Abstract
We report the cloning and sequencing of the gene containing cytochrome c′ (cycP) from the photosynthetic purple bacterium Rhodobacter capsulatus and the regions flanking that gene. Mutant strains unable to synthesize cytochrome c′ had increased sensitivity to nitrosothiols and to nitric oxide (which binds to the heme moiety of cytochrome c′).
The physiological function of periplasmic bacterial cytochrome c′ has eluded definition for over 40 years since its first discovery (27), despite extensive biochemical and biophysical analysis. The amino acid sequences of cytochromes c′ from metabolically diverse proteobacteria reveal these cytochromes to be entirely distinct from the class I c-type cytochromes that include the mitochondrial cytochrome c (1). Most striking is that the motif CXXCH, at which the heme is covalently bound, is located toward the C terminus of the polypeptide. Three-dimensional structures of cytochromes c′ from a number of different bacteria (9, 11, 22, 25, 29) show that the heme iron lacks an amino acid ligand at its sixth coordination site, consistent with the high-spin–intermediate-spin state of the heme iron and unusual optical spectral features (17). The hydrophobic environment in this unoccupied pocket explains why the binding of ligands to cytochrome c′ is limited to small, predominantly uncharged ligands, particularly nitric oxide (NO) and carbon monoxide (CO).
There are some data available which indicate that the physiological function of cytochrome c′ involves binding NO in vivo. Electron paramagnetic resonance spectroscopy of intact cells of denitrifying bacteria which produce cytochrome c′ has shown that these organisms contain a heme-nitrosyl after growth under denitrifying conditions, whereas organisms that do not synthesize cytochrome c′ do not give rise to a spectral feature due to a heme-nitrosyl (30, 31). The possibility cannot be excluded, however, that the spectral feature is due to some chromophore other than cytochrome c′. Work in our laboratory has shown that cytochrome c′ added to a suspension of denitrifying bacteria binds to NO which is produced as a freely diffusible intermediate during denitrification, indicating that the cytochrome is indeed capable of binding NO at the concentrations it achieves in vivo during denitrification (20). NO is a free radical that is capable of damaging cellular material, particularly by reaction with thiols and transition metals in proteins, and hence inhibiting normal metabolism. Furthermore, under aerobic conditions, NO reacts with the superoxide anion to form peroxynitrite (ONOO−), which may also be a potent agent of oxidative damage. Cytochrome c′ may function to prevent the accumulation of NO and hence protect the bacterial cell against the harmful effects of NO and nitrosative stress.
Cloning and sequencing of cycP.
To investigate the function of cytochrome c′, we amplified the gene encoding cytochrome c′, cycP, from the photosynthetic bacterium Rhodobacter capsulatus PAS100 by PCR using primers which had been designed to the cytochrome c′ amino acid sequence from R. capsulatus SP7 (1). Degeneracy in the oligonucleotide primers was biased according to the probable codon usage as determined for R. capsulatus by Armstrong et al. (3). Primer 1 was designed to an amino acid sequence toward the N terminus (VLEAREA), 5′-GTSCTKGARGCSCGSGARGC-3′; primer 2 was designed to be complementary to sequence toward the C terminus (CKACHDD), 5′-TCRTCRTGGCASGCYTTGCA-3′ (where S = G/C, K = G/T, R = G/A, and Y = C/T). Thermal cycling using these oligonucleotide primers with R. capsulatus chromosomal DNA as a template (35 cycles of 30 s of denaturation at 94°C, 60 s of annealing at 60°C, and 90 s of extension at 72°C) gave a 350-bp product which was confirmed as cycP by DNA sequencing using an ABI 373A DNA sequencer (Applied Biosystems). Digoxigenin-labeled deoxynucleoside triphosphate mix was used to amplify a labeled cycP product which was used as a hybridization probe. To identify a suitable fragment of R. capsulatus PAS100 chromosomal DNA which contained cycP, a Southern hybridization of DNA digested with a range of restriction enzymes was undertaken. A SalI fragment of 6 kb hybridized with the digoxigenin-labeled cycP probe. A library was constructed by cloning R. capsulatus PAS100 DNA SalI fragments of around 6 kb into the vector pZErO and maintaining the clones in Escherichia coli TOP10F′. A colony blot identified those transformants which contained the cloned cycP fragment. The plasmid was purified from a culture of one of these colonies and designated pCP101. The insert of pCP101 was sequenced using a primer walking approach, starting with primers designed to the central region of cycP, identified by sequencing of the initial 350-bp amplification product. Sequence was compiled using Staden, and further analysis of the sequence utilized the Wisconsin GCG package available from the SEQNET facility at the Daresbury Laboratory and sequence analysis program available on the Internet at http://www.expasy.hcuge.ch.
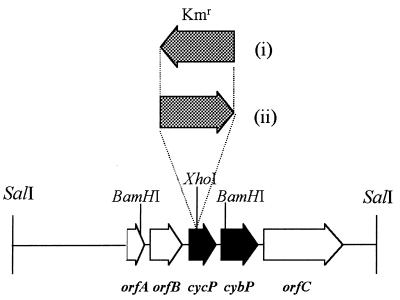
A map of the region containing cycP and flanking genes is shown in Fig. 1. cycP is predicted to encode a protein with 150 amino acids. From amino acid 22 onward, the sequence is identical to that of cytochrome c′ determined from the X-ray crystal structure of the cytochrome from R. capsulatus strain St. Louis (25). There are seven amino acid differences between these two identical sequences and the sequence determined biochemically for cytochrome c′ from R. capsulatus SP7 (1). These differences are indicative of minor genetic variation within the species R. capsulatus.
FIG. 1.
Map of cycP and flanking regions. Physical map of the R. capsulatus region containing cycP (encoding cytochrome c′), cybP (encoding a putative membrane-bound b-type cytochrome), and three open reading frames with no homology to genes of known function. Constructs were generated in this work containing a Kmr cassette inserted in the XhoI site within cycP in the opposite direction from cycP transcription (plasmids pCP201 and pCP301 and R. capsulatus strain MC101) (i) and in the same direction as cycP transcription (plasmids pCP202 and pCP302 and R. capsulatus strain MC111) (ii).
The N-terminal 21 amino acids which are not found in the mature protein sequence of cytochrome c′ are predominantly hydrophobic, except for two basic residues toward the N-terminal end. Signal peptide prediction programs TopPred2 and SOSUI strongly predict that this N-terminal region is a periplasmic leader sequence, and they predict a proteolytic cleavage site between amino acids 21 and 22. This is entirely in keeping with the fact that cytochrome c′ has been shown by cell fractionation studies to be a periplasmic protein (14) and that the biochemically determined N terminus is equivalent to amino acid 22 of the sequence predicted from the gene.
Database searches of the DNA sequence downstream of cycP revealed an open reading frame, cybP, with a deduced amino acid sequence which has 41% identity to that of an open reading frame found in the phototrophic purple sulfur bacterium Chromatium vinosum (10). In R. capsulatus, cybP is adjacent to, and transcribed in the same direction as, cycP. Interestingly, the gene in C. vinosum is also located downstream of the gene encoding cytochrome c′, but in this case the two genes are convergently transcribed. The possibility that there is functional interaction between cytochrome c′ and CybP in both bacteria is enticing. CybP is homologous to the cytochrome b-type subunit of Ni-Fe hydrogenases found in a range of hydrogen-utilizing bacteria (15). This family of b-type cytochromes consists of a core structure containing four putative transmembrane-spanning helices and two b-type hemes, ligated via conserved His residues within the membrane-spanning regions of the proteins. These conserved His residues are also conserved in CybP from R. capsulatus and C. vinosum.
In R. capsulatus, the predicted translational start site for cybP lies 60 bases downstream of the translational stop within cycP. Downstream of the coding region of cycP is a putative stem-loop (stability of formation, ΔG = −19.7 kcal, as calculated using GCG program Mfold) followed by a T-rich region which may together constitute the elements necessary to stop transcription. There is a purine-rich putative ribosome binding site just upstream from the predicted start site of cybP but no obvious promoter features. It is likely therefore that the expression of cybP relies upon read-through from cycP, and therefore the level of expression of the membrane cytochrome is low compared to that of cytochrome c′.
In order to gauge the distribution of cytochrome c′, a TBLASTN search was executed against a National Center for Biotechnology Information database containing all complete and partially complete eubacterial and archaeal genome sequences (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html). Genes with significant homology to cycP were identified in the genomes of Pseudomonas aeruginosa, Bordetella pertussis, and Neisseria meningitidis. Part of the gene for cytochrome c′ from N. meningitidis had previously been identified in a project to identify its neighboring gene lst, which encodes lipopolysaccharide α-2,3-sialyltransferase (13). In each case, the gene consists of an open reading frame whose predicted amino acid sequence bears significant similarity to the mature cytochrome c′ (including the conservation of the c-heme covalent attachment motif CXXCH toward the C terminus) and an N-terminal predicted periplasmic leader sequence. Although organisms of significantly variable metabolic diversity which contain cytochrome c′ have been identified, all those so far identified are members of the α-, β-, or γ-Proteobacteria.
Our analysis showing that the pathogen N. meningitidis is capable of synthesizing a cytochrome c′ is of interest, since this organism needs to withstand environments in which NO is produced specifically as a toxic agent, during its pathogenic life cycle. Macrophages produce NO as a consequence of bacterial infection causing induction of inducible NO synthase. The capacity of the organisms to withstand NO may be an important virulence factor. A further twist is that the production of NO may damage the blood-brain barrier, thus allowing N. meningitidis to enter the meninges and cause the disease state of meningitis (6).
Insertional mutagenesis of cycP.
Mutant strains in which cycP was disrupted were generated by inserting the kanamycin resistance gene derived from Tn903 into the XhoI site located within cycP. The kanamycin resistance gene was inserted in both orientations in order to ensure that polar effects of the insertion could be discounted. (The kanamycin resistance gene lacks transcriptional terminators [21], and therefore read-through into cybP should occur in mutant strains in which the resistance cassette is oriented in the same direction as transcription of cycP and cybP, but not when the cassette is oriented oppositely.)
The insert from pCP101 was excised with NsiI and ligated into pGEM-3Zf(+), which had been digested with PstI, to produce a plasmid with a unique XhoI site within cycP. A SalI restriction fragment containing the Tn903-derived kanamycin resistance cartridge from pUC4-K was inserted into the XhoI site, yielding plasmids pCP201 and pCP202 (Fig. 1 and Table 1). pCP201 and pCP202 were digested with KpnI, and the sticky ends were rendered blunt with T4 polymerase. The resultant 7-kb fragments containing disrupted copies of cycP were ligated into the vector pRVS1 (26) (a mobilizable vector capable of replicating within an E. coli host but not within R. capsulatus), which had been linearized with SmaI, to produce pCP301 and pCP302. These plasmids were transformed into E. coli S17-1 (24). E. coli S17-1(pCP301) and E. coli S17-1(pCP302) were allowed to undergo a conjugative mating with R. capsulatus PAS100. For the mating, bacterial strains were grown to mid-log phase, harvested and washed, mixed in a 2:1 ratio of R. capsulatus to E. coli, and maintained on 0.22-μm-pore-size nitrocellulose filters on RCV (28) agar plates for 6 h. R. capsulatus transconjugants in which the kanamycin resistance encoded within plasmids pCP301 and pCP302 had become incorporated into the chromosome by homologous recombination were selected on RCV-malate plates containing kanamycin and rifampin (R. capsulatus PAS100 is resistant to rifampin, the antibiotic being included to counterselect against the E. coli donor strain). Of the resultant transconjugants, 5% were spectinomycin sensitive, indicating that the plasmid DNA containing the spectinomycin resistance gene had become lost. Southern analysis confirmed that double crossovers had occurred such that R. capsulatus MC111 and MC101 each contained a single, disrupted copy of cycP with the kanamycin resistance cassette inserted so as to be transcribed in the same orientation as (MC111) and opposite orientation from (MC101) cycP.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype and description | Source or reference |
|---|---|---|
| R. capsulatus | ||
| PAS100 | Rfr Smr; str-2 hsd-1 restriction-deficient strain derived from wild-type B10 | 28 |
| MC101 | Rfr Kmr; derived from PAS100; Kmr cartridge inserted in cycP and transcribed in the opposite direction from cycP | This study |
| MC111 | As MC101, but Kmr gene transcribed in same direction as cycP | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco BRL |
| TOP10F′ | F′ (lacIq Tetr) mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL endA1 nupG | Invitrogen |
| S17-1 | thi pro hsdR recA RP4-2 integrated Tc::Mu Km::Tn7 (mobilizable vector) | 24 |
| Plasmids | ||
| pZErO | Kmr | Invitrogen |
| pGEM-3Zf(+) | Apr | Promega |
| pUC4-K | Apr Kmr; source of Tn903-derived Kmr cartridge | Pharmacia |
| pRVS1 | Spr Apr; mobilizable vector | 26 |
| pCP101 | Kmr; 6-kb SalI fragment containing cycP in pZErO | This study |
| pCP201 | pCP101 with Kmr cartridge in XhoI site within cycP; Kmr inserted in opposite direction from transcription of cycP | This study |
| pCP202 | As pCP201 with Kmr cartridge inserted in same direction as transcription of cycP | This study |
| pCP301 | cycP::Kmr insert from pCP201 cloned into pRVS1 | This study |
| pCP302 | cycP::Kmr insert from pCP202 cloned into pRVS1 | This study |
| pRK415 | Broad-host-range, mobilizable vector; Tcr | 16 |
| pRKMC401 | pRK415 containing BamHI fragment containing cycP | This study |
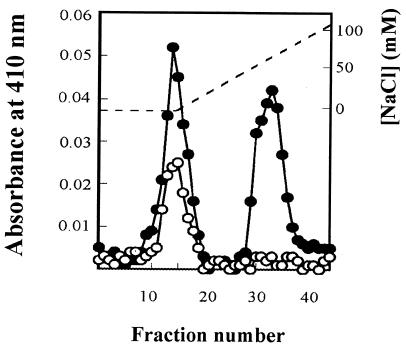
In order to confirm the mutant construction biochemically, periplasmic extracts from R. capsulatus PAS100 and the cycP mutants were prepared by harvesting 1 liter of cells; resuspending the resultant pellets in 25 ml of buffer containing 500 mM sucrose, 100 mM Tris-HCl (pH 8), and 3 mM EDTA; and incubating them with lysozyme (25 mg) at 30°C for 1 h. Spheroplasts and periplasm were separated by centrifugation at 17,500 × g for 5 min. Ion-exchange column profiles for R. capsulatus PAS100 and MC101 (Fig. 2) show that a cytochrome peak is absent from periplasm isolated from the cycP mutant strain. Spectroscopic analysis of peak fractions using a World Precision Instruments S2000 fiber optic spectrophotometer confirmed that MC101 synthesizes cytochrome c2 but not cytochrome c′ whereas the wild type synthesizes both cytochromes. Similar results were found for mutant MC111.
FIG. 2.
Separation of periplasmic cytochromes of R. capsulatus PAS100 and MC101. One-liter cultures of R. capsulatus PAS100 and MC101 were grown photosynthetically in RCV-malate, and the periplasmic extract from each culture was isolated. Each of these extracts was loaded onto a 1.7- by 15-cm DEAE-Sepharose CL6B anion-exchange column, and the columns were developed with an NaCl gradient in 100 mM Tris-HCl (pH 8). The figure shows the cytochrome absorbance (A410) profile during the elution of proteins from columns loaded with each of these periplasmic extracts, PAS100 (filled circles) and MC101 (open circles). [NaCl] is marked by a dotted line. The first cytochrome to elute is cytochrome c2, which is found in both strains. A second cytochrome, which has the spectral properties of cytochrome c′ (data not shown), eluted with 70 mM NaCl from columns loaded with PAS100 extract but not extracts from the cycP mutant strain MC101.
Growth rates of R. capsulatus strains.
R. capsulatus PAS100 (wild type) and MC101 and MC111 (cycP mutant strains) were grown under anaerobic photoheterotrophic conditions, chemoheterotrophic aerobic conditions, and chemoheterotrophic anaerobic conditions with 60 mM dimethyl sulfoxide (DMSO) as respiratory electron acceptor in the dark in RCV medium (28) supplemented with 25 mM malate. Under aerobic conditions, the wild-type and mutant strains grew at similar rates, as would be expected given that cytochrome c′ is expressed mainly under anaerobic conditions (4). A more unexpected finding was that the mutant strains grew slightly faster than the wild-type strain under anaerobic conditions. This was particularly notable during growth in the dark with DMSO as respiratory electron acceptor (growth rate, μPAS100 = 0.0117 ± 0.0011 h−1, μMC111 = 0.0176 ± 0.0022 h−1, and μMC101 = 0.0200 ± 0.0006 h−1). Cytochrome c′ is a highly expressed protein and represents ca. 5 to 10% of the total periplasmic protein in the wild type after anaerobic growth. The absence of cytochrome c′ may allow a subsequently higher concentration of other proteins in the periplasm (e.g., binding proteins necessary for transport processes and other electron transport proteins), enabling more rapid metabolism and hence growth. This metabolic burden imposed by synthesis of the cytochrome is most noticeable under anaerobic conditions in the dark with DMSO, presumably because the bacteria grow exceedingly slowly under these conditions, and hence the metabolic burden is exaggerated.
Photoheterotrophic growth was monitored after the inclusion of a bolus of 100 μM NO in cultures at the beginning of growth. A long lag phase was observed in wild-type and mutant strains alike, after which time normal growth resumed in all strains (data not shown). The lag phase is due to the toxicity of NO. We suppose that eventually the bacteria are capable of overcoming the inhibitory effects of NO and resuming normal growth after having reduced NO to the relatively inert N2O via the activity of the NO reductase known to be synthesized by R. capsulatus (5).
Disk diffusion susceptibility assays.
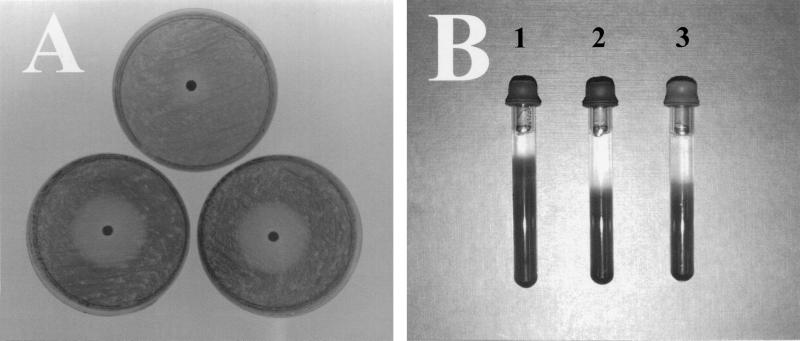
The toxicity of various compounds to strains of R. capsulatus was assessed on agar plates spread with a lawn of the bacteria. Whatman 3MM paper disks (4-mm diameter) soaked in a solution of test substance were carefully laid onto the center of the plates, and the plates were then incubated at 30°C overnight. A circular zone of clearing, within which there was no bacterial growth, occurred on the plates surrounding the filter disk which had been soaked in toxic chemical. The concentration of toxic compound decreases with distance from the disk since the concentration is dependent upon diffusion. The diameter of the clearance zone is a measure of the toxicity of the substance to a given R. capsulatus strain. Filters which were soaked with hydrogen peroxide (H2O2) or with methyl viologen or benzyl viologen, which are superoxide (O2−) releasers, gave similar clearance zones for mutant and wild-type strains (data not shown). On the other hand, soaking filter disks with S-nitrosoglutathione (GSNO) and S-nitrosopenicillamine (SNAP) (both of which can release NO on homolysis or act as nitrosating agents, transferring NO+) gave rise to a larger zone of clearance for the cycP mutant than for the wild type, indicating that the mutant is sensitive to NO or nitrosative stress (Fig. 3A).
FIG. 3.
(A) Disk diffusion susceptibility assays. Four-millimeter paper disks soaked with 15 μl of 125 mM GSNO were placed on RCV-malate plates on which a lawn of bacteria had been spread in order to examine the effect of NO on growth of R. capsulatus strains. The photograph shows R. capsulatus PAS100 (top), MC101 (lower left), and MC111 (lower right) demonstrating the greater degree of susceptibility of the cycP mutant strains to the NO releaser. (B) Growth of R. capsulatus strains under an NO headspace. NO gas (100 μl) was injected into the headspace of tubes containing R. capsulatus strains suspended in 0.3% agar which were subsequently incubated in the light for 24 h. Depth above which growth is not observed toward the top of the agar is shallower in PAS100 (tube 1) than in MC101 and MC111 (tubes 2 and 3, respectively), indicative of the greater capacity of PAS100 to withstand the toxic effects of NO than of either of the cycP mutants.
Effect of NO gas on growth rate.
The finding that the NO releasers and nitrosating agents GSNO and SNAP were less toxic to a strain of R. capsulatus expressing cytochrome c′ than to strains which could not express the cytochrome suggests that the cytochrome serves to detoxify NO in vivo. However, given the possibility that the effect of these chemicals is independent of NO, we devised a method to directly test the toxicity of NO. Growth of R. capsulatus strains was carried out in agar, in glass tubes with a 9-mm internal diameter. Suspensions of R. capsulatus were mixed with RCV-malate containing either 0.3 or 1.0% agar at 42°C, poured into glass tubes, and allowed to set. The tubes were fitted with gastight Suba-Seals, and the headspace was sparged with nitrogen gas. Subsequently, known volumes of NO gas (obtained from Aldrich) were injected into the headspace using a Hamilton syringe in order to assess the effect of the gas on growth of R. capsulatus strains. During the subsequent incubation, NO diffused from the headspace through the agar, and a zone of clearance formed at the top of the tubes due to the toxicity of NO gas. The depth of the clearance zone is a measure of the toxicity of NO to a given strain. Figure 3B shows typical NO tubes and demonstrates clearly that the wild type is significantly less susceptible to the toxicity of NO than is either of the cycP mutant strains. In order to exclude the possibility that the clearance toward the top of the tube was due to NO acting as a chemorepellant and that the bacteria were moving down the tubes by chemotaxis, the experiments were repeated with suspensions of bacteria maintained in 1% agar (in which motility is inhibited), yielding results similar to those shown in Fig. 3B.
Effect of NO on oxygen uptake.
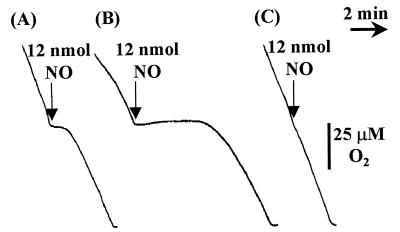
In R. capsulatus, oxygen utilization is performed by heme-copper oxidases, enzymes which are sensitive to NO (7). To test whether the presence of cytochrome c′ confers NO resistance by binding the ligand, the rates of oxygen uptake by R. capsulatus wild-type and cycP mutant strains were monitored in a Clark-type O2 electrode (Rank Brothers, Bottisham, Cambridge, United Kingdom). The inhibitory effect of NO was assessed by injecting volumes of an NO-saturated solution into the vessel containing the respiring cells. Figure 4 shows that NO has a greater inhibitory effect on oxygen respiration in the cycP mutant strains than in the wild type.
FIG. 4.
Effect of NO on oxygen respiration. R. capsulatus strains were grown under photoheterotrophic conditions, harvested, and resuspended in fresh RCV-malate growth medium. The rate of oxygen respiration by 2 mg (dry weight) of R. capsulatus contained in a volume of 3 ml was monitored in a Clark-type electrode maintained at 30°C. (A) Inhibitory effect of NO on R. capsulatus PAS100. (B) Inhibitory effect of NO on R. capsulatus MC111. (C) No discernible inhibition of oxygen respiration by 12 nmol of NO added to a suspension of R. capsulatus PAS100 containing plasmid pRKMC401. In each case, NO additions were made at [O2] = 100 μM.
A BamHI fragment containing cycP, but not cybP (Fig. 1), was cloned into broad-host-range plasmid pRK415 (16), yielding plasmid pRKMC401. Conjugative transfer of pRKMC401 into R. capsulatus PAS100 produced a strain that contained multiple copies of cycP. Figure 4 shows that in this cytochrome c′-overproducing strain oxygen respiration is less susceptible to NO than is that of the wild type, further demonstrating that the cytochrome confers increased resistance to NO.
Cytochrome c′ confers increased resistance to NO.
Since cytochrome c′ is known to bind NO, and it has been speculated that its physiological function is related to this property (20, 30, 31), the influence of NO on the growth of wild-type and cycP mutant R. capsulatus was examined by a number of methods. The introduction of NO into cultures of R. capsulatus as a bolus had no noticeably differential effect on growth rates of wild-type versus mutant strains (see above). However, convincing effects on wild-type and cycP mutant R. capsulatus were achieved either indirectly via NO releasers SNAP and GSNO or directly by supplying NO to the headspace above immobilized R. capsulatus. Clearly, the mutant is more susceptible to toxicity caused by NO than is the wild-type strain (Fig. 3). Other heme proteins have been found to function in detoxification, notably, flavohemoglobin from E. coli and Salmonella enterica serovar Typhimurium (12, 18) and hemoglobin from the nematode Ascaris lumbricoides (19).
Natural environments contain detectable levels of NO, which are generated as a result of disproportionation of nitrite under acidic conditions (8) and biologically via nitrification and denitrification (2). The synthesis of cytochrome c′ presumably protects R. capsulatus (and other organisms possessing the cytochrome) from the damaging effects of the radical in these environments. Bulk [NO] in soils has been measured at concentrations of up to 10−7 M (23), which is sufficient to cause potent inhibition of heme-copper oxidases (7). The capacity of cytochrome c′ to bind NO and hence allow oxygen respiratory metabolism to continue may be a key factor in the selection of bacteria that synthesize cytochrome c′. This latter supposition is clearly supported by the finding that oxygen consumption is significantly impaired in mutant strains unable to synthesize cytochrome c′ compared to wild-type strains (Fig. 4).
In conclusion, we have been able to demonstrate for the first time that cytochrome c′ has a physiological function and that this is to alleviate the toxic effects caused by a constant supply of NO to bacterial cultures.
Nucleotide sequence accession number.
The nucleotide sequence of the gene for cytochrome c′ and flanking genes has been deposited in the GenBank database under GenBank accession no. AF147705.
Acknowledgments
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grant P08290, awarded to J.W.B.M. and R.K.P.
REFERENCES
- 1.Ambler R P, Bartsch R G, Daniel M, Kamen M D, McLellan L, Meyer T E, Vanbeeumen J. Amino-acid sequences of bacterial cytochromes c′ and c556. Proc Natl Acad Sci USA. 1981;78:6854–6857. doi: 10.1073/pnas.78.11.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson I C, Levine J S. Relative rates of nitric oxide and nitrous oxide production by nitrifiers, denitrifiers, and nitrate respirers. Appl Environ Microbiol. 1986;51:938–945. doi: 10.1128/aem.51.5.938-945.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong G A, Alberti M, Leach F, Hearst J E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 4.Bartsch R G, Ambler R P, Meyer T E, Cusanovich M A. Effect of aerobic growth-conditions on the soluble cytochrome content of the purple phototrophic bacterium Rhodobacter sphaeroides: induction of cytochrome c554. Arch Biochem Biophys. 1989;271:433–440. doi: 10.1016/0003-9861(89)90293-2. [DOI] [PubMed] [Google Scholar]
- 5.Bell L C, Richardson D J, Ferguson S J. Identification of nitric oxide reductase activity in Rhodobacter capsulatus: the electron transport pathway can either use or bypass both cytochrome c2 and the cytochrome bc1 complex. J Gen Microbiol. 1992;138:437–443. doi: 10.1099/00221287-138-3-437. [DOI] [PubMed] [Google Scholar]
- 6.Boje K M K. Inhibition of nitric oxide synthase attenuates blood-brain barrier disruption during experimental meningitis. Brain Res. 1996;720:75–83. doi: 10.1016/0006-8993(96)00142-4. [DOI] [PubMed] [Google Scholar]
- 7.Brown G C, Cooper C E. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 8.Chalk P M, Smith C J. Chemodenitrification. Dev Plant Soil Sci. 1983;9:65–89. [Google Scholar]
- 9.Dobbs A J, Anderson B F, Faber H R, Baker E N. Three-dimensional structure of cytochrome c′ from two Alcaligenes species and the implications for four-helix bundle structures. Acta Crystallogr. 1996;D52:356–368. doi: 10.1107/S0907444995008328. [DOI] [PubMed] [Google Scholar]
- 10.Even M T, Kassner R J, Dolata M, Meyer T E, Cusanovich M A. Molecular-cloning and sequencing of cytochrome c′ from the phototrophic purple sulfur bacterium Chromatium vinosum. Biochim Biophys Acta. 1995;1231:220–222. doi: 10.1016/0005-2728(95)00101-n. [DOI] [PubMed] [Google Scholar]
- 11.Finzel B C, Weber P C, Hardman K D, Salemme F R. Structure of ferricytochrome c′ from Rhodospirillum molischianum at 1.67 Å resolution. J Mol Biol. 1985;186:627–643. doi: 10.1016/0022-2836(85)90135-4. [DOI] [PubMed] [Google Scholar]
- 12.Gardner P R, Gardner A M, Martin L A, Salzman A L. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert M, Watson D C, Cunningham A M, Jennings M P, Young N M, Wakarchuk W W. Cloning of the lipooligosaccharide alpha-2,3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J Biol Chem. 1996;271:28271–28276. doi: 10.1074/jbc.271.45.28271. [DOI] [PubMed] [Google Scholar]
- 14.Gilmour R, Goodhew C F, Pettigrew G W. Cytochrome c′ of Paracoccus denitrificans. Biochim Biophys Acta. 1991;1059:233–238. doi: 10.1016/s0005-2728(05)80208-7. [DOI] [PubMed] [Google Scholar]
- 15.Gross R, Simon J, Lancaster C R D, Kroger A. Identification of histidine residues in Wolinella succinogenes hydrogenase that are essential for menaquinone reduction by H2. Mol Microbiol. 1998;30:639–646. doi: 10.1046/j.1365-2958.1998.01100.x. [DOI] [PubMed] [Google Scholar]
- 16.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 17.Maltempo M M, Moss T H. The spin 3/2 state and quantum spin mixtures in haem proteins. Q Rev Biophys. 1976;9:181–215. doi: 10.1017/s0033583500002407. [DOI] [PubMed] [Google Scholar]
- 18.Membrillo-Hernandez J, Coopamah M D, Channa A, Hughes M N, Poole R K. A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohaemoglobin) gene by the 'NO releaser,‘ S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the glyA-hmp intergenic region. Mol Microbiol. 1998;29:1101–1112. doi: 10.1046/j.1365-2958.1998.01000.x. [DOI] [PubMed] [Google Scholar]
- 19.Minning D M, Gow A J, Bonaventura J, Braun R, Dewhirst M, Goldberg D E, Stamler J S. Ascaris haemoglobin is a nitric oxide-activated ‘deoxygenase.’. Nature. 1999;401:497–502. doi: 10.1038/46822. [DOI] [PubMed] [Google Scholar]
- 20.Moir J W B. Cytochrome c′ from Paracoccus denitrificans: spectroscopic studies consistent with a role for the protein in nitric oxide metabolism. Biochim Biophys Acta. 1999;1430:65–72. doi: 10.1016/s0167-4838(98)00276-3. [DOI] [PubMed] [Google Scholar]
- 21.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 22.Ren Z, Meyer T, McRee D E. Atomic-structure of a cytochrome c′ with an unusual ligand-controlled dimer dissociation at 1.8 Å resolution. J Mol Biol. 1993;234:433–445. doi: 10.1006/jmbi.1993.1597. [DOI] [PubMed] [Google Scholar]
- 23.Rudolph J, Conrad R. Flux between soil and atmosphere, vertical concentration profiles in soil, and turnover of nitric oxide. 2. Experiments with naturally layered soil cores. J Atmos Chem. 1996;23:275–300. [Google Scholar]
- 24.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 25.Tahirov T H, Misaki S, Meyer T E, Cusanovich M A, Higuchi Y, Yasuoka N. High-resolution crystal structures of two polymorphs of cytochrome c′ from the purple phototrophic bacterium Rhodobacter capsulatus. J Mol Biol. 1996;259:467–479. doi: 10.1006/jmbi.1996.0333. [DOI] [PubMed] [Google Scholar]
- 26.VanSpanning R J M, Wansell C W, Reijnders W N M, Harms N, Ras J, Oltmann L F, Stouthamer A H. A method for introduction of unmarked mutations in the genome of Paracoccus denitrificans: construction of strains with multiple mutations in the genes encoding periplasmic cytochrome c550, cytochrome c551i, and cytochrome c553i. J Bacteriol. 1991;173:6962–6970. doi: 10.1128/jb.173.21.6962-6970.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernon L P, Kamen M D. Hematin compounds in photosynthetic bacteria. J Biol Chem. 1954;211:643–662. [PubMed] [Google Scholar]
- 28.Weaver P F, Wall J D, Gest H. Characterisation of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 29.Yasui M, Harada S, Kai Y, Kasai N, Kusunoki M, Matsuura Y. 3-Dimensional structure of ferricytochrome c′ from Rhodospirillum rubrum at 2.8 Å resolution. J Biochem. 1992;111:317–324. doi: 10.1093/oxfordjournals.jbchem.a123756. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura T, Iwasaki H, Shidara S, Suzuki S, Nakahara A, Matsubara T. Nitric oxide complex of cytochrome c′ in cells of denitrifying bacteria. J Biochem. 1988;103:1016–1019. doi: 10.1093/oxfordjournals.jbchem.a122372. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura T, Shidara S, Ozaki T, Kamada H. 5 coordinated nitrosylhemoprotein in the whole cells of denitrifying bacterium, Achromobacter xylosoxidans NCIB 11015. Arch Microbiol. 1993;160:498–500. [Google Scholar]