Abstract
The chemoreceptor Tcp mediates taxis to citrate. To identify citrate-binding residues, we substituted cysteine for seven basic or polar residues that are chosen based on the comparison of Tcp with the well-characterized chemoreceptors. The results suggest that Arg-63, Arg-68, Arg-72, Lys-75, and Tyr-150 (and probably other unidentified residues) are involved in the recognition of citrate.
The closely related enteric bacteria Escherichia coli and Salmonella enterica serovar Typhimurium have multiple transmembrane receptors that mediate chemotactic responses to amino acids, non-PTS sugars, and other attractants and repellents (2, 11, 25, 26, 34, 35). Some receptors (Tar for aspartate, Tsr for serine, and Trg for ribose and galactose) are found in both species. Others are species specific. Belonging to the latter class is the Salmonella serovar Typhimurium-specific chemoreceptor Tcp, which mediates taxis to citrate and a divalent cation-citrate complex and away from phenol (39). Salmonella serovar Typhimurium, but not E. coli, can utilize citrate as a sole carbon source.
Ligand recognition by Tar and Tsr has been studied extensively using mutagenesis (13, 21, 22, 30, 38), chemical modification (14, 15, 17), X-ray crystallography (28, 40), and computer simulation (19). Mutations at the aspartate-binding residues of Tar cause defects in the aspartate-sensing ability without affecting the repellent-sensing ability or other receptor functions (13, 21, 30, 38), although in some cases the maltose-sensing ability is also affected due to a partial overlap of the binding sites for aspartate and the complex of maltose and maltose-binding protein (13). Tcp is homologous to Tar and Tsr, but it recognizes the non-amino acid ligand citrate. Identification of the citrate-binding residues of Tcp should further our understanding of the molecular logic underlying ligand recognition in this family of receptors.
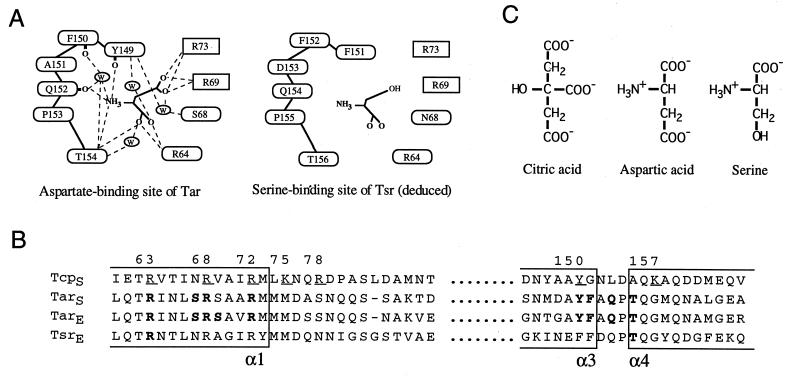
The residues involved in ligand binding in Tar and Tsr (Fig. 1A and B) are located in the two helices (α1 and α4) that extend through the cytoplasmic membrane as the first and the second transmembrane regions (TM1 and TM2), respectively (28, 40). In Tar, three Arg residues (residues 64, 69, and 73) within the helix α1 interact with the α- or β-carboxyl groups of aspartate (40) (Fig. 1A and B). The corresponding Arg residues of Tsr are predicted to interact with the α-carboxyl or the hydroxyl group of serine (19, 22) (Fig. 1A and B). This triplet is perfectly conserved in Tcp as residues 63, 68, and 72 (Fig. 1B), which are expected to interact with the carboxyl groups or the hydroxyl group of citrate. Tyr-149 of Tar, which is located near the top of the helix α4 and interacts with the carboxyl groups of the ligand via water molecules (40), is also conserved in Tcp as Tyr-150. Because citrate has no amino group (Fig. 1C), it is reasonable that the Thr residue (154 in Tar and 156 in Tsr), which interacts with the amino group of aspartate or serine, is not conserved in Tcp (39). Since citrate has a polar hydroxyl group and three carboxyl groups (Fig. 1C), it is likely that additional polar or positively charged residues in Tcp are involved in the interaction with citrate.
FIG. 1.
Ligand-binding sites of the bacterial chemoreceptors. (A) Schematic illustration of the ligand-binding residues of Tar and Tsr. The hydrogen bonding pattern of the aspartate binding site of Tar was drawn according to Yeh et al. (40). It should be noted that the backbone carbonyl atoms of Gln-152, Phe-150, and Tyr-149 are involved in hydrogen bonding. The serine-binding site of Tsr was deduced from the sequence similarity with Tar. Residues shown in ovals are from one subunit and those in boxes are from the other. W, a bound water molecule. (B) Alignment of the amino acid sequences of the bacterial chemoreceptors of E. coli (indicated with the subscript E) and Salmonella serovar Typhimurium (indicated with the subscript S). Bold letters indicate the residues involved in ligand recognition in Tar and Tsr. Underlined residues of Tcp indicate those replaced by Cys in this study. Numbering of residues is for Tcp. (C) Structures of the attractants specific for Tar, Tsr, or Tcp. The attractants are shown in their fully charged forms.
As an initial attempt to understand the citrate-recognition mechanism, we substituted Cys for seven polar or basic residues (Arg-63, Arg-68, Arg-72, Lys-75, Arg-78, Tyr-150, and Lys-157). Plasmids encoding the resulting mutant proteins were introduced into the E. coli strain HCB339 (ΔMCP) (37), which lacks all four chemoreceptors.
We first examined general receptor functions of the Cys-replaced Tcp proteins by swarming assay (Fig. 2). HCB339 cells expressing Tcp swarm in tryptone semisolid agar which does not contain citrate (32). Although the actual stimulus to which they respond is unknown, mutations in the C-terminal methyltransferase-binding sequence of Tcp affect swarming without impairing the citrate-sensing ability (32), demonstrating that this swarming requires the normal function of Tcp. HCB339 cells expressing any mutant Tcp receptor swarmed as fast as those expressing wild-type Tcp, suggesting that all of the mutant receptors retain general receptor functions including signaling and adaptation.
FIG. 2.
Swarming abilities of HCB339 (ΔMCP) cells expressing wild-type (WT) and mutant Tcp receptors in tryptone semisolid agar which does not contain citrate. Aliquots (1 μl each) of fresh overnight cultures were inoculated onto tryptone semisolid agar (1% tryptone, 0.5% NaCl, 0.3% agar) supplemented with 25 μg of chloramphenicol per ml, and then the plate was incubated at 30°C for 14 h.
We then examined receptor capabilities of the Cys-replaced Tcp proteins by temporal stimulation assay as described previously (31). Without chemotactic stimulation, cells expressing wild-type or any mutant Tcp swam smoothly. When 15% glycerol was added, cells expressing each mutant Tcp showed tumbling responses similar to those of cells expressing wild-type Tcp (data not shown), indicating that all of these proteins are expressed and retain the ability to mediate repellent responses to glycerol. Fig. 3A shows the citrate-sensing properties of the mutant Tcp proteins. The R78C and K157C receptors conferred the same citrate-sensing ability as wild-type Tcp. The R72C, K75C, or Y150C receptor mediated responses to citrate that were weaker than those mediated by wild-type Tar: 50 mM citrate was required for the 50% smooth-swimming fraction of cells expressing any of these mutant receptors, whereas a concentration of 1 mM is enough for that of cells expressing wild-type Tcp. HCB339 cells expressing Tcp-R63C or Tcp-R68C showed no response to citrate up to a concentration of 50 mM. These results suggest that the residues Arg-63, Arg-68, Arg-72, Lys-75, and Tyr-150 are important for sensing citrate.
FIG. 3.
Citrate-sensing abilities of the Cys-replaced Tcp receptors. (A) Temporal stimulation assay of cells expressing the Cys-replaced Tcp receptors. HCB339 (ΔMCP) cells expressing wild-type or mutant Tcp as the sole chemoreceptor were treated with 15% glycerol and then stimulated with various concentrations of citrate. The fraction of smooth-swimming cells after 30 s was measured at 25°C as described previously (31). Symbols: open circles, wild-type Tcp; closed circles, Tcp-R63C; open triangles, Tcp-R68C; closed triangles, Tcp-R72C; open squares, Tcp-K75C; closed squares, Tcp-R78C; open diamonds, Tcp-Y150C; closed diamonds, Tcp-K157C. (B) Immunoblotting analysis of methylation patterns of the Cys-replaced Tcp receptors. HCB339 (ΔMCP) cells expressing wild-type or mutant Tcp were incubated with 15% glycerol (Glyc), distilled water (None), 10 mM citrate (Cit-10), or 50 mM citrate (Cit-50) at 25°C for 30 min. Samples were subjected to SDS-PAGE followed by immunoblotting with anti-Tsr serum (18), which cross-reacts with Tcp, as described previously (32).
We next examined the methylation patterns of the mutant receptors by immunoblotting with anti-receptor serum (Fig. 3B) as described previously (32). Tcp is methylated at multiple residues in the cytoplasmic domain, and its methylation level increases and decreases in response to citrate and glycerol, respectively, to result in adaptation (39). Methylation and demethylation of a receptor can be detected as mobility shifts of the protein in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE): the more the receptor is methylated, the faster it migrates in the gel (3, 4, 9, 10). Tcp-R63C migrated a little faster than wild-type Tcp and the other mutant Tcp receptors. However, the receptor does not seem to be proteolytic fragments but seems to be a full-length receptor: (i) Tcp-R63C expressed in HCB339 appeared as multiple bands corresponding to differential levels of methylation (Fig. 3B), whereas the same receptor expressed in a ΔMCP strain lacking the methyltransferase CheR and the methylesterase/deamidase CheB appeared as a single band with a mobility faster than that of wild-type Tcp (data not shown); (ii) Tcp-R63C was detected with antiserum raised against the C-terminal 20-amino-acid sequence of Tar (data not shown), whereas the mutant Tcp receptor lacking the C-terminal residue (Phe-547) was not (H. Okumura, M. Homma, and I. Kawagishi, unpublished results); and (iii) the corresponding mutant (R64C) Tar protein also migrates faster than wild-type Tar (data not shown).
In the absence of citrate, all of the mutant proteins showed methylation patterns similar to that of wild-type Tcp. All of the mutant receptors were demethylated in response to the addition of the repellent glycerol. This result indicates that all of the mutant receptors retain general signaling and adaptation abilities. In the presence of citrate, the receptors which mediated attractant responses to citrate (Tcp-R78C and Tcp-K157C) showed elevated methylation levels. In contrast, citrate did not influence the methylation levels of Tcp-R63C and Tcp-R68C, which failed to mediate responses to citrate and increased only marginally those of Tcp-R72C, Tcp-K75C, and Tcp-Y150C, which mediated weaker responses. These results indicate that the R63C, R68C, R72C, K75C, and Y150C receptors are fully or partially defective in citrate-stimulated methylation, corresponding well to the defects in behavioral responses to citrate, and that the latter defects are not indirect results from elevated levels of methylation, which would bias the unstimulated behavior toward tumbling.
Two SH groups in close vicinity can form a disulfide bond under moderately oxidizing conditions. Indeed, in some Cys-replaced mutants of Tar, the two subunits of the homodimer are cross-linked by a spontaneously formed disulfide bond (27). Thus, the defects in citrate sensing could be due to indirect effects of disulfide cross-linking. We therefore looked for intersubunit disulfide bonds in the Cys-replaced Tcp proteins. After nonreducing SDS-PAGE followed by immunoblotting, bands with apparent molecular masses of about 60 kDa were detected for all samples (Fig. 4A). In addition, with the R68C, K75C, and R78C receptors, we found additional bands with apparent molecular masses of about 130 kDa (Fig. 4A). These bands disappeared by the treatment of the samples with 10% 2-mercaptoethanol prior to SDS-PAGE (Fig. 4A). Therefore, we concluded that these bands represent disulfide-cross-linked homodimers of the mutant Tcp proteins, suggesting that residues 68, 75, and 78 of one subunit of the Tcp homodimer are located near the same residues of the partner subunit. This result corresponds well to a configuration in Tar revealed by a comprehensive survey of disulfide cross-linking in Cys-scanned mutant proteins (7): Ser-68 and Met-75 of the Tar homodimer lie at the interface of helices α1 and α1′. In Tcp, the positional equivalents are Asn-67 and Leu-74, respectively. If Tcp has a similar helical structure, residues 68, 75, and 78 would be located at positions adjacent to the interface, and residue 72 would be in the opposite faces. Moreover, in the case of Tcp-R78C, which had normal sensing abilities, both the cross-linked and the non-cross-linked species showed increases and decreases in the methylation level in response to citrate and glycerol, respectively (Fig. 4B). Even in the case of R68C and K75C, which were defective in citrate sensing, the methylation levels of the cross-linked dimers decreased in response to glycerol (Fig. 4B). This finding suggests that Tcp forms a functional homodimer regardless of ligand occupancy states, as demonstrated for Tar (7, 8, 12, 24, 29), Tsr (23), and Trg (1, 16, 20).
FIG. 4.
Detection of disulfide-cross-linked dimers of the mutant Tcp proteins in nonreducing SDS-PAGE followed by immunoblotting. (A) Detection of cross-linked dimers of Tcp. Ice-cold 5% trichloroacetic acid was added to HCB339 (ΔMCP) cells expressing wild-type (WT) or mutant Tcp proteins pretreated with or without 10 mM N-ethylmalemide (NEM). NEM was added to prevent disulfide formation during sample preparation. The NEM-pretreated or untreated samples were collected by centrifugation and were dissolved in nonreducing SDS loading buffer supplemented with 10 mM NEM (lanes labeled with NEM) or 10% 2-mercaptoethanol (lanes labeled with 2ME), respectively. These samples were subjected to nonreducing SDS-PAGE followed by immunoblotting. (B) Methylation patterns of cross-linked and uncross-linked Tcp proteins. HCB339 (ΔMCP) cells expressing wild-type or mutant Tcp were incubated with 15% glycerol (Glyc), distilled water (None), 10 mM citrate (Cit-10), or 50 mM citrate (Cit-50) at 25°C for 30 min. After stimulation, the samples were treated with 10 mM NEM and 5% trichloroacetic acid and were further treated as described above.
For R68C, K75C, and R78C, the fractions of the cross-linked dimers were 0.2, 0.6, and 0.5, respectively. Thus, substantial amounts of uncross-linked homodimers are always available, even for mutant receptors that undergo cross-linking. Moreover, the addition of dithiothreitol (up to 50 mM) did not improve citrate responses of HCB339 cells expressing any of these receptors (data not shown). Therefore, it is likely that loss of the positively charged side chains of Arg-68 and Lys-75 itself impairs the ligand-binding affinity of Tcp for citrate. Taken together, the data suggest that the residues Arg-63, Arg-68, Arg-72, Lys-75, and Tyr-150 are involved in the recognition of citrate.
Tar has two rotationally symmetrical, antiparallel, nonoverlapping ligand-binding sites at the subunit interface (28, 40). Arg-64 in one subunit of the Tar homodimer interacts with the α-carboxyl group of aspartate, and Tyr-149 in the same subunit interacts with the α- and β-carboxyl groups via water molecules. Arg-69 and Arg-73 in the other subunit interact with the β-carboxyl group of aspartate. Presumably, some of the three carboxyl groups or the hydroxyl group of citrate may interact with Arg-68 and Arg-72 in one subunit of the Tcp homodimer and with Arg-63 and Tyr-150 in the other subunit. Lys-75 may also interact with some of the carboxyl groups and/or the hydroxyl group of citrate and may be one of the residues responsible for the ligand specificity, because this lysine is not conserved in Tar or Tsr. In contrast, the closely located basic residue Arg-78 does not seem to be involved in ligand recognition. Another candidate for a determinant of ligand specificity is Lys-157. In Tar, the amino group of aspartate interacts with Thr-154, which is not conserved in Tcp. Instead, Tcp has the basic residue Lys-157 in this region. However, the substitution of Cys for Lys-157 did not affect citrate sensing at all.
Binding of aspartate to Tar does not cause a large rearrangement between TM1-α1 and TM1′-α1′ (5, 6, 7, 16, 20, 27) or between TM1-α1 and α4′-TM2′ (36) but triggers a slight axial movement of α4-TM2 relative to TM1-α1 and TM1′-α1′ (6, 16, 33). It is this movement that transmits information about the extracellular binding event to the cytoplasmic signaling domain. Based on the homology of Tcp with Tar, it is assumed that residues 63, 68, 72, and 75 of Tcp are located at the apex of α1 and that residue 150 is near the apex of α4. Tcp would seem to transduce signals via a similar process. However, our results also imply a possible difference between Tcp and Tar. The critical movement of α4 in Tar presumably involves Thr-154 (Thr-156 in Tsr), since it is a major contact with the ligand in α4. Lys-157 would play a similar role in Tcp, but unexpectedly the K157C mutant was normal for citrate taxis.
In this study, we targeted several residues in the putative ligand-binding regions for Cys replacement. The mutants can be further characterized by chemical modification as has been successfully applied to Tar (14, 15) and Tsr (17, 18). These polar and positively charged residues were chosen for mutagenesis, based on the homology of Tcp with the well-characterized chemoreceptors Tar and Tsr. Among the residues at which Cys substitutions disrupted citrate taxis, Arg-63, 68 and 72 are conserved in Tar and Tsr and Tyr-150 is conserved in Tar. Lys-75 is the only residue unique to Tcp. It is likely that some other residues also interact with citrate. Further experiments, such as random mutagenesis, are needed to elucidate the precise molecular mechanism underlying the recognition of citrate, including discrimination between citrate and a metal ion-citrate complex.
Acknowledgments
We thank Michael D. Manson of Texas A & M University for critically reading the manuscript.
This work was supported in part by grants-in-aid for scientific research to I.K. from the Ministry of Education, Science, Sports and Culture of Japan and from the Takeda Science Foundation.
REFERENCES
- 1.Baumgartner J W, Hazelbauer G L. Mutational analysis of a transmembrane segment in a bacterial chemoreceptor. J Bacteriol. 1996;178:4651–4660. doi: 10.1128/jb.178.15.4651-4660.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 3.Boyd A, Simon M I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980;143:809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelsky D, Dahlquist F W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci USA. 1980;77:2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chervitz S A, Falke J J. Locked on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J Biol Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chervitz S A, Falke J J. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chervitz S A, Lin C M, Falke J J. Transmembrane signaling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danielson M A, Bass R B, Falke J J. Cysteine and disulfide scanning reveals a regulatory α-helix in the cytoplasmic domain of the aspartate receptor. J Biol Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFranco A L, Koshland D E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci USA. 1980;77:2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engström P, Hazelbauer G L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980;20:165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- 11.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falke J J, Koshland D E., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 13.Gardina P, Conway C, Kossmann M, Manson M. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J Bacteriol. 1992;174:1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomi S, Lee L, Iwama T, Imae Y. Inhibition of aspartate chemotaxis of Escherichia coli by site-directed sulfhydryl modification of the receptor. J Biochem. 1993;133:208–213. doi: 10.1093/oxfordjournals.jbchem.a124027. [DOI] [PubMed] [Google Scholar]
- 15.Gomi S, Lee L, Iwama T, Imae Y, Kawagishi I. Ligand recognition mechanism of bacterial chemoreceptors revealed by site-specific sulfhydryl modification. In: Kurihara K, Suzuki N, Ogawa H, editors. Olfaction and taste XI. Tokyo, Japan: Springer-Verlag; 1994. pp. 210–214. [Google Scholar]
- 16.Hughson A G, Hazelbauer G L. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc Natl Acad Sci USA. 1996;93:11546–11551. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwama T, Kawagishi I, Gomi S, Homma M, Imae Y. In vivo sulfhydryl modification of the ligand-binding site of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1995;177:2218–2221. doi: 10.1128/jb.177.8.2218-2221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwama T, Homma M, Kawagishi I. Uncoupling of ligand-binding affinity of the bacterial serine chemoreceptor from methylation- and temperature-modulated signaling states. J Biol Chem. 1997;272:13810–13815. doi: 10.1074/jbc.272.21.13810. [DOI] [PubMed] [Google Scholar]
- 19.Jeffery C J, Koshland D E., Jr Three-dimensional structural model of the serine receptor ligand-binding domain. Protein Sci. 1993;2:559–566. doi: 10.1002/pro.5560020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G F, Burrows G G, Lebert M R, Dutton D P, Hazelbauer G L. Deducing the organization of a transmembrane domain by disulfide cross-linking. The bacterial chemoreceptor Trg. J Biol Chem. 1994;269:29920–29927. [PubMed] [Google Scholar]
- 21.Lee L, Imae Y. Role of threonine residue 154 in ligand recognition of the Tar chemoreceptor in Escherichia coli. J Bacteriol. 1990;172:377–382. doi: 10.1128/jb.172.1.377-382.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee L, Mizuno T, Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J Bacteriol. 1988;170:4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Li G, Weis R M. The serine receptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 24.Lynch B A, Koshland D E., Jr Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macnab R M. Motility and chemotaxis. In: Neidhardt F C, Ingraham J, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 732–759. [Google Scholar]
- 26.Manson M D. Bacterial motility and chemotaxis. Adv Microb Physiol. 1992;33:277–346. doi: 10.1016/s0065-2911(08)60219-2. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama I N, Mikawa Y G, Maruyama H I. A model for transmembrane signalling by the aspartate receptor based on random-cassette mutagenesis and site-directed disulfide cross-linking. J Mol Biol. 1995;253:530–546. doi: 10.1006/jmbi.1995.0571. [DOI] [PubMed] [Google Scholar]
- 28.Milburn M V, Prive G G, Milligan D L, Scott W G, Yeh J, Jancarik J, Koshland D E, Jr, Kim S-H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 29.Milligan D L, Koshland D E., Jr Site-directed cross-linking: establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 30.Mowbray S L, Koshland D E., Jr Mutations in the aspartate receptor of Escherichia coli which affect aspartate binding. J Biol Chem. 1990;265:15638–15643. [PubMed] [Google Scholar]
- 31.Nishiyama S, Nara T, Imae Y, Homma M, Kawagishi I. Thermosensing properties of mutant aspartate receptors having methyl-accepting sites substituted multiply or singly with alanine. J Bacteriol. 1997;179:6573–6580. doi: 10.1128/jb.179.21.6573-6580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumura H, Nishiyama S-I, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxyl-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ottemann K M, Xiao W, Shin Y-K, Koshland D E., Jr A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 34.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 35.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingram J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 36.Umemura T, Tatsuno I, Shibasaki M, Homma M, Kawagishi I. Intersubunit interaction between transmembrane helices of the bacterial aspartate chemoreceptor homodimer. J Biol Chem. 1998;273:30110–30115. doi: 10.1074/jbc.273.46.30110. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff C, Parkinson J S. Aspartate taxis mutants of the Escherichia coli Tar chemoreceptor. J Bacteriol. 1988;170:4509–4515. doi: 10.1128/jb.170.10.4509-4515.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto K, Imae Y. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci USA. 1993;90:217–221. doi: 10.1073/pnas.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh J I, Biemann H P, Privé G G, Pandit J, Koshland D E, Jr, Kim S-H. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]