Abstract
The RIP homotypic interaction motif (RHIM) is a conserved protein domain that is approximately 18–22 amino acids in length. In humans, four proteins carrying RHIM domains have been identified: receptor-interacting serine/threonine protein kinase (RIPK) 1, RIPK3, Z-DNA-binding protein 1 (ZBP1), and TIR domain-containing adapter-inducing IFN-β (TRIF), which are all major players in necroptosis, a distinct form of regulated cell death. Necroptosis is mostly presumed to be a fail-safe form of cell death, occurring in cells in which apoptosis is compromised. Upon activation, RIPK1, ZBP1, and TRIF each hetero-oligomerize with RIPK3 and induce the assembly of an amyloid-like structure of RIPK3 homo-oligomers. These act as docking stations for the recruitment of the pseudokinase mixed-lineage kinase domain like (MLKL), the pore-forming executioner of necroptosis. As RHIM domain interactions are a vital component of the signaling cascade and can also be involved in apoptosis and pyroptosis activation, it is unsurprising that viral and bacterial pathogens have developed means of disrupting RHIM-mediated signaling to ensure survival. Moreover, as these mechanisms play an essential part of regulated cell death signaling, they have received much attention in recent years. Herein, we present the latest insights into the supramolecular structure of interacting RHIM proteins and their distinct signaling cascades in inflammation and infection. Their uncovering will ultimately contribute to the development of new therapeutic strategies in the regulation of lytic cell death.
Keywords: immunogenic cell death, necroptosis, RHIM domain
Introduction
RHIM (RIP homotypic interaction motif), an amino acid sequence of approximately 18–22 residues in length [1], is a phylogenetically old motif. Proteins interacting via RHIM domains (Figure 1A) have been described in several metazoan species, with RHIM-like domains found in fungi and even some prokaryotes [2]. The metazoan RHIM domain is structurally and functionally related to the prion-forming domain of the HET-s protein expressed in filamentous fungi [3] (Figure 1B), which oligomerizes and forms fibrils as part of the heterokaryon incompatibility system. Orthologs to mammalian RHIM proteins were discovered in Drosophila and Branchiostoma, where they are involved in innate immunity signaling pathways [3,4], underlining the conserved nature of this motif.
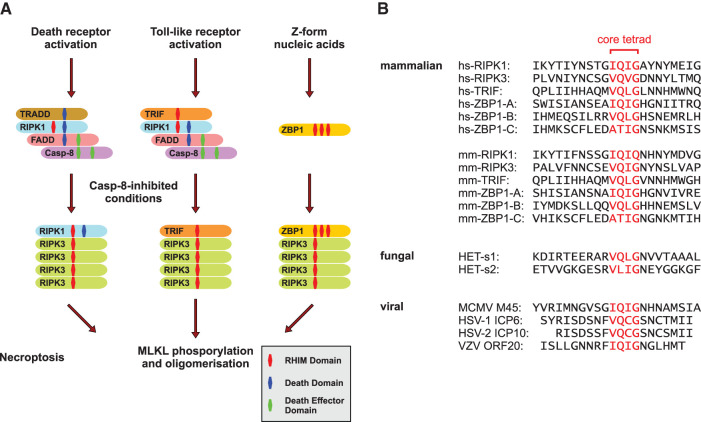
Figure 1. The conserved RHIM domain and its interactions in necroptosis.
(A) Schematic presentation of the established necroptosis pathways. In response to death receptor signaling, TRADD–RIPK1–FADD–caspase-8 complex assembly is initiated, mediated by death domain and death effector domain interactions. TLR signaling can also directly trigger cell death via assembly of the TRIF–RIPK1–FADD–caspase-8 complex. In caspase-8-inhibited conditions, RIPK1 or TRIF oligomerize with RIPK3 and serve as ‘seeds’ for RIPK3 homo-oligomer formation. Upon sensing intracellular Z-form nucleic acids, e.g. viral RNA, ZBP1 recruits RIPK3 and initiates oligomer formation. The amyloid-like RIPK3 homo-oligomers constitute in each case the platform for MLKL activation, a mandatory step for the execution of necroptotic cell death. (B) Alignment of the RHIM amino acid sequence of Homo sapiens (hs), Mus musculus (mm), the Podospora anserina proteins HET-s1 and HET-s2, and the viral RHIM domains of MCMV M45, HSV-1 ICP6, HSV-2 ICP10, and VZV ORF20. The conserved core tetrad is highlighted in red.
In mammals, the RHIM domain was originally identified as a short amino acid sequence in the intermediate domain of RIPK1 (receptor-interacting serine/threonine protein kinase 1), and near the RIPK3 C-terminus that was pivotal to protein interactions and the promotion of necroptosis [5,6], a form of regulated necrosis triggered in ischemia-reperfusion injury (IRI), systemic inflammation, autoimmunity, and neurodegenerative diseases, among others [7–16]. Also known as TICAM1, TRIF (TIR domain-containing adapter-inducing interferon (IFN)-β) was the third protein identified to carry a functional RHIM domain and induces a necroptosis-initiating complex with RIPK3 in response to TLR3 (Toll-like receptor 3) and TLR4 signaling [17,18]. The fourth and last protein demonstrated to be able to interact with RIPK3 via a RHIM domain to induce necroptosis is ZBP1, also known as the DNA-dependent activator of interferon (IFN) regulatory factors (DAI) [19–21]. The RHIM domain in these proteins is defined by a core tetrad with the consensus sequence (V/I)-Q-(V/I/L/C)-G, which is essential for intermolecular protein interaction and signal transduction [5,18,19,21–23]. Mutating all four conserved residues to alanine is a common laboratory approach for abolishing RHIM domain signaling and necroptosis without disrupting other functions of the protein [5,19,22,24], whereas mutation of a single valine residue of the RIPK3 core tetrad (mouse RIPK3 V448P corresponding to human RIPK3 V480P) is sufficient to rescue cells in vitro from canonical necroptosis [25]. The engagement with RIPK3 via RHIM domain interactions is essential for inducing activation of the effector MLKL (mixed-lineage kinase domain like) [5,19,22,26]. In the following execution phase of necroptosis, MLKL oligomerizes to permeabilize the cell membrane by a mechanism distinct among species that has been investigated in depth in recent years but is not fully understood [27–30]. Subsequently, cell lysis occurs and damage-associated molecular patterns (DAMPs) are released into the extracellular space [31–33]. The importance of RHIM interactions for the promotion of necroptosis signaling is underlined by the finding that while kinase activity of RIPK3 is dispensable for MLKL activation under RIPK3-overexpressed conditions, the RIPK3 RHIM-deficient mutant cannot induce necroptosis despite the fact that it still phosphorylates MLKL. In humans, a core oligomer of at least four RIPK3 protomers interacting via their RHIM domains appears essential to enable MLKL oligomer formation [26].
Identifying the precise order of the necroptosis signaling cascades and the composition of the signaling complexes in response to specific triggers has been the main challenge in past years. Canonically, necroptosis is triggered by death receptor signaling when caspases are inhibited and apoptosis is blocked. Under these circumstances, RIPK3 interacts with activated RIPK1 to initiate necroptosis [6,24], but non-canonical pathways involving RHIM interactions have been discovered. One of these involves the activation of some TLRs. Unlike several other TLRs, both TLR3 and TLR4 can directly initiate necroptotic cell death upon ligand binding via TRIF and do not rely on endocrine or paracrine TNF-dependent mechanisms [17,23]. Most studies have reported that TRIF transitions into a complex with RIPK1, caspase-8, FADD, and cFLIP to initiate apoptosis and interacts with RIPK3 to initiate necroptosis when this pro-apoptotic complex fails [17,34,35]. However, a model for Yersinia pseudotuberculosis infection by TLR4 activation under TAK1-inhibited conditions described a RHIM-dependent complex of TRIF, RIPK1, ZBP1, FADD, and caspase-8 termed the TRIFosome, which facilitated a pyroptosis-like form of regulated necrosis [36]. Interestingly, recent studies also uncovered mechanisms in which RIPK3 triggers RIPK1-dependent apoptosis, often as part of the induction of multiple regulated cell death pathways in parallel in response to infection [37–40]. While in vitro stimulation using defined cytokines and inhibitors to induce necroptosis has proven valuable to the initial description of RHIM function, we now need to understand these more diverse complexes and their role in inflammation and immunity.
Host immune evasion by RHIM domain interactions
Viruses infecting mammalian cells often need to dodge detection and triggering the host cell death machinery for long enough to enable efficient replication and spread to other cells [41]. Necroptosis plays a prominent role in the clearance of infected cells and appears to have developed as a backup mechanism to execute cell death when apoptosis fails. However, viral pathogens have co-evolved with the host-regulated cell death machinery and developed mechanisms to ensure cell survival during infection [19,41–43]. In particular, the primate genes encoding RIPK3 and MLKL are in an evolutionary arms race with viral-encoded pathogenicity factors containing functional RHIM domains [34,44]. Specifically, the Betaherpesvirinae, a family of large DNA viruses, utilize an elaborate system to evade several host immune response mechanisms and persist as latent infection [45]. Their mouse-specific member murine cytomegalovirus (MCMV), probably the best studied viral RHIM protein, expresses four different proteins to sequentially block caspase-8-mediated extrinsic apoptosis, RIPK3-mediated necroptosis, and BCL2 family-mediated mitochondrial cell death [42]. By interacting with ZBP1, RIPK1, and RIPK3, MCMV abolishes regulated cell death and TNF-mediated activation of NF-κB and MAPK [46,47]. Infection with an engineered MCMV variant with a mutational disruption of the viral M45 RHIM domain core tetrad led to ZBP1- and RIPK3-dependent but RIPK1-independent necroptosis [20,48], which emphasizes the enormous contribution of this protein to immune evasion and latent infection.
The Alphaherpesvirinae herpes simplex virus (HSV)-1 and HSV-2, both human pathogens, disrupt host cell necroptosis signaling by expressing the RHIM proteins infected cell protein (ICP)6 and ICP10, respectively [49]. As ICP6 simultaneously functions as a suppressor of caspase-8-mediated apoptosis and RIPK3-dependent necroptosis, it is central for establishing HSV infection [49–51]. In the execution of cell death, ICP6 hinders the translocation of the RIPK1–RIPK3–MLKL complex to caveolin-1-associated detergent-resistant membrane (DRM) vesicles, ensuring cell survival in human-derived HT-29 cells [43]. Consistent with that finding, HSV-1 carrying a RHIM-mutated ICP6 failed to evade the cell death of infected human cells and readily induced ZBP1- and RIPK3-dependent necroptosis [52]. Notably, the anti-necroptotic effects mediated by ICP6 are highly host-specific: in mouse cells, it induced RIPK1 and RIPK3 activation and promoted cell death, a finding that can be interpreted as a species barrier contributing to HSV-1 specificity for the natural human host organism [53]. In 2020, another herpesvirus protein with an RHIM domain, ORF20, which is expressed by varicella-zoster virus (VZV) and serves as a decoy for ZBP1, was discovered [37]. RHIM domain mutation inhibited viral spread in vitro; however, this mechanism relies on caspase activation rather than canonical necroptosis [37]. Inhibiting RHIM protein-mediated cell death appears to be particularly important in herpesvirus infections and is probably the main contributor to the typical persistent latent infection of these pathogens [45,51,54]. As the reactivation of these infections poses a major challenge in immunocompromised patients [55,56], overcoming pathogen persistence would be a substantial therapeutic advantage: the viral RHIM proteins are promising target structures in this process.
The expression of an interacting RHIM protein is not the only mechanism pathogens utilize to hinder necroptosis in the cells of mammalian host organisms. Many Gram-negative enteric pathogens have evolved bacterial factors that can be delivered to the cytosolic compartment of the infected host's cells via a type III secretion system (T3SS), which manipulates the immunological response by inhibiting inflammatory signaling and immunogenic cell death (reviewed in [57] and [58]). Both enteropathogenic Escherichia coli (EPEC) and Shigella express a protease delivered by the T3SS that specifically recognizes RHIM sequences in its hosts and thereby cleaves mammalian RHIM proteins [59,60]. The EPEC protease EspL recognizes the RHIM domain conserved core tetrad and cleaves RIPK1, RIPK3, TRIF, ZBP1, and even MCMV M45 [59]. OspD3, the EspL homolog in Shigella, also specifically cleaves the RHIM domain of human RIPK1 and RIPK3, extending the survival of infected host cells and thereby protecting the cytosolic replicative niche of the bacterium [60]. The physiological relevance of this pathogenicity factor was experimentally validated in a mouse model of infection with the EPEC-like mouse pathogen Citrobacter rodentium. During the resolving stage of the inflammation, EspL-proficient bacteria were more prevalent in the feces of infected animals as compared with an espL deletion mutant [59]. This suggested that inhibiting necroptosis additionally promotes bacterial persistence in the gut by limiting the extent of immunogenic cell death. Furthermore, the EspL cleavage site of the RIPK3 RHIM domain in primates was recognized as a rapidly evolving site likely owing to genetic selection [44]. Therefore, it can be assumed that the inhibition of RHIM protein-mediated cell death is a significant virulence factor that contributes to the highly elaborate mechanism utilized by these bacteria to dampen and delay the host immune response [57,59,60].
The depletion of host RIPK3 as a mechanism to enable longer persistence is also utilized by several orthopoxviruses: cowpox virus (CPXV), variola virus, ectromelia virus, and monkeypox virus (currently a worldwide concern); each encodes a viral inducer of RIPK3 degradation (vIRD) [61]. CPXV vIRD binds to the RIPK3 RHIM domain mediated by its N-terminal ankyrin repeats and promotes K48-linked RIPK3 ubiquitination, priming it for proteasomal degradation. In a mouse model of CPXV infection, wild-type virus killed the mice within 5 days while a vIRD-deficient variant was efficiently cleared [61]. However, further investigations will be needed to fully understand how these RIPK3-depleting mechanisms promote pathogenicity and whether they can be targeted in clinical settings to support host defense.
The RHIM domain assembly — an exceptional structure
It has been established for two decades that the RHIM domain facilitates protein–protein interactions [5], but the ongoing characterization of the nature of these interfaces at the molecular level only began more recently. Surprisingly, it was determined that the motif facilitates the formation of amyloid-like structures defined by fibrils of β-sheet-stabilized protein stacks [62,63]. For the longest time, such amyloid structures in mammals were only associated with pathological settings such as degenerative and prion diseases, but newer evidence showed that amyloids can form as a part of functional signal transduction [64]. Furthermore, several signalosomes involving homotypic interaction with high relevance in cell death and inflammation have been described [62].
The first major issue to understanding RHIM domain function was to determine its molecular structure in the native monomeric proteins. Nuclear magnetic resonance (NMR) studies on the C-terminal domain of human RIPK3 revealed that a sequence spanning 22 amino acids (P448–Q469) forms an S-shaped structure defined by three interacting β-sheets, with the VQVG consensus motif being the central part [65]. Upon homo-oligomerization, this structure assembled into fibrils with the S-shaped core region rotating in each layer, which might be necessary to sterically enable RIPK3 kinase domain positioning and activity [65]. All four human RHIM-expressing proteins have the capacity to self-assemble into such homotypic oligomers with distinct structural characteristics [66,67].
The first stable hetero-amyloid complex identified by structural biology was a heterocomplex of the human-derived RIPK1 and RIPK3 RHIM domains [68]. While the core tetrad was pivotal to proper amyloid formation, several surrounding residues stabilize the defined structure of this complex by their interactions, such as RIPK1 N535 and Q540 with RIPK3 N454 and Q459 [68]. However, Wu et al. showed that the central β-sheet containing the human RIPK3 core tetrad was sufficient to interact with the RIPK1 RHIM motif [65]. It is suggested that the RIPK1/RIPK3 hetero-amyloid core region in humans and mice acts as a ‘seed’ for further RIPK3 homo-amyloid fibrils as amplifiers [1,26,68]. RIPK3-dependent signal transduction appeared to be highly reliant on the correct formation of these RHIM complexes, and substitution mutants of RHIM domain residues substantially impaired downstream signaling [69]. Interestingly, RHIM-deficient human RIPK3 retained the ability to phosphorylate (activate) MLKL but failed to induce MLKL oligomer insertion into the membrane and thereby necroptosis [43]. Again, this underlines the essential role of correct RHIM domain alignment in physiological necroptosis signaling.
As RHIM interactions are highly complex and specifically defined, viral RHIM proteins can efficiently interfere with this system [34]. To understand their mode of action, MCMV M45 protein complexes involving both RIPK1 and RIPK3 have been investigated at the molecular level. In canonical necroptosis signaling, M45 inserts itself into hetero-oligomers with human-derived RIPK3, which results in deformed amyloid-like signaling filaments [69]. When comparing human-derived mixed RIPK3–RIPK1 and RIPK3–M45 hetero-oligomers under conditions allowing fibril assembly, the RIPK3–RIPK1 hetero-oligomer mostly formed single fibrils up to ∼200 nm in length whereas RIPK3–M45 oligomers formed extensive fibrillar networks [66]. HSV-1 ICP6 RHIM interactions with mammalian RHIM proteins have been characterized in more detail: the main interaction partners for ICP6 were identified to be ZBP1 and RIPK3 [52,70] and structural analysis of human-derived recombinant proteins revealed that ICP6–ZBP1 hetero-oligomers were more stable than ZBP1 homo-oligomers, forming longer and thicker fibrils. At the same time, the ICP6–RIPK3 hetero-oligomer was less stable and assembled in shorter fibrils than the RIPK3 homo-oligomer [70]. Studies on these distinct RHIM interactions emphasize the importance of the flanking residues around the core tetrad in shaping the structure of the fibril and defining its activity [66,70,71]. These tightly defined interactions have been positioned as promising targets for synthetic RHIM peptides with the potential to interfere with amyloid assembly to prevent necroptosis in human pathologies. A recent approach to designing synthetic peptides containing the M45 RHIM domain fused to the HIV TAT protein as a delivery system regrettably failed to inhibit necroptosis; on the contrary, the peptides killed the targeted cells by self-assembling into amyloid-like fibrils [72]. However, a 90-amino acid truncated MCMV M45 protein containing the viral RHIM domain associated with RIPK3 more efficiently than RIPK1 and prevented necroptosis in vitro [66]. Other than their assembly, the breakdown of higher-order RHIM domain amyloid-like structures are also not fully understood. In 2019, it was reported that autophagy defects led to an accumulation of high-molecular mass structures of TRIF, RIPK1, and RIPK3 — potentially oligomers — and sensitized macrophages to necroptosis [73], but much work remains to be done to understand the underlying process completely.
ZBP1 — a key mediator in cell fate decision
In recent years, much attention has focused on the role of ZBP1 in the fight against infection due to its role as an intracellular sensor of Z-form nucleic acids, and ZBP1 expression is tightly connected with inflammatory, especially IFN, signaling [74,75]. ZBP1-deficient mice were significantly protected in the setting of systemic inflammatory response syndrome (SIRS) induced by TNF and IFN-γ [75]. As part of influenza A virus (IAV) infection, inflammatory stimuli such as IFNs up-regulate the expression of ZBP1, which is able to interact with RIPK3 after sensing newly synthesized nuclear viral RNA, allowing infected cells to initiate pyroptosis, necroptosis, and apoptosis, an inflammatory pathway for which the term PANoptosis has been defined [76–78]. This multimodal cell death in response to IAV infection involves the activation of regulated cell death pathways via the ZBP1–RIPK3 axis, a complex involving RIPK3 fibril formation to induce necroptosis [77]. Furthermore, caspase-6 associates with this complex, enhancing RIPK3 interaction with ZBP1 and thereby facilitating activation of the NLRP3 inflammasome, a key structure in pyroptosis [79]. The ZBP1–RIPK1–RIPK3 interactions also triggered apoptotic and necroptotic cell death simultaneously in HSV-1-infected glial cells [80], suggesting a global role of ZBP1 for inducing multiple forms of regulated cell death. As IAV infection demonstrates several similarities with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in terms of cytokine signaling and cell death, the involvement of ZBP1 in COVID-19 is currently being considered [76,81]. These novel findings highlight the importance of RHIM-mediated signaling as part of inflammatory signaling during infectious diseases.
As part of inflammation, particularly sterile inflammation, necroptosis is a progressive process for which it is imperative to identify inhibitors that limit its pathological extent [7,8,11,82]. In cancer, however, targeted activation of immunogenic regulated cell death supported classical tumor therapy. As ZBP1 promoted necroptosis in response to the sensing of endogenous double-stranded RNA in RIPK1-deficient or caspase-8/FADD-inhibited conditions [83], ZBP1 activation presents opportunities for exploitation in this context. A recent study successfully explored a means of using a Z-DNA formation-triggering small molecule in a cancer model to sensitize tumor cells to ZBP1 activation [39]. Therein, the authors were able to induce necroptosis and apoptosis in vitro and re-sensitize tumor cells to treatment with checkpoint inhibitors in vivo [39]. In line with these observations, defects in the function of the RNA-editing enzyme adenosine deaminase acting on RNA 1 (ADAR1) induces ZBP1 activation via the accumulation of endogenous double-stranded RNA which, led to pathological cell death and autoinflammation [39,84–86]. Other newly discovered non-canonical activators of RIPK3-mediated cell death are heat stress and osmotic stress [87,88]. In this context, the ZBP1 RHIM-A domain was identified as essential to ZBP1 oligomerization and association with RIPK3 in response to heat shock [87]. However, osmotic stress induced by excess extracellular NaCl or sucrose could also trigger rapid RIPK3-dependent MLKL activation and lytic cell death in human and murine cells but required only an active RIPK3 kinase domain and did not involve RHIM domain interactions [88].
All these novel findings on the role of ZBP1 as a key player in innate immunity present promising opportunities to interfere with inflammatory cell death to improve treatment options in infectious diseases and cancer and suggest the ZBP1 RHIM domains (Figure 1B) as promising targets to mediate cellular processes in a variety of pathologies.
Conclusions
In this article, the novel insights presented surrounding the RHIM domain, its amyloid-like structures, and its manifold interactions with pathogens all corroborated the importance of this short amino acid sequence to mammalian immunity. In addition, future efforts will aid the discovery of how this highly conserved and specific domain can be exploited as a target structure to therapeutically interfere with immunogenic cell death signaling in necroptosis-associated morbidities and improve the therapeutic options against viral and bacterial pathogens in sterile inflammatory diseases.
Perspectives
RHIM-mediated signaling by RIPK1, TRIF, ZBP1, and RIPK3 culminating in necroptosis is a key pathway utilized by cells in inflammation and infection settings. As the RHIM structure is highly conserved and exclusive to regulated cell death signal transduction, it presents a promising target structure for regulating interference.
The structures of distinct RHIM domains are currently characterized in-depth at the molecular level. Understanding the exact features of these structures will aid the design of RHIM-mediating drugs for therapeutic interventions.
The nucleic acid sensor ZBP1 is emerging as a promiscuous regulator of cell death in response to virus infection. We are more than aware of the importance of the latter, especially since the outbreak of the infectious disease COVID-19. In particular, ZBP1 signaling could be a target for therapeutic exploitation in infectious diseases in the near future.
Abbreviations
- CPXV
cowpox virus
- DAMPs
damage-associated molecular patterns
- HSV
herpes simplex virus
- IAV
influenza A virus
- ICP
infected cell protein
- MCMV
murine cytomegalovirus
- MLKL
mixed-lineage kinase domain-like
- RHIM
RIP homotypic interaction motif
- RIPK
receptor-interacting serine/threonine protein kinase
- T3SS
type III secretion system
- TLR3
Toll-like receptor 3
- TRIF
TIR domain-containing adapter-inducing IFN-β
- vIRD
viral inducer of RIPK3 degradation
- ZBP1
Z-DNA-binding protein 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was funded by a grant from Dr. Werner Jackstädt-Stiftung (to SK).
Author Contributions
T.R. and S.K. drafted the manuscript. All authors reviewed and approved the final version.
References
- 1.Wu, X.L., Hu, H., Dong, X.Q., Zhang, J., Wang, J., Schwieters, C.D.et al. (2021) The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis. Nat. Commun. 12, 1627 10.1038/s41467-021-21881-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyrka, W., Coustou, V., Daskalov, A., Lends, A., Bardin, T., Berbon, M.et al. (2020) Identification of NLR-associated amyloid signaling motifs in bacterial genomes. J. Mol. Biol. 432, 6005–6027 10.1016/j.jmb.2020.10.004 [DOI] [PubMed] [Google Scholar]
- 3.Kajava, A.V., Klopffleisch, K., Chen, S. and Hofmann, K. (2014) Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci. Rep. 4, 7436 10.1038/srep07436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang, M., Yuan, S., Huang, S., Li, J., Xu, L., Huang, H.et al. (2011) Characterization of bbtTICAM from amphioxus suggests the emergence of a MyD88-independent pathway in basal chordates. Cell Res. 21, 1410–1423 10.1038/cr.2011.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun, X., Yin, J., Starovasnik, M.A., Fairbrother, W.J. and Dixit, V.M. (2002) Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 277, 9505–9511 10.1074/jbc.M109488200 [DOI] [PubMed] [Google Scholar]
- 6.Peltzer, N. and Walczak, H. (2019) Cell death and inflammation - a vital but dangerous liaison. Trends Immunol. 40, 387–402 10.1016/j.it.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 7.Riebeling, T., Jamal, K., Wilson, R., Kolbrink, B., von Samson-Himmelstjerna, F.A., Moerke, C.et al. (2021) Primidone blocks RIPK1-driven cell death and inflammation. Cell Death Differ. 28, 1610–1626 10.1038/s41418-020-00690-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moerke, C., Jaco, I., Dewitz, C., Müller, T., Jacobsen, A.V., Gautheron, J.et al. (2019) The anticonvulsive Phenhydan® suppresses extrinsic cell death. Cell Death Differ. 26, 1631–1645 10.1038/s41418-018-0232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lalaoui, N., Boyden, S.E., Oda, H., Wood, G.M., Stone, D.L., Chau, D.et al. (2020) Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 10.1038/s41586-019-1828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zelic, M., Pontarelli, F., Woodworth, L., Zhu, C., Mahan, A., Ren, Y.et al. (2021) RIPK1 activation mediates neuroinflammation and disease progression in multiple sclerosis. Cell Rep. 35, 109112 10.1016/j.celrep.2021.109112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degterev, A., Ofengeim, D. and Yuan, J. (2019) Targeting RIPK1 for the treatment of human diseases. Proc. Natl Acad. Sci. U.S.A. 116, 9714–9722 10.1073/pnas.1901179116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linkermann, A., Hackl, M.J., Kunzendorf, U., Walczak, H., Krautwald, S. and Jevnikar, A.M. (2013) Necroptosis in immunity and ischemia-reperfusion injury. Am. J. Transplant. 13, 2797–2804 10.1111/ajt.12448 [DOI] [PubMed] [Google Scholar]
- 13.He, S. and Wang, X. (2018) RIP kinases as modulators of inflammation and immunity. Nat. Immunol. 19, 912–922 10.1038/s41590-018-0188-x [DOI] [PubMed] [Google Scholar]
- 14.Yuan, J., Amin, P. and Ofengeim, D. (2019) Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 20, 19–33 10.1038/s41583-018-0093-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nailwal, H. and Chan, F.K. (2019) Necroptosis in anti-viral inflammation. Cell Death Differ. 26, 4–13 10.1038/s41418-018-0172-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zelic, M., Roderick, J.E., O'Donnell, J.A., Lehman, J., Lim, S.E., Janardhan, H.P.et al. (2018) RIP kinase 1-dependent endothelial necroptosis underlies systemic inflammatory response syndrome. J. Clin. Invest. 128, 2064–2075 10.1172/JCI96147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, W.J., Sridharan, H., Huang, C., Mandal, P., Upton, J.W., Gough, P.J.et al. (2013) Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 288, 31268–31279 10.1074/jbc.M113.462341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Martinon, F., Kelliher, M.et al. (2004) RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5, 503–507 10.1038/ni1061 [DOI] [PubMed] [Google Scholar]
- 19.Rebsamen, M., Heinz, L.X., Meylan, E., Michallet, M.C., Schroder, K., Hofmann, K.et al. (2009) DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 10, 916–922 10.1038/embor.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upton, J.W., Kaiser, W.J. and Mocarski, E.S. (2012) DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 10.1016/j.chom.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, W.J., Upton, J.W. and Mocarski, E.S. (2008) Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 181, 6427–6434 10.4049/jimmunol.181.9.6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, W.J. and Offermann, M.K. (2005) Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 174, 4942–4952 10.4049/jimmunol.174.8.4942 [DOI] [PubMed] [Google Scholar]
- 23.Berghe T., V., Hassannia, B. and Vandenabeele, P. (2016) An outline of necrosome triggers. Cell Mol. Life Sci. 73, 2137–2152 10.1007/s00018-016-2189-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho, Y.S., Challa, S., Moquin, D., Genga, R., Ray, T.D., Guildford, M.et al. (2009) Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, H., Wu, X., Li, X., Li, M., Li, F., Wang, L.et al. (2020) Crucial roles of the RIP homotypic interaction motifs of RIPK3 in RIPK1-dependent cell death and lymphoproliferative disease. Cell Rep. 31, 107650 10.1016/j.celrep.2020.107650 [DOI] [PubMed] [Google Scholar]
- 26.Chen, X., Zhu, R., Zhong, J., Ying, Y., Wang, W., Cao, Y.et al. (2022) Mosaic composition of RIP1-RIP3 signalling hub and its role in regulating cell death. Nat. Cell Biol. 24, 471–482 10.1038/s41556-022-00854-7 [DOI] [PubMed] [Google Scholar]
- 27.Ros, U., Peña-Blanco, A., Hänggi, K., Kunzendorf, U., Krautwald, S., Wong, W.W.et al. (2017) Necroptosis execution is mediated by plasma membrane nanopores independent of calcium. Cell Rep. 19, 175–187 10.1016/j.celrep.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores-Romero, H., Ros, U. and Garcia-Saez, A.J. (2020) Pore formation in regulated cell death. EMBO J. 39, e105753 10.15252/embj.2020105753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samson, A.L., Zhang, Y., Geoghegan, N.D., Gavin, X.J., Davies, K.A., Mlodzianoski, M.J.et al. (2020) MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 11, 3151 10.1038/s41467-020-16887-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethi, A., Horne, C.R., Fitzgibbon, C., Wilde, K., Davies, K.A., Garnish, S.E.et al. (2022) Membrane permeabilization is mediated by distinct epitopes in mouse and human orthologs of the necroptosis effector, MLKL. Cell Death Differ. 10.1038/s41418-022-00965-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samson, A.L., Garnish, S.E., Hildebrand, J.M. and Murphy, J.M. (2021) Location, location, location: a compartmentalized view of TNF-induced necroptotic signaling. Sci Signal. 14, eabc6178 10.1126/scisignal.abc6178 [DOI] [PubMed] [Google Scholar]
- 32.Kolbrink, B., Riebeling, T., Kunzendorf, U. and Krautwald, S. (2020) Plasma membrane pores drive inflammatory cell death. Front. Cell Dev. Biol. 8, 817 10.3389/fcell.2020.00817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, L., Wang, H., Wang, Z., He, S., Chen, S., Liao, D.et al. (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 34.Baker, M., Shanmugam, N., Pham, C.L.L., Strange, M., Steain, M. and Sunde, M. (2020) RHIM-based protein:protein interactions in microbial defence against programmed cell death by necroptosis. Semin. Cell Dev. Biol. 99, 86–95 10.1016/j.semcdb.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 35.He, S., Liang, Y., Shao, F. and Wang, X. (2011) Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl Acad. Sci. U.S.A. 108, 20054–20059 10.1073/pnas.1116302108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muendlein, H.I., Connolly, W.M., Magri, Z., Smirnova, I., Ilyukha, V., Gautam, A.et al. (2021) ZBP1 promotes LPS-induced cell death and IL-1β release via RHIM-mediated interactions with RIPK1. Nat. Commun. 12, 86 10.1038/s41467-020-20357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steain, M., Baker, M., Pham, C.L.L., Shanmugam, N., Gambin, Y., Sierecki, E.et al. (2020) Varicella zoster virus encodes a viral decoy RHIM to inhibit cell death. PLoS Pathog. 16, e1008473 10.1371/journal.ppat.1008473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogusa, S., Thapa, R.J., Dillon, C.P., Liedmann, S., Oguin, III, T.H., Ingram, J.P.et al. (2016) RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza A virus. Cell Host Microbe 20, 13–24 10.1016/j.chom.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, T., Yin, C., Fedorov, A., Qiao, L., Bao, H., Beknazarov, N.et al. (2022) ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 606, 594–602 10.1038/s41586-022-04753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shubina, M., Tummers, B., Boyd, D.F., Zhang, T., Yin, C., Gautam, A.et al. (2020) Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J. Exp. Med. 217, e20191259 10.1084/jem.20191259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tummers, B. and Green, D.R. (2022) The evolution of regulated cell death pathways in animals and their evasion by pathogens. Physiol. Rev. 102, 411–454 10.1152/physrev.00002.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandal, P., Nagrani, L.N., Hernandez, L., McCormick, A.L., Dillon, C.P., Koehler, H.S.et al. (2021) Multiple autonomous cell death suppression strategies ensure cytomegalovirus fitness. Viruses 13, 1707 10.3390/v13091707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali, M., Roback, L. and Mocarski, E.S. (2019) Herpes simplex virus 1 ICP6 impedes TNF receptor 1-induced necrosome assembly during compartmentalization to detergent-resistant membrane vesicles. J. Biol. Chem. 294, 991–1004 10.1074/jbc.RA118.004651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, S.N., Chappidi, S., Pinkham, C. and Hancks, D.C. (2021) Evolutionary profile for (host and viral) MLKL indicates its activities as a battlefront for extensive counteradaptation. Mol. Biol. Evol. 38, 5405–5422 10.1093/molbev/msab256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picarda, G. and Benedict, C.A. (2018) Cytomegalovirus: shape-shifting the immune system. J. Immunol. 200, 3881–3889 10.4049/jimmunol.1800171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Upton, J.W., Kaiser, W.J. and Mocarski, E.S. (2008) Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J. Biol. Chem. 283, 16966–16970 10.1074/jbc.C800051200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mack, C., Sickmann, A., Lembo, D. and Brune, W. (2008) Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl Acad. Sci. U.S.A. 105, 3094–3099 10.1073/pnas.0800168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Upton, J.W., Kaiser, W.J. and Mocarski, E.S. (2010) Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 10.1016/j.chom.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo, H., Omoto, S., Harris, P.A., Finger, J.N., Bertin, J., Gough, P.J.et al. (2015) Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 17, 243–251 10.1016/j.chom.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mostafa, H.H., Thompson, T.W., Konen, A.J., Haenchen, S.D., Hilliard, J.G., Macdonald, S.J.et al. (2018) Herpes simplex virus 1 mutant with point mutations in UL39 is impaired for acute viral replication in mice, establishment of latency, and explant-induced reactivation. J. Virol. 92, e01654-17 10.1128/JVI.01654-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He, S. and Han, J. (2020) Manipulation of host cell death pathways by herpes simplex virus. Curr. Top. Microbiol. Immunol. 10.1007/82_2020_196 [DOI] [PubMed] [Google Scholar]
- 52.Guo, H., Gilley, R.P., Fisher, A., Lane, R., Landsteiner, V.J., Ragan, K.B.et al. (2018) Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1. Cell Death Dis. 9, 816 10.1038/s41419-018-0868-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang, Z., Wu, S.Q., Liang, Y., Zhou, X., Chen, W., Li, L.et al. (2015) RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 17, 229–242 10.1016/j.chom.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 54.Gershon, A.A., Breuer, J., Cohen, J.I., Cohrs, R.J., Gershon, M.D., Gilden, D.et al. (2015) Varicella zoster virus infection. Nat. Rev. Dis. Primers 1, 15016 10.1038/nrdp.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fishman, J.A. (2013) Overview: cytomegalovirus and the herpesviruses in transplantation. Am. J. Transplant. 13, quiz 10.1111/ajt.12002 [DOI] [PubMed] [Google Scholar]
- 56.Gianella, S. and Letendre, S. (2016) Cytomegalovirus and HIV: a dangerous Pas de Deux. J. Infect Dis. 214, S67–S74 10.1093/infdis/jiw217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan, J., Giogha, C. and Hartland, E.L. (2021) Molecular mechanisms employed by enteric bacterial pathogens to antagonise host innate immunity. Curr. Opin. Microbiol. 59, 58–64 10.1016/j.mib.2020.07.015 [DOI] [PubMed] [Google Scholar]
- 58.Santos, A.S. and Finlay, B.B. (2015) Bringing down the host: enteropathogenic and enterohaemorrhagic Escherichia coli effector-mediated subversion of host innate immune pathways. Cell Microbiol. 17, 318–332 10.1111/cmi.12412 [DOI] [PubMed] [Google Scholar]
- 59.Pearson, J.S., Giogha, C., Mühlen, S., Nachbur, U., Pham, C.L., Zhang, Y.et al. (2017) Espl is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat. Microbiol. 2, 16258 10.1038/nmicrobiol.2016.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashida, H., Sasakawa, C. and Suzuki, T. (2020) A unique bacterial tactic to circumvent the cell death crosstalk induced by blockade of caspase-8. EMBO J. 39, e104469 10.15252/embj.2020104469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu, Z., Nailwal, H., Rector, J., Rahman, M.M., Sam, R., McFadden, G.et al. (2021) A class of viral inducer of degradation of the necroptosis adaptor RIPK3 regulates virus-induced inflammation. Immunity 54, 247–258.e7 10.1016/j.immuni.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nanson, J.D., Kobe, B. and Ve, T. (2019) Death, TIR, and RHIM: self-assembling domains involved in innate immunity and cell-death signaling. J. Leukoc. Biol. 105, 363–375 10.1002/JLB.MR0318-123R [DOI] [PubMed] [Google Scholar]
- 63.Li, J., McQuade, T., Siemer, A.B., Napetschnig, J., Moriwaki, K., Hsiao, Y.S.et al. (2012) The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 10.1016/j.cell.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otzen, D. and Riek, R. (2019) Functional amyloids. Cold Spring Harb. Perspect. Biol. 11, a033860 10.1101/cshperspect.a033860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, X., Ma, Y., Zhao, K., Zhang, J., Sun, Y., Li, Y.et al. (2021) The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3. Proc. Natl Acad. Sci. U.S.A. 118, 14 10.1073/pnas.2022933118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pham, C.L., Shanmugam, N., Strange, M., O'Carroll, A., Brown, J.W., Sierecki, E.et al. (2019) Viral M45 and necroptosis-associated proteins form heteromeric amyloid assemblies. EMBO Rep. 20, e46518 10.15252/embr.201846518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker, M., Shanmugam, N., Pham, C.L.L., Ball, S.R., Sierecki, E., Gambin, Y.et al. (2022) The RHIM of the immune adaptor protein TRIF forms hybrid amyloids with other necroptosis-associated proteins. Molecules 27, 3382 10.3390/molecules27113382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mompeán, M., Li, W., Li, J., Laage, S., Siemer, A.B., Bozkurt, G.et al. (2018) The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell 173, 1244–1253.e10 10.1016/j.cell.2018.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu, H., Wu, X., Wu, G., Nan, N., Zhang, J., Zhu, X.et al. (2021) RIP3-mediated necroptosis is regulated by inter-filament assembly of RIP homotypic interaction motif. Cell Death Differ. 28, 251–266 10.1038/s41418-020-0598-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shanmugam, N., Baker, M., Sanz-Hernandez, M., Sierecki, E., Gambin, Y., Steain, M.et al. (2021) Herpes simplex virus encoded ICP6 protein forms functional amyloid assemblies with necroptosis-associated host proteins. Biophys. Chem. 269, 106524 10.1016/j.bpc.2020.106524 [DOI] [PubMed] [Google Scholar]
- 71.Mompeán, M., Bozkurt, G. and Wu, H. (2019) Mimicry by a viral RHIM. EMBO Rep. 20, e47433 10.15252/embr.201847433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolbrink, B., Riebeling, T., Teiwes, N.K., Steinem, C., Kalbacher, H., Kunzendorf, U.et al. (2022) TAT-RHIM: a more complex issue than expected. Biochem. J. 479, 259–272 10.1042/BCJ20210677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim, J., Park, H., Heisler, J., Maculins, T., Roose-Girma, M., Xu, M.et al. (2019) Autophagy regulates inflammatory programmed cell death via turnover of RHIM-domain proteins. eLife 8, e44452 10.7554/eLife.44452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kesavardhana, S., Kuriakose, T., Guy, C.S., Samir, P., Malireddi, R.K.S., Mishra, A.et al. (2017) ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 214, 2217–2229 10.1084/jem.20170550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, D., Liang, Y., Zhao, S., Ding, Y., Zhuang, Q., Shi, Q.et al. (2020) ZBP1 mediates interferon-induced necroptosis. Cell. Mol. Immunol. 17, 356–368 10.1038/s41423-019-0237-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Basavaraju, S., Mishra, S., Jindal, R. and Kesavardhana, S. (2022) Emerging role of ZBP1 in Z-RNA sensing, influenza virus-induced cell death, and pulmonary inflammation. mBio 13, e0040122 10.1128/mbio.00401-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, T., Yin, C., Boyd, D.F., Quarato, G., Ingram, J.P., Shubina, M.et al. (2020) Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e13 10.1016/j.cell.2020.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng, M. and Kanneganti, T.D. (2020) The regulation of the ZBP1-NLRP3 inflammasome and its implications in pyroptosis, apoptosis, and necroptosis (PANoptosis). Immunol. Rev. 297, 26–38 10.1111/imr.12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng, M., Karki, R., Vogel, P. and Kanneganti, T.D. (2020) Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell 181, 674–687.e13 10.1016/j.cell.2020.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeffries, A.M., Suptela, A.J. and Marriott, I. (2022) Z-DNA binding protein 1 mediates necroptotic and apoptotic cell death pathways in murine astrocytes following herpes simplex virus-1 infection. J. Neuroinflammation. 19, 109 10.1186/s12974-022-02469-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flerlage, T., Boyd, D.F., Meliopoulos, V., Thomas, P.G. and Schultz-Cherry, S. (2021) Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 19, 425–441 10.1038/s41579-021-00542-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weisel, K., Berger, S., Papp, K., Maari, C., Krueger, J.G., Scott, N.et al. (2020) Response to inhibition of receptor-interacting protein kinase 1 (RIPK1) in active plaque psoriasis: a randomized placebo-controlled study. Clin. Pharmacol. Ther. 108, 808–816 10.1002/cpt.1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiao, H., Wachsmuth, L., Kumari, S., Schwarzer, R., Lin, J., Eren, R.O.et al. (2020) Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580, 391–395 10.1038/s41586-020-2129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Reuver, R., Verdonck, S., Dierick, E., Nemegeer, J., Hessmann, E., Ahmad, S.et al. (2022) ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607, 784–789 10.1038/s41586-022-04974-w [DOI] [PubMed] [Google Scholar]
- 85.Hubbard, N.W., Ames, J.M., Maurano, M., Chu, L.H., Somfleth, K.Y., Gokhale, N.S.et al. (2022) ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 607, 769–775 10.1038/s41586-022-04896-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiao, H., Wachsmuth, L., Wolf, S., Lohmann, J., Nagata, M., Kaya, G.G.et al. (2022) ADAR1 averts fatal type I interferon induction by ZBP1. Nature 607, 776–783 10.1038/s41586-022-04878-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yuan, F., Cai, J., Wu, J., Tang, Y., Zhao, K., Liang, F.et al. (2022) Z-DNA binding protein 1 promotes heatstroke-induced cell death. Science 376, 609–615 10.1126/science.abg5251 [DOI] [PubMed] [Google Scholar]
- 88.Zhang, W., Fan, W., Guo, J. and Wang, X. (2022) Osmotic stress activates RIPK3/MLKL-mediated necroptosis by increasing cytosolic pH through a plasma membrane Na(+)/H(+) exchanger. Sci. Signal. 15, eabn5881 10.1126/scisignal.abn5881 [DOI] [PubMed] [Google Scholar]