Abstract
Expression of the Escherichia coli mutA mutator phenotype requires recA, recB, recC, ruvA, and ruvC gene, but not recD, recF, recO, or recR genes. Thus, the recBCD-dependent homologous recombination system is a component of the signal pathway that activates an error-prone DNA polymerase in mutA cells.
DNA replication fidelity can be transiently reduced in response to environmental and physiological stimuli. In addition to the well-known Escherichia coli SOS system, emerging evidence suggests the existence of a number of such pathways in E. coli (7). One of the more intriguing newly recognized mutagenic pathways is the one elicited in mutA cells (7, 16, 27), in which the expression of an altered glyV glycine tRNA gene results in a strong mutator phenotype (27) characterized by elevation of transversions. In the mutA allele, the normal 3′-CCG anticodon is mutated to a 3′-CUG anticodon such that the mutant tRNA misreads the aspartate codon 5′-GAU/C as glycine at a low efficiency.
Expression of the mutA phenotype is constitutive and requires the recA and recB genes, but not umuD, umuC, dinB, or other lexA-repressible functions (16, 23), and thus represents a novel inducible mutagenic pathway termed “translational stress-induced mutagenesis” (TSM) (7).
The unexpected requirement for recA (in a non-SOS role) and recB genes in this pathway suggested that the mutA phenotype is homologous recombination dependent, since the RecA protein and RecBCD nuclease are principal components of the major homologous recombination pathway in E. coli (10). Whereas recA and recBCD functions are required for initiation of homologous recombination, ruvA and ruvB functions act together to catalyze branch migration of the Holliday junction, and ruvC encodes a Holliday junction-specific exonuclease (30).
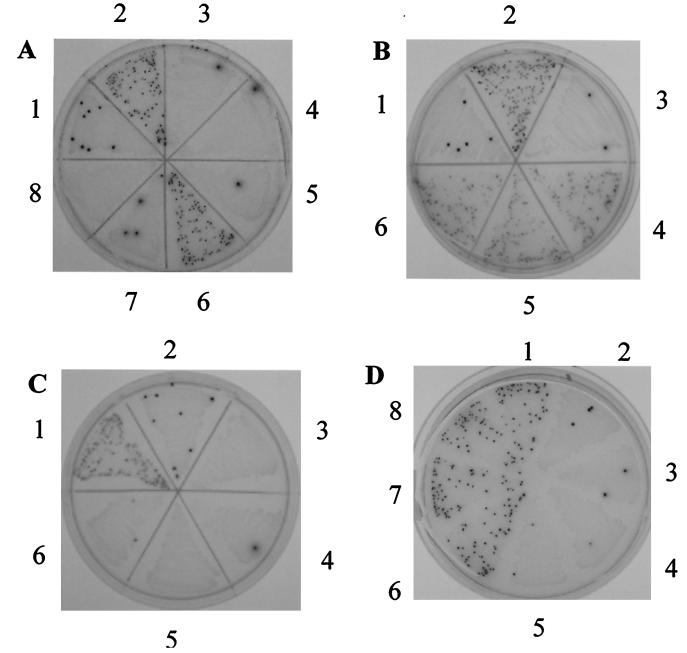
To detect the mutator phenotype as elevated background mutagenesis, a colony papillation assay based on reversion of a lacZ mutant allele to lacZ+ status, as described in detail by Miller and coworkers (14, 15), was used. The strains and plasmids used in this study are listed in Table 1. In this assay, lacZ mutant colonies are grown on minimal A agar plates containing limiting amounts of glucose on which they form colorless (white) colonies. After exhausting glucose as the carbon source in the medium, the colony stops growing. However, the P-Gal (phenyl-β-d-galactoside) in the medium can be utilized as a carbon source by any lacZ+ revertant cells present within the lacZ mutant colonies. As a result, the lacZ+ cells continue to divide to form microcolonies (papillae) within the larger growth-arrested lacZ mutant colony. For ease of observation, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), which is hydrolyzed to an insoluble blue dye by β-galactosidase (encoded by the lacZ+ gene), is included on the papillation plates so that the papillae stain dark blue and hence become easier to detect (15). This papillation assay was originally used to identify mutA and mutC cells (14). An example of the effect of the mutA allele on colony papillation can be seen in Fig. 1A, sector 1, which shows that a streak of CC105 (wild-type) cells contains only a small number of papillae, reflecting a normal background level of mutagenesis. In contrast, sector 2 shows that a streak of CC105mutA cells contains numerous blue papillae, reflecting elevated background mutagenesis. The mutA phenotype is abolished in LR600 cells (CC105mutA recC [sector 4]) and in LR140 (CC105mutA ruvC [sector 8]) cells, but not in LR800 (CC105mutA recD [sector 6]) cells. The mutA phenotype is restored when complemented for recBCD genes on a multicopy plasmid (Fig. 1B, sectors 4 to 6). In contrast, expression of the mutA phenotype does not require recD (Fig. 1A, sector 6), as expected, because recD-defective cells remain recombination proficient (10). Figure 1C shows that the ruvA gene is also required (sectors 5 and 6) for the mutA phenotype. Figure 1D shows that the mutA phenotype is unaffected in cells defective for recR (sector 6), recO (sector 7), and recF (sector 8) genes, suggesting that in contrast to the recBCD-dependent homologous recombination pathway, a functional recFOR-dependent recombinational repair pathway (10, 28, 29) is not required for the mutA phenotype.
TABLE 1.
Bacterial and plasmid strains used in this study
| E. coli strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| AB1157 | argE3 hisG4 leuB6 proA2 thr-1 ara-14 galK2 lacY1 mtl-1 xyl-1 thi-1 ara-1 rpsL31 supE44 tsx-33 | B. Bachman (4) |
| AK3 | recD1903::mini-tet | A. Kuzminov (11) |
| AM115 | CC105 recF322::Tn3 (Apr) | P1.JAS34 X CC105 to Apr; screen for UVS |
| AM116 | CC105mutA recF322::Tn3 (Apr) | P1.JAS34 X CC105mutA to Apr; screen for UVS |
| AM117 | CC105 recO1504::Tn5 (Kanr) | P1.JAS20 X CC105 to Kanr; screen for UVS |
| AM118 | CC105mutA recO1504::Tn5 (Kanr) | P1.JAS20 X CC105mutA to Kanr; screen for UVS |
| AM119 | CC105 recR252::Tn10-9 (Kanr) | P1.JAS31 X CC105 to Kanr; screen for UVS |
| AM120 | CC105mutA recR252::Tn10-9 (Kanr) | P1.JAS31 X CC105mutA to Kanr; screen for UVS |
| CC105 | [ara Δ(lac proB) xiii] F′ lacI Z proB+ | J. Miller (14) |
| CC105mutA | mutA590C in CC105 | J. Miller (14) |
| GS1481 | ΔruvC64::Kan (Kanr) in AB1157 | R. G. Lloyd (13) |
| JAS34a | recF322::Tn3(Apr) | J.A. Sawitzke (25) |
| JAS20a | recO1504::Tn5 (Kanr) | J. A. Sawitzke (25) |
| JAS31a | recR252::Tn10-9 (Kanr) | J. A. Sawitzke (25) |
| KH2R | Δ(srlR-recA)306::Tn10 (Tetr) in KH2 | This laboratory (20) |
| LR300 | recB268::Tn10 (Tetr) in CC105 | This laboratory (23) |
| LR400 | recB268::Tn10 (Tetr) in CC105mutA | This laboratory (23) |
| LR500 | recC266::Tn10 (Tetr) in CC105 | P1.N2103 X CC105 to Tetr; screen for UVS |
| LR600 | recC266::Tn10 (Tetr) in CC105mutA | P1.N2103 X CC105mutA to Tetr; screen for UVS |
| LR700 | recD1903::mini-tet (Tetr) in CC105 | P1.AK3 X CC105 to Tetr; screen for absence of Exo V activity |
| LR800 | recD1903::mini-tet (Tetr) in CC105mutA | P1.AK3 X CC105mutA to Tetr; screen for absence of Exo V activity |
| LR110 | ruvA60::Tn10 (Tetr) in CC105 | P1.N2507 X CC105 to Tetr; screen for UVS |
| LR120 | ruvA60::Tn10 (Tetr) in CC105mutA | P1.N2507 X CC105mutA to Tetr; screen for UVS |
| LR130 | ΔruvC64::Kan (Kanr) in CC105 | P1.GS1481 X CC105 to Kanr; screen for UVS |
| LR140 | ΔruvC64::Kan (Kanr) in CC105mutA | P1.GS1481 X CC105mutA to Kanr; screen for UVS |
| N2103 | recC266::Tn10 (Tetr) in AB1157 | R. G. Lloyd (12) |
| N2507 | ruvA60::Tn10 (Tetr) in AB1157 | R. G. Lloyd (26) |
| Plasmids | ||
| pBR322 | Apr Tetr (vector) | R. Brent (2) |
| pDWS2 | pBR322 derivative harboring recB, recC, and recD genes | A. Kuzminov (22) |
Other markers used were recB21 recC22 sbcB15 sbcC201sup0 hsdR ara-14 Δ(gpt-proA)62 lacY1 tsx-33 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 Rac− F− λ−.
FIG. 1.
Requirement for recombination genes required for the mutA phenotype detected by a colony papillation assay based on reversion of the lacZ mutant to a lacZ+ phenotype as described in detail elsewhere (15). Strains were streaked on minimal agar-based indicator plates (papillation plates) and incubated for 5 days at 37°C before observation. (A) Effect of recC, recD, and ruvC mutations on expression of the mutA mutator phenotype. Sectors: 1, E. coli CC105 (wild-type) control cells; 2, CC105mutA cells showing characteristically high papillation; 3, E. coli LR500 (CC105 recC); 4, E. coli LR600 (CC105mutA recC); 5, E. coli LR700 (CC105 recD); 6, E. coli LR800 (CC105mutA recD); 7, E. coli LR130 (CC105 ruvC); 8, E. coli LR140 (CC105mutA ruvC). (B) Overexpression of recBCD genes restores the mutA phenotype in mutA recC cells. Sectors: 1, CC105 control; 2, CC105mutA, showing high papillation; 3, LR500 (CC105 recC)/pDWS2(recB+ -C+ -D+); 4, 5, and 6, LR600 (CC105mutA recC [three isolates])/pDWS2(recB+ -C+ -D+) showing restoration of papillation. (C) Effect of ruvA mutation on the expression of the mutA mutator phenotype. Sectors: 1, CC105mutA showing high papillation characteristic of mutA cells; 2, CC105 control with few papillae; 3 and 4, E. coli LR110 (CC105 ruvA [two isolates]) controls; 5 and 6, E. coli LR120 (CC105mutA ruvA [two isolates]) showing that the mutA phenotype is abolished in ruvA cells. (D) Effect of recF, recO, and recR mutations on the expression of the mutA mutator phenotype. Sectors: 1, CC105mutA showing characteristically high papillation; 2, CC105 control, showing few papillae; 3, E. coli AM115 (CC105 recF); 4, E. coli AM117 (CC105 recO) showing few papillae; 5, E. coli AM119 (CC105 recR) showing few papillae; 6, E. coli AM120 (CC105mutA recR) showing that the high papillation characteristic of mutA cells is unaffected in recR cells; 7, E. coli AM118 (CC105mutA recO) showing that the high papillation characteristic of mutA cells is unaffected in recO cells; 8, E. coli AM116 (CC105mutA recF) showing that the high papillation characteristic of mutA cells is unaffected in recF cells.
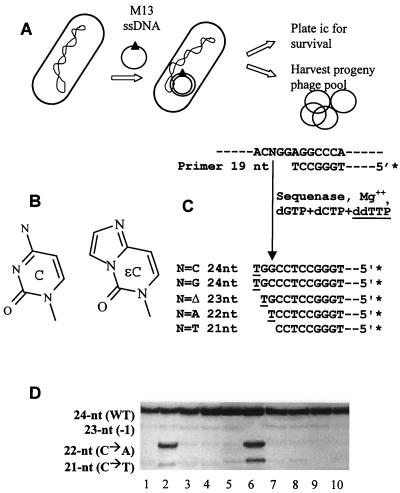
The mutA phenotype is manifested not only as an elevation in background mutagenesis at apparently undamaged DNA sites, as detected by the papillation assay, but also as a significant elevation in mutagenesis at the mutagenic exocyclic DNA lesion ɛC (see Fig. 2B for chemical structure) borne on M13 single-stranded DNA (ssDNA) vectors transfected into E. coli cells (16, 23). In this assay, M13 ssDNA bearing a single site-specific lesion (ɛC-ssDNA) is transfected into an appropriate strain, and the resulting progeny phage are analyzed for mutations at the ɛC site by a quantitative multiplex sequence analysis procedure summarized in Fig. 2C (16, 17, 19, 21). The assay depends on limited elongation of a prelabeled primer to characteristic lengths, depending on the base replacing the lesion upon replication.
FIG. 2.
(A) Summary of methodology used to analyze survival effects and mutagenesis at a site-specific ɛC residue (solid triangle) borne on M13 ssDNA. Procedures for transfection and measurement of survival (as infectious centers [ic]) and mutagenic effects have been described in detail elsewhere (17, 19–21) and in Materials and Methods. (B) Chemical structure of ɛC, shown alongside that of normal cytosine for comparison. (C) Principles of multiplex sequence analysis as previously described in detail (17, 21). Five micrograms of pooled progeny phage DNA (∼2 pmol) was annealed to ∼1 pmol of 5′-32P-end-labeled 19-mer primer. Approximately 0.2 pmol of the annealed template was incubated with approximately 0.5 U of T7 DNA polymerase devoid of 3′-to-5′ exonuclease activity (Sequenase 2.0; U.S. Biochemicals) in the presence of 1 μM (each dCTP and dGTP, 10 μM dideoxythymidine-5′-triphosphate (ddTTP), and 20 mM MgCl2 in buffer (40 mM Tris-HCl [pH 7.6], 50 mM NaCl, 10 mM dithiothreitol). Under these conditions, limited primer extension occurs, such that elongation on each of the four species of template DNA (i.e., wild type, C→T transitions, C→A transversions, and 1-nt deletions) results in a product of a different length. Note that C→G transversions are not induced by ɛC at significant levels (8, 18, 20) and are therefore not separately measured in the assay. The elongation products were fractionated on high-resolution 16% polyacrylamide–8 M urea gels, and the proportion of each product was determined from densitometric analyses of autoradiographs as described previously (17–19). Every elongation assay was monitored by parallel elongation of standard template DNA mixes containing known proportions of authentic mutant and wild-type DNAs. Mutation frequency was calculated by dividing the signal in each mutant band by the sum of signals in all bands. (D) Examples of multiplex sequence analyses of mutagenesis at the ɛC lesion. The elongation products are identified to the left of the autoradiograph. WT, wild type. Lanes: 1, E. coli CC105 (barely detectable signal in C→A and C→T bands); 2, CC105mutA (strong signal in C→A and C→T bands); 3, LR300 (CC105 recB); 4, LR400 (CC105mutA recB); 5, LR700 (CC105 recD); 6, LR800 (CC105mutA recD); 7, LR110 (CC105 ruvA); 8, LR120 (CC105mutA ruvA); 9, LR130 (CC105 ruvC); 10, LR140 (CC105mutA ruvC).
An example of the effect of the mutA allele on mutation fixation at ɛC can be seen in Fig. 2D, in which lane 1 shows low mutagenesis (i.e., low intensity of 22- and 21-nucleotide [nt] bands corresponding to C→A and C→T mutants, respectively) in CC105 (wild-type) cells, whereas lane 2 shows elevated mutagenesis (significantly increased signal in C→A and C→T mutant bands) in CC105mutA cells. In quantitative terms, mutagenesis at ɛC in CC105 (wild-type) cells is about 5% (Table 2), whereas in CC105mutA cells, it is about 45% (Table 2). As shown in Fig. 2, in cells defective for recB (lane 4), ruvA (lane 8), or ruvC (lane 10), the mutA phenotype is abolished, whereas it is unaffected in cells defective for recD (lane 6), in complete agreement with the results obtained with the papillation assay. These observations are quantitatively expressed in Table 2.
TABLE 2.
Effect of recB, recD, ruvA, and ruvC defects on the mutA phenotype detected as mutation fixation at an ɛC residue borne on transfected M13 ssDNA
| E. coli strain | Mean ± SD survivala | Mean ± SD % mutation frequencyb
|
||
|---|---|---|---|---|
| Total | C→A | C→T | ||
| CC105 | 630 ± 185 | 5 ± 1 | 2 ± 0 | 3 ± 1 |
| CC105mutA | 800 ± 220 | 45 ± 3 | 38 ± 2 | 7 ± 1 |
| LR300 (CC105 recB) | 650 ± 132 | 5 ± 2 | 2 ± 1 | 3 ± 1 |
| LR400 (CC105mutA recB) | 270 ± 65 | 6 ± 2 | 3 ± 1 | 3 ± 1 |
| LR700 (CC105 recD) | 840 ± 92 | 6 ± 2 | 3 ± 1 | 3 ± 1 |
| LR800 (CC105mutA recD) | 910 ± 164 | 47 ± 4 | 36 ± 2 | 11 ± 2 |
| LR110 (CC105 ruvA) | 680 ± 130 | 5 ± 2 | 2 ± 1 | 3 ± 1 |
| LR120 (CC105mutA ruvA) | 320 ± 40 | 7 ± 2 | 2 ± 1 | 5 ± 1 |
| LR130 (CC105 ruvC) | 720 ± 190 | 2 ± 2 | 1 ± 1 | 1 ± 1 |
| LR140 (CC105mutA ruvC) | 350 ± 36 | 2 ± 2 | 1 ± 1 | 1 ± 1 |
Values represent numbers of infectious centers per transfection (per 50 ng of ssDNA) and are averages of results from three independent transfections of ɛC-ssDNA.
Multiplex sequence analysis data shown were averaged from three to six independent elongation assays. Numbers are rounded to the nearest integer.
The requirement for recA, recB, recC, ruvA, and ruvC genes (but not the recD gene) allows the conclusion that a functional recBCD-dependent homologous recombination system is indeed required for the expression of the mutA phenotype. While this finding is intriguing, it is not immediately apparent why a functional recBCD-mediated recombination system is required for the expression of the mutA phenotype. Even though it is tempting to propose that the special features of recombination-mediated initiation of a replication fork on the bacterial chromosome (9) might account for the involvement of recombination in the mutator phenotype, it does not readily account for several observations. (i) An error-prone DNA polymerase is found in cell extracts from mutA cells, implying the modification of an existing DNA polymerase or the induction of a normally repressed polymerase (1). (ii) In the in vivo ɛC mutagenesis assay, mutation fixation occurs during the conversion of the transfected ɛC-ssDNA to the parental double-stranded replicative form DNA; it is possible that blocked elongation at the lesion site mimics a recombination-mediated initiation event, but this possibility by itself cannot explain mutation elevation at undamaged sites (1, 16). (iii) The requirement not only for recombination-initiation functions, such as recA, recB, and recC, but also for those required for its completion, such as ruvA and ruvC, suggest that the ability to conclude recombination is as important as the initiation process.
Exposed ssDNA regions at the sites of replication arrest are thought to be the signal required for SOS induction. Formation of specific DNA structures during homologous recombination (such as the cross-strand Holliday junction) may similarly act as a signal for TSM induction. However, the requirement for ruvC, the Holliday junction resolvase, suggests that the junction by itself probably does not constitute the signal, although other interpretations cannot be ruled out. Rather, the nucleoprotein complex containing the Holliday junction, as well as ruvA-, ruvB-, and ruvC-encoded proteins, may constitute the signal.
It is interesting that the so-called adaptive mutagenesis phenomenon (for recent reviews, see references 3, 5, 6, and 24) is similar to the TSM pathway in its genetic requirements and the fact that mutagenesis is elevated in a lacZ marker gene on the F′ episome. In adaptive mutagenesis, −1-bp deletions appear to be increased in the stationary phase, and this increase is partially suppressed by mutations in cells defective for recA, recBC, and ruvAB genes. However, the TSM pathway differs from adaptive mutagenesis in several regards: TSM is manifested in growing cells, mainly induces base substitutions, elevates mutation fixation at a DNA lesion, and increases mutagenesis not only in marker genes carried on the F′ episome, but also on the chromosome, as evidenced by the elevation in forward mutagenesis to rifampin resistance in mutA cells (16). Furthermore, mutagenesis is also elevated on a transfected M13 viral genome, and, finally, an error-prone DNA polymerase activity is expressed in TSM-induced cells (1).
Acknowledgments
We thank the individuals identified in Table 1, especially J. A. Sawitzke, R. G. Lloyd, and A. Kuzminov, for the bacterial and plasmid strains.
This study was supported in part by United States Public Health Research Service grants awarded by the National Cancer Institute (R01 CA73026) and the National Institutes of General Medical Sciences (R01 GM58253).
REFERENCES
- 1.Al Mamun A A M, Rahman M S, Humayun M Z. Escherichia coli cells bearing mutA, a mutant glyV tRNA gene, express a recA-dependent error-prone replication activity. Mol Microbiol. 1999;33:732–740. doi: 10.1046/j.1365-2958.1999.01520.x. [DOI] [PubMed] [Google Scholar]
- 2.Brent R, Ptashne M. The lexA gene product represses its own promoter. Proc Natl Acad Sci USA. 1980;77:1932–1936. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns J. Mutation and cancer: the antecedents to our studies of adaptive mutation. Genetics. 1998;148:1433–1440. doi: 10.1093/genetics/148.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Witt S K, Adelberg E A. The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics. 1962;47:577–586. doi: 10.1093/genetics/47.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster P L. Adaptive mutation: has the unicorn landed? Genetics. 1998;148:1453–1459. doi: 10.1093/genetics/148.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster P L, Rosche W A. Increased episomal replication accounts for the high rate of adaptive mutation in recD mutants of Escherichia coli. Genetics. 1999;152:15–30. doi: 10.1093/genetics/152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humayun M Z. SOS and Mayday: multiple inducible mutagenic pathways in Escherichia coli. Mol Microbiol. 1998;30:905–910. doi: 10.1046/j.1365-2958.1998.01120.x. [DOI] [PubMed] [Google Scholar]
- 8.Jacobsen J S, Perkins C P, Callahan J T, Sambamurti K, Humayun M Z. Mechanisms of mutagenesis by chloroacetaldehyde. Genetics. 1989;121:213–222. doi: 10.1093/genetics/121.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzminov A, Stahl F W. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J Bacteriol. 1997;179:880–888. doi: 10.1128/jb.179.3.880-888.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd R G, Buckman C, Benson F E. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- 13.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaels M L, Cruz C, Miller J H. mutA and mutC: two mutator loci in Escherichia coli that stimulate transversions. Proc Nat Acad Sci USA. 1990;87:9211–9215. doi: 10.1073/pnas.87.23.9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 16.Murphy H S, Humayun M Z. Escherichia coli cells expressing a mutant glyV (glycine tRNA) gene have a UVM-constitutive phenotype: implications for mechanisms underlying the mutA or mutC mutator effect. J Bacteriol. 1997;179:7507–7514. doi: 10.1128/jb.179.23.7507-7514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palejwala V A, Pandya G A, Bhanot O S, Solomon J J, Murphy H S, Dunman P M, Humayun M Z. UVM, an ultraviolet-inducible RecA-independent mutagenic phenomenon in Escherichia coli. J Biol Chem. 1994;269:27433–27440. [PubMed] [Google Scholar]
- 18.Palejwala V A, Rzepka R W, Humayun M Z. UV irradiation of Escherichia coli modulates mutagenesis at a site-specific ethenocytosine residue on M13 DNA. Evidence for an inducible recA-independent effect. Biochemistry. 1993;32:4112–4120. doi: 10.1021/bi00066a037. [DOI] [PubMed] [Google Scholar]
- 19.Palejwala V A, Rzepka R W, Simha D, Humayun M Z. Quantitative multiplex sequence analysis of mutational hot spots. Frequency and specificity of mutations induced by a site-specific ethenocytosine in M13 viral DNA. Biochemistry. 1993;32:4105–4111. doi: 10.1021/bi00066a036. [DOI] [PubMed] [Google Scholar]
- 20.Palejwala V A, Simha D, Humayun M Z. Mechanisms of mutagenesis by exocyclic DNA adducts. Transfection of M13 viral DNA bearing a site-specific adduct shows that ethenocytosine is a highly efficient RecA-independent mutagenic noninstructional lesion. Biochemistry. 1991;30:8736–8743. doi: 10.1021/bi00100a004. [DOI] [PubMed] [Google Scholar]
- 21.Palejwala V A, Wang G, Murphy H S, Humayun M Z. Functional recA, lexA, umuD, umuC, polA, and polB genes are not required for the Escherichia coli UVM response. J Bacteriol. 1995;177:6041–6048. doi: 10.1128/jb.177.21.6041-6048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponticelli A S, Schultz D W, Taylor A F, Smith G R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- 23.Ren L, Al Mamun A A M, Humayun M Z. The mutA mistranslator tRNA-induced mutator phenotype requires recA and recB genes, but not derepression of lexA-regulated functions. Mol Microbiol. 1999;32:607–616. doi: 10.1046/j.1365-2958.1999.01378.x. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg S M, Thulin C, Harris R S. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics. 1998;148:1559–1566. doi: 10.1093/genetics/148.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawitzke J A, Stahl F W. Roles for lambda Orf and Escherichia coli RecO, RecR and RecF in lambda recombination. Genetics. 1997;147:357–369. doi: 10.1093/genetics/147.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shurvinton C E, Lloyd R G, Benson F E, Attfield P V. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol Gen Genet. 1984;194:322–329. doi: 10.1007/BF00383535. [DOI] [PubMed] [Google Scholar]
- 27.Slupska M M, Baikalov C, Lloyd R, Miller J H. Mutator tRNAs are encoded by the Escherichia coli mutator genes mutA and mutC: a novel pathway for mutagenesis. Proc Natl Acad Sci USA. 1996;93:4380–4385. doi: 10.1073/pnas.93.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng Y C, Hung J L, Wang T C. Involvement of RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat Res. 1994;315:1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 29.Webb B L, Cox M M, Inman R B. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 30.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]