Abstract
A physical map of the apple proliferation phytoplasma strain AT chromosome was constructed from genomic DNA extracted from diseased tobacco plants. The map was generated with single and double digestions of the chromosome with BssHII, SmaI, MluI, and ApaI restriction endonucleases and resolving the fragments by pulsed-field gel electrophoresis. Partial digestion and Southern blot analysis were used to assist in the arrangement of the 14 contiguous restriction fragments obtained. From the restriction fragments generated by double digestions, the size of the circular chromosome was calculated to be approximately 645 kb. Locations of the two rRNA operons, the operon including the fus and tuf genes, and three other genes were placed on the map. Genome sizes and BssHII restriction profiles of apple proliferation strain AP15 and the pear decline and European stone fruit yellows phytoplasmas were different from that of strain AT.
Phytoplasmas are a large and diverse group of wall-less prokaryotes of the class Mollicutes (trivial name, mycoplasmas). They infect several hundred plant species and cause many severe diseases of crop plants (18). Since phytoplasmas cannot be cultured under axenic conditions, our knowledge about these microorganisms is unsatisfactory. Only recently have the phylogenetic relationships of phytoplasmas within the mollicutes been established by sequence analysis of genes encoding ribosomal RNAs and other conserved genes (see reference 18 for a review). Furthermore, detection and classification of phytoplasmas could be significantly improved by the use of molecular methods. However, most genetic, physiological, and pathological aspects are still poorly understood.
One of the economically important phytoplasmoses is apple proliferation (AP), a disease found only in Europe. The causal agent, the AP phytoplasma, is a member of the phylogenetic AP phytoplasma group, which includes other temperate fruit tree pathogens, such as pear decline (PD), European stone fruit yellows (ESFY), and peach yellow leaf roll phytoplasmas (18). The AP agent is one of the most intensively studied phytoplasmas, of which several genes have been identified and sequenced. The G+C content of the AP phytoplasma chromosome was estimated to be as low as 23.7 mol% (11). In order to enhance further molecular research on the AP phytoplasma, we constructed a physical map of its chromosome, employing an approach similar to that used by Firrao et al. (7) to generate the first physical map of a phytoplasma chromosome, that of the western X-disease (WX) agent. This strategy is based on the fact that restriction endonucleases with GC-rich recognition sequences produce a limited number of usually large restriction fragments from AT-rich phytoplasma chromosomal DNA. These fragments differ significantly in size from fragments obtained from contaminating host plant DNA which show higher G+C contents (11). Following the separating and sizing of the phytoplasma restriction fragments under appropriate pulsed-field gel electrophoresis (PFGE) conditions, the fragments were arranged by a combinatorial approach that included double and partial digestions and Southern blot hybridization.
German AP phytoplasma strain AT, which was previously transmitted to Catharanthus roseus (periwinkle) via dodder bridges (15), was used for mapping. In the course of this work, this strain was transmitted by dodder to Nicotiana occidentalis and from there via grafting to Nicotiana tabacum (tobacco) cv. Samsun and other solanaceous plants. Other phytoplasmas used in this study were periwinkle-maintained strains AP15 of the AP phytoplasma from Italy (5), PD1 of the PD phytoplasma, and GSFY2 of the ESFY phytoplasma (13).
Leaf midribs, cortical stem tissue, or stem phloem preparations were taken for phytoplasma extraction. Phloem was prepared by peeling off 2- to 3-mm-wide strips of cortical stem tissue and removing the fine layer of vascular phloem with a scalpel. Samples were prepared for PFGE as described by Firrao et al. (7). Restriction digestions were carried out for 20 to 24 h with 50 to 100 U per block according to the directions of the manufacturer (MBI Fermentas, Vilnius, Lithuania). Double digestions were carried out sequentially in the same block. In addition, most major restrictions fragments were excised from the gel and digested with the second enzyme individually. Blocks were embedded in 1% (wt/vol) SeqPlaque (FMC Bioproducts, Rockland, Maine) agarose gel, and PFGE was carried out as described by Firrao et al. (7). Concatameric lambda ladder DNA, Saccharomyces cerevisiae chromosomes (both from Bio-Rad Laboratories, Hercules, Calif.), and Marker XV (Roche Diagnostics, Mannheim, Germany) were used as size markers. Third- to fifth-degree polynomial equations were generated by multilinear regression from molecular weight migration distances for each PFGE gel and used to determine fragment sizes. Chromosomal DNA of the AP phytoplasma, either enriched by CsCl density gradient centrifugation (12) or recovered from the PFGE gel by agarase (Roche Diagnostics) digestion, and cloned DNA fragments were used to probe Southern blots of phytoplasmal DNA.
In attempts to extract strain AT chromosomal DNA, the quantities obtained from midribs, cortical tissue, and phloem preparations of periwinkle, N. occidentalis, Nicotiana clevelandii, Nicotiana quadrivalvis, and tomato were too low for restriction mapping. However, phloem preparations from tobacco cv. Samsun produced higher DNA yields and were thus used further. The size of the high-molecular-weight DNA forming a discrete band in the PFGE gel was estimated to be about 645 kb, thus confirming the value previously obtained by Marcone et al. (14) in work with periwinkle. Southern blot hybridization using CsCl-purified AP phytoplasma DNA as a probe resulted in a strong reaction with the 645-kb band, indicating that it represents full-length, linearized chromosomal DNA of strain AT (not shown).
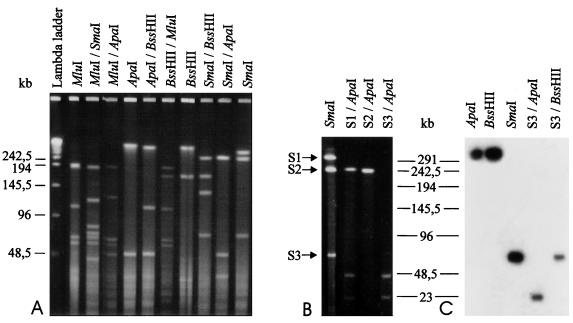
Chromosomal DNA of strain AT from tobacco was digested individually with 13 different restriction enzymes possessing GC-rich recognition sequences. Restriction enzymes NotI, BglI, Cfr42I, and RsrII failed to cleave the chromosomal DNA, whereas BamHI, Kpn2I, Eco52I, and SalI yielded eight or more fragments when separated by PFGE and were thus not used for the mapping studies. No consistent results were obtained with XhoI. Restriction enzymes BssHII, SmaI, ApaI, and MluI produced two, three, four, and five discrete restriction fragments (Table 1; Fig. 1A), respectively, and were chosen to construct the map. Whole PFGE-purified chromosomal DNA of strain AT was used to probe Southern blots of PFGE-separated BssHII, SmaI, ApaI, and MluI restriction fragments; the hybridization pattern revealed two, three, four, and five bands for BssHII, SmaI, ApaI, and MluI, respectively, as seen in ethidium bromide-stained gels. Reciprocal double digestions resulted in restriction fragments with sizes between 7 and 411 kb (Table 1; Fig. 1A). Small DNA fragments obtained by single and double digestion which could not be distinguished from bands arising from plant DNA contamination in the ethidium bromide-stained PFGE gel were identified by Southern blot analysis employing a probe derived from PFGE-purified chromosomal DNA (not shown). Analysis of the restriction data indicate that there must be, in addition to the clearly identified fragments, one BssHII-SmaI, one BssHII-ApaI, and one SmaI-MluI fragment with sizes between 2 and 5 kb, which were too small to be detected due to insufficient amounts of phytoplasmal DNA in the gel (see Table 1). No discrete DNA bands larger than 40 kb were detected in ethidium bromide-stained gels of preparations from healthy plants (not shown).
TABLE 1.
Number and average size of fragments obtained by single and double restriction digestions of the chromosome of strain AT of the AP phytoplasma
| Fragment | Average size (kb) of each fragmenta
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BssHII | SmaI | MluI | ApaI | BssHII-SmaI | BssHII-MluI | BssHII-ApaI | SmaI-MluI | SmaI-ApaI | MluI-ApaI | |
| A | 473 | 314 | 192 | 543 | 245 | 192 | 411 | 186 | 250 | 188 |
| B | 168 | 247 | 186 | 47 | 170 | 168 | 107 | 121 | 248 | 128 |
| C | 68 | 113 | 47 | 140 | 113 | 47 | 86 | 47 | 68 | |
| D | 72 | 22 | 78 | 72 | 47 | 78 | 47 | 66 | ||
| E | 65 | 5* | 65 | 22 | 72 | 24 | 60 | |||
| F | 27 | 2* | 65 | 22 | 51 | |||||
| G | 8 | 40 | 7 | 50 | ||||||
| H | 3* | 33 | ||||||||
| I | 10 | |||||||||
| Total size | 641 | 629 | 628 | 659 | 638 | 645 | 636 | 651 | 645 | 654 |
The mean of the sums of the restriction fragments generated from the double digests was 645 kb (standard deviation, 9 kb). *, fragments evidenced by restriction and alignment of larger fragments but not identified in PFGE gels.
FIG. 1.
PFGE of chromosomal DNA of the AP phytoplasma prepared from diseased tobacco. (A) Ethidium bromide-stained gel of single and double digests with various rare-cutting enzymes. Concatameric lambda DNA was used as a size marker. PFGE parameters were 0.7 to 12.5 s, 6 V/cm, 20 h. (B) Ethidium bromide-stained gel of three SmaI fragments individually digested with ApaI, showing that the SmaI-ApaI digest results in three double bands. (C) Southern blot hybridization of various fragments obtained by single and double digestions with gene probe IMP, revealing the position of one of the two small SmaI-ApaI fragments. PFGE parameters for panels B and C were 0.7 to 15 s, 6 V/cm, 19 h.
Different pulse and total electrophoresis times were evaluated to determine the size of the restriction fragments greater than 5 kb. Each value was established from a minimum of three different gels. The mean values of restriction fragments are listed in Table 1. From the sums of restriction fragments generated from all double digests, the size of the strain AT chromosome was calculated to be 645 kb (standard deviation, 9 kb) (Table 1; Fig. 2). This value is slightly larger than the mean calculated from single digests (639 kb) and similar to the size estimation from the entire chromosome (see above).
FIG. 2.
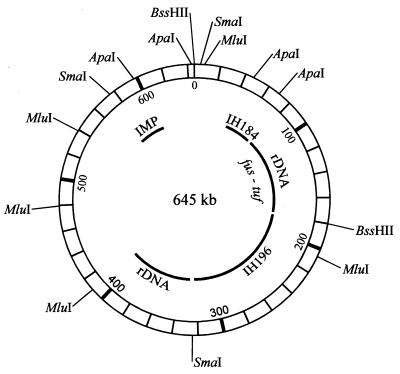
Physical map of the chromosome of the AP phytoplasma. Restriction sites for BssHII, SmaI, MluI, and ApaI are indicated. Loci of the two rRNA operons, the operon containing fus and tuf, an IMP gene, a putative nitroreductase (IH196) gene, and randomly cloned DNA fragment IH184 (3) were also mapped.
To lay out the map after blotting, restriction sites were positioned after subjective correlation of all data on fragment sizes from single and double digestions. Construction of the map shown in Fig. 2 started with double digestion of strain AT chromosome with BssHII and SmaI, which showed that two of the three SmaI sites are located in the large BssHII fragment, whereas the third must be close to one of the BssHII sites. One of these BssHII sites was arbitrarily chosen as the start point of the map. Single digestion of the chromosome with MluI and double digestions with SmaI-MluI and BssHII-MluI allowed positioning of the SmaI and BssHII sites and four of the five MluI sites on the map. The location of the fifth MluI site, position 483, was determined by partial digestion of the 253-kb SmaI fragment between positions 327 and 580. ApaI sites were determined by digesting BssHII, SmaI, and MluI fragments with ApaI (results not shown). As there was evidence that ApaI digestion resulted in two fragments with sizes of 47 kb, their presence could be verified by digesting individual SmaI and BssHII fragments with ApaI (Fig. 1B). The positions of three ApaI sites were confirmed by Southern hybridization of ApaI fragments with probes IMP (immunodominant membrane protein) (1) (Fig. 1C) and IH184 (3). Southern blot hybridization with the other gene probes used confirmed the positions of other sites on the map. The alignment of the restriction fragments revealed that the chromosome of strain AT is circular, like that of all other mycoplasmas previously examined.
Several gene loci were mapped by Southern blot hybridization of restriction fragments with probes from the AP phytoplasma (Fig. 1C and 2). It could be shown that the two rRNA operons, which seem to be present in all phytoplasmas (17), are unlinked. A similar chromosomal arrangement is known from the WX phytoplasma (7) and some culturable mycoplasmas (6, 16). In the sweet potato little leaf phytoplasma, one of the rRNA operons is inverted (A. C. Padovan and K. S. Gibb, Proc. 12th Conf. Int. Org. Mycoplasmol., p. 130–131, 1998). The locus of the linked genes encoding elongation factors G (fus) and Tu (tuf) (2) was identified on the same fragment as one of the rRNA operons. However, the positions of the fus and tuf genes relative to the position of the rRNA operon was not determined. Other mapped loci were those of a gene encoding an immunodominant protein (1) and a putative nitroreductase gene (9).
As for strain AT, chromosomes of strains AP15, PD1, and GSFY2 were cut twice by BssHII. However, the sizes of the restriction fragments differed from that of strain AT and each other (data not shown). Also, based on BssHII restriction fragments, the sizes of the entire chromosomes of the other strains examined were different from the size of strain AT, being approximately 685, 675, and 635 kb for strains AP15, PD1, and GSFY2, respectively (data not shown). These values are close to those of size estimations from the entire chromosomes, which were 690, 660, and 630 kb, respectively (14). Whether and how the differences in chromosome sizes are linked to phenotypical traits is unknown. The two AP phytoplasma types represented by strains AT and AP15 are common in natural populations and can be distinguished by Southern blot analysis using randomly cloned DNA fragments as probes (3, 10). The PD and ESFY phytoplasma strains differ from each other and the AP agent not only in host specificity; they are also transmitted by different insect vectors (4). In clinical microbiology, analysis of chromosomal DNA restriction patterns by PFGE is increasingly included for the typing of bacterial pathogens (for references, see reference 8). This method may also be suitable for distinguishing phytoplasmas with differences in phenotypical traits. One such trait would be virulence, in which phytoplasma strains may differ considerably. For Mycoplasma gallisepticum, an avirulent strain has a smaller genome than virulent strains, possibly due to deletion of virulence factors (19).
The restriction enzymes we used differ from those employed in WX agent chromosome mapping in that NotI, SalI, and RsrII, which were employed in WX phytoplasma mapping, either did not cut or cut too often the strain AT chromosome. Also, the number and sizes of fragments obtained in digestions with BssHII or SmaI differed considerably. In addition, the restriction sites mapped on the WX phytoplasma chromosome are evenly distributed, whereas a pronounced clustering was observed with strain AT. Nine of the 14 restriction sites are located in a 180-kb region flanking the arbitrary start point. A particularly high concentration of restriction sites was identified in the immediate vicinity of the start point. This clustering may indicate differences in the overall base pair composition of the genome, that is, that the region flanking the start point is richer in G+C than other portions of the genome. Differences in the distribution of GC-rich restriction sites are known from distributions of other mycoplasmas. For the genomes of Ureaplasma urealyticum and Mycoplasma mycoides, it is noted that the smaller region bound by the rRNA operons is much richer in GC-rich restriction sites than the larger portion of the chromosome (6, 16). However, the region of the strain AT chromosome in which the GC-rich restriction sites are concentrated does not include fragments in which the rRNA operons are located.
Acknowledgments
We thank Mike Rott for correcting the English text and Felix Hergenhahn for transmitting the AP phytoplasma from periwinkle to the Nicotiana species.
The work was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Berg M, Davies D L, Clark M F, Vetten H J, Maier G, Marcone C, Seemüller E. Isolation of a gene encoding an immunodominant membrane protein of the apple proliferation phytoplasma, and expression and characterization of the gene product. Microbiology. 1999;145:1937–1943. doi: 10.1099/13500872-145-8-1937. [DOI] [PubMed] [Google Scholar]
- 2.Berg M, Seemüller E. Chromosomal organization and nucleotide sequence of the genes coding for the elongation factors G and Tu of the apple proliferation phytoplasma. Gene. 1999;226:103–109. doi: 10.1016/s0378-1119(98)00552-6. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet F, Saillard C, Kollar A, Seemüller E, Bové J M. Detection and differentiation of the mycoplasmalike organism associated with apple proliferation disease using cloned DNA probes. Mol Plant-Microbe Interact. 1990;3:438–443. [Google Scholar]
- 4.Carraro L, Osler R, Loi N, Ermacora P, Refatti E. Transmission of European stone fruit yellows phytoplasma by Cacopsylla pruni. J Plant Pathol. 1998;80:233–239. [Google Scholar]
- 5.Carraro L, Osler R, Refatti E, Poggi Pollini C. Transmission of the possible agent of apple proliferation to Vinca rosea by dodder. Riv Patol Veg. 1988;24:43–52. [Google Scholar]
- 6.Cocks B G, Pyle L E, Finch L R. A physical map of the genome of Ureaplasma urealyticum 960T with ribosomal RNA loci. Nucleic Acids Res. 1989;16:6713–6719. doi: 10.1093/nar/17.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firrao G, Smart C D, Kirkpatrick B C. Physical map of the western X disease phytoplasma chromosome. J Bacteriol. 1996;178:3985–3988. doi: 10.1128/jb.178.13.3985-3988.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hielm S, Björkroth J, Hyytia E, Korkeala H. Genomic analysis of Clostridium bolutinum group II by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1998;64:703–708. doi: 10.1128/aem.64.2.703-708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarausch W, Saillard C, Dosba F, Bové J M. Differentiation of mycoplasmalike organisms (MLOs) in European fruit trees by PCR using specific primers derived from the sequence of a chromosomal fragment of the apple proliferation MLO. Appl Environ Microbiol. 1994;60:2916–2923. doi: 10.1128/aem.60.8.2916-2923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kison H, Schneider B, Seemüller E. Restriction fragment length polymorphism within the apple proliferation mycoplasmalike organism. J Phytopathol. 1994;141:395–401. [Google Scholar]
- 11.Kollar A, Seemüller E. Base composition of the DNA of mycoplasmalike organisms associated with various plant diseases. J Phytopathol. 1989;127:177–186. [Google Scholar]
- 12.Kollar A, Seemüller E, Bonnet F, Saillard C, Bové J M. Isolation of the DNA of various plant pathogenic mycoplasmalike organisms from infected plants. Phytopathology. 1990;80:233–237. [Google Scholar]
- 13.Marcone C, Hergenhahn F, Ragozzino A, Seemüller E. Dodder transmission of pear decline, European stone fruit yellows, rubus stunt, picris echioides yellows and cotton phyllody phytoplasmas to periwinkle. J Phytopathol. 1999;147:187–192. [Google Scholar]
- 14.Marcone C, Neimark H C, Ragozzino A, Lauer U, Seemüller E. Chromosome sizes of phytoplasmas composing major phylogenetic groups and subgroups. Phytopathology. 1999;89:805–810. doi: 10.1094/PHYTO.1999.89.9.805. [DOI] [PubMed] [Google Scholar]
- 15.Marwitz R, Petzold H, Özel M. Untersuchungen zur Übertragbarkeit des möglichen Erregers der Triebsucht des Apfels auf einen krautigen Wirt. Phytopathol Z. 1974;81:85–91. [Google Scholar]
- 16.Pyle L E, Taylor T, Finch L R. Genomic map of some strain within the Mycoplasma mycoides cluster. J Bacteriol. 1990;172:7265–7268. doi: 10.1128/jb.172.12.7265-7268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider B, Seemüller E. Presence of two sets of ribosomal genes in phytopathogenic mollicutes. Appl Environ Microbiol. 1994;60:3409–3412. doi: 10.1128/aem.60.9.3409-3412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seemüller E, Marcone C, Lauer U, Ragozzino A, Göschl M. Current status of molecular classification of the phytoplasmas. J Plant Pathol. 1998;80:3–26. [Google Scholar]
- 19.Tigges E, Minion C. Physical map of Mycoplasma gallisepticum. J Bacteriol. 1994;176:4157–4159. doi: 10.1128/jb.176.13.4157-4159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]