Abstract
The Bacillus subtilis merodiploid strain GSY1127 contains a large nontandem duplication of a portion of its chromosome within its left (anticlockwise) replication segment. This causes displacement of the replication terminus region to a noticeably asymmetric location relative to oriC. The utilization of the subsidiary replication terminators, TerIII and TerV, in the merodiploid strain has been compared with that in B. subtilis 168. It is shown that TerIII is utilized to a significant extent in GSY1127 and that TerV is used only marginally at the most. Neither of these terminators is used to a measurable extent in the 168 strain. It is concluded that TerIII and TerV do indeed function as backups to the major terminator TerI, as has been generally thought. It is further concluded that, in the 168 strain, the vast majority of clockwise forks are arrested at the highly efficient TerI terminator, with fork fusion between the approaching forks occurring frequently while the clockwise fork is stationary at TerI.
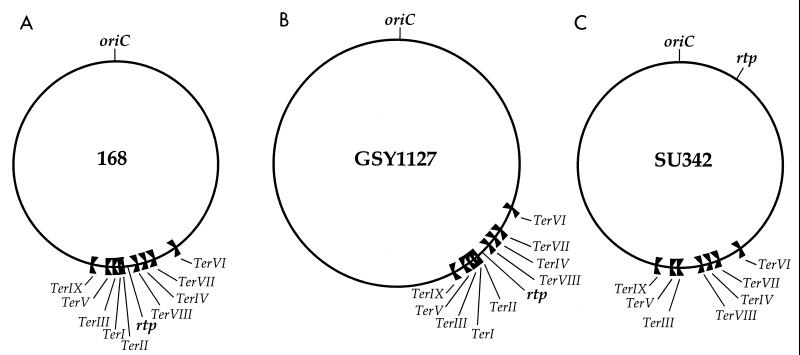
In the circular chromosomes of Bacillus subtilis and Escherichia coli the approach of replication forks towards the end of a round of replication is modulated by the presence of DNA replication terminators. Such terminators are short DNA sequences that bind a cognate terminator protein (RTP [for replication terminator protein] in B. subtilis and Tus [for terminus utilization substance] in E. coli). A terminator-terminator protein complex has the ability to arrest (or cause a severe pausing of) a replication fork in a polar manner. Both organisms contain multiple terminators (9 in B. subtilis and 10 in E. coli) in the terminus region, and they are organized as two opposed groups in each case (2, 6). Figure 1A shows the location and orientation of the terminators (TerI to TerIX) in the B. subtilis 168 chromosome. Ter1, -III, -V, and -IX are oriented to arrest the clockwise-moving fork, while TerII, -VIII, -IV, -VII, and -VI arrest the anticlockwise fork. The gene for RTP (rtp) lies between TerII and TerVIII, and its expression is autoregulated through a promoter that overlaps TerI (1). It has been generally agreed that the multiplicity of terminators reflects the use of the outer (subsidiary) ones as backups to those more centrally located within the terminus region. Thus, if the clockwise fork passed through TerI, its movement out of the terminus region would be subsequently impeded when it reached TerIII. Consistent with this concept was the observation that some of the outer terminators in E. coli operated only when the inner ones were deleted (7). The opposed arrangement of two groups of terminators in the chromosome has been referred to as constituting a “replication fork trap” (8).
FIG. 1.
Location of DNA replication terminators in the chromosomes of various B. subtilis strains. TerI, -III, -V, and -IX are oriented to block movement of the clockwise fork generated at oriC. The others are oriented to block the anticlockwise fork. The position of the rtp gene is also shown in each case.
In the present work, and with knowledge of the full complement of terminus region terminators in B. subtilis, we have used a new approach to demonstrate directly that the outer terminators do in fact function as backups in order to prevent passage of a fork out of the terminus region. We took advantage of the availability of a stable merodiploid strain of B. subtilis, GSY1127 (11). This strain contains a nontandem duplication of a 932-kb segment of the left replication half of the chromosome, which causes displacement of the terminus region to a markedly asymmetric location relative to oriC (Fig. 1B). The orientation of the duplication within the enlarged oriC-to-terminus segment is the same as in the primary segment. (It should be noted that the progenitor of GSY1127 and B. subtilis 168 were both derived from the B. subtilis Marburg strain.) TerI appears to be the most frequently used terminator in B. subtilis 168, and this probably reflects its asymmetric positioning at ∼172° on the 360° map (9). Clearly, one would expect TerIII and TerV, if they were needed as backups, to be utilized more frequently in GSY1127 than in B. subtilis 168. It will be shown that this is indeed the case. Also, it can be firmly concluded that very few, if any, of the clockwise forks in B. subtilis 168 pass through TerI.
B. subtilis 168 trpC2 and the GSY1127 strain (hisH2 ilvC1/ilvC+) were obtained from E. W. Nester (Stanford collection) and C. Anagnostopoulos, respectively. DNA was extracted from mid-exponential-phase Penassay broth cultures at 37°C using procedures described previously (13). After being cut with the appropriate restriction enzyme, the DNA was examined for the presence of forked molecules arrested at the terminator being investigated by Southern transfer and hybridization using an appropriate 32P-labeled probe (4). In all cases two completely independent experiments were performed, and they gave identical results.
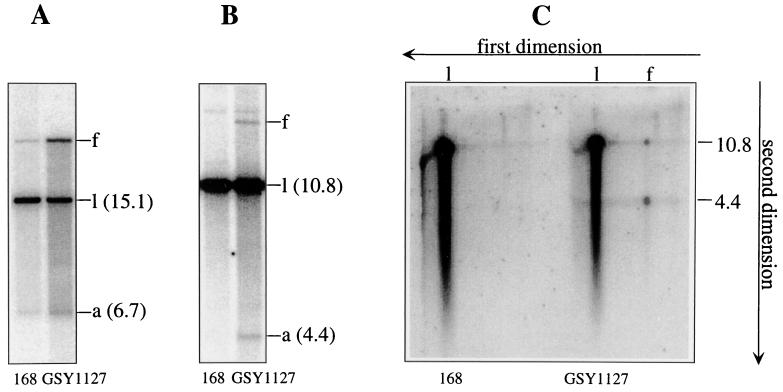
Figure 2A shows the result of testing for fork arrest at TerI. This terminator lies within a 15.1-kb NcoI fragment. The forked molecule, indicative of arrest at TerI, migrates more slowly than the linear form; the faster-migrating species (6.7 kb) represents an arm released from the fork (13). It is clear that, as expected, the level of arrested fork in GSY1127 relative to the linear form is significantly higher (∼5-fold) than in the 168 strain. In the case of the merodiploid, the ratio of the fork (after correcting for breakdown) to the linear form was quite high (∼0.3). Obviously, TerI is a highly efficient terminator.
FIG. 2.
Clockwise replication fork arrest at TerI and TerIII in B. subtilis 168 and GSY1127. (A and B) Results of neutral-gel assays (4, 13) for arrest at TerI and TerIII, respectively. (C) Results of analysis of the DNA samples analyzed in panel B by 2-D (neutral-alkaline) gel electrophoresis (14). B. subtilis 168 and GSY1127 were grown in Penassay broth at 37°C, and samples were collected at mid-exponential phase for DNA extraction and analysis as described previously (4, 13). For assays of fork arrest at TerI the DNA was cut with NcoI, and for TerIII assays SalI was used. The 32P-labeled probes used corresponded to a 0.53-kb segment of the 6.7-kb arm of the fork arrested at TerI and a 5.1-kb segment covering the complete arm (4.4 kb) plus part of the stem of the fork arrested at TerIII.
TerIII lies only 16.4 kb from TerI and within a 10.8-kb SalI fragment. Figure 2B shows the results of Southern analysis of a SalI digest of the DNA samples examined in Fig. 2A using an appropriate TerIII probe. Clearly there has been measurable fork arrest at TerIII in GSY1127, indicated by the presence of forked DNA, a significant portion of which has broken down to yield the 4.4-kb arm. But the TerIII-related fork is completely absent from the 168 strain. That the band labeled “f” in the GSY1127 lane was indeed a forked molecule was established by two-dimensional (2-D) (neutral-alkaline) gel electrophoresis (Fig. 2C, right section). In the second (alkaline) dimension the forked molecule was resolved into its expected component single strands of 4.4 and 10.8 kb. The left section of Fig. 2C confirmed that the fork was undetectable in the 168 strain. The ratio of forked DNA at TerIII in GSY1127 (corrected for breakdown) relative to the linear (10.8-kb) species was ∼0.05. Even in examining more intense phosphorimager images, no TerIII-derived forked molecules could be detected in DNA from the 168 strain. It is concluded that a small but significant fraction of the clockwise replication forks in GSY1127 pass through TerI to be arrested at TerIII, but this is not so for the 168 strain.
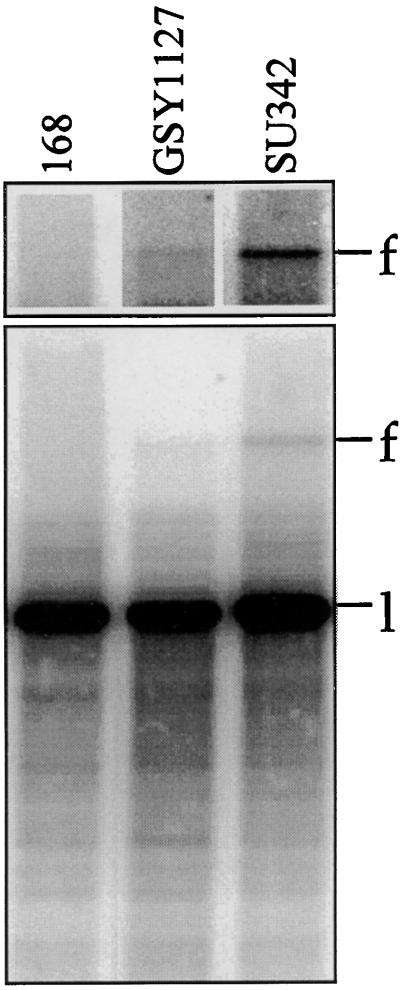
TerV lies 117 kb from TerIII and within a 20.3-kb NcoI fragment. Results of an examination of the DNA samples from the 168 and GSY1127 strains for forks arrested at TerV are shown in Fig. 3. The position expected for an arrested fork at TerV was established by examining DNA from the SU342 strain (Fig. 3), from which TerI and TerII had been deleted and rtp placed at amyE under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac-1 gene (3). The upper section of Fig. 3 shows the area of the forked molecule at a higher phosphorimager intensity. There was possibly a trace amount of forked DNA in GSY1127, but it was completely missing from the 168 strain. The ratio of forked to linear DNA in GSY1127 was <0.002. The band identified in SU342 as the TerV-related fork was too faint to establish its validity by 2-D electrophoresis. This was achieved by comparing DNA from SU342 and SU341 (parent of SU342 without rtp) and finding that, after cutting of the DNA individually with five different restriction enzymes, the putative forked molecule in SU342 (absent from SU341) in every case was of the expected apparent size (approximately the sum of the stem plus two arms).
FIG. 3.
Assays for fork arrest at TerV in various B. subtilis strains. The DNA samples were obtained and assayed, after being cut with NcoI, as described in the legend to Fig. 2. The 32P-labeled probe corresponded to a 1.5-kb segment (labeling by PCR) from within the 12.0-kb arm of the fork arrested at TerV.
The relative strength of individual terminators within their chromosomal context and at normal cellular levels of RTP is not known. However, we have previously shown that both TerIII and TerV function very efficiently and similarly to TerI in the presence of overproduced RTP in a plasmid assay system (4). The results described here show clearly that in the merodiploid strain GSY1127, TerIII is utilized to a small but significant extent and TerV is used only marginally at the most. (The potential usage of the next terminator, TerVI, was not investigated, but it would be expected to be negligible in both strains studied here.) On the other hand, neither TerIII nor TerV was utilized to a measurable extent in the 168 strain. Thus, in the 168 strain the vast majority of clockwise forks are effectively arrested at TerI and do not pass through the additional 16.4 kb to reach TerIII. This is completely consistent with TerIII and TerV functioning as backup terminators, being utilized only if some of the clockwise replication forks pass through TerI. This would be likely to occur more frequently in the merodiploid because of the clockwise fork entering the displaced terminus region well in advance of the anticlockwise fork.
TerI is a very efficient terminator, as evidenced by its presence as forked DNA to such a large extent in GSY1127. Under the conditions of growth used here, approximately 20% of all replicating chromosomes in GSY1127 would have contained a clockwise fork arrested at TerI, reflecting arrest of each individual fork for a significant portion of a round of replication. It is thus not surprising that no detectable clockwise forks pass through TerI in the 168 strain in the relatively short time available before arrival of the anticlockwise fork. Appropriate strains to test the behavior of the anticlockwise fork through the same type of approach are not available. While we have not tested directly for the level of fork arrest at TerII in the 168 chromosome, it is likely that on most occasions, the clockwise fork would encounter TerI (at 172°) in advance of the anticlockwise fork encountering TerII. Thus, in B. subtilis 168 it is highly likely that the majority of approaching forks fuse in the close vicinity of TerI and that in many instances fork fusion would take place while the clockwise fork was stationary.
Some years ago the approach of replication forks in the terminus region of GSY1127 was investigated by the use of outgrowing spores of this strain in conjunction with a density shift approach (10). The data showed severe blockage of the clockwise fork in the vicinity of gltA, but it could not be decided on what side of gltA the block was more likely to have taken place. gltA is now known to reside <4 kb from TerI, which is clearly the major site of arrest. One potentially interesting aspect of the behavior of GSY1127 should be mentioned. In this strain the clockwise fork appears to remain arrested at TerI for a significant portion of each round of replication. In an earlier study in which TerI was placed at metD (100°) in the 168 strain and well outside the normal terminus region (to yield SU227), the clockwise fork was estimated to be arrested at this site for the order of 15 min at 37°C (5). This caused the two forks to fuse, on the average, in the vicinity of 145°. Noticeable elongation of the cells was evident, with a small portion becoming filamentous. It was concluded that the cell elongation was most likely mediated through at least partial SOS induction as a result of the clockwise forks being arrested for such a long time. In spite of arrest of the clockwise fork for an extended period of time in GSY1127, this strain gave no indication of filamentation. In GSY1127 most of the approaching forks would still meet within the normal terminus region, in contrast to SU227, and this might be the causative factor in avoiding filamentation. Of course it is possible that GSY1127 carries a mutation suppressing any filamentation. The nonfilamentous behavior of GSY1127 deserves further investigation.
Acknowledgments
This work has been supported by the Australian Research Council.
REFERENCES
- 1.Ahn K S, Malo M S, Smith M T, Wake R G. Autoregulation of the gene encoding the replication terminator protein of Bacillus subtilis. Gene. 1993;132:7–13. doi: 10.1016/0378-1119(93)90508-z. [DOI] [PubMed] [Google Scholar]
- 2.Coskun-Ari F F, Hill T M. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem. 1997;272:26448–26456. doi: 10.1074/jbc.272.42.26448. [DOI] [PubMed] [Google Scholar]
- 3.Duggin I G, Andersen P A, Smith M T, Wilce J A, King G F, Wake R G. Site-directed mutants of RTP of Bacillus subtilis and the mechanism of replication fork arrest. J Mol Biol. 1999;286:1325–1335. doi: 10.1006/jmbi.1999.2553. [DOI] [PubMed] [Google Scholar]
- 4.Franks A H, Griffiths A A, Wake R G. Identification and characterization of new DNA replication terminators in Bacillus subtilis. Mol Microbiol. 1995;17:13–23. doi: 10.1111/j.1365-2958.1995.mmi_17010013.x. [DOI] [PubMed] [Google Scholar]
- 5.Franks A H, Wake R G. Replication fork arrest at relocated replication terminators on the Bacillus subtilis chromosome. J Bacteriol. 1996;178:4258–4265. doi: 10.1128/jb.178.14.4258-4265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths A A, Andersen P A, Wake R G. Replication termination protein-based replication fork-arrest systems in various Bacillus species. J Bacteriol. 1998;180:3360–3367. doi: 10.1128/jb.180.13.3360-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill T M. Features of the chromosomal terminus region. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1602–1614. [Google Scholar]
- 8.Kuempel P L, Pelletier A J, Hill T M. Tus and the terminators: the arrest of replication in prokaryotes. Cell. 1989;59:581–583. doi: 10.1016/0092-8674(89)90001-9. [DOI] [PubMed] [Google Scholar]
- 9.Kunst F, Ogasawara N, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 10.O'Sullivan M A, Anagnostopoulos C. Replication terminus of the Bacillus subtilis chromosome. J Bacteriol. 1982;151:135–143. doi: 10.1128/jb.151.1.135-143.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider A-M, Gaisne M, Anagnostopoulos C. Genetic structure and internal rearrangements of stable merodiploids from Bacillus subtilis strains carrying the trpE26 mutation. Genetics. 1982;101:189–210. doi: 10.1093/genetics/101.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith M T, Wake R G. Definition and polarity of action of DNA replication terminators in Bacillus subtilis. J Mol Biol. 1992;227:648–657. doi: 10.1016/0022-2836(92)90214-5. [DOI] [PubMed] [Google Scholar]
- 13.Weiss A S, Wake R G. A unique DNA intermediate associated with termination of chromosome replication in Bacillus subtilis. Cell. 1984;39:683–689. doi: 10.1016/0092-8674(84)90475-6. [DOI] [PubMed] [Google Scholar]
- 14.Yasbin R E, Firshein W, Laffan J, Wake R G. DNA repair and DNA replication in Bacillus subtilis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 295–326. [Google Scholar]