tandem duplications: an insertion (≥6 bp) that is an exact duplicate of the DNA sequence immediately flanking it. Especially Cas9 nickase enzymes, combined with two sgRNAs targeting opposite strands, produce DNA breaks with protruding ends resulting in this type of genetic alterations (15–18). Finally, SIQ is also able to detect precise gene editing outcomes by matching the reads to a supplied repair template; such events are classified as homology-directed repair. In addition to event classification, additional metadata such as event location with respect to cut site, size and microhomology usage are determined. SIQ can process multiple sequence files and targets simultaneously: even on a regular computer, SIQ can analyse millions of reads per minute. To inspect whether SIQ correctly determines mutagenic outcomes, we validated SIQ using an in silico-generated dataset of 200 target sites (Figure 1C). For each target site, we generated 11 sets of 10 000 reads with a mutation frequency ranging from 0 to 100% with 10% incremental steps. The mutations ranged in size between −25 bp (deletions) and +25 bp (insertions) at the cut site (see also ‘Materials and Methods’ section). Running this dataset through SIQ resulted in a perfect match between the in silico-generated and detected mutation frequencies (Figure 1C). For reference purposes, we assayed two other available mutation detection tools and found a similar performance for SIQ and CRISPResso2 (6), whereas both performed somewhat better than AmpliCan (7).
The output of SIQ is an Excel table, which can be analysed directly, or be processed through a dedicated web tool called SIQPlotteR, which we made publicly available (https://siq.researchlumc.nl/SIQPlotteR/). SIQPlotteR can also be installed locally. We created SIQPlotteR as we experienced that the amount of data produced by targeted sequence experiments requires condensed data visualizations as well as an interactive environment to allow researchers to explore the data from different angles. To capture the entire spectrum of mutational outcomes, we developed a novel visualization that we termed ‘tornado plot’, which shows in a single graph the repair outcome type, the weight of each mutation, the extent of microhomology at the junctions and the location of the event with respect to the target site (Figure 1B).
Mutation analysis on cells treated with CRISPR–Cas9 variants
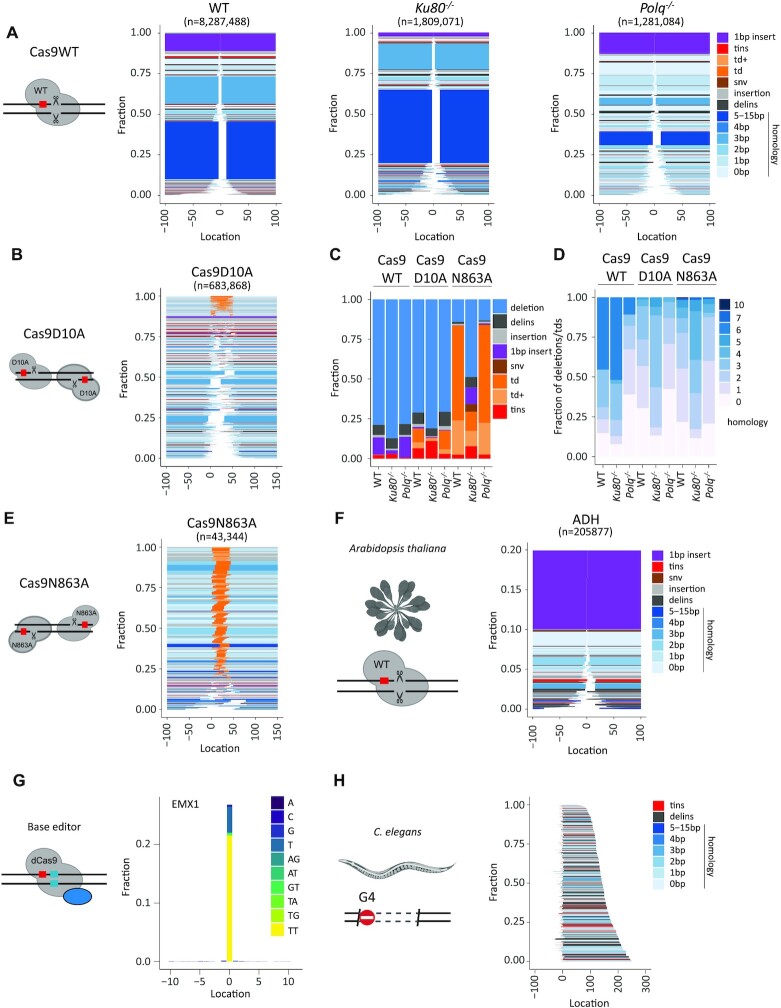
To showcase SIQ, we processed a series of experiments. We induced DSBs with different configurations in mES cells: blunt DSBs using wild-type Cas9, and DSBs with 5′ and 3′ protruding overhangs using Cas9 nickases (Cas9D10A and Cas9N863A, respectively). In addition to wild-type mES cells, we included cells with a deficiency in Polθ, which is critically important for TMEJ, and cells with a deficiency in Ku80, which is a key factor in non-homologous end joining (NHEJ). To generate datasets rich with different variants, we targeted the selectable gene Hprt, which when mutated confers resistance to treatment with 6-TG. DNA from 6-TG-resistant cells was isolated for each cell line and amplified using specific primers (Supplementary Table S1). Deep sequencing was performed on these amplicons and the data were analysed by SIQ, and in parallel by CRISPResso2, AmpliCan and ScarMapper (19) to compare the performances on real data (Supplementary Figure S2). Importantly, the cellular selection we applied (i.e. 6-TG selection) is not a prerequisite for establishing detailed mutation profiles as NGS data from pools of unselected cells also produce a wide spectrum of mutational outcomes, even if they contain up to 90% wild-type reads, which can be filtered out in SIQPlotteR (Figure 1B).
Figure 2 demonstrates that SIQPlotteR visualizes SIQ-processed NGS data in intuitively interpretable formats, which can be adapted in several dimensions (types of outcome to visualize, scales, colour coding and sorting; Supplementary Figure S4). Furthermore, the data obtained with SIQ analysis recapitulate the repair profiles that have been previously found for the tested conditions (15,17,20–22). Cas9WT and Cas9 nickase variants produce entirely different mutation profiles: Cas9WT-induced blunt DSBs create small deletions that, in mES cells, are characterized by microhomology and 1 bp insertions; Cas9 nickase-induced DSBs with 3′ overhangs (Cas9N863A) predominantly give rise to tandem duplications, whereas DSBs with 5′ overhangs (Cas9D10A) mostly produce deletions in which the 5′ protruding sequence has been lost, but also tandem duplications, in which fill-in has occurred (Figure 2A, B and E; Supplementary Figure S5) (1,18). Overtly different mutation profiles are produced in cells that contain DNA repair deficiencies, such as in TMEJ- and NHEJ-deficient cells. Confirming published work, Figure 2 shows a prominent role for Polθ in mutagenic repair of DSBs induced by Cas9WT, leading to a characteristic microhomology-mediated repair profile: the two dominant microhomology-mediated outcomes that are present in wild-type cells are almost completely absent in cells with a deficiency in TMEJ, which is accompanied by an overall reduction in homology usage (i.e. the two blue blocks; Figure 2A and D, Cas9WT: WT versus Polq−/−) (18,21). The action of NHEJ can also be observed in wild-type cells, as it is reflected by the presence of 1 bp insertions (Figure 2A and D, purple) that are absent in NHEJ-deficient cells. In addition, Figure 2C further highlights the following genetic requirements: (i) a Polθ dependence for deletions containing templated insertions, which are increasingly manifest in Ku80−/− cells; (ii) a Ku80 dependence for tandem duplications at DSBs having 5′ protruding ends; and (iii) also a Ku80 dependence for tandem duplications at DSBs having 3′ protruding ends. Note that SIQ refrains from classifying events as being the product of NHEJ or TMEJ action based on, for example, degree of junctional microhomology or presence of inserts (of a given size or configuration) as we have observed that overt differences exist between mutation profiles in different contexts (e.g. DNA context, cell type, species), but also that NHEJ and TMEJ can produce identical mutations. Inferring the potential contribution of a specific EJ pathway in mutation profiles thus ideally requires genetic support.
Figure 2.
Showcasing SIQ capabilities. (A) Tornado plot representation of mutation profiles at Cas9-induced DSBs in mES cells of the indicated genotypes, which were transfected with Cas9WT and HPRT_Ex3.1 sgRNA. Data are plotted relative to the Cas9WT break site (at 0) and sorted by deletion size. Tins (templated insertions; see the ‘Materials and Methods’ section), 1 bp insertions, insertions, deletions and deletion insertions (delins) are colour coded. The degree of blue colouring reflects the extent of microhomology found at the deletion junction. (B) Similar to panel (A), but for wild-type cells transfected with Cas9D10A in combination with two sgRNAs targeting opposite strands of the DNA to create a break with a 5′ overhang of 50 bp. Td and td+ (tandem duplication and tandem duplication compound; see the ‘Materials and Methods’ section) are depicted in orange and indicate DNA that has been duplicated. (C) Fraction of mutation types identified for each Cas9 variant in mES cells of indicated genotype. (D) The degree of microhomology that is found at the junctions of deletions and tandem duplications for each Cas9 variants in mES cells of indicated genotype. (E) Similar to panel (A), but for wild-type cells transfected with Cas9N863A in combination with two sgRNAs targeting opposite strands of DNA to create a break with a 3′ overhang of 43 bp. (F) Tornado plot representing the mutation profile at the ADH locus in Arabidopsis thaliana. Only mutagenic events are shown. Data are plotted relative to the Cas9WT break site (at 0) and sorted by deletion size. Tins (templated insertions; see the ‘Materials and Methods’ section), 1 bp insertions, insertions, deletions and deletion insertions (delins) are colour coded. The degree of blue colouring reflects the extent of microhomology found at the deletion junction. (G) An SNV alteration plot that displays base editing at an EMX1 site in data obtained from (23). Data are plotted relative to the Cas9 base editor target site (at 0); the y-axis reflects the fraction of total reads with an alteration. Dinucleotide SNVs are detected as delins (del = 2, ins = 2) and can be optionally displayed in SIQPlotteR. (H) Tornado plot (inverted) representing the mutation profile at a G4 site in DNA extracted from DOG-1-deficient C. elegans. The PCR strategy chosen generates an ∼350 bp PCR product, which excludes wild-type events from being detected (reads are 2 × 150 bp). Deletion products are ordered on end position. Tins (templated insertions; see the ‘Materials and Methods’ section), insertions, deletions and deletion insertions (delins) are colour coded. The degree of blue colouring reflects the extent of microhomology found at the deletion junction.
To illustrate the ability of SIQ to determine mutation spectra that are the consequence of repair of other types of DNA damage than nuclease-induced DBSs, we analysed three datasets. First, we analysed data from an experiment in A. thaliana where the ADH locus was targeted by expressing Cas9 and an sgRNA against ADH in root explants (see the ‘Materials and Methods’ section). About 20% of the reads contained a mutation of which 1 bp insertions were the dominant repair outcome. The remainder of the mutation profile consists primarily of deletions with limited microhomology (Figure 2F). Second, we used published data from another research group that used base-editing CRISPR technology to induce specific base substitutions at the EMX1 locus in HEK293T cells (23). Figure 2G shows the output of SIQPlotteR that visualizes the presence of base alterations at a given target, in this case the result of base editing EMX1 in HEK293T cells, which are dominated by mutations to TT at the target site. Third, we used data derived from C. elegans FancJ mutants in which DSBs spontaneously occur at G-rich sequences, as these can form stable DNA secondary structures called G-quadruplexes (G4s), which impede ongoing DNA replication (24). In the absence of FANCJ/DOG-1 in C. elegans, genomic deletions arise that have lost a G4 motif as well as 50–200 bases of downstream sequence (25–27). Performing SIQ on NGS data of targeted sequencing around such G4 sites in worm populations produces G4 deletion spectra that recapitulate repair of G4-induced DSBs at an unprecedented scale (Figure 2H). These examples illustrate the utility of SIQ to provide detailed insight into mutation profiles in a range of species (worms, plants, mouse and human cells), and in very different experimental set-ups.
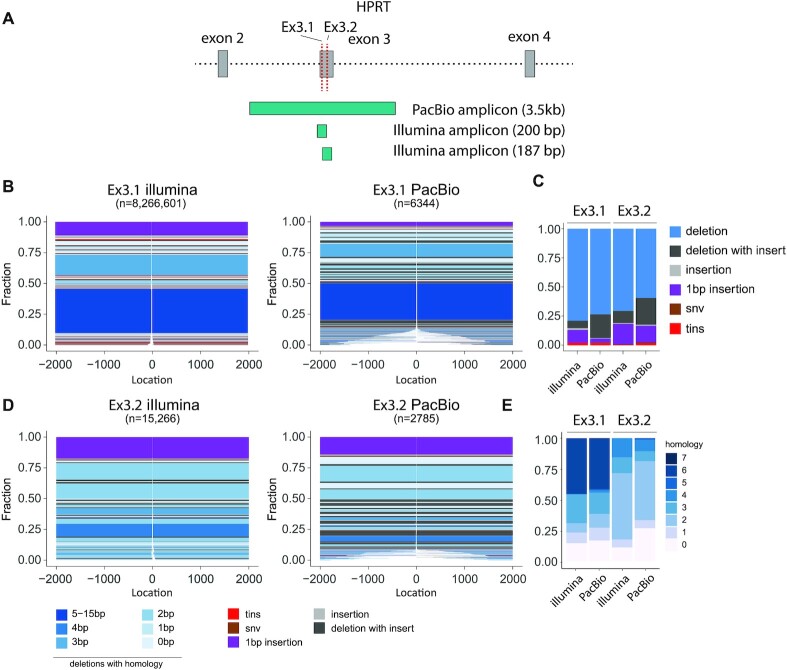
SIQ on long-read PacBio data
While short-read sequencing (e.g. by Illumina platforms) is often informative and affordable, the use of long-read sequencing also starts to gain momentum as it allows for inclusion of large structural variants in the analysis. Yet also for their output, easy processing tools for user-friendly quantification and inspection are grosso modo missing. Therefore, we designed the current version of SIQ to also create mutation profiles from long-read NGS data (i.e. PacBio data). To generate proof of concept, we isolated DNA from cells that were transfected with Cas9WT and either of two different sgRNAs that induce DSBs in exon 3 of the Hprt gene. We designed primers to produce amplicons of 270 bp and 3 kb (Figure 3A) to compare short- and long-read sequencing. PCR products were obtained and sequenced on a PacBio SequelII and on an Illumina HiSeq. In the size range covered by both technologies (0–200 bp), we find SIQ to produce comparable spectra (Supplementary Figure S4C–E). Using PacBio sequencing, we can detect mutations that otherwise would be missed in Illumina sequencing as those events remove either one or both of the primers used for amplification, which constituted 8% and 12% of events, respectively (Figure 3B and D). In terms of mutation types (Figure 3C) and homology at the junctions (Figure 3E), the two sequence methods generate comparable footprints, with the exception of deletions with insertions (delins), which are more frequently found in PacBio data (Figure 3C). While the additional mutations detected in PacBio versus Illumina sequencing on these CRISPR sites in mES cells may appear relatively modest, research has shown that such large deletions may occur more frequently in certain cell types and species and that long-read sequencing provides a powerful method to detect undesired genome modifications (28).
Figure 3.
SIQ on PacBio data. (A) PCR strategy for Illumina and PacBio sequencing of the mouse HPRT gene targeted at exon 3 with either HPRT_Ex3.1 or Ex3.2 sgRNA. (B) Tornado plot representation of mutation profiles at Cas9-induced DSBs in mES cells, which were transfected with Cas9WT and HPRT_Ex3.1 sgRNA and sequenced with Illumina or PacBio. Data are plotted relative to the Cas9WT break site (at 0) and sorted by deletion size. Tins (templated insertions; see the ‘Materials and Methods’ section), 1 bp insertions, insertions, deletions and deletion insertions (delins) are colour coded. The degree of blue colouring reflects the extent of microhomology found at the deletion junction. (C) Fraction of mutation types identified for HPRT_Ex3.1 and HPRT_Ex3.2 for the indicated sequencing strategy. (D) As in panel (B), but here for HPRT_Ex3.2. (E) The degree of microhomology that is found at the deletion junctions for HPRT_Ex3.1 and HPRT_Ex3.2, for the indicated sequencing strategy.
DISCUSSION
Advantages of SIQ
Here, we have developed user-friendly software to translate complex NGS outcomes into an Excel file format that allows for multifactorial data mining, and into intuitive and easily amendable graphics to facilitate interpretation. Because SIQ only needs NGS output and a reference target that is suspected of having mutations, it can be used to create mutation profiles for a wide range of experimental approaches that apart from the now common CRISPR/Cas9 technology include targeting by base editors, TALENs, endonucleases, (plasmid-based) DNA cross-links and replication blocks (e.g. via G4s).
SIQ is designed to facilitate researchers who do not have in-depth knowledge on how to handle NGS data, or are not skilled in programming: a user can simply select the amplicon NGS files to be analysed and input the DNA reference. Once the target location(s) are set and the primers used are added (optionally), analysis can commence. When analysis is complete, the resulting Excel table can be data-mined via the numerous parameters that are annotated to each mutational outcome. The Excel table can also be directly uploaded in SIQPlotteR to analyse data quality as well as to generate various interactive data visualizations. We have included visualizations that show quality control, targeting frequency, repair-type classification, size alteration, microhomology, SNV alteration and the insightful tornado plots. For all of these plots, we allow users to filter experiments based on the number of reads or event type, select and sort samples, choose colours and finally export their plots to a PDF format.
Comparison to other methods
In recent years, several tools have been created to analyse amplicon data, such as AmpliCan (7), CRISPResso2 (6), CrispRVariants (29), ScarMapper (19) and CRISPAltRations (30). We found SIQ to perform on par with CRISPResso2 and AmpliCan, and both to perform slightly better than ScarMapper on Cas9-induced breaks. For Cas9 nickase experiments, we observed larger differences between the tools, likely due to the inability to set multiple target sites or the limited quantification window around the target site (Supplementary Figure S2). In general, these tools have been designed to analyse a specific type of CRISPR editing. Some tools, such as CRISPAltRations, are specifically trained to detect CRISPR edits in a limited window around the break site, precluding detection of other types of events, such as large deletions, or its use in analysing experiments that employ other means of creating DNA alterations. While most tools provide basic classification of events, such as deletions and insertions, none of these report tandem duplications or templated insertions. Apart from SIQ generating multidimensional output visualizations that can easily be modified, another major difference to the now available tools is the ease of installation and usage. Some of the current tools require additional software dependences to be installed, or cannot be run from Windows or MacOS. We feel that in most cases (bio)informatic expertise is needed or nearby experts are required to install the software and run analyses. To optimally facilitate unrestricted data processing, without restrictions on accessibility, file size and number limits, we developed SIQ to not depend on websites, but instead operate on a local computer, and to implement it in Java to allow researchers to simply launch SIQ upon download.
DATA AVAILABILITY
The latest versions of SIQ and SIQPlotteR are available in the GitHub repository: https://github.com/RobinVanSchendel/SIQ/releases/latest. Since this software is commonly used in our lab, we expect to develop and extend it further in the future.
ACCESSION NUMBERS
The raw targeted sequencing data generated in this study have been deposited in the NCBI SRA database under accession number PRJNA802705. The base editor data for EMX1 used for Figure 2G were downloaded from accession number SRR3305545. CAS9D10A data were previously generated and can be found in accession numbers SRR12079930, SRR12079938 and SRR12079923, and Cas9N863A in wild-type cells in accession number SRR12079956.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Tijsterman lab for critical testing of SIQ and SIQPlotteR and for critical reading of the manuscript. We thank Dr Bert van de Kooij for critical reading of the manuscript.
Contributor Information
Robin van Schendel, Human Genetics, Leiden University Medical Center, Leiden, The Netherlands.
Joost Schimmel, Human Genetics, Leiden University Medical Center, Leiden, The Netherlands.
Marcel Tijsterman, Human Genetics, Leiden University Medical Center, Leiden, The Netherlands; Institute of Biology Leiden, Leiden University, Leiden, The Netherlands.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Dutch Cancer Society [2020-1/12925 to J.S., 11251/2017-2 to M.T.]; Holland Proton Therapy Centre [2019020-PROTON-DDR to M.T.]; Netherlands Organization for Scientific Research for Earth and Life Sciences (NWO) [OP.393 to M.T.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schimmel J., Muñoz-Subirana N., Kool H., van Schendel R., Tijsterman M. Small tandem DNA duplications result from CST-guided Pol α-primase action at DNA break termini. Nat. Commun. 2021; 12:4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014; 42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloh K., Kanchana R., Bialk P., Banas K., Zhang Z., Yoo B.C., Kmiec E.B. Deconvolution of Complex DNA Repair (DECODR): establishing a novel deconvolution algorithm for comprehensive analysis of CRISPR-edited Sanger sequencing data. CRISPR J. 2021; 4:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conant D., Hsiau T., Rossi N., Oki J., Maures T., Waite K., Yang J., Joshi S., Kelso R., Holden K. et al. Inference of CRISPR edits from Sanger trace data. CRISPR J. 2022; 5:123–130. [DOI] [PubMed] [Google Scholar]
- 5. Huang W., Li L., Myers J.R., Marth G.T. ART: a next-generation sequencing read simulator. Bioinformatics. 2011; 28:593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Labun K., Guo X., Chavez A., Church G., Gagnon J.A., Valen E. Accurate analysis of genuine CRISPR editing events with ampliCan. Genome Res. 2019; 29:843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fauser F., Schiml S., Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014; 79:348–359. [DOI] [PubMed] [Google Scholar]
- 9. Lazo G.R., Stein P.A., Ludwig R.A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (NY). 1991; 9:963–967. [DOI] [PubMed] [Google Scholar]
- 10. den Dulk-Ras A., Hooykaas P.J. Electroporation of Agrobacterium tumefaciens. Methods Mol. Biol. 1995; 55:63–72. [DOI] [PubMed] [Google Scholar]
- 11. Vergunst A.C., de Waal E.C., Hooykaas P.J. Root transformation by Agrobacterium tumefaciens. Methods Mol. Biol. 1998; 82:227–244. [DOI] [PubMed] [Google Scholar]
- 12. de Pater S., Neuteboom L.W., Pinas J.E., Hooykaas P.J., van der Zaal B.J. ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 2009; 7:821–835. [DOI] [PubMed] [Google Scholar]
- 13. Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schimmel J., van Schendel R., den Dunnen J.T., Tijsterman M. Templated insertions: a smoking gun for polymerase theta-mediated end joining. Trends Genet. 2019; 35:632–644. [DOI] [PubMed] [Google Scholar]
- 15. Bothmer A., Phadke T., Barrera L.A., Margulies C.M., Lee C.S., Buquicchio F., Moss S., Abdulkerim H.S., Selleck W., Jayaram H. et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 2017; 8:13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen P.J., Hussmann J.A., Yan J., Knipping F., Ravisankar P., Chen P.F., Chen C., Nelson J.W., Newby G.A., Sahin M. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021; 184:5635–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schiml S., Fauser F., Puchta H. Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proc. Natl Acad. Sci. U.S.A. 2016; 113:7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schimmel J., Kool H., van Schendel R., Tijsterman M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017; 36:3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng W., Simpson D.A., Cho J.E., Carvajal-Garcia J., Smith C.M., Headley K.M., Hathaway N., Ramsden D.A., Gupta G.P. Marker-free quantification of repair pathway utilization at Cas9-induced double-strand breaks. Nucleic Acids Res. 2021; 49:5095–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell. 2016; 63:633–646. [DOI] [PubMed] [Google Scholar]
- 21. Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P., Ramsden D.A. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell. 2016; 63:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018; 563:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002; 31:405–409. [DOI] [PubMed] [Google Scholar]
- 25. Koole W., van Schendel R., Karambelas A.E., van Heteren J.T., Okihara K.L., Tijsterman M. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014; 5:3216. [DOI] [PubMed] [Google Scholar]
- 26. Kruisselbrink E., Guryev V., Brouwer K., Pontier D.B., Cuppen E., Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 2008; 18:900–905. [DOI] [PubMed] [Google Scholar]
- 27. van Schendel R., Romeijn R., Buijs H., Tijsterman M. Preservation of lagging strand integrity at sites of stalled replication by Pol α-primase and 9-1-1 complex. Sci. Adv. 2021; 7:eabf2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Höijer I., Emmanouilidou A., Östlund R., van Schendel R., Bozorgpana S., Tijsterman M., Feuk L., Gyllensten U., Hoed den, Ameur A CRISPR–Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 2022; 13:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindsay H., Burger A., Biyong B., Felker A., Hess C., Zaugg J., Chiavacci E., Anders C., Jinek M., Mosimann C. et al. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 2016; 34:701–702. [DOI] [PubMed] [Google Scholar]
- 30. Kurgan G., Turk R., Li H., Roberts N., Rettig G.R., Jacobi A.M., Tso L., Sturgeon M., Mertens M., Noten R. et al. CRISPAltRations: a validated cloud-based approach for interrogation of double-strand break repair mediated by CRISPR genome editing. Mol. Ther. Methods Clin. Dev. 2021; 21:478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The latest versions of SIQ and SIQPlotteR are available in the GitHub repository: https://github.com/RobinVanSchendel/SIQ/releases/latest. Since this software is commonly used in our lab, we expect to develop and extend it further in the future.