Abstract
RNA plays a critical role in many biological processes, and the structures it adopts are intimately linked to those functions. Among many factors that contribute to RNA folding, van der Waals interactions between adjacent nucleobases stabilize structures in which the bases are stacked on top of one another. Here, we utilize fluorescence-detected circular dichroism spectroscopy (FDCD) to investigate base-stacking heterogeneity in RNA labeled with the fluorescent adenine analogue 2-aminopurine (2-AP). Comparison of standard (transmission-detected) CD and FDCD spectra reveals that in dinucleotides, 2-AP fluorescence is emitted almost exclusively by unstacked molecules. In a trinucleotide, some fluorescence is emitted by a population of stacked and highly quenched molecules, but more than half originates from a minor ∼10% population of unstacked molecules. The combination of FDCD and standard CD measurements reveals the prevalence of stacked and unstacked conformational subpopulations as well as their relative fluorescence quantum yields.
The functions of biological macromolecules are intimately linked to their structures. Ribonucleic acid (RNA) can fold into a wide variety of 3-dimensional structures that endow it with unique catalytic and regulatory properties. RNA structure is determined by several different types of interactions.1 Cations such as Na+ and Mg2+ enable RNA to adopt compact conformations by neutralizing the negatively charged sugar–phosphate backbone and, in some cases, binding at specific sites.2,3 Hydrogen bonding between nucleobases gives rise to the phenomenon of Watson–Crick base pairing, in which adenine (A) residues base-pair with uracil (U), and guanine (G) base-pairs with cytosine (C). Many types of non-Watson–Crick base pairs also occur in RNA.4 In addition, van der Waals interactions between the aromatic nucleobases help to stabilize structures in which the bases stack on top of one another.5 Base stacking contributes significantly to the stability of both RNA and DNA structures,6 and stacking between nucleobases and aromatic amino acids plays a role in RNA binding by certain proteins.7,8 Bases fluctuate between stacked and unstacked conformations on a microsecond time scale,9,10 and these transient structures may provide targets that are kinetically trapped by ligand or protein binding.11−13 Base stacking heterogeneity therefore plays a crucial role in nucleic acid structure and function.
A powerful approach to monitoring base stacking is the use of fluorescent base analogues (FBAs). These are variants of the natural bases that are chemically modified in order to render them fluorescent, with the goal of exerting minimal perturbations on the nucleic acid they are used to study. Many FBAs have been developed,14−20 but the adenine analogue 2-aminopurine (2-AP) is by far the most widely utilized.21 When free in solution, 2-AP riboside (Figure 1A; R = ribose) exhibits red-shifted absorption relative to the natural bases and strong fluorescence with a quantum yield of ∼0.68 and a lifetime of ∼10 ns (Figure 1B,C).21−23 Upon incorporation into nucleic acids, it is quenched up to 100-fold in a sequence- and structure-dependent manner.21,24,25 This change can be ascribed to alterations in the relative energies of bright π–π* and dark n−π* transitions that result from stacking interactions and charge transfer with neighboring bases.26,27 Incorporation into RNA and DNA also leads to complex decays in time-resolved fluorescence experiments that can be fit with multiple exponential decay components,28 continuous lifetime distributions,29 or a combination thereof,30 indicating the presence of base stacking heterogeneity.
Figure 1.
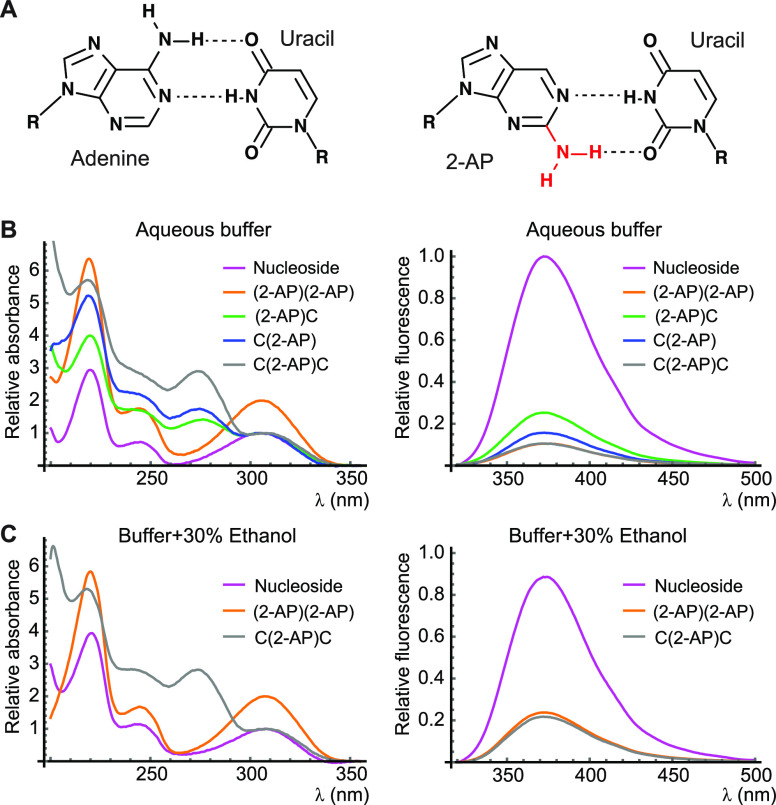
(A) Watson–Crick base pairs between adenine and uracil (left) and 2-AP and uracil (right). Chemical modification is indicated in red. (B and C) Absorbance (left) and fluorescence emission (right) spectra of the samples used in this study in aqueous phosphate buffer (B) or phosphate buffer containing 30% ethanol (C). Absorbance spectra have been scaled so that the longest-wavelength absorbance peak has a value of 2 for 2-AP dinucleotide (orange) and 1 for all other samples. Fluorescence spectra report the intensity per 2-AP residue and have been scaled such that free 2-AP riboside in aqueous buffer has a peak intensity of 1.
In this work, we use standard (transmission-detected) and fluorescence-detected circular dichroism spectroscopy (CD and FDCD, respectively) and time-correlated single-photon counting (TCSPC) to probe base stacking in 2-AP-labeled RNA oligonucleotides. In dinucleotides containing 2-AP, we detect fluorescence only from unstacked conformations with spectra that are nearly identical to isolated 2-AP riboside. In contrast, fluorescence is observed from stacked conformations in a trinucleotide, enabling us to determine the populations and relative fluorescence quantum yields of stacked and unstacked conformational states using a parameter-free model. This work provides a unique window into the distribution of conformations that are present in solution, as well as the effects of oligonucleotide structure on 2-AP photophysics.
We first recorded fluorescence, CD, and FDCD spectra of 2-AP riboside and dinucleotide. 2-AP dinucleotide exhibited 9.7-fold fluorescence quenching relative to 2-AP riboside (Figures 1B and S1), and significant dark (τ < 200 ps) and partially quenched (τ = 2 ns) populations were observed through TCSPC (Table 1 and Figure S2). As previously observed for 2-AP (deoxyribo)dinucleotide,31 2-AP (ribo)dinucleotide exhibited a sigmoidal CD spectrum arising from coupling between strong π–π* transitions on each base,22,32 with features centered around its electronic transition bands at 220 and 310 nm (Figure 2C). In contrast, 2-AP riboside exhibited a weak CD signal that was nonzero only at wavelengths <240 nm, arising from coupling between high-energy electronic transitions on the ribose sugar and the base (Figure 2B).32 Surprisingly, the FDCD spectra of both samples exhibited extremely similar lineshapes (Figure S3). This led us to hypothesize that fluorescence from 2-AP dinucleotide was produced almost entirely by emitting species that are unstacked (Figure 2A). The CD spectra of these conformations would contain only contributions similar to those observed in the nucleoside.
Table 1. Observed Average Lifetime (⟨τ⟩obs) and Populations (αic) and Lifetimes (τi) of Individual Decay Components Obtained through TCSPCa.
| sample | solvent | ⟨τ⟩obs | α0 (%) | α1c | τ1 (ns) | α2c | τ2 | α3c | τ3 |
|---|---|---|---|---|---|---|---|---|---|
| 2-AP riboside | B | 9.17 | 0 | 2.9 | 2.58 | 97.1 | 9.37 | ||
| 2-AP riboside | B+E | 8.40 | 0 | 10.9 | 1.25 | 89.1 | 9.28 | ||
| (2-AP)(2-AP) | B | 2.95 | 68.6 | 27.5 | 2.03 | 3.9 | 9.46 | ||
| (2-AP)(2-AP) | B+E | 4.16 | 42.3 | 47.7 | 3.55 | 10.0 | 7.05 | ||
| (2-AP)C | B | 5.02 | 54.3 | 4.0 | 0.29 | 28.7 | 3.65 | 13.0 | 9.49 |
| C(2-AP) | B | 2.51 | 42.0 | 11.5 | 0.23 | 41.3 | 2.43 | 5.2 | 8.35 |
| C(2-AP)C | B | 4.11 | 77.0 | 15.2 | 2.59 | 7.8 | 7.11 | ||
| C(2-AP)C | B+E | 5.14 | 55.8 | 15.1 | 2.77 | 29.1 | 6.36 |
α0 is a dark population whose decays are not resolved (τ < 200 ps). The populations αic have been corrected to account for the presence of the dark population (see Experimental Methods for details). “B”, aqueous buffer; “B+E”, buffer containing 30% ethanol.
Figure 2.
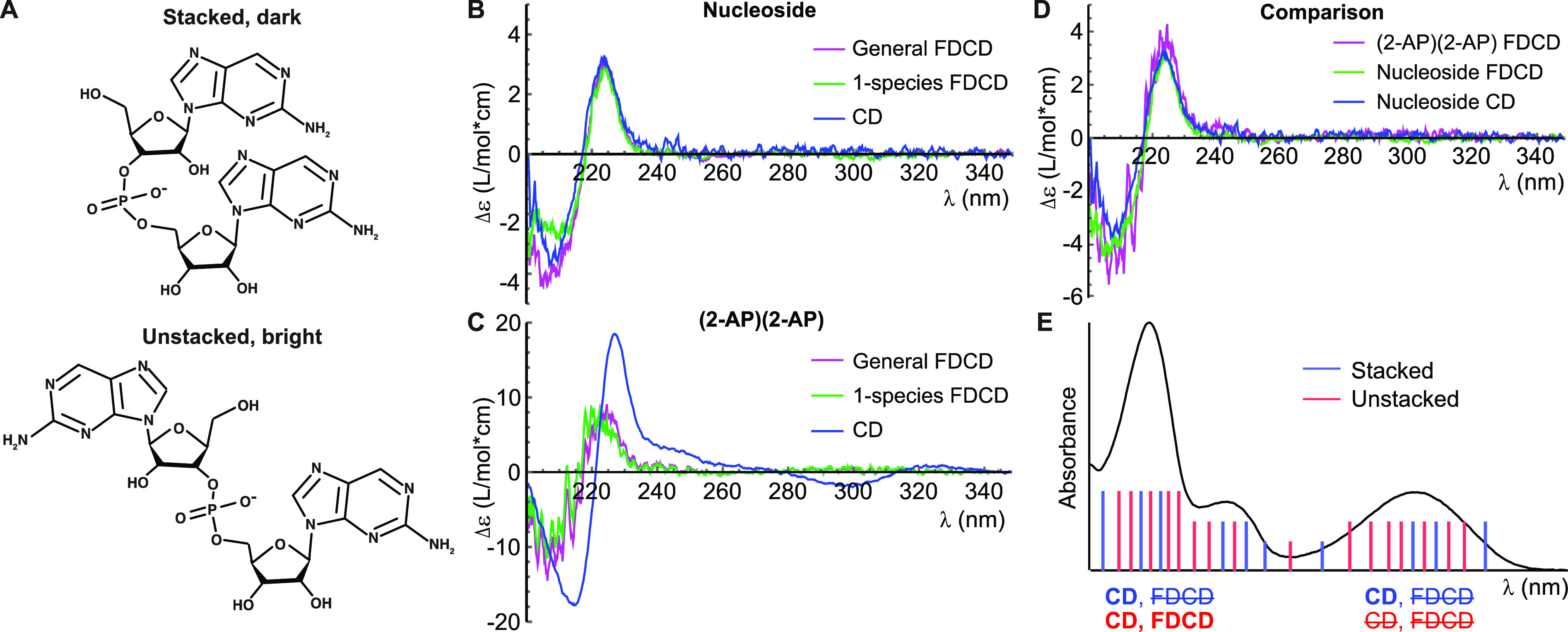
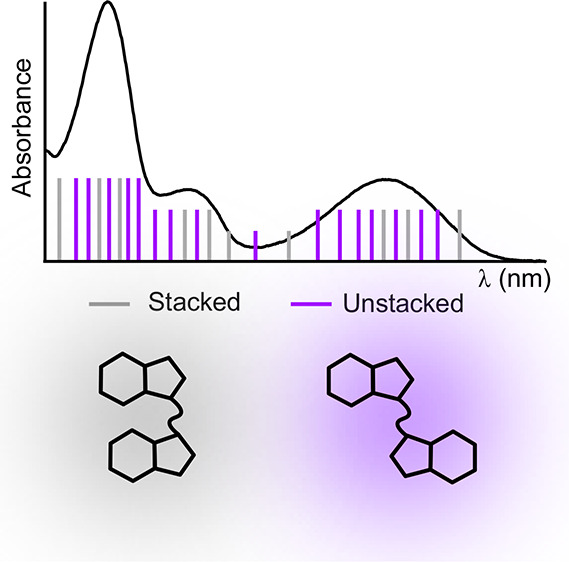
(A) Schematic of 2-AP dinucleotide in a stacked conformation (top) in which it is highly quenched and a brighter unstacked conformation (bottom). (B) Spectra of 2-AP riboside. (C) Spectra of 2-AP dinucleotide. Magenta, FDCD spectra processed with eq 1; green, FDCD spectra processed with eq 2; blue, standard CD spectra. (D) Magenta: FDCD spectrum of 2-AP dinucleotide processed using eq 1 under the assumption that all observed fluorescence comes from unstacked “nucleoside-like” structures. This spectrum shows strong agreement with the nucleoside FDCD (green) and CD (blue) spectra. (E) Illustration of how CD and FDCD report on different conformational subpopulations. The inhomogeneously broadened absorbance spectrum of 2-AP dinucleotide contains contributions (sticks) from stacked (blue) and unstacked (red) conformations. Labels below absorption bands indicate whether the stacked and/or unstacked conformations contribute to that band’s FDCD and/or CD signal. Nonzero CD is observed when 2-AP has optical activity, while nonzero FDCD is observed when 2-AP is fluorescent and optically active.
To test this hypothesis quantitatively, we used established theory33 to convert our FDCD spectra into predicted CD spectra of the fluorescent species in the sample: ΔεF = εF,L – εF,R, where εF,L(R) is the extinction coefficient of the fluorophore only under excitation with left (right)-handed circularly polarized light. These predictions will henceforth be called “processed” FDCD spectra. ΔεF can be predicted using eq 1, termed the “general FDCD equation”, where εF is the polarization-independent extinction coefficient of the fluorophore only, θ the FDCD signal in units of millidegrees, CD the total CD signal (including both fluorescent and nonfluorescent species) in units of millidegrees, and A the total absorbance:
| 1 |
The right-hand side of eq 2 will henceforth be abbreviated as “FDCDadj”, as it contains the raw FDCD signal θ adjusted to account for the overall absorbance and CD of the sample. If there are no species present that absorb but are nonfluorescent, then ΔεF/εF = ΔA/A. Given that CD = ΔεF·32980·c·l, where c is the concentration of the fluorophore and l is the path length, eq 1 can be modified to eq 2, termed the “1-species FDCD equation”:
| 2 |
The measured CD spectrum of 2-AP riboside agrees closely with the processed FDCD spectrum, with both the lineshape and the intensity being accurately predicted (Figure 2B). In contrast, the CD and processed FDCD spectra bear little resemblance for 2-AP dinucleotide (Figure 2C), indicating the presence of multiple structures that interconvert on a time scale slower than the fluorescence lifetime. If, as hypothesized above, the “fluorophore” in the dinucleotide sample is a “nucleoside-like” unstacked species (Figure 2A), then eq 2 must be altered such that εF is that of the nucleoside rather than the dinucleotide. When this adjustment is made, the processed FDCD spectra of both samples agree closely with each other and with the nucleoside’s measured CD spectrum (Figure 2D), indicating that the fluorescent species in the dinucleotide sample has the same CD spectrum as the nucleoside. A similar conclusion was previously drawn in an FDCD study of dinucleotides containing the fluorophore 1,N6-ethenoadenosine.34 We confirmed the chemical purity of our samples using thin-layer chromatography, demonstrating that the fluorescence arises from an unstacked conformational subpopulation rather than monomers generated by chemical degradation of 2-AP dinucleotide (Figure S4).
We next investigated a series of dinucleotides and trinucleotides in which 2-AP was placed adjacent to cytosine residues. Cytosine was chosen to avoid the complicating factors of energy transfer from adjacent bases to 2-AP, which is most efficient for adenine,35 and quenching of 2-AP fluorescence due to charge transfer to neighboring bases, which is most efficient for guanine.36,37 The dinucleotide (2-AP)C exhibited 4.0-fold fluorescence quenching relative to free 2-AP riboside and a strong CD signal across the UV, including at wavelengths >300 nm where only 2-AP absorbs directly (Figure 3A). C(2-AP) exhibited 6.3-fold quenching and minimal CD signal >300 nm (Figure 3B). For both dinucleotides, FDCD spectra processed using eq 1 predicted that the CD spectra of the fluorescent species in the sample were nearly identical to that of the free nucleoside (Figure 3D). In 2-AP dinucleotide, (2-AP)C and C(2-AP), TCSPC measurements identified populations with lifetimes of 2–4 ns and 8–10 ns, as well as a significant (40–70%) dark population with lifetime <200 ps (Table 1). The FDCD spectra suggest that even the partially quenched (τ = 2–4 ns) population is sufficiently unstacked to have a CD spectrum that lacks contributions from base–base coupling. We conclude that in all three dinucleotides, fluorescence comes almost exclusively from an ensemble of nucleoside-like unstacked conformations that includes both unquenched and partially quenched subpopulations.
Figure 3.
FDCD and CD spectra of (2-AP)C (A), C(2-AP) (B), and C(2-AP)C (C). Magenta, FDCD spectra processed with eq 1; green, standard CD spectra. (D) Overlay of FDCD spectra for all aqueous samples, processed with eq 1.
In the trinucleotide C(2-AP)C, fluorescence was quenched to a greater degree (9.7-fold relative to 2-AP riboside), indicating a shift in the conformational ensemble toward stacked structures. In addition, a nonzero FDCD signal was observed in the lowest-energy absorption band of 2-AP around 310 nm, indicating that some fluorescence originated from conformations with optical activity at that wavelength (Figure 3C). However, the FDCD spectrum predicts a CD intensity that is smaller than what is observed experimentally. To determine the implications of the discrepancy between the trinucleotide’s CD and FDCD spectra, we assumed that it exists in equilibrium between an ensemble of unstacked conformations with the same fluorescence quantum yield and CD spectrum as 2-AP riboside and an ensemble of stacked conformations (Figure 4A). In aqueous buffer, we observed 9.7-fold fluorescence quenching in the trinucleotide compared to the nucleoside, allowing us to write
where cnuc, cun, and cst are the concentrations of the nucleoside, stacked population, and unstacked population, respectively, and ϕnuc, ϕun, and ϕst are their fluorescence quantum yields. Assuming that the quantum yield of the unstacked population is equal to that of the nucleoside, and accounting for the fact that the sum of the concentrations of stacked and unstacked populations is equal to the total sample concentration, we obtain eq 3:
| 3 |
where ctot is the total sample concentration and Rϕ = ϕnuc/ϕst is the ratio of the quantum yields of the nucleoside and stacked conformation.
Figure 4.
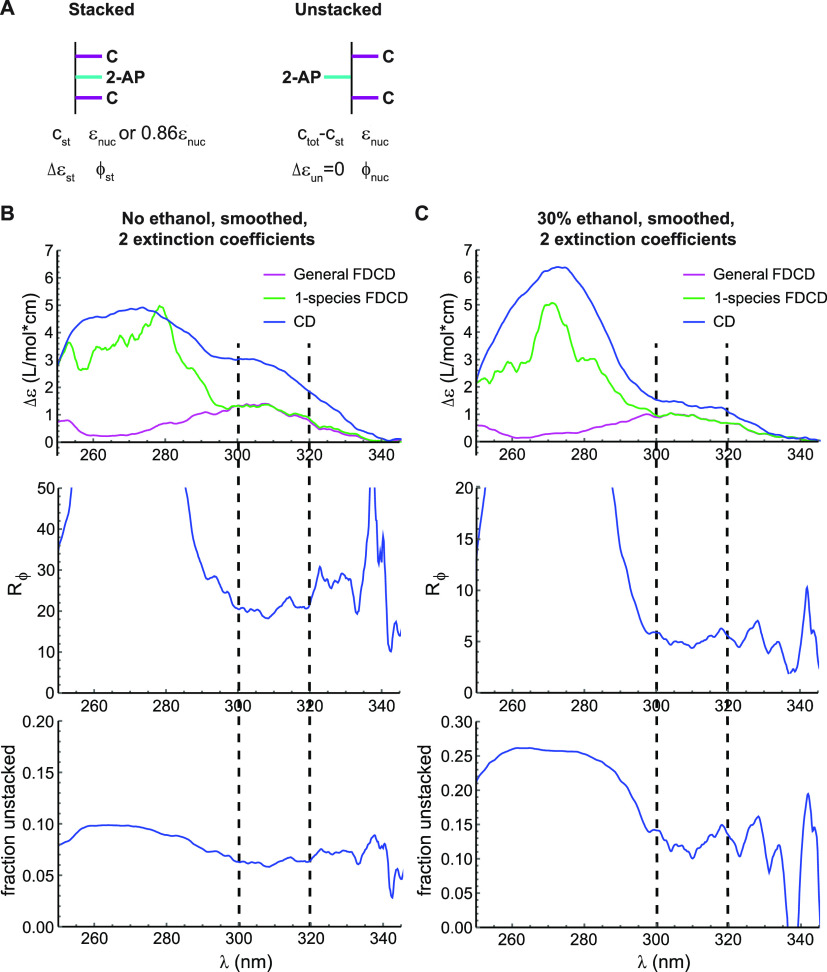
Fluorescence quenching and stacking heterogeneity in C(2-AP)C. (A) Model used for data analysis. In the unstacked (“un”) conformation, 2-AP has properties characteristic of the free nucleoside “nuc”, while the stacked conformation (“st”) has a different CD signal and fluorescence quantum yield. c = concentration, ε = extinction coefficient, Δε = εL – εR, ϕ = quantum yield. (B) Results of the “smoothed, 2 extinction coefficient” model for C(2-AP)C in aqueous buffer. Top: Close-up of the long-wavelength region of Figure 3C. Magenta, FDCD spectra processed with eq 1; green, FDCD spectra processed with eq 2; blue, standard CD spectra. Middle: Value of the quenching ratio Rϕ obtained at each wavelength by solving the system of eqs 3 and 7. Bottom: Value of the fraction unstacked fun = (ctot – cst)/ctot obtained at each wavelength. Dashed lines indicate the wavelength range over which parameter values were quantified. (C) Analogous plots for C(2-AP)C in buffer containing 30% v/v ethanol.
At wavelengths longer than ∼300 nm, native base absorption is negligible and ΔεF/εF = ΔA/A could potentially apply. Indeed, the 1-species and general equations agree with each other at these wavelengths, indicating that the signals are not impacted by direct absorbance by the C residues. 2-AP riboside exhibits no CD signal at these wavelengths (Figure 2B), so the CD of the trinucleotide comes entirely from the stacked population (eq 4). This step sorts the continuum of structures potentially adopted by an oligonucleotide into an ensemble of unstacked conformations with zero CD signal at the wavelengths being considered and an ensemble of stacked conformations that contribute the entire CD signal.
| 4 |
The FDCD spectra of samples containing multiple fluorescence species can be processed using eq 5, where i counts over all fluorescent species, whose contributions are weighted by their quantum yields ϕi and concentrations ci:
| 5 |
Plugging in the properties of the stacked and unstacked populations into eq 5 yields eq 6:
| 6 |
Noting that Δεnuc = 0 at the wavelengths being considered and plugging in eq 4 for cstΔεst (λ), we obtain eq 7:
| 7 |
One obtains estimates of Rϕ and cst by solving the system of eqs 3 and 7 at each wavelength based on the experimental measurements of CD, A, θ, and ε (Figure 4 and Table 2), with no free parameters required. We investigated a model in which the extinction coefficient of the nucleoside was used for both the stacked and unstacked populations (εnuc = εun = εst) and one in which they have different extinction coefficients (Figures S5 and S6). For the latter, the difference in extinction coefficient between the two states was estimated by measuring absorbance of 2-AP riboside and C(2-AP)C as a function of temperature, which yielded εst = 0.86εnuc = 0.86εun (Figure S7; see Experimental Methods for details). For C(2-AP)C, the resulting values of Rϕ and cst are stable across the range of wavelengths in which the stated assumptions hold and the sample exhibits appreciable signal (300–320 nm) and are largely independent of whether data smoothing is used and whether one or two extinction coefficients are used (Table 2). For example, with data smoothed using a 5 nm rolling average and εst = 0.86εun, we obtain Rϕ = 23 ± 1 and an unstacked population of 6.3 ± 0.2% of the total (mean ± standard deviation across all data points from 300 to 320 nm). This indicates that the stacked conformation is quenched to a greater extent than one might assume based on the overall 9.7-fold fluorescence intensity ratio (free nucleoside:trinucleotide) and that a minor unstacked population of only 6.3% of the total gives rise to 61% of the fluorescence emitted by the sample.
Table 2. Quenching Ratios and Unstacked Populations for C(2-AP)Ca.
| buffer |
30% ethanol |
||||||
|---|---|---|---|---|---|---|---|
| smoothing | ε | Rϕ | % un | % fl un | Rϕ | % un | % fl un |
| none | 1 | 22 ± 4 | 5.9 ± 0.7 | 58 | 6 ± 1 | 11 ± 4 | 42 |
| 5 nm | 1 | 22 ± 1 | 6.0 ± 0.3 | 59 | 5.7 ± 0.5 | 11 ± 2 | 42 |
| none | 2 | 23 ± 4 | 6.2 ± 0.6 | 61 | 6 ± 1 | 12 ± 3 | 48 |
| 5 nm | 2 | 23 ± 1 | 6.3 ± 0.2 | 61 | 6.2 ± 0.5 | 12 ± 1 | 47 |
Results are shown for models with and without smoothing by a 5 nm rolling average, and with (ε = 2) and without (ε = 1) accounting for hypochromicity of 2-AP in the stacked conformation. Rϕ = quenching ratio (ϕun/ϕst); % un = percent unstacked; % fl un = % of total fluorescence that comes from the unstacked population. Mean ± standard deviation of the results obtained at all data points in the range of 300–320 nm.
We repeated these measurements on 2-AP dinucleotide and C(2-AP)C in phosphate buffer containing 30% v/v ethanol (mole fraction ∼0.1), which is thought to shift population away from stacked conformations.31 Addition of ethanol at this concentration increases the viscosity of pure H2O from 0.89 × 103 Pa·s to approximately 2 × 103 (ref (38)) and decreases its dielectric constant from 78.41 to approximately 66.39 It has previously been shown that a decrease in dielectric constant of this magnitude impacts the morphology of double-stranded DNA condensates40 and promotes folding of certain RNA species,41 potentially by enhancing counterion condensation around the phosphate backbone. This is not expected to be a dominant factor for our short single-stranded oligonucleotides, which have only 1–2 phosphate groups per molecule.
As expected from the known effects of ethanol on base stacking, the oligonucleotides were quenched to a lesser degree in 30% ethanol (3.5-fold for 2-AP dinucleotide and 3.7-fold for C(2-AP)C relative to 2-AP riboside in the same solvent; Figures 1C and S1). In TCSPC measurements, the slowest decay component shortened and increased in amplitude, while the dark population decreased (Table 1). The CD spectrum of 2-AP dinucleotide decreased in intensity, and its lineshape changed at long wavelengths, whereas the FDCD spectrum remained nearly unchanged (Figure S8). With fluorescence coming almost exclusively from unstacked conformations even in an aqueous buffer, it was unsurprising that the same observation held when ethanol was used to shift the stacking equilibrium. The CD spectrum of C(2-AP)C showed more complex changes when ethanol was added (Figures 4 and S8). Our model suggested that the quenching ratio Rϕ decreases significantly to only 6, while the unstacked population doubles to 12% of the total, resulting in 47% of fluorescence being emitted by the unstacked population (Table 2). The increase in unstacked population was expected, but the large decrease in the quenching ratio suggests that ethanol alters the structure of the stacked population, mitigating its quenching. An alteration of the stacked structure is consistent with the complex changes in the CD spectrum that are observed upon addition of ethanol to C(2-AP)C.
2-AP-labeled dinucleotides and trinucleotides have previously been investigated by CD and time-resolved fluorescence measurements,24,30,31,42,43 although the DNA versions of these small model systems have been far more studied than their RNA counterparts. Two decay components (8.1 and 1.1 ns) were resolved using frequency-domain fluorescence lifetime measurements for 2-AP deoxyribodinucleotide [d(2-AP)d(2-AP)]. The slower (8.1 ns) and predominant (∼64%) component approached the 9.0 ns lifetime of free 2-AP determined in that study.31 We similarly observed decays of 9.5 and 2.0 ns in 2-AP ribodinucleotide, but the shorter (2.0 ns) one was predominant, a pattern that was recapitulated in (2-AP)C and C(2-AP). Similar behavior was previously observed in d(2-AP)dC43 and dCd(2-AP),24 each of which exhibits a minor population (∼2% at 20 °C) with a lifetime of about 8 ns, a larger population (∼36%) with a lifetime of about 2.5 ns, and two subnanosecond decay components. These results show that the members of this family of deoxyribo- and ribodinucleotides have minor unquenched conformational states that persist for at least the fluorescence lifetime, in addition to larger partially quenched populations. Significantly, our FDCD results demonstrate that these partially quenched populations are still highly unstacked, with CD spectra that are nearly identical to that of free 2-AP riboside. Consequently, the ensemble of conformations that gives rise to the CD signal at wavelengths >240 nm has minimal overlap with the ensemble that gives rise to fluorescence.
Three exponential decay components were observed in 2-AP-labeled DNA trinucleotides dXd(2-AP)dX, but the slowest was only ∼3.5 ns at 20 °C, significantly shorter than the lifetime of free 2-AP. It was concluded that 2-AP persists in a fully unstacked conformation for less than 10 ns, with fluctuations back to stacked conformations “gating” decay to the ground state.42 Slower stacking dynamics on the time scale of microseconds have been observed through single-molecule FRET9 and temperature-jump infrared spectroscopy.10 Gating appears to be less significant in C(2-AP)C, which exhibits a slowest decay component of 7.1 ns in an aqueous buffer. The lifetime of this component decreases to 6.4 ns in 30% ethanol despite an increase in viscosity that would be expected to slow the stacking/unstacking kinetics, suggesting that gating is not responsible for the reduction in lifetime. Furthermore, because FDCD is a fluorescence-detected excitation spectroscopy, the spectral intensity and lineshape reflect species that are present on the time scale of absorption rather than emission (fs rather than ps–ns). Dynamics occurring on time scales faster than the fluorescence lifetime would alter the apparent quantum yields of different conformations but would not alter their excitation spectra. Our results suggest that there is a population present in 2-AP-labeled RNA dinucleotides and trinucleotides that is sufficiently unstacked to lack CD contributions from coupling between 2-AP and the adjacent bases.
The 2-state model presented here sorts intermediate structures into stacked or unstacked ensembles based on whether they contribute to the CD spectrum. In the dinucleotides studied here, the “zero CD” ensemble includes both unquenched and partially quenched subpopulations identified through TCSPC. In contrast, in C(2-AP)C, the prevalence of the least quenched subpopulation alone is comparable to (in buffer) or larger than (in 30% EtOH) the entire “zero CD” population (Tables 1 and 2). Free energies determined through NMR44,45 and temperature-dependent UV spectroscopy46 show that stacking is slightly disfavorable in rArC and rCrA, while molecular dynamics simulations predict it to be slightly favorable.47 Assuming that 2-AP exhibits stacking thermodynamics similar to A, this is consistent with the assertion above that the ensemble of unstacked conformations encompasses more structures and is more populated than the minor unquenched conformation observed in lifetime measurements. For C(2-AP)C, the free energy change resulting from a transition from the stacked to the unstacked structure depicted in Figure 4A can be very roughly estimated by adding together the enthalpies of unstacking of rArC and rCrA, and averaging their entropies of unstacking and multiplying by 1.5 (because complete trinucleotide unstacking liberates three bases rather than two). Using values from ref (46), ΔG = ΔH – TΔS yields a rough estimate of ΔGunstack = +2.1 kcal/mol at 298 K, corresponding to an unstacked population of 3%. This suggests that our model’s estimate of 6% is a reasonable value based on stacking energetics.
Base stacking plays a critical role in determining the structural free energy landscapes of RNA and DNA. The work presented here provides insight into the thermodynamics of base stacking in RNA and the photophysical behavior of the FBAs that are widely used to study it. Future work will include FDCD measurements on more complex RNA systems and FDCD measurements with FBAs other than 2-AP.
Experimental Methods
Materials. RNA di- and trinucleotides were purchased from Dharmacon (Horizon Discovery) and deprotected and desalted by the manufacturer. 2-AP riboside was purchased from TriLink Biotechnologies and 2-AP nucleobase was purchased from Sigma-Aldrich. Except where otherwise noted, all measurements were performed in a buffer containing 20 mM NaiPO4 and 100 mM NaCl at pH 7.5.
Spectroscopy. FDCD and CD measurements were performed on a Jasco J1500 circular dichroism spectrometer with the sample in a 1 cm path length cylindrical cell inserted into a Jasco FDCD-551 cell holder. This holder envelops the cell in an elliptical mirror,48 which directs fluorescence into a PMT (Hamamatsu R374) at 90° to the excitation light. A long-pass colored glass filter with a 380 nm cutoff was placed in front of the fluorescence detector to remove scattered light. Scans were performed with an excitation bandwidth of 4 nm, an integration time of 4 s, and a scan speed of 20 nm/min. The spectra that are shown are the average of 10 scans for FDCD and 5 scans for CD, and samples were measured at a concentration of 5 μM. The measurement channels “FDCD” and “DC” were both recorded, with the former yielding signal proportional to FL – FR and the latter yielding signal proportional to FL + FR (essentially, the fluorescence excitation spectrum without correction for lamp intensity at different wavelengths) (Figure S3). FDCD baselines were recorded individually for each sample using the achiral 2-AP nucleobase (Figure 1A, R = H) at a concentration that yielded the same brightness as the sample when excited at 305 nm. CD, absorbance, and fluorescence baselines were recorded using buffer.
Thermal ramps were carried out at a concentration of 20 μM, ramping from 7 to 80 °C at a rate of 1 °C per minute while monitoring absorbance at 308 nm. Fluorescence emission spectra were recorded on a Molecular Devices SpectraMax I3 fluorometer using a plate reader attachment with excitation at 295 nm and the sample at a concentration of 500 nM. Fluorescence lifetime measurements were performed on a Horiba Fluoromax-5 with a TemPro lifetime system using a NanoLED340 pulsed light source and the same long-pass filter used for FDCD. Sample concentrations were 1 μM for 2-AP riboside and 5 μM for all other samples. Instrument response functions were collected using a dilute suspension of Ludox. The full width at half-maximum (FWHM) of the instrument response function (IRF) was approximately 900 ps, and the bin width was 55 ps.
Thin-Layer Chromatography. TLC experiments were performed using silica gel 60G F254 plates and a solvent system consisting of 50:40:3:15 n-butanol:acetone:33% ammonia:water, as previously described.49 One nanomole of nucleoside or dinucleotide was spotted, and after development, the plate was imaged on a 302 nm ultraviolet transilluminator.
Data Analysis. Most data processing and analysis was performed in Mathematica (Wolfram Alpha). The FDCD spectrum of 2-AP nucleobase was subtracted from the FDCD spectrum of the sample, and the DC spectrum of the solvent was subtracted from the DC spectrum of the sample. θ was then calculated by dividing the FDCD spectrum by the DC spectrum (Figure S3). We utilized the absorption spectra of 2-AP nucleoside in pure H2O as a proxy for the absorption spectra of the fluorophore for all aqueous samples containing a single 2-AP residue. Its spectrum was scaled to a value of 6 000 L/mol·cm at its longest-wavelength absorption peak, generating εF, which was then utilized in processing of FDCD spectra.33 The absorption spectrum of 2-AP dinucleotide in pure H2O was used to generate its own εF spectrum by scaling to a peak value of 12 000 L/mol·cm. Likewise, absorption spectra of 2-AP nucleoside and dinucleotide in 30% ethanol were used to generate εF spectra for analysis of the corresponding FDCD spectra.
The relative hypochromicity of the stacked conformation of C(2-AP)C was estimated by calculating the difference in 308 nm absorbance between C(2-AP)C and 2-AP nucleoside at the minimum and maximum temperatures sampled (7 and 80 °C, respectively; Figure S7). The difference at 80 °C was then subtracted from the difference at 7 °C, and the resulting value was divided by the absorbance of the trinucleotide at 7 °C. This yielded a value of 0.16, so we concluded that the extinction coefficient of the unstacked conformation was 1.16 times that of the stacked conformation. Taking εun = εnuc, we thus used εst = 0.86εnuc in our 2-extinction coefficient models. The system of eqs 3 and 7 has two solutions when separate extinction coefficients are used for the stacked and unstacked conformations, but only one is physically meaningful (the other has a stacked concentration that exceeds the overall sample concentration).
Fluorescence lifetime data were analyzed in Horiba Decay Analysis Software v6.6. Three-exponential models were used for all samples, and inclusion of additional exponentials yielded negligible improvement in the χ2 value and residuals (fitting parameters are in Table S1). Because of the 900 ps IRF width, decays with apparent time constants below 200 ps were considered unreliable and were subsumed into the dark population in downstream analysis. With <200 ps components excluded, the average observed lifetime was calculated according to ⟨τ⟩obs = Σαiτi, where τi is the time constant of exponential decay component i and αi is its amplitude. The dark population of molecules with lifetimes that were too short to resolve, α0, was calculated according to α0 = 1 – ⟨τnuc⟩/(⟨τsample⟩R), where ⟨τnuc⟩ is the average lifetime of 2-AP riboside, ⟨τsample⟩ is the average lifetime of the sample, and R is the ratio of the steady-state fluorescence intensity of 2-AP riboside to the sample.50 The amplitudes of the decay components were then corrected to account for the dark population according to αic = αi(1 – α0).
Acknowledgments
This work was funded by NIH R00 GM120457 to J.R.W.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.2c01778.
The authors declare no competing financial interest.
Supplementary Material
References
- Butcher S. E.; Pyle A. M. The Molecular Interactions That Stabilize RNA Tertiary Structure: RNA Motifs, Patterns, and Networks. Acc. Chem. Res. 2011, 44, 1302–1311. 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- Lipfert J.; Doniach S.; Das R.; Herschlag D. Understanding Nucleic Acid–Ion Interactions. Annu. Rev. Biochem. 2014, 83, 813–841. 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D. E. A Guide to Ions and RNA Structure. RNA 2004, 10, 335–343. 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K.; Li S.; Kaukonen T.; Colasanti A. V.; Xin Y.; Lu X.-J. Effects of Noncanonical Base Pairing on RNA Folding: Structural Context and Spatial Arrangements of G·A Pairs. Biochemistry 2019, 58, 2474–2487. 10.1021/acs.biochem.9b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala A.; Ali Z.; Bhattacharya S.; Halder A.; Mitra A.; Sharma P. On the Nature of Nucleobase Stacking in RNA: A Comprehensive Survey of Its Structural Variability and a Systematic Classification of Associated Interactions. J. Chem. Inf. Model. 2021, 61, 1470–1480. 10.1021/acs.jcim.0c01225. [DOI] [PubMed] [Google Scholar]
- Yakovchuk P.; Protozanova E.; Frank-Kamenetskii M. D. Base-Stacking and Base-Pairing Contributions into Thermal Stability of the DNA Double Helix. Nucleic Acids Res. 2006, 34, 564–574. 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N.; Allers J.; Myers J.; Shamoo Y. Protein–RNA Interactions: Exploring Binding Patterns with a Three-Dimensional Superposition Analysis of High Resolution Structures. Bioinformatics 2006, 22, 2746–2752. 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- Koh Y. Y.; Wang Y.; Qiu C.; Opperman L.; Gross L.; Hall T. M. T.; Wickens M. Stacking Interactions in PUF–RNA Complexes. RNA 2011, 17, 718–727. 10.1261/rna.2540311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israels B.; Albrecht C. S.; Dang A.; Barney M.; von Hippel P. H.; Marcus A. H. Submillisecond Conformational Transitions of Short Single-Stranded DNA Lattices by Photon Correlation Single-Molecule Förster Resonance Energy Transfer. J. Phys. Chem. B 2021, 125, 9426–9440. 10.1021/acs.jpcb.1c04119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik A. L.; Brauns E. B. Rearrangement of Partially Ordered Stacked Conformations Contributes to the Rugged Energy Landscape of a Small RNA Hairpin. Biochemistry 2008, 47, 10834–10840. 10.1021/bi801170c. [DOI] [PubMed] [Google Scholar]
- Krishnan R.; Blanco M. R.; Kahlscheuer M. L.; Abelson J.; Guthrie C.; Walter N. G. Biased Brownian Ratcheting Leads to Pre-mRNA Remodeling and Capture Prior to First-Step Splicing. Nat. Struct. Mol. Biol. 2013, 20, 1450–1457. 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala K. C.; Wang J.; Hou Q.; Walter N. G. Mg2+ Shifts Ligand-Mediated Folding of a Riboswitch from Induced-Fit to Conformational Selection. J. Am. Chem. Soc. 2015, 137, 14075–14083. 10.1021/jacs.5b09740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y. A.; Meyer I. M. Four RNA Families with Functional Transient Structures. RNA Biol. 2015, 12, 5–20. 10.1080/15476286.2015.1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bood M.; Füchtbauer A. F.; Wranne M. S.; Ro J. J.; Sarangamath S.; El-Sagheer A. H.; Rupert D. L. M.; Fisher R. S.; Magennis S. W.; Jones A. C.; Pentacyclic Adenine M.; et al. Pentacyclic Adenine: A Versatile and Exceptionally Bright Fluorescent DNA Base Analogue. Chem. Biol. 2018, 9, 3494–3502. 10.1039/C7SC05448C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin P.; Börjesson K.; Li H.; Maartensson J.; Brown T.; Wilhelmsson L. M.; Albinsson B. Characterization and Use of an Unprecedentedly Bright and Structurally Non-Perturbing Fluorescent DNA Base Analogue. Nucleic Acids Res. 2008, 36, 157–167. 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson L. M.; Sandin P.; Holmén A.; Albinsson B.; Lincoln P.; Nordén B. Photophysical Characterization of Fluorescent DNA Base Analogue, tC. J. Phys. Chem. B 2003, 107, 9094–9101. 10.1021/jp034930r. [DOI] [Google Scholar]
- Wilhelmsson L. M.; Holmén A.; Lincoln P.; Nielsen P. E.; Nordén B. A Highly Fluorescent DNA Base Analogue That Forms Watson–Crick Base Pairs with Guanine. J. Am. Chem. Soc. 2001, 123, 2434–2435. 10.1021/ja0025797. [DOI] [PubMed] [Google Scholar]
- Hawkins M. E. Fluorescent Pteridine Nucleoside Analogs. Cell. Biochem. Biophys. 2001, 34, 257–281. 10.1385/CBB:34:2:257. [DOI] [PubMed] [Google Scholar]
- Shin D.; Sinkeldam R. W.; Tor Y. Emissive RNA Alphabet. J. Am. Chem. Soc. 2011, 133, 14912–14915. 10.1021/ja206095a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A. R.; Fin A.; Tor Y. Chemical Mutagenesis of an Emissive RNA Alphabet. J. Am. Chem. Soc. 2015, 137, 14602–14605. 10.1021/jacs.5b10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. C.; Reich E. Fluorescence Studies of Nucleotides and Polynucleotides. J. Biol. Chem. 1969, 244, 1228–1237. 10.1016/S0021-9258(18)91833-8. [DOI] [PubMed] [Google Scholar]
- Holmén A.; Nordén B.; Albinsson B. Electronic Transition Moments of 2-Aminopurine. J. Am. Chem. Soc. 1997, 119, 3114–3121. 10.1021/ja9635600. [DOI] [Google Scholar]
- Neely R. K.; Magennis S. W.; Dryden D. T. F.; Jones A. C. Evidence of Tautomerism in 2-Aminopurine from Fluorescence Lifetime Measurements. J. Phys. Chem. B 2004, 108, 17606–17610. 10.1021/jp0490857. [DOI] [Google Scholar]
- Somsen O. J. G.; Hoek van A.; Amerongen van H. Fluorescence Quenching of 2-Aminopurine in Dinucleotides. Chem. Phys. Lett. 2005, 402, 61–65. 10.1016/j.cplett.2004.11.122. [DOI] [Google Scholar]
- Jones A. C.; Neely R. K. 2-Aminopurine as a Fluorescent Probe of DNA Conformation and the DNA-Enzyme Interface. Q. Rev. Biophys. 2015, 48, 244–279. 10.1017/S0033583514000158. [DOI] [PubMed] [Google Scholar]
- Jean J. M.; Hall K. B. 2-Aminopurine Fluorescence Quenching and Lifetimes: Role of Base Stacking. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 37–41. 10.1073/pnas.98.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J. M.; Hall K. B. 2-Aminopurine Electronic Structure and Fluorescence Properties in DNA. Biochemistry 2002, 41, 13152–13161. 10.1021/bi020308y. [DOI] [PubMed] [Google Scholar]
- Gelot T.; Tourón-Touceda P.; Crégut O.; Léonard J.; Haacke S. Ultrafast Site-Specific Fluorescence Quenching of 2-Aminopurine Studied by Femtosecond Down-Conversion. J. Phys. Chem. A 2012, 116, 2819–2825. 10.1021/jp212187m. [DOI] [PubMed] [Google Scholar]
- Bharill S.; Sarkar P.; Ballin J. D.; Gryczynski I.; Wilson G. M.; Gryczynski Z. Fluorescence Intensity Decays of 2-Aminopurine Solutions: Lifetime Distribution Approach. Anal. Biochem. 2008, 377 (2), 141–149. 10.1016/j.ab.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty A. C.; Jones A. C.; Camp P. J. Extraction of Lifetime Distributions from Fluorescence Decays with Application to DNA-Base Analogues. Phys. Chem. Chem. Phys. 2011, 13, 3819–3830. 10.1039/c0cp01779e. [DOI] [PubMed] [Google Scholar]
- Johnson N. P.; Baase W. A.; von Hippel P. H. Low-Energy Circular Dichroism of 2-Aminopurine Dinucleotide as a Probe of Local Conformation of DNA and RNA. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 3426–3431. 10.1073/pnas.0400591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K. E.; Johnson W. C.; Ho P. S.. Principles of Physical Biochemistry, 2nd ed.; Prentice Hall, 2006. [Google Scholar]
- Tinoco I. Jr; Turner D. H. Fluorescence Detected Circular Dichroism. Theory. J. Am. Chem. Soc. 1976, 98, 6453–6456. 10.1021/ja00437a003. [DOI] [Google Scholar]
- Reich C.; Tinoco I. Fluorescence-Detected Circular Dichroism of Dinucleoside Phosphates. A Study of Solution Conformations and the Two-State Model. Biopolymers 1980, 19, 833–848. 10.1002/bip.1980.360190409. [DOI] [Google Scholar]
- Xu D.-G.; Nordlund T. M. Sequence Dependence of Energy Transfer in DNA Oligonucleotides. Biophys. J. 2000, 78, 1042–1058. 10.1016/S0006-3495(00)76663-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill M. A.; Dohno C.; Barton J. K. Direct Chemical Evidence for Charge Transfer between Photoexcited 2-Aminopurine and Guanine in Duplex DNA. J. Am. Chem. Soc. 2004, 126, 1316–1317. 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]
- Wan C.; Fiebig T.; Schiemann O.; Barton J. K.; Zewail A. H. Femtosecond Direct Observation of Charge Transfer between Bases in DNA. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 14052–14055. 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattab I. S.; Bandarkar F.; Fakhree M. A. A.; Jouyban A. Density, Viscosity, and Surface Tension of Water + Ethanol Mixtures from 293 to 323K. Korean J. Chem. Eng. 2012, 29, 812–817. 10.1007/s11814-011-0239-6. [DOI] [Google Scholar]
- Moriyoshi T.; Ishii T.; Tamai Y.; Tado M. Static Dielectric Constants of Water + Ethanol and Water + 2-Methyl-2-Propanol Mixtures from 0.1 to 300 MPa at 298.15 K. J. Chem. Eng. Data 1990, 35, 17–20. 10.1021/je00059a005. [DOI] [Google Scholar]
- Arscott P. G.; Ma C.; Wenner J. R.; Bloomfield V. A. DNA Condensation by Cobalt Hexaammine(III) in Alcohol–Water Mixtures: Dielectric Constant and Other Solvent Effects. Biopolymers 1995, 36, 345–364. 10.1002/bip.360360309. [DOI] [PubMed] [Google Scholar]
- Gunawardhana S. M.; Holmstrom E. D. Apolar Chemical Environments Compact Unfolded RNAs and Can Promote Folding. Biophys. Rep. 2021, 1, 100004. 10.1016/j.bpr.2021.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J. M.; Hall K. B. Stacking–Unstacking Dynamics of Oligodeoxynucleotide Trimers. Biochemistry 2004, 43, 10277–10284. 10.1021/bi049701p. [DOI] [PubMed] [Google Scholar]
- Somsen O. J. G.; Keukens L. B.; de Keijzer M. N.; van Hoek A.; van Amerongen H. Structural Heterogeneity in DNA: Temperature Dependence of 2-Aminopurine Fluorescence in Dinucleotides. ChemPhysChem 2005, 6, 1622–1627. 10.1002/cphc.200400648. [DOI] [PubMed] [Google Scholar]
- Ezra F. S.; Lee C.-H.; Kondo N. S.; Danyluk S. S.; Sarma R. H. Conformational Properties of Purine-Pyrimidine and Pyrimidine-Purine Dinucleoside Monophosphates. Biochemistry 1977, 16, 1977–1987. 10.1021/bi00628a035. [DOI] [PubMed] [Google Scholar]
- Lee C.-H.; Ezra F. S.; Kondo N. S.; Sarma R. H.; Danyluk S. S. Conformational Properties of Dinucleoside Monophosphates in Solution: Dipurines and Dipyrimidines. Biochemistry 1976, 15, 3627–3639. 10.1021/bi00661a034. [DOI] [PubMed] [Google Scholar]
- Frechet D.; Ehrlich R.; Remy P.; Gabarro-Arpa J. Thermal Perturbation Differential Spectra of Ribonucleic Acids. II. Nearest Neighbour Interactions. Nucleic Acids Res. 1979, 7, 1981–2001. 10.1093/nar/7.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatshahi H. S.; Henriksen N. M.; Cheatham T. E. Consensus Conformations of Dinucleoside Monophosphates Described with Well-Converged Molecular Dynamics Simulations. J. Chem. Theory Comput. 2018, 14, 1456–1470. 10.1021/acs.jctc.7b00581. [DOI] [PubMed] [Google Scholar]
- Nehira T.; Tanaka K.; Takakuwa T.; Ohshima C.; Masago H.; Pescitelli G.; Wada A.; Berova N. Development of a Universal Ellipsoidal Mirror Device for Fluorescence Detected Circular Dichroism: Elimination of Polarization Artifacts. J. Appl. Spectrosc. 2005, 59, 121–125. 10.1366/0003702052940459. [DOI] [PubMed] [Google Scholar]
- Kolassa N.; Roos H.; Pfleger K. A Separation of Purine Derivatives by Thin-Layer Chromatography on Silica Gel Plates Suitable for Metabolic Studies. J. Chromatogr. A 1972, 66, 175–177. 10.1016/S0021-9673(01)82946-0. [DOI] [PubMed] [Google Scholar]
- Greiner V. J.; Kovalenko L.; Humbert N.; Richert L.; Birck C.; Ruff M.; Zaporozhets O. A.; Dhe-Paganon S.; Bronner C.; Mély Y. Site-Selective Monitoring of the Interaction of the SRA Domain of UHRF1 with Target DNA Sequences Labeled with 2-Aminopurine. Biochemistry 2015, 54, 6012–6020. 10.1021/acs.biochem.5b00419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.