Abstract
The linked resistance to nickel and cobalt of Ralstonia eutropha-like strain CH34 (Alcaligenes eutrophus CH34) is encoded by the cnr operon, which is localized on the megaplasmid pMOL28. The regulatory genes cnrYXH have been cloned, overexpressed, and purified in Escherichia coli. CnrY fractionated as a 10.7-kDa protein in in vitro translation assays. CnrX, a periplasmic protein of 16.5 kDa, was overproduced and purified as a histidine-tagged fusion protein in E. coli. His-CnrX was found to posses a secondary structure content rich in alpha-helical and beta-sheet structures. CnrH, a sigma factor of the extracytoplasmic function family, was purified as an N-terminally histidine-tagged fusion. In gel shift mobility assays, His-CnrH, in the presence of E. coli core RNA polymerase enzyme, could retard at least two different promoter DNA targets, cnrYp and cnrHp, localized within the cnrYXH locus. These promoters and their transcription start sites were confirmed by primer extension. Purified His-CnrX did not inhibit the DNA-binding activity of His-CnrH and is therefore unlikely to be an anti-sigma factor, as previously hypothesized (EMBL M91650 description entry). To study the transcriptional response of the regulatory locus to metals and to probe promoter regions, transcriptional fusions were constructed between fragments of cnrYXH and the luxCDABE, luciferase reporter genes. Nickel and cobalt specifically induced the cnrYXH-luxCDABE fusion at optimal concentrations of 0.3 mM Ni2+ and 2.0 mM Co2+ in a noncomplexing medium for metals. The two promoter regions PY (upstream cnrY) and PH (upstream cnrH) were probed and characterized using this vector and were found to control the nickel-inducible regulatory response of the cnr operon. The cnrHp promoter was responsible for full transcription of the cnrCBA structural resistance genes, while the cnrYp promoter was necessary to obtain metal-inducible transcription from the cnrHp promoter. The zinc resistance phenotype (ZinB) of a spontaneous cnr mutant strain, AE963, was investigated and could be attributed to an insertion of IS1087, a member of the IS2 family of insertion elements, within the cnrY gene.
The resistance of Ralstonia eutropha-like strain CH34 (Alcaligenes eutrophus CH34) against multiple heavy metals can be regarded as a phenomenon in its own right (15) and has found increasing applications within the field of environmental technology (29). The metal resistance determinants, which are localized predominantly on either of the two indigenous megaplasmids pMOL28 (15, 28) and pMOL30 (15), have been recently reviewed by Taghavi et al. (27). One important application has been the development of metal-specific biosensors based on in vivo and in vitro gene fusions of CH34 heavy metal resistance determinants. This was possible only through a detailed knowledge of the regulation of the metal resistances at the molecular level (2).
The cnr operon, located on pMOL28 (13, 28), encodes a phenotype of inducible resistance to 5 mM Co2+ and 3 mM Ni2+ in minimal medium. Resistance is mediated by an intricate coordination between the genes of the regulatory locus, cnrYXH, located upstream of the structural locus, cnrCBA (13). The latter encodes a three-component cation/proton antiporter (4), whose topological orientation in the membrane has been elucidated largely by comparison with the well-studied czc (cadmium-zinc-cobalt resistance) system (3, 17, 21), with which it shares close homology at the protein level (18, 20).
Despite the strong similarity between the structural resistance genes of the czc (17, 18), czr (9), sil (8), cnr (13), and ncc (25) operons, the regulation of both cnr and ncc (nickel resistance determinants) differs greatly from that of the other three-component-system-based heavy metal resistance operons. The cnr and ncc operons share an average of 66% identity in their regulatory loci and 79% in their structural loci. For cnr, regulation requires the activities of at least three genes, cnrYXH. Until now, the gene products of the regulatory locus have remained elusive. Only CnrH shares a close similarity (32%) with a known gene product, namely, ςE of Escherichia coli, an alternative ς70-like sigma factor of the extracytoplasmic function (ECF) family (14, 16, 22). CnrY and CnrX, however, show no homology to known proteins, and therefore their functional roles in vivo have remained open to debate. In this paper, we report the preliminary characterization of the Cnr regulatory proteins, through cloning and isolation from E. coli overexpression systems, and the molecular characterization of a spontaneous cnr mutant harboring zinc resistance. Furthermore, we identified promoter regions within the regulatory locus by use of transcriptional fusions with reporter bioluminescence genes and examined their heavy metal-dependent activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. eutropha-like strains were grown in either 869 medium (15) or Tris-buffered 284 medium with 0.2% (wt/vol) gluconate (15) as a carbon source. Bioluminescence assays were carried out in either 284-gluconate liquid medium or RM medium (284 medium supplemented with 0.1% [wt/vol] gluconate, MOPS [morpholinepropanesulfonic acid], and beta-glycerol phosphate replacing Tris and inorganic phosphate, respectively) (2). For E. coli, 869 medium supplemented with antibiotics was used for propagation, while Luria broth (LB) supplemented with antibiotics and/or 2% (wt/vol) glucose was used in protein overexpression cultures. Antibiotic concentrations were 20 μg of tetracycline (TET) per ml and 100 μg of ampicillin (AMP) per ml. NiCl2 and CoCl2 were used for lux induction experiments. R. eutropha and E. coli strains were grown at 30 or 37°C, respectively, unless stated otherwise.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| R. eutropha strains | ||
| CH34 | Wild type, containing pMOL28 and pMOL30, czc cnr chr mer cop pbr | 15 |
| AE126 | pMOL28, cnr chr mer | 15 |
| AE104 | Plasmid free, metal sensitive | 15 |
| AE963 | pMOL29, cnrY963::IS1087 chr mer, ZinB phenotype (due to derepression of cnr) | 1 |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ− rpsL nupG | GIBCO BRL |
| CM1446 | DH10B containing pMOL877 | 30 |
| JM105 | F′ traD36 lacIqΔ(lacZ)M15 proA +B+/thi rpsL (Strr) endA sbcB15 sbcC hsdR4 (rK− mK+) Δ(lac-proAB) | 32 |
| BL21(DE3) | F−dcm ompT hsdS (rB− mB−) galDE3 | Stratagene |
| TOP10 F′ | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH5α | F− φ80d lacZΔM15 Δ(lacZyA-argF)U169 deoR recA1 endA1 phoA hsdR17 (rK− mK+) supE44 λ−thi-1 gyrAg6 relA1 | GIBCO BRL |
| pMOL877-based plasmids | ||
| pMOL877 | luxCDABE, Tetr | 30 |
| pMOL1550 | cnrYXH-luxCDABE, Tetr | This study |
| pMOL1551 | cnrYXHC′-luxCDABE, Tetr | This study |
| pMOL1583 | cnrYp-luxCDABE, Tetr | This study |
| pMOL1561 | cnrYXa H-luxCDABE, Tetr | This study |
| pMOL1586 | cnrYp-cnrYX-cnrHp-luxCDABE, Tetr | This study |
| pMOL1587 | cnrHp-luxCDABE, Tetr | This study |
| pMOL1588 | cnrHp-cnrH-luxCDABE, Tetr | This study |
| pMOL1591 | cnrX-cnrHp-luxCDABE, Tetr | This study |
| pMOL1592 | cnrYp-cnrY-luxCDABE, Tetr | This study |
| pMOL1593 | (cnrY-cnrYp) -cnrX-cnrHp-luxCDABEb, Tetr | This study |
| pMOL1596 | ivrHC-luxCDABE, Tetr | This study |
| Other plasmids | ||
| pTrc99A | Ampr | Pharmacia Biotech Ltd. |
| pTrcCnrX | Ampr, cnrX in pTrc99A | This study |
| pTrcCnrYa | Ampr, cnrYa in pTrc99A | This study |
| pTrcCnrYb | Ampr, cnrYb in pTrc99A | This study |
| pCnrXphoA1 | Ampr Kmr, pTrc-cnrX′ in pGV4218 | This study |
| pCnrXphoA2 | Ampr Kmr, cnrX′-pTrc in pGV4218 | This study |
| pGV4218 | Ampr Kmr | 5, 19 |
| pVDZ′2-cnr | cnr as EcoRI-PstI fragment in pVDZ′2 | 13 |
| pRSETA | Ampr | Invitrogen |
| pRSETCnrX | Ampr, cnrX in pRSETA | This study |
| pRSETCnrH | Ampr, cnrH in pRSETA | This study |
| pMOL29 | pMOL28 containing cnrY963::IS1087 | 1 |
Point mutations in PH region.
cnrY-cnrYp region cloned in the opposite orientation as cnrX-cnrHp-luxCDABE.
Genetic techniques.
Standard molecular genetic techniques were used (23). The pMOL28 plasmid (15) was used as the source for the cnr genes and was extracted from AE126 by the procedure of Taghavi et al. (26). Electroporation (26) was used to introduce plasmid DNA into R. eutropha and E. coli.
Construction of Cnr protein overexpression clones.
The cnrY gene was cloned in its two possible open reading frames (ORFs) (13). The cnrYa ORF (positions 970 to 1266, according to the numbering of Liesegang et al. [13]) was PCR amplified as an NcoI-BamHI fragment from pMOL28, using 5′-GATCTCCATGGAGGTTTGCCACGGC-3′ as the upstream primer and 5′-GCGGCAAGGATCCTGTCAGC-3′ as the downstream primer. The resulting fragment spanned the region from position 962 to 1747 of the cnr sequence. The cnrYb ORF (positions 982 to 1270) was amplified as an NcoI-HindIII fragment with the upstream and downstream primers 5′-GTTTGCCATGGCAGACGTGGAAGA-3′ and 5′-ATGGCAAGCTTGCCGACGCC-3′, respectively, spanning positions 981 to 1516. The inserts were cloned into the expression vector pTrc99A, linearized with either NcoI-BamHI or NcoI-HindIII. The cnrX gene (positions 1272 to 1727) was cloned as an RcaI-BamHI fragment using the primers 5′-CGGTAAATCATGAAATCTCGT-3′ (upstream) and 5′-GCGGCAAGGATCCTGTCAGC-3′ (downstream) into pTrc99A, linearized with NcoI and BamHI. Following ligation, plasmid constructs were transformed into E. coli JM105 and selected on LB-AMP. Positive transformants were sequenced to ensure the in-frame insertion of the cloned fragments within the expression vector.
Expression of cnr regulatory genes in a cell-free system.
The protein expression of pTrcCnrX, pTrcCnrYa, and pTrcCnrYb in E. coli JM105 was tested in a cell-free system. Plasmid DNAs (4 μg) of the clones were used as templates (in both linearized and circularized forms) in an in vitro coupled transcription-translation protocol, using the Promega (Madison, Wis.) E. coli S30 Extract System For Linear Templates kit, as specified by the manufacturer. l-[4,5-3H]-Leucine (Amersham Pharmacia Biotech, Rainham, United Kingdom) was used to label nascent polypeptides. The translation products were separated on a 16.5% T, 6% C Tricine sodium dodecyl sulfate (SDS)-polyacrylamide gel (24) and visualized by autoradiography. 14C-labeled protein markers (Amersham Pharmacia Biotech) were included as standards.
Expression and production of cnrX gene products in E. coli.
Overnight cultures of E. coli(pTrcCnrX) were diluted 100-fold in 5 ml of LB-AMP and grown to an optical density at 600 nm (OD600) of 1.0. IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) was added to induce protein overexpression for 4 to 5 h. The cells were harvested and lysed by sonication in lysis solution (25 mM Tris [pH 7.5], 0.1 M NaCl, 1 mM EDTA, 15% [vol/vol] glycerol) with 0.1 mg of phenylmethylsulfonyl fluoride added as a protease inhibitor. The pellet fraction was collected following centrifugation at 18,000 rpm for 10 min (Sorvall centrifuge, SS34 rotor) and solubilized in 0.25% (wt/vol) Thesit (Roche Diagnostics, Brussels, Belgium) plus 10 mM EDTA, followed by incubation at 4°C for 30 min. The samples were centrifuged at 40,000 rpm for 2 h at 4°C, using a Beckman ultracentrifuge. The supernatant was collected, mixed with Laemmli loading buffer, fractionated on a 16.5% Tricine SDS-polyacrylamide gel (24), and visualized with Coomassie blue R-250.
Construction of PhoA (alkaline phosphatase) fusions with CnrX.
A gene fusion between the nucleotide sequence of cnrX′ encoding the first 27 amino acids of CnrX (MMKSRT . . . . . AAWL) and that of the ORF encoding the mature form of alkaline phosphatase (phoA) was constructed in vitro. In this construct, transcription of cnrX and that of phoA are coupled and directed from the pTrc promoter, resulting in a hybrid CnrX-PhoA fusion protein. The pTrc promoter together with cnrX were obtained by PCR amplification of pTrcCnrX using the downstream primer 5′-CGCGAATGGATCCGCTCATGTTTGAC-3′ (BamHI site) and the upstream primer 5′-TGCGAGTAGATCTGCCATGCCGC-3′ (BglII site). The cnrX′ fragment was cloned as a BamHI-BglII insert in the BglII-linearized and dephoshorylated plasmid pGV4218 (5, 19). The hybrid plasmid was used to transform E. coli DH5α. Transconjugants were selected directly on LB plates with 25 μg of kanamycin per ml, 100 μg of AMP per ml, 1 mM IPTG, and 0.5 mg of XP (5-bromo-4-chloro-3-indolylphosphate, 4-toluidine salt) (Boehringer Mannheim) per ml. This resulted in two plasmids, pCnrXphoA1 and pCnrXphoA2, having the pTrc-cnrX′ fragment cloned with phoA in the same and opposite orientations, respectively. The orientations of the cloned inserts were verified by PCR (5), while the structural integrity of the constructs was confirmed by sequence analysis. Constructs with plasmid pCnrXphoA1 formed small, dark blue colonies (PhoA+), while PhoA− transconjugants with the insert in the opposite orientation formed larger, light green colonies with a white halo or simply uniform white colonies.
Construction of histidine-tagged fusion proteins.
Both cnrX and cnrH were cloned as tagged fusions incorporating an N-terminal His tag. For the construction of pRSETCnrX, the cnrX ORF was amplified from pMOL28 as a BamHI-PstI fragment with downstream and upstream primers 5′-TTTGGATCCATGATGAAATCTCGTACCCGACGG-3′ and 5′-TTTCCTGCAGCGTCTTCCGGATTCACTGCGAGCCGCG-3′, which includes the cnrX region from position 1260 to 1738. The cnrH ORF was amplified as a BamHI-PstI fragment with primers 5′-TTTGGATCCAATCCGGAAGACGCTGACAGAATCC-3′ and 5-TTTGACTGCAGACTTATTTTTCCGAGTCAGCATCCAGC-3′, which includes the cnrH region from position 1711 to 2296. The pRSETA vector was isolated from E. coli TOP10 F′ using a Plasmid Midi-prep kit for high-copy-number plasmids and then linearized with BamHI and PstI. Following ligation of the vector and insert, plasmid constructions were transformed into electrocompetent E. coli BL21(DE3), and transconjugants were selected on LB-AMP plates.
Production and purification of His-CnrX and His-CnrH in E. coli.
Overnight cultures of E. coli BL21(DE3) bearing pRSETCnrH were diluted 100-fold into 3.3 liters of LB plus 2% (wt/vol) glucose and AMP and grown to an OD660 of 0.5. The cells were collected by centrifugation and washed once with fresh LB-AMP medium. The cells were suspended in the same volume of fresh LB-AMP. For induction of protein overexpression, 1 mM IPTG was added and the cells were incubated for 16 h at 28°C with shaking. Subsequently, the cells were harvested by centrifugation and suspended in 50 ml of lysis buffer (500 mM NaCl, 50 mM Tris-Cl, pH 8) plus the protease inhibitors 4-(2-aminoethyl)benesulfonyl fluoride (AEBSF) (0.1 mg/ml) and leupeptin (1 μg/ml). Cells were lysed by a double passage through a French pressure disruption cell. The cell pellet was recovered following centrifugation at 12,000 rpm (Sorvall centrifuge, SS34 rotor) at 4°C for 30 min. The recovered pellet was suspended in 30 ml of immobilized metal chromatography (IMAC) buffer G (6 M GdCl2, 50 mM Tris [pH 7.9], 0.5 M NaCl) and left on a rotary mixer for 1 h at 4°C. The denatured proteins were recovered in the supernatant fraction following centrifugation at 18,000 rpm at 4°C for 30 min using a Beckman centrifuge. The guanidine HCl-solubilized supernatant fraction was mixed with 6 ml of Ni-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN, Westburg B. V., The Netherlands) in a Falcon tube and left to mix on a rotary mixer for 1 h at 4°C. The Ni-NTA resin–His-CnrH mix was packed in batch fashion into a 5-ml column (Amersham Pharmacia Biotech). The purification was carried out using the Pharmacia fast protein liquid chromatography system at 4°C. The column was washed with IMAC buffer G for 10 min at 1 ml/min until the OD280 attained the baseline value. The adsorbed, denatured protein was allowed to slowly renaturate by the application of a 6 to 0 M GuCl2 gradient, which was achieved by applying a continuous gradient beginning with IMAC buffer G and ending with IMAC buffer B (50 mM Tris-Cl [pH 8.0], 1 M NaCl, 20% [vol/vol] glycerol) over 4 h at 1 ml/min. His-CnrH was eluted with 10 column volumes (40 ml) of imidazole-containing IMAC buffer I (0.5 M imidazole, 1 M NaCl, 50 mM Tris-Cl, pH 7.6) at 1 ml/min. Two-milliliter fractions were collected. Fractions containing His-CnrH were pooled and further purified by gel filtration chromatography, using a Superdex 75 (16/60) column (Amersham Pharmacia) equilibrated with gel filtration buffer (50 mM Tris-Cl [pH 8.0], 200 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% [vol/vol] glycerol). The pure protein was stored in aliquots in 50% (vol/vol) glycerol at −80°C. A similar purification protocol was followed for the pRSETCnrX overexpression constructs. The following buffers replaced the equivalents used for His-CnrH: IMAC buffer Ix (0.5 M imidazole, 1 M NaCl, 50 mM Tris-Cl [pH 7.6], 0.1% [wt/vol] Thesit) and gel filtration buffer X (50 mM Tris-Cl [pH 8.0], 200 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.1% [wt/vol] Thesit).
Gel shift mobility assays.
DNA targets were obtained by PCR amplification from pMOL28. The amplified products corresponded to the following targets: PY fragment (positions 820 to 1028 according to Liesegang et al. [13]), PH fragment (positions 1600 to 1800), and ivrHC fragment, containing the intervening region between cnrH and cnrC (positions 2270 to 2482). The DNA was end labeled with [γ-32P]dATP using polynucleotide kinase (GIBCO BRL, Life Technologies N. V., Merelbeke, Belgium), according to the manufacturer's specifications. The labeled DNA was recovered using a MicroSpin column (Amersham Pharmacia Biotech) as specified by the manufacturer. The protocol of Landini et al. (12) was used as the basis for the gel shift mobility assays. Purified His-CnrH (see above) was concentrated to approximately 0.2 mg/ml (≈9 μM) using a Centricon instrument (Millipore, Bedford, Mass.). The protein concentration was determined spectrophotometrically at 280 nm. Reconstitution of His-CnrH with the E. coli RNA polymerase core enzyme (Biozym, Landgraaf, The Netherlands) was allowed to occur by incubation of various concentrations of His-CnrH with the core enzyme (50 nM) in vitro at 37°C for 20 min in the presence of binding buffer (50 mM Tris-Cl [pH 8], 1 mM EDTA, 1 mM dithiothreitol). The reconstituted mixture was allowed to interact with 1.5 pmol of labeled DNA (10 ng) according to the protocol of Landini et al. (12) (37°C for 30 min). Subsequently, the reaction mixture was separated on a 5% nondenaturing acrylamide gel, using 0.5× Tris-borate-EDTA as the running buffer. DNA bands were visualized using autoradiography.
Primer extension for determination of mRNA transcription start sites.
Total RNA extraction from the R. eutropha CH34 derivatives AE104, AE963, and AE126 was carried out using the RNeasy purification kit (QIAGEN), following the manufacturer's guide, with minor modifications for cell lysis. The CH34 derivative strains were grown in 284-gluconate medium to stationary phase and then diluted 100-fold in fresh medium and grown to an OD660 of 0.5. The cells were induced with NiCl2 to a final concentration of 0.4 mM for 20 min and immediately chilled on ice. The pellet was recovered by centrifugation and then resuspended in Insta-Pure solution (EUROGENTEC S. A., Seraing, Belgium) as specified by the manufacturer. Chloroform was added to 1/10 volume, and the mixture was centrifuged at 3000 rpm for 15 min (Sorvall centrifuge, Sg3 rotor). Subsequent purification steps were as specified in the QIAGEN RNeasy purification kit manual.
For the determination of transcription start sites, 40 pmol of each of the oligonucleotides, cnrYp (5′-GGGCCGGCTCTGCACTGATG-3′), cnrHp (5′-CGCCACAAGTTGGCCGAATG-3′), and cnrCp (5′-GCTGCAACAGGTTCGG-3′), was labeled using 5 μl of [γ-32P]dATP (3,000 mCi/mmol) (Amersham) and polynucleotide kinase (New England Biolabs) by standard protocols. The final oligonucleotide concentration was adjusted to 0.8 pmol/μl. For primer extension experiments, a modification of the Promega protocol was used. Hybridization was carried out using 2 pmol of the labeled primer and 10 to 20 μg of total RNA extract at 45°C for 3 h in 30 μl of hybridization buffer (40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid] [pH 6.4], 400 mM NaCl, 80% formamide). The extension reaction was carried out in a 50-μl volume of extension mix, containing 10 mM dithithreitol, 1× First Strand Synthesis Buffer (GIBCO BRL), and 1 mM deoxynucleoside triphosphates. Superscript reverse transcriptase (GIBCO BRL, Life Technologies) (200 U) was added for extension, which was carried out for 1 h at 42°C. Following heat denaturation of the enzyme and RNase treatment, the extension products were recovered by ethanol precipitation, resuspended in formamide loading buffer, and separated on a 5% acrylamide sequencing gel next to a sequencing ladder. Sequencing reactions were carried out using the Pharmacia Amersham Biotech Thermosequenase kit according to the manufacturer's protocol for dITP cycle sequencing: the sequencing ladder of pUC18 DNA generated using the forward universal primer (5′-GTTTTCCCAGTCACGACGTTGTA-3′) was used to as a reference to mark the positions of the primer extension products, in base pairs, from the site of primer hybridization.
Construction of transcriptional fusions.
Different regions of the cnr regulatory locus were amplified by PCR. EcoRI sites were introduced into the primer sequences to clone the inserts into the unique EcoRI site of pMOL877. The inserts were obtained by PCR amplification using pMOL28 DNA as a template (see Table 2). For the construction of point mutations within cnrYXH, a strand overlap extension protocol was used. Two primary PCRs were carried out on cnr DNA using the primer pairs CNRLUX1-PXLUX1 and CNRLUX2-PXLUX2; the resulting amplicons corresponded to the 5′ and 3′ ends of cnrYXH, respectively. The point mutations incorporated in PXLUX1 and PXLUX2 affected the positions 1657 (G to A), 1659 (A to G), and 1662 (T to C). This changed the presumed cnrHp promoter sequence (based on the AAC consensus for RpoE-like ECF factors) from 5′-CCGGAACATCG-3′ to 5′-CCGAAGCACCG-3′. The second PCR step used the products of the first PCR step as the DNA template and the primer pair CNRLUX1-CNRLUX2. Plasmid pMOL877 was isolated from CM1446 using the GIBCO BRL Plasmid Midi-prep kit for low-copy-number plasmids. Constructs were transformed into E. coli DH10B by electroporation, and transconjugants were selected on 869 medium supplemented with 20 μg of TET per ml. Plasmid DNA of transconjugants was isolated and subsequently electroporated into the R. eutropha strains AE104 and CH34. Transconjugants were selected on 869 medium supplemented with 20 μg of TET per ml. Ninety-six clones were selected and inoculated onto different Ni induction plates (0.3 mM Ni), which were examined using autoradiography. Those constructs showing Ni-responsive bioluminescence (black spots on X-ray film) were further characterized.
TABLE 2.
Probing promoters cnrYXHa
| Plasmid | Cloned fragment | Maximum fold induction with nickelbd | Transcription | Relative signal strength (%)cd |
|---|---|---|---|---|
| pMOL1551 (1–2418) | 1,027 (11) | Inducible | 100 (0.7) | |
| pMOL1550 (1–2298) | 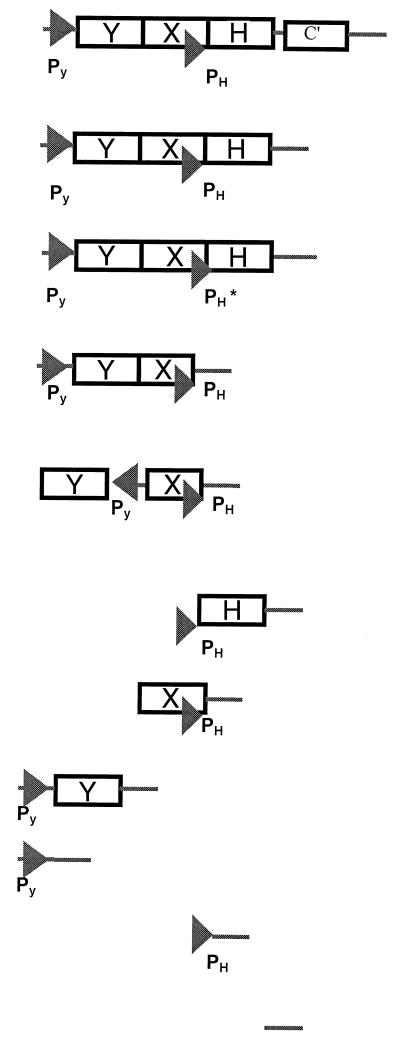 |
1,036 (25) | Inducible | 100 |
| pMOL1561 (1–2298) | <2 | Constitutive | 13 (2.0) | |
| pMOL1586 (870–1720) | 210 (12) | Inducible | 23 (3.5) | |
| pMOL1593 (843–1256), (1256–1720) | <2 | Constitutive | 15 (3.8) | |
| pMOL1588 (1579–2298) | <2 | Constitutive | 58 (9.2) | |
| pMOL1591 (1253–1720) | <2 | Constitutive | 17 (2.8) | |
| pMOL1592 (870–1280) | <2 | Constitutive | 11 (0.7) | |
| pMOL1583 (870–1000) | <2 | Constitutive | 13 (2.8) | |
| pMOL1587 (1579–1720) | <2 | Constitutive | 17 (3.5) | |
| pMOL1596 (2280–2418) | <2 | Constitutive | 0.4 (0.07) | |
| pMOL877 | 0 | Inactive | 0.0 |
Transcriptional fusions between different regions of the cnr regulatory locus and the luxCDABE genes were tested for their metal-responsive activity in vivo in CH34.
The metal response to 0.3 mM nickel was quantitated as the fold induction compared to noninduced cells and is expressed as the signal-to-noise ratio. A value of <2 is considered constitutive transcription.
Maximum light production (in ALU) divided by the OD660nm of the culture, as compared to the transcription of the complete cnr regulatory locus from pMOL1550.
Standard deviations are indicated in parentheses and are based on the results from three independent experiments.
Luminometry assays.
The bioluminescence of cnr-lux constructs was measured with a LUCY1 luminometer (Anthos Labtech B.V., Heerhugowaard, The Netherlands) at 23°C. Cells were grown overnight in 869 medium plus 20 μg of TET per ml to an OD660 of 1.0. The cells were harvested by centrifugation, washed once with RM-gluconate (0.1%, wt/vol) plus MOPS and beta-glycerol phosphate, and suspended in fresh medium at an OD660 of 0.3. Subsequently, 20 μl of the metal salt solutions was added to the microtiter plates at 10× the working concentrations immediately before 180 μl of the test cultures was added to the well. Water was used as a negative control. The bioluminescence emitted in absolute light units (ALU) was measured over 16 h at 30-min intervals, using the MIKROWIN software of the Anthos LUCY1 luminometer. Data processing was carried out using EXCEL 7.0 (Microsoft). Constructs harboring pMOL877 in CH34 gave a noise level of approximately 100 ALU.
Nucleotide sequence accession number.
The sequence of IS1087 has been entered in the EMBL database under accession no. AJ243722.
RESULTS
In vitro transcription-translation of cnrY.
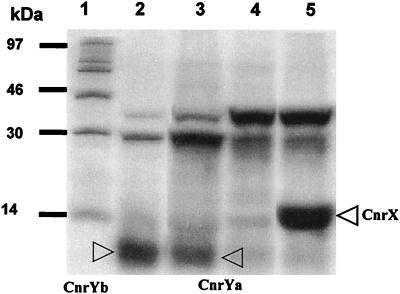
Based on sequence data, two possible ORFs have been hypothesized for cnrY (13). In nccY, which is closely related to cnr, only the ORFb reading frame is present. Translational fusions of the ORFa and ORFb reading frames were constructed in the E. coli expression vector pTrc99A, resulting in pTrcCnrYa and pTrcCnrYb, respectively, and subsequently tested in an in vitro coupled transcription-translation system. As shown in Fig. 1, both could be expressed in vitro. However, the protein product corresponding to the ORFb coding frame appeared more abundant (also by comparing the intensity of the 28-kDa internal control). Since in the homologous nccYXH system ORFa does not exist, in all likelihood the CnrY protein is translated from ORFb in vivo. It was not possible to overproduce the CnrY protein in E. coli cultures. Sequence analysis confirmed that both pTrcCnrYa and pTrcCnrYb had the correct structural integrity, and both plasmids were stably maintained in E. coli. Therefore, protein instability due to toxicity may have been the basis of this result.
FIG. 1.
Coupled in vitro transcription-translation of cnrY and cnrX. Plasmids pTrcCnrX, pTrcCnrYa, and pTrcCnrYb were used as DNA templates in an in vitro transcription-translation reaction. The templates were linearized or circularised to maximize efficient transcription from the pTrc promoter; a linearized template with a predominant 30-kDa band suggests inefficient transcription-translation coupling. The protein products are indicated. Lane 1, size marker; lane 2, pTrcCnrYb, linearized template; lane 3, pTrcCnrYa, linearized template; lane 4, pTrc99A; circularized template; lane 5, pTrcCnrX, circularized template.
Purification and characterization of CnrX.
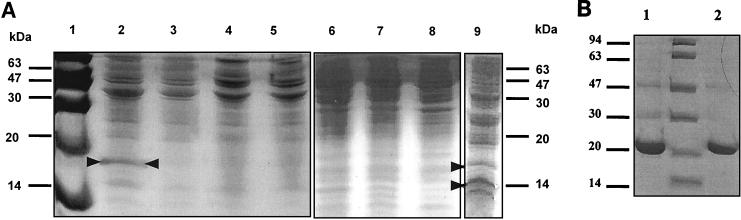
The cnrX gene was cloned in the pTrc99A expression vector, and after induction with IPTG, CnrX was over expressed in E. coli as a 16.5-kDa protein which was localized in the membrane fractions (Fig. 2A, compare membrane fractions in lanes 2 and 3 with soluble fractions in lanes 6 and 7, respectively). In whole-cell preparations of IPTG-induced cultures, a lower-molecular-mass band of approximately 14 kDa was also observed to co-migrate in SDS-polyacrylamide gels (Fig. 2A, lane 9), which suggested that CnrX may exist as processed and nonprocessed forms, with the latter being located in the membrane fractions. The existence of a membrane-translocating leader sequence at the N terminus of CnrX, as suggested on the basis of the amino acid sequence (MMKSRTRRLSLSTLFGALLGVSVAAA | WLY [the bar represents the cleavage site, and hydrophobic residues are in boldface]), was supported by studies with pTrc-cnrX′–phoA fusions. A fusion between the N-terminal 27 amino acids of CnrX and a phoA reporter system where PhoA lacks its own signal sequence (19) resulted in strong PhoA activity; such transformants formed small, dark blue colonies when plated on selective medium (see Materials and Methods). Transformants having the pTrcCnrX′ fused to phoA in the opposite orientation (pCnrXphoA2) did not show PhoA activity. These data are consistent with those in the accompanying study by Grass et al. (6).
FIG. 2.
Overproduction of CnrX and purification of His-CnrX. (A) Constructs bearing pTrcCnrX expressed CnrX as a 16.5-kDa protein fractionating in membrane components of cell lysates. Lane 1, size marker; lanes 2 and 3, protein extracts from induced and noninduced cultures of pTrcCnrX, respectively; lanes 4 and 5, protein extracts from induced and noninduced cultures, respectively, of the pTrc99A control with no insert; lanes 6 to 8, soluble fractions of induced pTrcCnrX, noninduced pTrcCnrX, and control pTrc99A cultures, respectively; lane 9, whole-cell preparation showing two forms of CnrX. (B) His-CnrX was overproduced as a 21-kDa protein and purified by Ni-NTA IMAC and gel filtration chromatography. Lane 1, pooled Ni-NTA IMAC fractions; lane 2, pooled gel filtration fractions.
The levels of overproduction of CnrX in E. coli (Fig. 2A) were considered too low for isolation purposes. To improve yields, CnrX was cloned and overproduced as a histidine-tagged fusion (pRSETCnrX), purified by use of Ni-NTA IMAC followed by gel filtration chromatography (Fig. 2B). The structural integrity of the purified protein was examined by secondary structure determination using circular dichroism (unpublished data), which revealed a high alpha-helical content (37.3%) and a relatively high content of antiparallel beta-sheets (21.4%), features which indicate the correct folding of the protein.
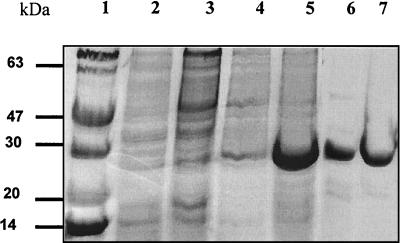
Purification and characterization of CnrH.
CnrH was overproduced in E. coli(pRSETCnrH) with an N-terminal histidine tag (Fig. 3), and the resulting inclusion bodies were purified under denaturing conditions by Ni-NTA IMAC, with protein renaturation prior to elution (Fig. 3, lane 5). Gel filtration was used as a final purification step (Fig. 3, lane 6). The purity of the protein was quantified to be greater than 95% (as determined by reverse-phase high-pressure liquid chromatography) (data not shown). The structural integrity of the purified protein was verified by the determination of its secondary structure, which was found to include an alpha-helical content exceeding 40% (unpublished data).
FIG. 3.
Overproduction and purification of His-CnrH. Constructs bearing pRSETCnrH overproduced His-CnrH as a 28-kDa protein in inclusion bodies. Lane 1, molecular mass marker; lanes 2 and 3, supernatant and pellet protein fractions of noninduced cultures, respectively; lanes 4 and 5, supernatant and pellet protein fractions of induced cultures, respectively. His-CnrH inclusion bodies were purified by Ni-NTA IMAC followed by gel filtration chromatography. Lane 6, pooled IMAC fractions; lane 7, gel filtration fraction.
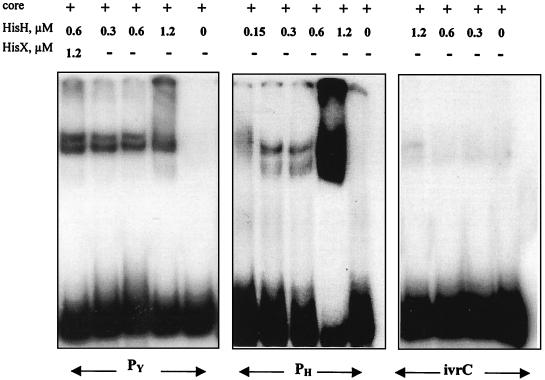
CnrH is thought to be a member of the ECF ς70 family. As such, CnrH should interact with a specific DNA target in the presence of core RNA polymerase. The close similarity with RpoE of E. coli prompted us to test the DNA-binding activity of His-CnrH using the core RNA polymerase of E. coli. Possible DNA targets were identified following a systematic analysis of the cnr sequence, both for regions homologous to the presumed cnrYp promoter sequence (13) and based on the consensus sequence of RpoE promoters (14). Three fragments were identified for further studies: the PY fragment (containing the putative cnrYp ς70-like promoter), the PH fragment (containing the putative cnrHp RpoE-like promoter), and the intervening region between cnrH and cnrC (ivrHC), showing similarity to the sequence around the presumed cnrYp promoter (6). The three potential targets were used for in vitro gel shift mobility assays. As shown in Fig. 4, His-CnrH could retard the PY and PH DNA fragments, but only when the core RNA polymerase enzyme of E. coli was included in the reactions. In the absence of the core RNA polymerase enzyme, no retardation was observed (results not shown). The addition of His-CnrX in the gel retardation reactions did not interrupt the interaction of His-CnrH with DNA (Fig. 4).
FIG. 4.
Gel shift mobility assays. Purified His-CnrH was tested for its DNA-binding activity in vitro in the presence of core RNA polymerase of E. coli. The interaction with PY, PH, and ivrHC with the indicated concentrations of His-CnrH, His-CnrX, and 50 nM core enzyme is shown.
Metal-responsive transcription from the regulatory locus, cnrYXHC′.
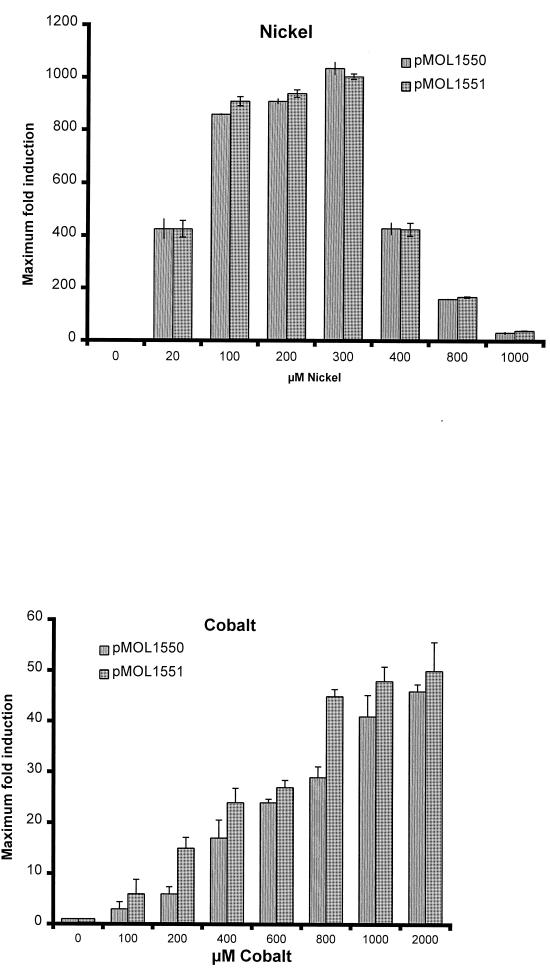
The transcriptional fusion of plasmid pMOL1551 was constructed by cloning the cnr regulatory locus, cnrYXHC′ (positions 1 to 2418 according to the numbering of Liesegang et al. [13]), upstream of the luxCDABE genes in pMOL877 (30). The transcriptional responses to nickel and cobalt were tested in the CH34 as well as the AE104 background. In the latter, no inducible bioluminescent response was observed. This indicates that additional regulatory functions are present in CH34 that are lacking in AE104. Also in E. coli, no transcription from the cnr regulatory locus was observed (data not shown). As shown in Fig. 5, transcription from cnrYXH was basal in the absence of Ni2+ and Co2+. The maximal transcriptional response was observed in RM medium with 0.3 mM nickel. At between 0 and 0.3 mM nickel (low levels), transcription was positively induced with increasing nickel concentrations. At higher concentrations (>0.4 mM), transcription became repressed with increasing metal concentration, and toxicity effects of the metal on the growth became apparent. The maximal transcription response was attained as a first peak during the mid-log phase and a second peak in the early stationary phase. The induction of cnr was also observed with cobalt, but the sensitivity differed from that of nickel. The maximum level of transcription was observed with cobalt concentrations of up to 2 mM.
FIG. 5.
Metal-inducible transcription from cnrYXH in the presence of nickel and cobalt. CH34 constructs bearing pMOL1550 and pMOL1551 were tested for their light production in the presence of various concentrations of nickel and cobalt at 23°C. The fold induction was calculated as the signal-to-noise ratio as a function of the biomass (OD660). The data presented are averages and standard deviations from three independently performed experiments.
Role of the intervening region between cnrH and cnrC (ivrHC) in metal-responsive transcription.
In order to examine the effect of the intervening region between cnrH and cnrC (ivrHC), plasmid pMOL1550 was constructed. It contains the fusion cnrYXH-luxCDABE and completely lacks the ivrHC region. The transcriptional responses of pMOL1550 and pMOL1551 were compared in strain CH34(pMOL28, pMOL30). The results are presented in Fig. 5. Nickel-dependent induction of lux in pMOL1550 and pMOL1551 showed similar kinetics. Therefore, it can be concluded that the ivrHC region is not essential for Ni-inducible regulation of transcription from the cnr regulatory locus. Neither Zn(II) nor Cr (VI) was able to induce cnrYXH transcription (data not shown). However, induction of cnr-derived transcription induced by low levels of Co2+ was more efficient with pMOL1551 than with pMOL1550, indicating that the ivrHC region might play a role in low-level Co2+ induction of cnrCBA transcription. However, with pMOL1596, which contains an ivrHC-lux fusion, no transcription of lux was observed in either the presence or the absence of Ni2+ and Co2+ (see Table 2). Thus, the ivrHC region is unlikely to contain a metal-inducible promoter.
Probing promoters and regulatory elements within the cnrYXH regulatory locus.
Transcriptional fusions between the cnr regulatory locus and luxCDABE were constructed as shown in Table 2. The selection of putative promoter fragments was based on the gel retardation data with CnrH. The metal responsiveness of the fusions was tested in the CH34 background, to ensure the presence of the cnrYXH genes in trans, and in the presence of 0.3 mM nickel. For comparison, transcription in the CH34 construct containing pMOL1550 was considered maximal (100% strength). As shown in Table 2, the promoter regions PY (in pMOL1583) and PH (in pMOL1587) were transcriptionally active, with relative signal strengths of 13 and 17%, respectively, compared to that of the complete cnrYXH fusion in pMOL1550. However, despite the presence of the CnrY, CnrX, and CnrH proteins in trans, transcription from these promoters was no longer Ni2+ inducible. Interestingly, the strain containing pMOL1586 (cnrYp-cnrYX-cnrHp-luxCDABE) showed metal induction kinetics for Lux comparable to that of pMOL1550. However, the maximum signal/noise ratio was approximately one-fifth of that attained with pMOL1550, probably due to a CnrH titration effect. In pMOL1588, where cnrH is transcribed from cnrHp, transcription was maintained for 20 h (until stationary phase) at a constant, high, constitutive level (maximum signal strength, 58%). Thus, despite the presence of CnrY and CnrX in trans, transcription from cnrHp was no longer metal regulated, indicating that an additional function was required in cis. Since cnrY and cnrX, together with the promoters cnrYp and cnrHp, were able to promote an Ni2+-responsive transcription, transcriptional coupling derived from cnrYp and cnrHp might additionally be necessary for metal-dependent regulation of the cnr operon. To test this hypothesis, the cnrYp-cnrY region was oriented in the opposite direction to the cnrX-cnrHp region (Table 2). Induction studies showed that the transcriptional response was no longer metal inducible (compare pMOL1586 and pMOL1593). These data indicate that both promoters cnrYp and cnrHp are required in cis and must be aligned in the same direction of transcription for a metal induction response of cnr. Therefore, cnr induction by nickel (and also cobalt) is achieved by the combined activities of these two promoters. In pMOL1561, point mutations were incorporated in the putative −35 region of cnrHp; this changed the CnrX codon usage without disturbing the CnrX protein sequence, while a wild-type copy was present in addition in trans on pMOL28. The Lux phenotype became constitutive and dropped to the same level as that with pMOL1593, where transcription is directed only from the cnrYp promoter. This suggests the removal of a promoter region, as compared to the case with pMOL1550.
Genetic analysis of the ZinB phenotype.
To complete the characterization of mutant phenotypes affected in cnr regulation, the genetic basis of the ZinB phenotype (1) was investigated. In strain AE963, nickel and cobalt resistances are constitutive and accompanied by a low-level zinc resistance that is probably due to overexpression of the cnr structural resistance proteins (1). Comparison of the EcoRI digests of pMOL28 from AE126 and pMOL29 from AE963 showed that in the latter, the EcoRI fragment bearing the cnr operon was approximately 1 kb larger than in the original pMOL28 (unpublished data). Further restriction analyses showed that an insertion in the cnrY gene was responsible for the observed ZinB mutant phenotype. Sequencing analysis revealed the presence of an IS2-type insertion element, which was designated IS1087.
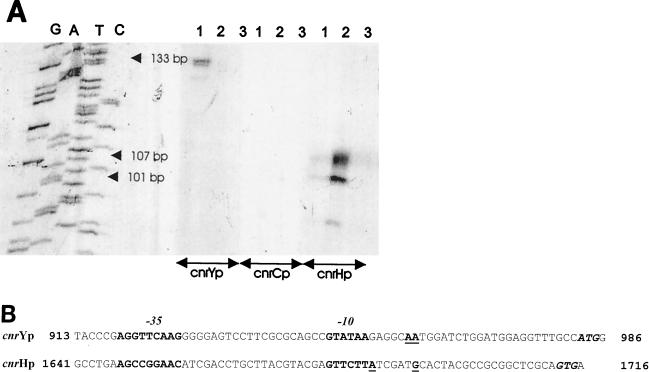
Transcription start sites.
The transcription start sites in cnr mRNA on induction with nickel were determined by primer extension. As shown in Fig. 6A, extension products were obtained in AE126 with both the cnrYp- and cnrHp-specific primers. No extension product with the ivrHC primer (corresponding to the presumed cnrCp promoter [6]) was observed. The cnrYp extension product corresponded to a distance of 133 nucleotides from the site where the pUC18 universal primer hybridized to its template. The deduced transcription start is therefore 5′-T949ATAAGAGGCAATGGATCTGGATG (the +1 nucleotide A959 is underlined). The cnrHp extension product corresponded to a distance of 101 nucleotides from the site where the primer hybridized, with the deduced start site being 5′-T1683CTTATCGATGC (the +1 G1693 is underlined). In AE963 (cnrY963::IS1087), no extension product was obtained with either the cnrYp or the ivrHC primers, while a strong signal was observed at the same position with the cnrHp primer as in AE126. An additional extension product was detected which would correspond to a transcription start 5′-T1683CTTATCGATGCACTACG. Since the cnrYp promoter is located upstream of the IS1087 insertion point in cnrY (position 1036), this also proves that no promoter from which transcription of the cnr structural genes occurs is present on IS1087. Therefore, the constitutive cnr expression in AE963 is due to transcription from the cnrHp promoter. No signals were obtained with the plasmid-free control strain, AE104.
FIG. 6.
Transcription start sites in the putative cnr promoters. Primer extension was used to determine the start sites of the potential cnr promoters for transcription under metal induction conditions (0.4 mM Ni2+). (A) A pUC18 DNA sequencing ladder was used as a reference to determine the sizes of the extension products. Lanes 1 to 3, primer extension products for mRNAs of nickel-induced cultures of AE126, AE963, and AE104, respectively. The primers used to identify transcripts from the presumed cnrYp, cnrCp, and cnrHp promoters are indicated under the respective lanes. The sizes of the extension products, calculated with the help of the pUC18 sequencing ladder, are also indicated. (B) Predicted structures of the cnrYp and cnrHp promoters. Based on the sizes of the primer extension products found in panel A, the transcriptional start sites of cnrYp and cnrHp were determined. The positions of the −35 and −10 regions (in boldface) and +1 transcriptional start sites (in boldface and underlined) are shown. Sequence numbering is according to Liesegang et al. (13). The translation start sites are indicated in boldface italics.
DISCUSSION
Since the early report on the nucleotide sequence of the cnr cobalt and nickel resistance operon of the R. eutropha-like strain CH34 by Liesegang et al. (13), the regulation of this resistance determinant has remained a mystery. In addition, the novelty of this regulatory locus, compared to well-characterized systems, at the genetic and protein levels has contributed to the slow advances made. In this paper, the roles of the individual components of the cnr regulatory region are examined.
The first determinant examined was cnrY, which is immediately preceded by the cnrYp promoter. Our findings point to ORFb (13) as being the correct ORF for cnrY, and this is further supported by the strong similarity of ORFb with nccY (where only one ORF exists). Furthermore, the ATG translation start of the putative protein ORFb is 6 nucleotides from the proposed ribosome-binding site, while no ribosome-binding site for ORFa translation could be identified (13). The CnrY protein is thought to have a downregulating role, based on cnrY963::IS1087, displaying a zinc-resistant (ZinB) phenotype. In this mutant, IS1087 is inserted at position 1036 in cnrY and disrupts the CnrY protein close to the N terminus. The resulting phenotype is low-level zinc resistance and elevated constitutive resistance to both cobalt and nickel, suggesting the inactivation of a repressor (1, 13). In primer extension assays with the cnrYp primer, no fragment was obtained with mRNA of strain AE963, showing that there is no promoter activity from IS1087 in the direction of cnrXH. It was not possible to overproduce the CnrY protein in vivo in E. coli; this is commonly encountered for negative regulators, which can confer toxicity to host cells on overproduction. CnrY, a 95-amino-acid protein (13) (for the revised sequence, see reference 6), shows no homology to well-characterized proteins in the database, except the homologous NccY. There are no obvious motifs in the primary amino acid structure of CnrY. However, the predicted secondary structure suggests two potential transmembrane domains towards the C terminus of the protein, a feature which was confirmed by use of phoA fusions (6). It is therefore possible that CnrY could be a negative regulator acting at the level of the cytoplasmic membrane. There are no obvious DNA-binding motifs in CnrY or obvious operator regions within cnrYXH, and therefore it is difficult to assign the protein a role as a DNA-binding repressor (13). Nevertheless, a putative role as an anti-sigma factor against CnrH can be envisaged, since many ECF sigma factors are controlled by a small, membrane-bound protein which is produced in stoichiometric proportions and which ensures a rapid on-off switch of sigma activity (10, 11). For cnrY963::IS1087 in AE963, transcription from cnrYp is disrupted. Therefore, high-level constitutive transcription in ZinB mutants must be due to transcription from a promoter downstream of cnrY, since there are no candidate promoter sequences within IS1087 in the direction of cnrXH, and no transcripts were identified by primer extension studies. In the accompanying study by Grass et al. (6), a similar (but not identical) constitutive, elevated resistance phenotype due to a frameshift mutation in cnrY resulting from a gene duplication of 14 bp mediated by an insertion sequence element is described. This supports the hypothesis that high-level transcription of cnrCBA is directed from another promoter downstream of cnrYp and that it is the combination of a disrupted CnrY protein and uncoupling of cnrYp and a downstream promoter which give rise to the constitutive, elevated nickel resistance. The localization of this promoter is discussed below.
CnrH, a 21-kDa protein, belongs to a family of ECF ς70-like proteins that direct the specific transcription of signals generated outside the cytoplasmic boundaries (14). The closest homologs of CnrH are the NccH (65% identity) and RpoE (32% identity) of E. coli. As with other ς70-like factors, there is a prominent helix-turn-helix motif at the carboxy terminus of CnrH (14). Purified His-CnrH, with an N-terminal histidine tag, in the presence of core RNA polymerase of E. coli could bind DNA targets containing the presumed cnrYp and cnrHp promoters. The DNA targets were confirmed to display promoter activities in lux-based transcription studies, and the corresponding promoter transcription start sites were identified using primer extension. It is interesting that the two promoter DNA targets have quite different sequences (13). In vitro, the PY promoter region was retarded by purified His-CnrH. Therefore, it is expected that in vivo CnrH would also transcribe cnr from the cnrYp promoter. Interestingly, the ivrHC region, whose sequence includes a region similar to the −10 region of cnrYp, was not retarded in in vitro DNA-binding assays. Furthermore cnr-lux transcriptional fusions incorporating (pMOL1551) and lacking (pMOL1550) this region showed similar Lux induction kinetics in the presence of Ni2+, while an ivrHC-lux fusion (pMOL1596) showed a background activity in strain CH34. Also, no promoter in this region could be identified by the primer extension experiments. The activity of cnrHp in the presence of metal was confirmed in primer extension assays, which contradicts the findings of Grass et al. (6), who were not able to detect a transcription start site from this region, although a downstream start site was reported originating from the ivrHC region (cnrCp promoter). Nonetheless, the observation of reverse transcriptase PCR products for the intervening region of cnrH and cnrC, as described by Grass et al. (6), also further supports a possible cnrHCBA transcript. The presumed cnrHp sequence resembles closely that of the RpoE promoter consensus and closely fits the common −35 . . . . . . AAC. . . . . . consensus proposed for the ECF family. The location of cnrHp within the cnrX coding region was confirmed by mutating the −35 . . . . . . AAC. . . . . . sequence without affecting the amino acid sequence of cnrX. A lux fusion of the mutated cnrHp no longer had a metal-inducible Lux+ phenotype, indicating that the mutation completely inactivated the promoter activity of the PH region. Thus, the . . . . . . AAC. . . . . . sequence is indeed part of the cnrHp promoter. It is not unknown for ECFs to recognize different promoter sequences. Since the assembly of the RNA polymerase holoenzyme involves a complex interaction between the sigma factor, core, and DNA, it is conceivable that CnrH is recruited by one type of core RNA polymerase to direct transcription from cnrYp and by another type for transcription from cnrHp.
The role of cnrX in regulation has been the most intriguing. Metal resistance in a CnrX− mutant was constitutively expressed, without affecting the level of resistance (13). It therefore appears that CnrX is not essential for the resistance phenotype (13). However the transcription data suggest that this locus is required for metal-inducible transcription from cnrYp and cnrHp. It has been postulated that CnrX functions as an anti-sigma factor on CnrH. However, our results do not support this hypothesis, since the binding of CnrH to the cnrYp fragment was not affected by the addition of purified His-CnrX. The N-terminal domain of CnrX is quite hydrophobic and has features of a leader peptide. CnrX overproduced in E. coli fractionated predominantly in the membrane fractions, although in whole-cell cultures a lower-molecular-weight form was also evident. This would indicate an inefficient processing of a signal sequence. PhoA fusions confirmed that the CnrX N terminus would function as a leader sequence. The topological orientation of the CnrX C terminus would therefore be periplasmic, which is corroborated by the findings of by Grass et al. (6), who found that the CnrX C terminus induced high alkaline phosphatase activities in phoA reporter fusions. CnrX contains six histidine residues, whose spatial arrangement in the primary amino acid sequence is identically conserved in NccX. The cluster of these residues together with the numerous glutamate residues may constitute important metal-binding sites in the protein. Therefore, CnrX may function as a periplasmic metal-sensing protein, interacting with CnrY, which is also localized at the membrane (6).
In conclusion, the cnr regulation appears to be effected by the activities of at least two promoters, cnrYp and cnrHp, whose presence was confirmed by gel retardation studies, primer extension, and transcriptional fusions. The cnrHp promoter, located within the cnrX coding region, is associated with high-level activity and is therefore likely to promote the high levels of transcription observed in the presence of metal. The cnrYp promoter is also transcriptionally active, but with a lower promoter strength than cnrHp. CnrH recognizes both promoter regions in complex with the core RNA polymerase, and the combined activities of both promoters are necessary for metal-responsive transcription of the cnr operon. ivrHC, which is not recognized by CnrH and seems indispensable for Ni-induced cnrCBA transcription, nevertheless seems to be required for optimal transcription when low levels of Co2+ are present. CnrY might function as a repressor or anti-sigma factor, and CnrX might function as a periplasmic sensor. Clearly, further studies on the in vivo preferences of the cnr promoters in the presence and absence of metal will be required if a model of regulation is to be hypothesized.
ACKNOWLEDGMENTS
We thank Albert Bossus and Philippe Corbisier for help on luminometry assays, Jon Hobman and Paolo Landini for help with DNA-binding studies, and Safieh Taghavi and Ann Provoost for help with DNA extraction procedures. C.T. thanks Joris Messens and Gaetan Muyldermans for help with protein purification, Yves Guenes for computer assistance, Henri De Greve for primer extension protocols, Viet Khong Nguyen for sequence analyses, and Maria Vanderveken for technical assistance. Dietrich Nies is acknowledged for fruitful and cooperative discussion.
This work was supported by a grant to C.T. from the Flemish Government and the VIB (Vlaamse Instelling voor Biotechnologie).
REFERENCES
- 1.Collard J M, Provoost A, Taghavi S, Mergeay M. A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J Bacteriol. 1993;175:779–784. doi: 10.1128/jb.175.3.779-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbisier P, van der Lelie D, Borremans B, Provoost A, de Lorenzo V, Brown N, Lloyd J, Hobman J, Csöregi E, Johansson G, Mattiasson B. Whole cell- and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal Chim Acta. 1999;387:235–244. [Google Scholar]
- 3.Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol. 1995;14:142–153. doi: 10.1007/BF01569896. [DOI] [PubMed] [Google Scholar]
- 4.Dong Q, Mergeay M. Czc/Cnr efflux: a three-component chemiosmotic antiport pathway with a 12-transmembrane-helix protein. Mol Microbiol. 1994;14:185–187. doi: 10.1111/j.1365-2958.1994.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 5.Gaballa A, Baysse C, Koedam N, Muyldermans S, Cornelis P. Different residues in periplasmic domains of the CcmC inner membrane protein of Pseudomonas fluorescens ATCC 17400 are critical for cytochrome c biogenesis and pyoverdine-mediated iron uptake. Mol Microbiol. 1998;30:547–555. doi: 10.1046/j.1365-2958.1998.01085.x. [DOI] [PubMed] [Google Scholar]
- 6.Grass G, Grosse C, Nies D H. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J Bacteriol. 2000;182:1390–1398. doi: 10.1128/jb.182.5.1390-1398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosse C, Grass G, Anton A, Franke S, Navarrete Santos A, Lawley B, Brown N L, Nies D H. Transcriptional organization of the czc heavy metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A, Matsui K, Lo J-F, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 9.Hassan, M., D. van der Lelie, D. Springael, U. Römling, N. Ahmed, and M. Mergeay. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene, in press. [DOI] [PubMed]
- 10.Helmann J D. Anti-sigma factors. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 11.Hughes K T, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 12.Landini P, Bown J A, Volkert M R, Busby S J. Ada protein-RNA polymerase sigma subunit interaction and alpha subunit-promoter DNA interaction are necessary at different steps in transcription initiation at the Escherichia coli ada and aidB promoters. J Biol Chem. 1998;273:13307–13312. doi: 10.1074/jbc.273.21.13307. [DOI] [PubMed] [Google Scholar]
- 13.Liesegang H, Lemke K, Siddiqui R A, Schlegel H G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993;175:767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sig E gene reveals a new sub-family of eubacterial RNA polymerase factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mergeay M, Nies D H, Schlegel H G, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 17.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 18.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattery T, Hernalsteens J-P, De Greve H. Identification and molecular characterization of a novel Salmonella enteritidis pathogenicity islet encoding an ABC transporter. Mol Microbiol. 1999;33:791–805. doi: 10.1046/j.1365-2958.1999.01526.x. [DOI] [PubMed] [Google Scholar]
- 20.Paulsen I T, Park J H, Cho P S, Saier M H., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 21.Rensing C, Pribyl T, Nies D H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouviere P E, De Las Penas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. RpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt T, Schlegel H G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghavi S, van der Lelie D, Mergeay M. Electroporation of Alcaligenes eutrophus with (mega)plasmids and genomic DNA fragments. Appl Environ Microbiol. 1994;60:3585–3591. doi: 10.1128/aem.60.10.3585-3591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taghavi S, Mergeay M, Nies D, van der Lelie D. Alcaligenes eutrophus as a model system for bacterial interactions with heavy metals in the environment. Res Microbiol. 1997;148:536–551. doi: 10.1016/S0923-2508(97)88361-1. [DOI] [PubMed] [Google Scholar]
- 28.Taghavi S, Mergeay M, van der Lelie D. Genetic and physical map of the Alcaligenes eutrophus CH34 megaplasmid pMOL28 and its derivative pMOL50 obtained after temperature induced mutagenesis and mortality. Plasmid. 1997;37:22–34. doi: 10.1006/plas.1996.1274. [DOI] [PubMed] [Google Scholar]
- 29.van der Lelie D. Biological interactions: the role of soil bacteria in the bioremediation of heavy metal-polluted soils. In: Vangronsveld J, Cunningham S, editors. Metal-contaminated soils: in situ inactivation and phytoremediation. Berlin, Germany: Landes Bioscience, Springer-Verlag; 1998. pp. 31–50. [Google Scholar]
- 30.van der Lelie D, Regniers L, Borremans B, Provoost A, Verschaeve L. The VITOTOX test, a SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat Res. 1997;389:279–290. doi: 10.1016/s1383-5718(96)00158-1. [DOI] [PubMed] [Google Scholar]
- 31.van der Lelie D, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies D H. Two-component regulatory system involved in transcriptional control of heavy-metal homeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]