BACKGROUND
The Fifth Biennial Conference on Bacterial Locomotion and Signal Transduction was held on 16 to 21 January 1999 in Cuernavaca, Mexico. The meeting brought together 202 researchers in the fields of bacterial motility, transcription control by environmental signals, and bacterial behavior and development. Many prominent advances were seen in the description of the transmembrane signaling pathway of chemotaxis in Escherichia coli to near atomic resolution, the revelation of the wide diversity of chemotactic signaling and behavioral responses in many nonenteric bacteria, the extension of bacterial-type signaling processes in eucarya, and the further dissection of action of two-component signaling systems in control of metabolism and gene expression. A growing consensus was reached on the molecular motions that occur during transmembrane signaling. Finally, structural information on the recently described PAS domain revealed novel and unexpected flexibility in its accommodation of different types of prosthetic groups and the different ways it can detect and decode specific stimuli.
This minireview summarizes information presented by the speakers at the meeting. An excellent introduction to these topics is in the minireview summarizing the previous meeting (25). Briefly, the meeting revolved around two-component regulatory systems that control chemotactic behavior and gene expression in response to environmental signals. Two-component systems comprise at least a sensor kinase and a response regulator. The sensor kinase catalyzes an autokinase reaction, or transfer of phosphate from ATP to a histidine residue, probably by intermolecular phosphorylation in a dimer. The protein structures and sequences of the histidine-protein kinases (termed HK) differ from those of other protein kinases. The phosphate is then transferred from the HK-histidine to a specific aspartate residue on the cognate response regulator protein (termed RR).
The activity of the RR protein, whether to influence the direction of flagellar rotation or to affect specific gene transcription, changes after phosphorylation. The effect of phosphorylation on RR structure is still incompletely defined but is likely to be widely conserved. In contrast, the effect of phosphorylation on RR activity appears to be quite varied. Phosphorylation can influence binding of the RR to its target, change its oligomeric state, or activate its function. Most HK proteins also possess a phosphatase activity to accelerate removal of phosphate from their cognate RR protein, often by reversal of the pathway of phosphate transfer. Changes in the ratio of autokinase and phosphatase activities are the primary mode of regulation by the appropriate environmental signal, which in many cases binds to the external, periplasmic domain of the HK. It is of interest how the cognate HK and RR recognize one another so efficiently and specifically, in light of the fairly high conservation in the sequences of their active sites.
Although HK proteins differ considerably in overall amino acid sequence from each other and from other protein kinases, they possess several conserved sequence motifs, separated by constant spacings. One motif, the H box, contains the phosphorylated histidine embedded in the dimerization domain in most HK. In contrast, the H box in CheA, the HK in chemotaxis, resides in a distinct location, called the P1 domain. Chemotactic signaling (diagrammed in Fig. 1) also differs from most other two-component systems because the kinase CheA is separate from the signal recognition proteins. This separation allows a single kinase to respond to and integrate the signals from multiple signal receptor proteins. Most chemotactic signaling proteins have two transmembrane segments (TMS) joining the ligand-binding sites on the periplasmic side of the cytoplasmic membrane to the highly conserved cytoplasmic portions which bind to CheA in complex with CheW. The mechanism whereby transmembrane signal transduction occurs is fairly well defined in the case of chemotactic signaling and may be a universal mechanism.
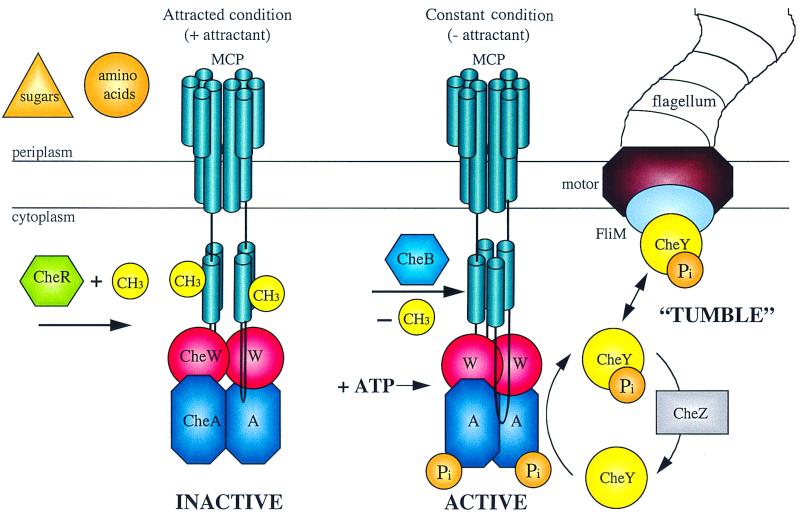
FIG. 1.
Schematic representation of the protein interactions and covalent modification during chemotaxis in enteric bacteria. Chemoeffector binding or release from the transmembrane MCP signal transducers results in a transmembrane conformational change that alters the activity of the CheA protein kinase and the susceptibility of the MCP protein to methylation or demethylation. Phosphorylated CheA can donate the phosphate to CheY, and P-CheY interacts with FliM in the flagellar basal body to influence the direction of flagellar rotation. (Figure provided by Jayna Ditta and Caroline Harwood.)
Unlike most signaling processes, chemotactic signaling in enteric bacteria exhibits adaptation. Bacterial swimming behavior changes quickly in response to addition or removal of a chemoeffector but then gradually returns to the original behavior even in the continued presence of the signal. This adaptation involves the reversible methylation of conserved glutamate residues in the cytoplasmic domain of the transducer proteins, the MCPs or methylated chemotaxis proteins. The changes in methylation are catalyzed by CheR methyltransferase and CheB methylesterase and are coupled to the same conformational changes that control CheA kinase activity. Chemotactic signaling is controlled so that addition of an attractant results in decreased activity of the CheA kinase and hence decreased phosphorylation of the RR, CheY. Phosphorylated CheY, P-CheY, favors clockwise (CW) flagellar rotation, so an increase in attractant concentration results in reduced CW events. P-CheY binds to signaling protein FliM in the flagellar basal body, but the manner in which the direction of flagellar rotation is determined is unknown. In enteric bacteria, CW flagellar rotation results in tumbling and reorientation of the cell in random directions. Counterclockwise (CCW) rotation allows the flagellar filaments on the cell to coalesce into a bundle which drives the cell in a fairly straight line. In nonenteric bacteria, different swimming behaviors are encountered depending on the number, location, and helical character of the flagellar filaments.
INTERCELLULAR COMMUNICATION AND BACTERIAL BEHAVIOR
Recent studies have uncovered many forms of communication between bacterial cells to measure their population density or to coordinate multicellular developmental processes. Upon starvation for readily utilizable nitrogen sources, the filamentous cyanobacterium Anabaena sp. strain PCC 7120 produces terminally differentiated nitrogen-fixing heterocysts, which arise from roughly every 10th cell in the filament. To elucidate how heterocyst spacing is regulated, James Golden (Texas A&M University) showed that pattern formation is controlled by PatS, a 17-amino-acid peptide produced by the cells that eventually differentiate into heterocysts (41). Release of the peptide, or more likely a processed product, from preheterocyst cells prevents adjacent cells from developing into heterocysts, as evidenced by the formation of chains of heterocysts when a patS deletion mutant is shifted to nitrogen-free medium. The last five amino acids of PatS are critical for heterocyst inhibition, and this pentapeptide suppressed heterocyst development when added to the patS deletion strain. The few heterocysts that formed were randomly spaced, presumably because the PatS peptide was not provided in a spatial gradient. Yet to be determined are the target of PatS, the basis for resistance of heterocysts to the peptide, and the nature of the in vivo processing of the peptide.
The cyanobacterium Synechococcus exhibits oscillations in biological functions, such as photosynthesis, with a periodicity of about 24 h, even in the absence of environmental cues. Most of the Synechococcus genome is transcribed with a circadian rhythm. Mutants with altered circadian timing can be identified by screening for changes in expression of specific promoters fused to the luciferase genes, luxAB. Susan Golden (Texas A&M University) and collaborators described the kaiABC pacemaker genes which appear to encode the primary internal clock (11). In kai null mutants the periodic expression of all reporter genes investigated is altered by a similar amount. The kai genes are not related to eukaryotic pacemaker genes, but homologues exist in other cyanobacteria and some archaea. Genes outside the kai cluster also affect circadian control of gene expression. These include a gene encoding a sigma factor which, when mutated, gives different periodicity in expression of different promoters. This suggests the possible presence of minor clocks that act with Kai to control the circadian rhythms of certain genes. In addition, mutants have been identified that have the same period length as wild-type cells, but altered trough and peak heights or offset phasing at different promoters (14).
Multiple signals are detected and integrated during fruiting body development in Myxococcus xanthus. Nutrient starvation is signaled by the generation of ppGpp, and the presence of sufficient cell numbers to allow proper fruiting body formation is detected from the level of amino acids released from cell surface proteins by the action of the asg-encoded proteases. Heidi Kaplan (University of Texas—Houston) discussed the cell density-responsive signal transduction pathway (13). Components of this signaling system were identified by analysis of mutants that bypass the usual requirement for nutrient starvation or high cell density. Three of the four proteins encoded by the sasB locus are required for development, and all control expression of a developmental reporter gene. The SasS and SasR proteins are an HK-RR pair that responds to cell density via A-signal amino acids, while the novel SasN and SasP proteins are proposed to influence SasS activity in response to nutrient status and cell density. Other mutations that bypass the requirement for A signal affect lipopolysaccharide O antigen. These mutants are also defective for fruiting body formation, sporulation, and motility and exhibit altered production of surface fibrils. These results point to a complex network of positive and negative regulatory elements that integrate early developmental signals in response to the integrity of the cell surface and perhaps the secreted fibrils.
Anke Treuner-Lange (D. Zusman's laboratory, University of California at Berkeley) identified in M. xanthus a serine/threonine protein phosphatase, Pph1, which belongs to the PP2C family of eukaryotic phosphatases. The pph1 gene is expressed at higher levels during development than during vegetative growth. The cellular substrate of the Pph1 phosphatase and its role in development are under investigation.
QUORUM SENSING
Quorum sensing refers to the mechanisms that alter gene expression in response to bacterial population density. In most cases, each bacterium in a population produces a constant amount of a low-molecular-weight signaling molecule, called an autoinducer, whose concentration reflects the total number of bacteria. The recognition of autoinducers can be specific and measure the number of a particular bacterial species, or it can be more generic and measure the total number of bacteria. Autoinducers include peptides in gram-positive bacteria and acylated homoserine lactones in gram-negative bacteria. The prototype quorum-sensing system controls the expression of bioluminescence in Vibrio fischeri. This marine bacterium produces a homoserine lactone autoinducer which activates bioluminescence lux genes upon binding to the LuxR regulatory protein. Only when cells reach a threshold density does the concentration of autoinducer suffice to activate lux gene transcription. The luxI product catalyzes autoinducer synthesis from S-adenosylmethionine and the appropriate acyl-acyl carrier protein.
Many gram-negative bacteria have quorum-sensing systems that involve LuxI-LuxR homologues for production and detection of acyl-homoserine lactones. Most of these organisms form pathogenic or symbiotic associations with plant or animal hosts. Some V. fischeri isolates colonize the light organ of the Hawaiian squid Euprymna scolopes and provide bioluminescence to the squid in exchange for a supply of nutrients. The squid are colonized by V. fischeri only after their birth, but nonmotile bacterial mutants are unable to colonize light organs. Karen Visick (Loyola University) reported that lux mutants defective for luciferase production, as well as luxI and luxR mutants, exhibited a three- to fourfold decrease in colonization of the light organ. This suggests that luminescence or the enzymatic reaction carried out by luciferase is required for normal symbiotic colonization, possibly because luciferase action lowers the oxygen tension in the light organ.
Bonnie Bassler (Princeton University) reported on the cell density-dependent induction of bioluminescence in Vibrio harveyi. This quorum-sensing system differs from others in gram-negative bacteria in that transcription is controlled through an HK-RR phosphorelay system rather that a luxI-luxR system. V. harveyi has two parallel phosphorelay systems: one that responds to autoinducer 1 (AI-1), a typical acyl-homoserine lactone, and one that responds to autoinducer 2 (AI-2), a compound of different but unknown structure. Many species of bacteria, including E. coli O157 and Salmonella enterica serovar Typhimurium 14028, produce compounds that induce bioluminescence in V. harveyi lacking the AI-1 response system and are therefore similar or identical to AI-2. In E. coli and Salmonella serovar Typhimurium, AI-2 is produced during the exponential phase and is degraded at the onset of stationary phase (35). An exciting challenge is to learn what genes respond to AI-2 and whether they are important for virulence. The Bassler group has cloned the luxS genes responsible for AI-2 production from V. harveyi, Salmonella serovar Typhimurium, and E. coli. luxS does not resemble any genes of known function, but homologues were found in at least 25 gram-positive and gram-negative bacteria.
BEHAVIOR ON SURFACES
The mechanism of gliding motility—the movement across solid surfaces by bacteria that lack flagella or any other apparent appendages—is still a mystery. Mark McBride (University of Wisconsin—Milwaukee) analyzed transposon insertion mutants of Flavobacterium johnsoniae to identify genes required for gliding motility. Some of these gene products include components of an ATP-binding cassette (ABC) transporter, a hemolysin, and proteins involved in synthesis of extracellular polysaccharides or lipopolysaccharides. Although it is not yet clear how these proteins contribute to movement, their properties should help test the two models for gliding. Lapidus and Berg (19) suggest that adhesion molecules in the outer membrane mediate cellular movement by moving along surface tracks anchored to the peptidoglycan. Movement of these adhesins is presumed to be driven by the proton motive force. Another model is that polysaccharide excretion propels cells by a conveyer belt mechanism whereby polysaccharide is secreted at one region of the cell surface and resorbed at another region.
Gliding of Myxococcus xanthus occurs either independently as single cells (adventurous motility) or coordinately as groups of cells (social motility). The frz genes (39) are required for chemotaxis and motility of single cells and have homology to the E. coli chemotaxis genes. Zhaomin Yang (W. Shi's laboratory, University of California at Los Angeles [UCLA]) reported on a new set of genes (named dif for defective in fruiting and fibrils) which encode a second set of homologues of MCPs, CheW, CheY, and CheA. The dif mutants are defective for social motility, but not for single-cell motility. Inactivation of dif genes also reduced production of an extracellular fibril material composed of polysaccharides and proteins and required for social motility and cell aggregation. It thus appears that M. xanthus uses the dif system to organize single cells into groups and coordinate cell movement within the group and uses the frz system to direct chemotactic movements of single cells and of cell groups into aggregation centers.
Some bacteria use a small number of flagella to swim in liquid media but differentiate into hyperflagellated and elongated swarmer cells when placed on solid surfaces. Bob Belas (University of Maryland) described precocious swarming mutants of Proteus mirabilis (6). These mutants are able to migrate on minimal agar and differentiate to the swarmer state earlier than the wild type when placed on a solid surface. A gene named rsbA for regulator of swarming behavior was identified in these mutants. RsbA resembles an HK and may control swarmer cell formation by monitoring some factor at the cell surface or restrict flagellar rotation. Azospirillum brasilense is a plant growth-promoting soil bacterium that produces two structurally different kinds of flagella: a polar flagellum for swimming and lateral flagella for swarming. Using a gusA reporter fusion, Sara Moens (Catholic University of Leuven) identified several regulatory genes that influence expression of laf1 encoding lateral flagellin. The lafR and flbT genes flank laf1 and seem to be positive regulators of its expression. LafR resembles an RR and affects laf1 expression only at low temperatures, but a cognate HK has not been identified.
FLAGELLAR STRUCTURE AND ASSEMBLY
The flagellar basal body is a complex structure which couples transmembrane ion movement to the rotation of the flagellar filament. It is also involved in the assembly of the entire structure, and some of its components control the direction of flagellar rotation. The synthesis of all flagellar components is orchestrated in several transcriptional cascades, which are controlled by environmental and physiological signals and include checkpoints that coordinate the transcription of the external flagellar components with their assembly into a functional structure.
The flagellar expression cascade begins with the master regulatory operon whose products, the FlhDC complex, activate transcription of the class II flagellar genes. Andres Campos (P. Matsumura's lab, University of Illinois—Chicago) described several novel aspects of FlhD action. In addition to its obligate role in flagellar synthesis, FlhD but not FlhC is also involved in regulation of cell division. FlhD mutants retain a high rate of cell division after entry into stationary phase, leading to formation of progressively smaller cells. This coupling to cell division may occur through polyamine levels, because genetic searches for FlhD target genes showed that FlhD activates expression of the polyamine-metabolizing enzymes encoded by speG (spermidine acetyltransferase) and cadA (lysine decarboxylase). FlhD is proposed to be the first bacterial example of combinatorial specificity, where the target specificity of a transcription activator is determined by its association with another protein. The crystal structure of FlhD revealed an N-terminal dimerization domain followed by a C-terminal domain predicted to form a flexible DNA-binding helix-turn-helix motif. Perhaps FlhC and the other presumptive partners of FlhD alter its target specificity by stabilizing the helix-turn-helix reading head in different orientations.
A current issue in flagellar assembly concerns the function and interactions of the components of the flagellum export apparatus. Only two flagellar proteins have typical N-terminal signal sequences and are translocated to their periplasmic site by the Sec pathway. Homologues of many components of the flagellar export pathway are found in type III secretion systems which inject bacterial virulence factors directly into eukaryotic cells. More than 10 proteins form the export apparatus residing in the C ring (28). One of those components, FliI, has ATPase activity and might be a selective gate for protein translocation from the cytoplasm into the export channel (8a). To support this hypothesis, Eugenia Silva-Herzog (G. Dreyfus's lab, Universidad Nacional Autónoma de México [UNAM]) reported that FliI binds flagellin and hook proteins, which activate FliI ATPase activity. Another component of the putative gate system is FliK, which controls hook length, perhaps by regulating the entry of substrates into the export pathway or by directly measuring hook length as a molecular ruler. Tohru Minamino (R. Macnab's lab, Yale University, in collaboration with S.-I. Aizawa's lab, Teikyo University) showed that FliK is secreted into the medium by wild-type cells and some mutants (27). This secretion of FliK is essential for hook length control, but the detailed mechanism remains unsolved.
In addition to these proteins of the export apparatus, the MS ring complex in the cytoplasmic membrane plays an important role in export, forming the channel through the membrane and defining sites for initiation of flagellar assembly. The central channel of the MS ring and export complex is normally closed. Hirofumi Suzuki (Keiichi Namba ERATO project, Japan) described computer-aided reconstitution of electron microscopy images of negatively stained MS ring complexes purified from FliF-overproducing strains (36). The presence of high salt concentrations allowed deeper penetration of stain into the central channel than in particles formed by removal of salt. Presumably some component of the flagellar export apparatus is responsible for the normal opening and closing of this channel.
In studies of the posttranslational targeting of flagellin in Helicobacter pylori, Christine Josenhans (Ruhr University, Bochum, Germany) introduced green fluorescent protein (GFP)-tagged flagellins from various species. One interesting observation was that GFP-flagellins accumulated at one cell pole only in late-log-phase cells, suggesting that the traffic to the pole was activated only in a portion of the cell cycle.
After passing through the export apparatus, flagellin molecules polymerize into a helical filament. The helical parameters are crucial for the flagellum to produce an effective thrust. In contrast, spirochetes have periplasmic endoflagellar filaments, whose rotation gives rise to counterrotation of the helical or wave-like cell body. Nyles Charon and colleagues (West Virginia University) have identified flagellar proteins (three types of flagellins and one sheath protein) of the spirochete Serpulina hyodysenteriae and created several flagellar mutants. Filaments from mutants lacking the sheath protein were thinner and had a shorter helical pitch and smaller helical diameter. These results indicate that the sheath protein influences the shape of the flagella.
The needle complex formed by the type III secretion apparatus of various pathogenic bacteria resembles a flagellar basal body (17), consistent with the hypothesis that these two structures have a common origin. Glenn Young (V. Miller's lab, Washington University) showed that the flagellar export apparatus can also mediate the release of a nonflagellar protein. Export of phospholipase A by Yersinia enterocolitica required the function of the flagellar regulatory proteins, FlhDC and FliA, as well as the proper assembly of the basal body (43). Export is independent of the Vir plasmid-encoded type III secretion system used by other virulence factors.
FLAGELLAR FUNCTION
A current goal of research on the mechanism of torque generation in the flagellar motor addresses the identity and relative orientation of the rotor and stator elements of the motor. It is controversial whether FliG rotates against MotA, FliM rotates against MotA, or FliG rotates against FliM and MotA. David Blair (University of Utah) provided support for the first possibility by showing that charged residues in FliG interact with charged residues in MotA. The FliG protein plays an important role in many aspects of flagellar function. Its N-terminal half is needed for flagellar assembly, and its C-terminal half is important for torque generation and motility. Changes in several charged residues in the C-terminal portion of FliG (Arg-281, Asp-288, and Asp-289) result in a paralyzed phenotype, which can be suppressed by changes in Arg-90 and Glu-98 of MotA. Frank Whitby (University of Utah) in collaboration with Blair's lab solved the crystal structure of the C-terminal domain of FliG by using the very stable domain from the hyperthermophilic bacterium Thermotoga maritima (22). This domain folded into six helices connected by loops, and the mutations conferring different phenotypes were mapped to different portions of the structure. For example, mutations resulting in a paralyzed Mot− phenotype affected the hydrophobic core of the domain and presumably interfere with domain stability. Separate surfaces were involved in FliM binding or in chemotactic signaling. The charged residues that may interact with MotA were exposed on the surface of an α-helix and nearby loops, which may form a ridge-like structure that determines the path of motion with respect to the stator MotA. A model where 30 FliG subunits form the circumference of the rotor was presented.
The stator function of MotA requires MotB, whose N-terminal TMS interacts with MotA and whose periplasmic C-terminal region has a peptidoglycan-binding motif for anchorage to the cell wall. MotB has dispensable regions throughout its length (29). Susan Van Way (M. Manson's lab, Texas A&M University) used amber mutations in motB to analyze the restoration of motility in response to different amino acid-inserting tRNA suppressor mutations and concluded that MotB must coinsert in proper stoichiometry with MotA for proper formation of the stator complex.
Genetic dissection of the pathway of ion movement through the flagellar motor may be simpler in systems driven by a gradient of sodium ions rather than of protons. In Vibrio alginolyticus, rotation of the polar flagellum is driven by the sodium gradient and is specifically inhibited by the sodium channel inhibitor phenamil. Seiji Kojima (M. Homma's lab, Nagoya University) analyzed phenamil-resistant motility mutants and identified the phenamil-binding sites in transmembrane portions of PomA and PomB (homologues of E. coli MotA and MotB) (15). Although the individual pomA and pomB mutants swam at almost normal rates, the double pomAB mutant showed greatly reduced motility and was resistant to phenamil but, surprisingly, was still susceptible to the more polar analog amiloride. The locations of the mutated sites near the cytoplasmic ends of the putative TMS imply that multiple residues from both proteins contribute to the high-affinity phenamil-binding site on the inner surface of the PomA-PomB complex.
DIVERSITY OF TAXIS PROCESSES
Sequence analyses of gene clusters involved in chemotactic signaling in nonenteric bacteria revealed additional protein components. Michael Saulmon (G. Ordal's lab, University of Illinois) noted that Bacillus subtilis has, besides the CheAWYRB components of the E. coli pathway, several additional components, including CheV (a CheW-CheY hybrid), CheC, and CheD. Tethered-cell analysis of the chemotactic responses of cheRBCD and cheRBCDV deletion strains suggested that CheAWCD and CheV interact with signal receptors in a more complex cluster than in the enteric bacteria. The suggestion was made that P-CheY might inhibit CheA activity.
Multiple copies of many chemotaxis genes are present in members of the alpha subgroup of Proteobacteria. These bacteria have a greater variety of behavioral responses than do enteric bacteria (2). Rhodobacter sphaeroides, Caulobacter crescentus, and several nitrogen-fixing rhizobacteria have multiple cheY genes and no apparent cheZ homologue. Mutational analyses suggest that one CheY functions as the primary motor-binding protein. R. sphaeroides has an extremely versatile metabolism and is able to grow aerobically, anaerobically, photosynthetically with complex carbon sources, or photosynthetically by fixing carbon dioxide. At least three operons encode multiple copies of chemotaxis gene homologues (to date, two cheA, one cheB, two cheR, three cheW, four cheY) and numerous MCP genes. These seemingly redundant genes may allow optimal chemotaxis in different life styles. Deepan Shah (J. Armitage's lab, University of Oxford) showed that CheY4 alone was sufficient for a normal chemotactic response, and its deletion resulted in a completely nonresponsive phenotype (31). The presence of che operon 1 without operon 2 resulted in an inverted tactic response.
Swarmer cells of C. crescentus have a single polar flagellum and reverse the direction of movement by changing the direction of flagellar rotation. Chemotactic ability is provided by the 12-gene mcpA operon, whose features include the absence of CheZ and the presence of three CheY homologs, only one of which is necessary for chemotaxis. Dickon Alley (Imperial College of Science, London, United Kingdom) reported that cheYII mutants were nonchemotactic, whereas cheY1 and cheYIII mutants were partially defective. The cheW product was not essential for chemotaxis, suggesting that at least one of its three homologues in the genome sequence can substitute. As expected, the CheA, CheB, and CheR proteins were essential for taxis. Several genes in the mcpA operon do not resemble enteric chemotaxis genes, and one was related to the B. subtilis cheD gene. Finally, the polar localization of McpA was only partially dependent on the presence of other mcpA operon components.
Chemotaxis in Sinorhizobium meliloti also differs from the enteric bacterial model. Instead of runs and tumbles, this organism performs runs and turns. Unlike the flexible, left-handed helices of enteric flagella, which form a bundle when rotating CCW and are disrupted when rotating CW, flagella in rhizobia are rigid and right-handed. CW rotation results in a run, but there is no reversal of rotation. Instead, removal of attractant results in the slowing of some flagella, so that the cell turns rather than tumbles. Furthermore, attractants produce an increase in the speed of flagellar rotation, unlike the constant speed in the enteric situation. This organism has more than six membrane transducers encoded in the 41-gene chemotaxis operon (33). Rudiger Schmitt (University of Regensberg) proposed that the absence of CheZ is compensated by the presence of a second CheY (CheY1), which has little signaling activity but may serve as a sink to help remove phosphate from P-CheY2 (32). This retrograde phosphorylation transfers phosphate from P-CheY2 back to CheA and thence to CheY1, without direct transfer between CheY1 and CheY2. This raises the question why CheA does not transfer phosphate to CheY1 during the initial chemotactic stimulus.
Multiple transducer genes are present in other nitrogen-fixing bacteria, but the chemoeffectors recognized by many of them have not been identified. Two laboratories showed that MCPs play a role in the establishment of root nodule symbioses. Rhizobium leguminosarum strain VF39 has at least 17 genes related to MCP genes (42), some of which are carried on large plasmids (100 to 800 kb) along with genes for catabolism, nitrogen fixation, and nodulation. Michael Hynes (University of Calgary) reported that at least two of these MCP genes (mcpB and mcpC) are required for competitive root nodulation. When tested in the laboratory, only the mcpB mutant has a defective Che phenotype. These mcp genes are constitutively expressed in free-living cells, but are down-regulated in bacteroids in the nodules. Albert Mendoza (J. Mora's lab, UNAM, Cuernavaca) identified an MCP gene from Rhizobium etli that complements E. coli tar and tsr mutants. An R. etli strain mutated in this gene is unable to switch from aerobic metabolism to the fermentative metabolism resembling that in root nodules. This mutant is also altered in its cellular aggregation behavior (clumping and poly-β-hydroxybutyrate synthesis).
Phototaxis in the light-harvesting archaeon Halobacterium salinarum responds to attractant orange light and repellent blue light through the action of two seven-helix retinal-containing pigments, sensory rhodopsins SR I and SR II. Each light-sensing pigment associates with an MCP-like signal-transducing Htr protein through membrane domain interactions (34). Bastianella Perazzona (J. Spudich's lab, University of Texas—Houston) showed that methylation of these proteins occurs on three glutamate residues in HtrI and one in HtrII. However, mutational alteration of these residues did not block phototaxis or adaptation to light stimuli. Repetitive stimulation of the unmethylatable form of the SRI-HtrI complex induced methyl group turnover in other transducers in the cell (30). This behavior may be due to global activation of the methylesterase. Alternatively, hetero-oligomers of MCPs may form, in which one molecule of HtrI specifies recognition of SRI, while the other promoter undergoes light-dependent methylation. These data support the idea that sensory signaling involves higher-order transducer clusters (21).
In addition, H. salinarum contains >10 chemotaxis transducer proteins in three structural classes: those typical of bacterial Tar protein, soluble transducer proteins, and those with small periplasmic domains and two or more TMS. Maia Kokoeva (D. Oesterhelt's lab, Max-Planck-Institute, Martinsried, Germany) described BasT, which has a topology and conserved signaling domain similar to Tar and is methylatable. Behavioral studies show that BasT is a chemoreceptor for branched-chain and sulfur-containing amino acids and functions together with a substrate-binding lipoprotein on the external face of the cytoplasmic membrane. It remains to be seen whether this binding protein interacts with the transducer directly or whether transport is required.
SIGNAL RECEPTION
The MCP transducer Tsr mediates a repellent response to weak acids, whereas Tsr mediates an opposite response. Tohru Umemura (Nagoya University) described the use of chimeric Tar-Tsr and Tsr-Tar proteins to localize the region responsible for sensing cytoplasmic pH to residues 256 to 309. Arginine-259 and serine-260 were identified as important for the pH response. The corresponding residues in Tsr are glycine and glutamate, respectively. Perhaps the pH affects the conformation of this region through interaction with other residues or the membrane bilayer.
PAS DOMAINS IN SIGNAL RECEPTION
PAS domains burst onto the scene as sensor modules present in all phylogenetic branches. Their name comes from the proteins in which they were first recognized: the Drosophila period clock protein, the vertebrate aryl hydrocarbon-receptor nuclear translocator, and the Drosophila single-minded protein. As reviewed in reference 37, PAS domains have a distinct α/β fold, designated the PAS core-helical connector-β-scaffold. Particularly fascinating is the finding that PAS domains accommodate a variety of prosthetic groups that recognize and allow response to a variety of signals. Some bound cofactors are heme, flavin adenine dinucleotide, flavin mononucleotide, and hydroxycinnamoyl moieties. So far in bacteria, PAS domains have been found in chemotaxis transducers and HK proteins of two-component systems that respond to light, oxygen, or redox potential. In eucarya, PAS domains have been found in clock proteins, hypoxia-inducible factors, voltage-gated ion channels, and other signaling proteins.
Several examples of PAS domains in bacterial signaling were described. The flavin adenine dinucleotide-containing Aer protein is a transducer that mediates E. coli aerotaxis by sensing the redox state of the electron transport system. The photoreceptor PYP carries a 4-hydroxycinnamoyl chromophore and mediates a photorepellent response in Ectothiorhodospira halophila. ArcB is an HK that controls expression of genes of anaerobic respiration and metabolism in response to the redox state, but no prosthetic group has been found yet. PAS domains have been found in KinA, which initiates the sporulation cascade in B. subtilis, and PhoR, which regulates phosphate-repressible genes, but the role of these domains has not been analyzed.
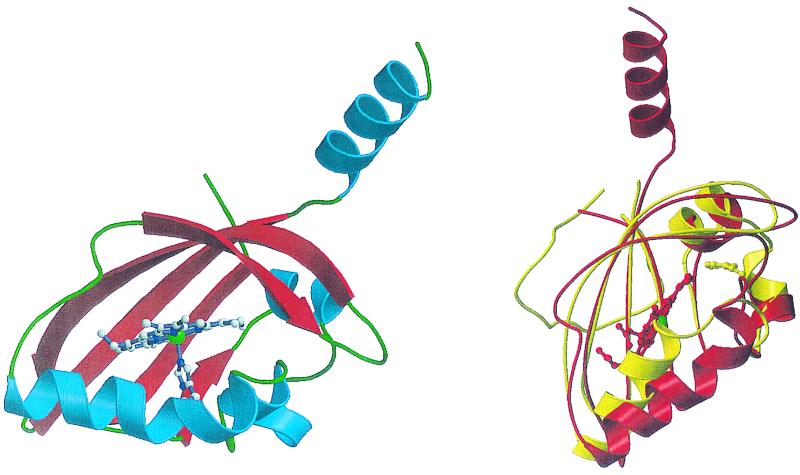
Expression of the nitrogenase pathway in plant-symbiotic bacteria is controlled by multiple factors. The FixL protein of Bradyrhizobium japonicum is an oxygen-sensing HK that prevents nitrogenase expression under aerobic conditions. Vondolee Delgado-Nixon (M.-A. Gilles-Gonzalez's lab, Ohio State University) described the crystal structure of its heme-containing PAS domain in the absence and presence of the ligand, CN (10) (Fig. 2). Ligand binding results in puckering of the porphyrin ring, which changes protein contacts of equatorial substituents along one side of the ring, rather than the expected changes in binding of axial ligands, as in other heme proteins. Gilles-Gonzalez and colleagues proposed that the conformational change induced in the prosthetic group upon signal reception is likely to involve different parts of the PAS domain in different proteins. Thus, the remarkable versatility of the PAS domain to accommodate different prosthetic groups may be mirrored in the use of different parts of the domain for signaling.
FIG. 2.
Structure of the heme-binding PAS domain from the FixL protein of Bradyrhizobium japonicum. (Left) A ribbons diagram with α-helices in blue and β-sheets in red. The helix pointing up leads to the HK domain. The heme moiety, with the heme iron in green, and its axial histidine ligand are shown. (Right) A comparison of the structures of the FixL heme-binding PAS domain in red and the PAS domain of the photoactive yellow protein PYP in yellow. The locations of the heme moiety of FixL, with the heme iron in green, and the p-hydroxycinnamate chromophore of PYP are indicated. Reprinted from reference 10 with permission of the publisher. (Figure provided by M.-A. Gilles-Gonzalez.)
RECEPTOR COMPLEXES
Considerable discussion focused on the role of receptor clustering and interactions to provide the high degree of signal gain that occurs in chemotactic sensing. Although flagella are inserted at random positions on the cell surface, the majority of MCP molecules in E. coli and other bacteria, including C. crescentus, M. xanthus, R. sphaeroides, and H. salinarum, are clustered in tight patches at the cell poles. Janine Maddock (University of Michigan) showed that CheA and CheW are clustered with the MCPs, and this clustering requires the presence of all three proteins (24). CheZ is also localized at the poles. The presence of the methyl-transferring enzymes CheB and CheR does not affect the clustering of the signaling complex, which might facilitate methyl transfer between MCP dimers and allow signal amplification (23). Dennis Bray (University of Cambridge) described computer modeling using Monte-Carlo simulations to examine how receptor clustering contributes to chemotactic signal gain. A chemotactic response can operate over a 5-log range of effector concentrations. Such a dynamic range cannot be obtained with independent receptor-binding kinetics but is described by coupling the activity of clustered receptors. The number of receptors in a fully coupled signaling complex was modeled to be at least 360. J. Parkinson commented that expression of even a few CCW-locked receptors interferes with signaling by wild-type receptors.
Tapu Shaikh (D. DeRosier's lab, Brandeis University) addressed biochemical characterization of a signaling complex containing a soluble form of Tar (sTar) with a leucine zipper dimerization domain in place of the transmembrane segments. This sTar dimer combined with the CheW and CheA proteins to form a large complex which was purified and visualized by electron microscopy and image analysis. The protein composition of the sTar-CheW-CheA complex was estimated to be 28:6:4. The complex was active for autokinase activity and responded to methylation. Using a similar approach, Peter Ames (University of Utah) described the properties of a stable Tsr-CheW-CheA complex whose kinase activity was regulated by appropriate signals. The Tsr/CheW/CheA stoichiometry was 16:1:1. The shortest fragment of Tsr that could interact with the CheA-CheW complex and specify CW signaling (activation of CheA kinase activity) contained residues 347 to 470, the same region identified by J. Falke's group for MCP coupling (see below). This fragment of Tsr binds to glutathione S-transferase (GST)-CheW with a Kd of 2 μM, and GST-CheW binds to CheA with Kd of 16 μM. The Tsr-binding region of CheA included the C-terminal M and C domains, whose removal eliminated CheW binding and control of kinase activity. Expression of the Tsr signaling fragment (residues 347 to 470) inhibited CheA kinase activity, even in the absence of CheW. The portion of Tsr needed for this squelching effect overlapped the region responsible for CheW binding but involved different amino acids. This result shows that Tsr can interact with CheA directly, as well as through CheW.
TRANSMEMBRANE SIGNALING
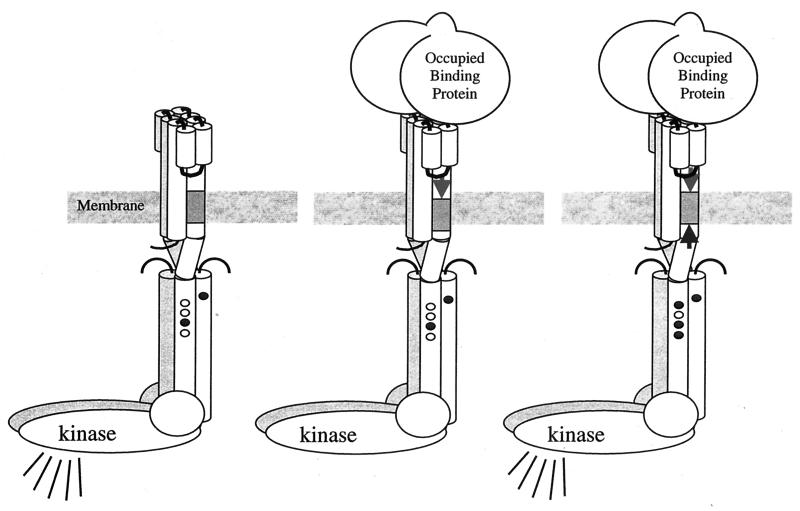
Chemotactic signaling involves a transmembrane signal transduction process, where occupancy of ligand-binding sites on the external periplasmic sector of the MCP transducer proteins results in intracellular changes in CheA autokinase activity and altered activity in methyl transfer reactions. A central issue is the nature of the coupled conformational changes between the two parts of the MCPs and how they are transmitted through the cytoplasmic membrane. The MCP dimer has four TMS. The arrangement of this four-helix bundle and the movements of its helices relative to one another in response to receptor occupancy have been probed by site-directed disulfide bond formation. A consensus opinion regarding the relative motions of the four helices now favors a piston model, in which one or more TMS moves vertically with respect to the other TMS, over a scissors model, in which the TMS twist or rotate relative to one another (Fig. 3).
FIG. 3.
Helical sliding mediates transmembrane signaling and compensatory adaptation. Cartoon representation of the receptor-kinase complex shows sites of adaptational methylation (small ovals) in methylated (shaded) and unmethylated (white) states. (Left) In the absence of ligand, complexed kinase is active (symbolized by broadcast lines). (Middle) Binding of ligand (e.g., sugar-occupied, sugar-binding protein) induces sliding of TM2 toward cytoplasm (arrow pointing down), resulting in kinase inhibition (no broadcast lines). (Right) Increased adaptational methylation slides TM2 back toward the periplasm (arrow pointing up), and kinase activity is restored. (Figure provided by Bryan Beel and Gerald Hazelbauer.)
The analysis of site-directed disulfide bond formation between a single cysteine or pairs of cysteine residues allows deduction of the contact surfaces of the transmembrane segments of the chemoreceptors. Studying the Trg chemoreceptor, Bryan Beel (G. Hazelbauer's lab, Washington State University) described the changes in the intrasubunit interfaces of TM1 to TM2 in response to ligand binding. There was no change in the intersubunit contacts between TM1 and TM1′. Furthermore, both signal binding and the adaptation resulting from receptor methylation lead to changes in the depth of penetration of TM2 in the membrane.
Using 165 individual cysteine residues substituted throughout regions of the Tar cytoplasmic domain, Randal Bass (J. Falke's lab, University of Colorado) measured their rates of reactivity with a fluorescent reagent to map patterns of surface exposure or burial indicative of local secondary structure. Moreover, the ability of engineered cysteine pairs to form disulfide bonds revealed intersegment contact surfaces. These results were interpreted as indicating that the cytoplasmic domain forms a coiled bundle arranged so an extended rod containing helices 6 and 7 is coiled with the 6′-7′ rod from the other subunit and with the antiparallel 8-9 and 8′-9′ segments. Several cysteine substitutions in the buried surfaces of helices 6, 7, and 8 resulted in a constitutively signaling state, suggesting that changes in the helix-helix interactions are directly linked to the signaling state of the receptor. The introduced cysteine residues were also useful for identification of surfaces on Tar that interact with CheA and CheW. A clever approach termed PICM (for protein interactions by chemical modification) tested the effect on CheW-coupled CheA kinase activity when specific cysteine residues were modified by a bulky sulfhydryl reagent. This method showed that conserved residues in the loop between helices 7 and 8 (residues 364 to 399) lie within the docking surface for CheA (3–5).
PHOSPHATE AND ACETATE TRANSFER IN CHEMOTAXIS
Phosphorylation of CheY is the primary signaling process in chemotaxis. Rina Barak and Michael Eisenberg (Weizmann Institute) have drawn attention to acetylation of CheY on lysine residues 92 and 109 by acetyl-AMP or acetyl coenzyme A (acetyl-CoA) as another signal that activates CheY. CheY can be acetylated directly by acetyl-CoA without enzymatic mediation. The role of CheY acetylation in chemotaxis was examined in mutants with decreased levels of acetyl-CoA. Decreased levels of acetyl-CoA had little effect on the response to attractants but markedly impaired the response to repellents. This finding is the first evidence that the attractant and repellent responses are not identical and that CheY acetylation might be important in the latter.
Rick Stewart (University of Maryland) measured the kinetics of the phosphotransfer reactions of CheA by rapid-quench analyses (26). Domain P2 is adjacent to the site of phosphorylation in the P1 domain and is involved in binding CheY and CheB. Deletion of P2 resulted in a 20- and 50-fold decrease in the rate of phosphate transfer to CheY and CheB, respectively. This slowed rate is no longer saturated by the level of either acceptor protein (i.e., increased Km). Even though the acceptor proteins can no longer dock with the P2-deleted form of CheA, phosphate transfer still occurs at a rate about 106 times faster than the transfer from low-molecular-weight phosphate donors, such as phosphoramidate. This suggests that the P1 domain around the phosphorylated histidine contributes substantially to the efficiency of phosphate transfer. The P2 domain could orient the acceptor molecules or induce a conformational change that augments CheA catalytic activity.
TWO-COMPONENT SYSTEMS IN METABOLIC CONTROL
In addition to their role in chemotaxis, two-component systems control transcription of a number of bacterial genes in response to environmental signals. NtrB-NtrC is a paradigm two-component system that responds to changes in the nitrogen supply. NtrB is the HK which transfers phosphate to the enhancer-binding protein NtrC (NRI) to activate it for transcription of ς54-dependent nitrogen-responsive gene promoters. Glutamine synthetase is the key enzyme in the assimilation of ammonium. Both the phosphorylation of NtrC and the adenylylation of glutamine synthase are regulated by the levels of glutamine (Gln, signaling sufficiency of fixed nitrogen) and 2-oxoglutarate (2OG) and are coordinated by the trimeric regulatory protein, PII. PII is itself subject to covalent modification by uridylyl groups at low levels of Gln. Alex Ninfa (University of Michigan) showed that the physiological behavior of the Ntr signal transduction pathway could be reproduced in vitro with purified components (12). In the presence of Gln, the unmodified PII trimer stimulates the adenylylation reaction that inhibits glutamine synthase activity, and it inhibits NtrB (NRII) kinase activity and activates its phosphatase activity. 2OG binds to PII with strong negative cooperativity. The PII-2OG1 complex promotes removal of adenylyl groups from glutamine synthase and increases the NtrB kinase activity and decreases its phosphatase activity. This demonstration revises the previous view that only the phosphatase activity of NtrB was regulated, unlike most other HK studied. The PII-2OG3 complex formed at higher levels of 2OG is inactive for control of NtrB activity or adenylylation of glutamine synthase. Thus, Gln and 2OG act at different steps to influence nitrogen metabolism.
Two-component systems control numerous virulence factors. Kurt Amrein (Hoffmann-La Roche, Basel, Switzerland) identified the set of two-component systems in the genome sequence of the major human pathogen Streptococcus pneumoniae. Thirteen linked RR-HK pairs in several structural classes were found. The role of each RR was examined by genetic disruption experiments. Of the 13 RRs, 2 were essential for growth on laboratory media. None of the viable deletions affected virulence upon intraperitoneal injection in mice. One HK was highly conserved in gram-positive bacteria, carried a PAS domain, and affected competence and virulence. Finally, the apparent sequence similarity of HK to DNA gyrases was supported by their susceptibility to inhibition by certain gyrase inhibitors (18).
Two-component systems also occur in eukaryotes, usually as hybrid proteins containing both histidine kinase and phosphorylation modules. One of these, the 1,670-residue DokA protein of Dictyostelium discoideum, affects sporulation and the response to high-osmolarity stress. Stephan Schuster (Max-Planck-Institute, Martinsried) showed that the receiver module of DokA, but not the HK activity, is crucial for production of cyclic AMP, a key developmental signal. Mutations in dokA interfere with sporulation by causing production of cyclic AMP at aberrant levels. The role of DokA in osmoregulation appears to involve phosphorylation on serine/threonine residues in the HK domain; phosphorylation on histidine had no obvious effect.
SENSOR KINASES
Regulation of more than 30 operons in response to changes of the cellular redox state during the switch from aerobic to anaerobic conditions is controlled by the ArcAB proteins, first identified by their role in anaerobic repression of genes of aerobic respiration. ArcA is an RR of the OmpR family, and ArcB is a tripartite sensor kinase. Upon stimulation, ArcB catalyzes a phosphorelay in which the phosphoryl group from ATP is sequentially transferred to the conserved His-292, Asp-576, and His-717 of ArcB before ending on the conserved Asp-54 of ArcA. O. Kwon (E. Lin's lab, Harvard Medical School) reported that, upon cessation of stimulation, signal decay occurs by dephosphorylation of Asp-54 of ArcA by reverse transfer of the phosphoryl group sequentially to His-717 and Asp-576 of ArcB. The phosphoryl group is then released as inorganic phosphate (9). ArcB is anchored to the cytoplasmic membrane at its N-terminal end via two TMS flanked by a short periplasmic loop. To test whether membrane anchorage specifically interacts with a membrane component, the TMS were replaced by a heterologous TMS. Interestingly, ArcB function was retained upon substitution of either TMS, but not when both were replaced. Perhaps both TMS carry redundant structural elements.
Expression of most terminal reductases is activated under anoxic conditions by FNR and also controlled by the presence of the electron acceptors, nitrate and nitrite. Their presence is detected by the action of two sensor kinases, NarX and NarQ, acting on the response regulators NarL and NarP, whose phosphorylated form activates or represses target genes. There are few systems in which regulation of HK activities can be studied. Angela Lee (R. Gunsalus' lab, UCLA) reported that membranes enriched for NarX displayed high levels of NarX autokinase activity which were stimulated five- or twofold by nitrate or nitrite, respectively. Signaling occurs in the same range of anion concentrations as are effective in cells. Detergent-solubilized NarX exhibited reduced autokinase activity and lost all response to nitrate, indicating the importance of the membrane environment for coupling ligand occupancy to kinase activity (20).
Most HK have high kinase and low phosphatase activities, which are reversed by their regulatory signals. Some HK, including UhpB, which controls expression of the sugar phosphate transport system, display the opposite behavior. Jesse Wright (R. Kadner's lab, University of Virginia) showed that, in the absence of inducer and the associated regulatory protein UhpC, UhpB lacks autokinase activity and has a strong dominant-negative effect when expressed in cells, consistent with its default state being kinase off. Mutations in various sites in the kinase portion of UhpB result in constitutive expression in vivo and restore kinase activity in vitro. The surprising inhibitory effect of UhpB on variants of the RR UhpA which are active without phosphorylation suggests that UhpB may also sequester UhpA.
PilS is an HK of Pseudomonas aeruginosa that, in concert with PilR, controls production of the polar type IV pili, which are responsible for twitching motility and adherence to epithelial cells. Unlike most HK, the N-terminal portion of PilS contains six TMS. Jessica Boyd (University of Calgary) showed that a fusion of PilS to GFP was strongly localized to the cell poles. The six-TMS domain was sufficient for membrane insertion, but polar localization also required the cytoplasmic linker region after the last TMS.
Guadalupe Espin and Miguel Castañeda (UNAM, Cuernavaca) described the role of AsrA, an HK similar to ArcB, in alginate production and encystment in Azotobacter vinelandii. Alginate production is essential for encystment and depends on the sigma factor AlgE. Action of AsrA is related to its activation of algD gene transcription.
Carey Waldburger (Columbia University) reported on the structural and genetic analyses of the periplasmic sensor domain of the HK PhoQ, which responds to divalent cation limitation and pH to activate transcription of magnesium transport genes in enteric bacteria and some virulence genes in Salmonella. The crystal structure of the E. coli protein showed that the dimeric periplasmic domain folded into a five-strand β-sheet core surrounded by helices. The helices forming most of the dimer interface appear to be continuations of the transmembrane helices. This suggests that ligand binding may cause a rearrangement of the dimer interface which is transmitted across the membrane to reduce net phosphoryl transfer to PhoP. Genetic studies defined the positions of potential salt bridges and examined the role of an acidic cluster, whose binding of Ni ions in the crystal may mimic the binding of the natural substrate, Mg. Changing the acidic residues to neutral homologs increased protein stability but reduced response to Mg depletion.
RESPONSE REGULATORS
OmpR, one of the most studied response regulators, controls two of its target genes, encoding the outer membrane porin channels, in opposite directions. Low levels of P-OmpR activate ompF gene transcription, whereas higher levels of P-OmpR result in activation of the ompC promoter and repression of ompF. Unphosphorylated OmpR is silent. Linda Kenney (Oregon Health Sciences University) showed that phosphorylation of OmpR results in a 15- to 25-fold increase in affinity for its specific DNA target. Typical of coupled processes, binding of OmpR to DNA strongly stimulated its ability to be phosphorylated by acetyl phosphate (1). The α3 of the phosphorylation module was proposed to play a central role in coupling OmpR phosphorylation and DNA binding. Both phosphorylation and DNA binding resulted in changes in the conformation of the linker between the phosphorylation and DNA-binding domains, as shown by changes in susceptibility to tryptic digestion.
To test the universality of the conformational changes that occur upon RR phosphorylation, Bill McCleary (Brigham Young University) constructed three chimeric CheY-PhoB molecules to examine whether phosphorylation of the CheY portion could affect PhoB-specific transcription activation by the DNA-binding domain. Although one hybrid protein was constitutively active, the other two were activated by phosphate transfer from CheA or acetyl phosphate. The ability to couple signaling in these chimeras suggests that a common activation mechanism transmits the phosphorylation status to the output domains.
STRUCTURES IN SIGNAL TRANSDUCERS
In Saccharomyces cerevisiae, osmotic stress activates the SLN1p-YPD1p-SSK1p phosphorelay to control a MAP kinase cascade that leads to production of the osmoprotectant glycerol. To study the structural basis for phosphate transfer in this pathway, Ann West (University of Oklahoma) determined the crystal structure of the histidine-containing protein YPD1p. The phosphorylated histidine residue is exposed on part of the central four-helix bundle, whose structure is conserved in the sensor kinases, ArcB, EnvZ, and the P1 domain of CheA. A model for the interaction of this domain with the other pathway components was supported by the properties of mutations that decrease phosphate transfer, presumably by interfering with RR domain interactions and/or stabilization of the transition state (40).
Kottayil Varaughese (Scripps Research Institute) reported on the crystal structures of the B. subtilis sporulation phosphorelay proteins, the aspartate-containing receiver Spo0F and the histidine-containing Spo0B, and of the complex of both proteins (38). Spo0F is a single-domain protein similar to CheY. Spo0B has two domains. The N-terminal helical hairpin dimerizes to form a four-helix bundle from which the carrier histidine protrudes, similar in essence but not in detail to the P1 and P2 domains of CheA. Docking of Spo0F into Spo0B brings the active-site aspartate and histidine into close proximity to allow phosphate transfer. Even though all four domains of the Spo0B dimer participate in binding of Spo0F, there are no major changes in the polypeptide backbones upon binding. The residues in Spo0F that are the contact points for docking to Spo0B are highly conserved in Spo0A, the ultimate phosphate acceptor in the phosphorelay, suggesting that it has a similar mode and specificity of recognition.
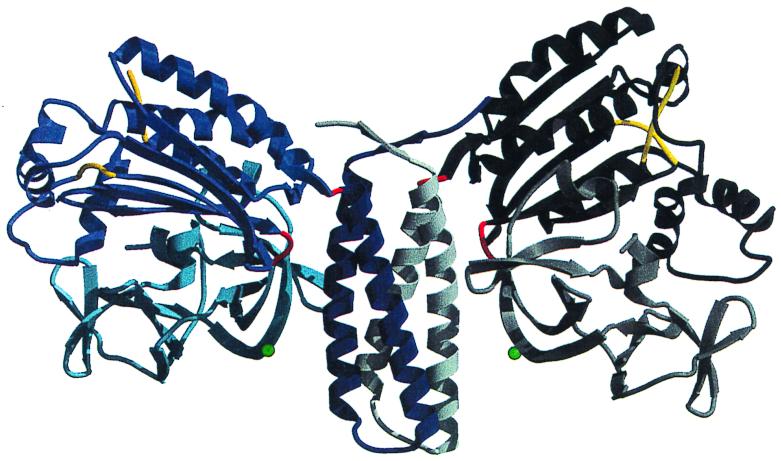
Alex Bilwes (M. Simon's lab, California Institute of Technology) described the structure of the kinase and regulatory portion of CheA from the hyperthermophilic bacterium Thermotoga maritima (7) (Fig. 4). The ATP-binding kinase domain differed in structure from known protein kinases but resembled that of DNA gyrase B, Hsp90, and MutL. The kinase site is incomplete, and the remainder of the catalytic portion may be carried by the P1 domain. The dimerization domain formed a four-helix bundle that provides an extensive and stable hydrophobic interface. The C-terminal regulatory domain resembles SH3 domains in structure but not in sequence. Surprisingly, the CheA dimer does not exhibit precise twofold symmetry, and the same residue in the two subunits can differ in relative location by as much as 6 Å. CheA is structurally related to the phosphorelay protein Spo0B, which also comprises a dimerization and a kinase-like domain, but is not a histidine kinase. Most HK differ from CheA by carrying the phosphorylated histidine on the dimerization domain rather than on a separate domain.
FIG. 4.
Crystal structure of the dimerization, ATP-binding, and regulatory domains of the histidine kinase CheA. CheA is the central component of the signaling pathway that controls bacterial chemotaxis. This is the first crystal structure of a histidine kinase. It reveals the modular composition of CheA, where the three functions of dimerization, ATP binding, and regulation are segregated to three separate domains. The domains are linked by hinges (in red) that allow movement. As a result, the interfaces between domains are different in the two subunits. Unexpectedly, the ATP-binding domain is different from other known kinases but resembles the ATP-binding domain of three ATPases, the topoisomerase gyrase B, the DNA repair enzyme MutL, and the chaperone Hsp90. Subunit A is shown in blue tones, whereas subunit B is shown in gray tones. The dimerization (middle of the figure) is a four-helix bundle where two helices are provided by each monomer (dark blue and white). The two ATP-binding domains (black and blue) are independent and are separated by 90 Å. Each regulatory domain (light gray and light blue) forms two β-barrels of topology reminiscent of SH3 domains. Reprinted from reference 7 with permission of the publisher. (Figure provided by A. Bilwes.)
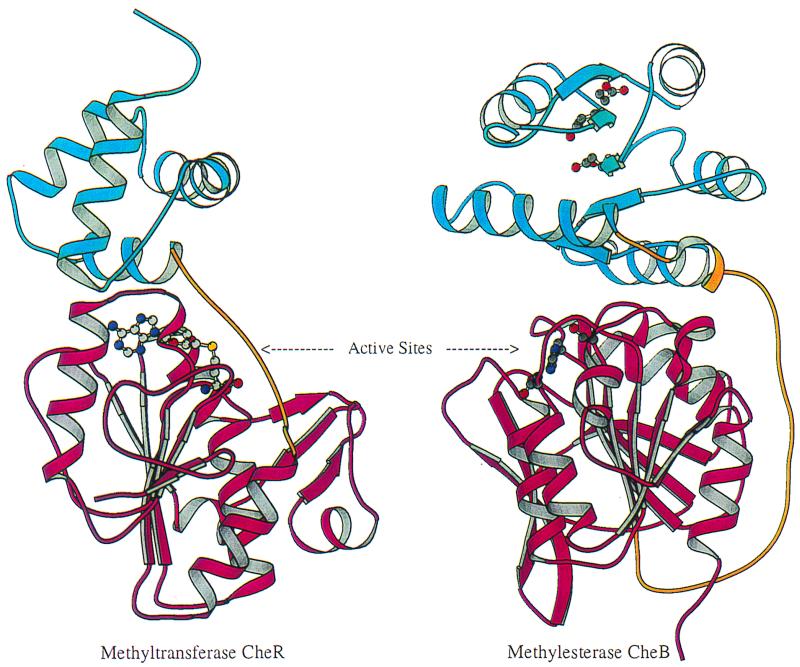
To investigate the specificity and control of the methylation reactions essential for the adaptation phase of chemotaxis, Ganesh Anand (A. Stock's lab, University of Medicine and Dentistry of New Jersey) compared the crystal structures of the methyltransferase CheR and the methylesterase CheB (8). Both proteins recognize the same protein substrates, the reversibly methylated glutamate or glutamine residues spaced along a helical face of the chemoreceptors. Despite a lack of sequence similarity, the structures of their catalytic domains are remarkably similar (Fig. 5). Mutational analyses were consistent with a role of positively charged residues in the active site of CheR in recognition of the methylation sites on the target proteins. The active site of methylesterase CheB is blocked by a receiver module. Phosphorylation results in strong activation of methylesterase activity, but this domain does not just block the active site. P-CheB is about 10 times more active than CheB lacking the phosphorylation module, suggesting that the phosphorylated N-terminal domain has a direct effect in stimulation of methylesterase catalysis. Additional regulation results from the tethering of CheB and CheR to the receptor clusters.
FIG. 5.
Structural comparison of receptor modification enzymes methyltransferase CheR and methylesterase CheB. Each protein contains two domains, an N-terminal domain (blue) and a C-terminal domain (magenta) connected by a linker (gold). The images are oriented to show the similar topologies of the C-terminal domains, the cofactor-binding domain of CheR, and the methylesterase catalytic domain of CheB. The S-adenosylmethionine cofactor in CheR and the active sites for phosphotransfer and methylesterase activities in CheB are depicted by ball-and-stick models (carbon, oxygen, nitrogen, and sulfur are shown in gray, red, blue, and yellow, respectively). Images were prepared using MOLSCRIPT (16). (Figure provided by Ann Stock.)
Megan McEvoy (Rick Dahlquist's lab, University of Oregon) mapped the sites on CheY for interaction with the phosphatase CheZ and the switch protein FliM by multidimensional nuclear magnetic resonance using peptides derived from both proteins. Binding to CheY required residues 196 to 214 of CheZ and residues 1 to 16 of FliM. P-CheY bound the peptides with about 20-fold-higher affinity than did CheY. P-CheY had about a threefold-higher affinity for CheA than for CheZ or FliM. The peptides from FliM and CheZ bound to nearby but different residues around the C-terminal portion of CheY, which includes the least conserved portion of the RR.
ACKNOWLEDGMENTS
Financial contributions from the National Science Foundation, Genencor, and Pfizer Co. are gratefully acknowledged.
REFERENCES
- 1.Ames S K, Frankema J, Kenney L J. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci USA. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations of a theme? Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 3.Bass R B, Coleman M D, Falke J J. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass R B, Falke J J. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Structure. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass R B, Falke J J. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J Biol Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belas R, Schneider R, Melch M. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180:6126–6139. doi: 10.1128/jb.180.23.6126-6139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 8.Djordjevic S, Stock A M. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J Struct Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 8a.Fan F, Macnab R M. Enzymatic characterization of FliI—an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis D, Kwon O, De Wulf P, Lin E C C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 10.Gong W, Hao B, Mansy S S, Gonzalez G, Gilles-Gonzalez M A, Chan M K. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA. 1998;96:15189–15193. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Ninfa A J. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J Bacteriol. 1999;181:1906–1911. doi: 10.1128/jb.181.6.1906-1911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan H B, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 14.Katayama M, Tsinoremas N, Kondo T, Golden S S. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by a mutation of putative channel component. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 17.Kubori T, Matsushima Y, Nakamura D, Urali J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S-I. The invasion-associated type III protein secretion system forms a supramolecular structure on the envelope of Salmonella typhimurium. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 18.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein K E. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 19.Lapidus I R, Berg H C. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee A, Delgado A, Gunsalus R P. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J Bacteriol. 1999;181:5309–5316. doi: 10.1128/jb.181.17.5309-5316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levit M N, Liu Y, Stock J B. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays. Mol Microbiol. 1998;30:459–466. doi: 10.1046/j.1365-2958.1998.01066.x. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd S A, Whitby F G, Blair D F, Hill C F. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 23.Lybarger S R, Maddock J R. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J Bacteriol. 1999;181:5527–5529. doi: 10.1128/jb.181.17.5527-5529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 25.Manson M D, Armitage J P, Hoch J A, Macnab R M. Bacterial locomotion and signal transduction. J Bacteriol. 1998;180:1009–1022. doi: 10.1128/jb.180.5.1009-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayover T L, Halkides C J, Stewart R C. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry. 1999;38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]
- 27.Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab R M. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 28.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muramoto K, Macnab R M. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol Microbiol. 1998;29:1191–1202. doi: 10.1046/j.1365-2958.1998.00998.x. [DOI] [PubMed] [Google Scholar]
- 30.Perrazona B, Spudich J L. Identification of methylation sites and effects of phototaxis stimuli on methylation in Halobacterium salinarum. J Bacteriol. 1999;181:5676–5683. doi: 10.1128/jb.181.18.5676-5683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah, D. S. H., S. L. Porter, D. C. Harris, G. H. Wadhams, P. A. Hamblin, and J. P. Armitage. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol. Microbiol., in press. [DOI] [PubMed]
- 32.Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;38:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 33.Sourjik V, Sterr W, Platzer J, Bos I, Haslbeck M, Schmitt R. Mapping of 41 chemotaxis, flagellar and motility genes to a single region of the Sinorhizobium meliloti chromosome. Gene. 1998;223:283–290. doi: 10.1016/s0378-1119(98)00160-7. [DOI] [PubMed] [Google Scholar]
- 34.Spudich J L. Variations on a molecular switch: transport and sensory signaling by archael rhodopsins. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 35.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J Struct Biol. 1998;124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 37.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varughese K I, Madhusadan, Zhou X Z, Whiteley J M, Hoch J A. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 39.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, West A H. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J Mol Biol. 1999;292:1039–1050. doi: 10.1006/jmbi.1999.3143. [DOI] [PubMed] [Google Scholar]
- 41.Yoon H S, Golden J W. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 42.Yost C K, Rochepeau P, Hynes M F. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology. 1998;144:1945–1956. doi: 10.1099/00221287-144-7-1945. [DOI] [PubMed] [Google Scholar]
- 43.Young G M, Schmiel D H, Miller V L. A new pathway for secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]