Abstract
Retinal ganglion cells (RGCs) are tasked with transmitting all light information from the eye to the retinal recipient areas of the brain. RGCs can be classified into many different types by morphology, gene expression, axonal projections, and functional responses to different light stimuli. Ultimately, these classification systems should be unified into a comprehensive taxonomy. Toward that end, we show here that nearly all RGCs express either Islet-2, Tbr2 or a combination of Satb1 and Satb2. We present gene expression data supporting the hypothesis that Satb1 and Satb2 are expressed in ON-OFF direction-selective (DS) RGCs, complementing our previous work demonstrating that RGCs that express Islet-2 and Tbr2 are non-DS and non-image-forming, respectively. Expression of these transcription factors emerges at distinct embryonic ages and only in postmitotic cells. Finally, we demonstrate that these transcription factor-defined RGC classes are born throughout RGC genesis.
Keywords: retinal ganglion cells, transcription factors, cell fate, RRIDs AB_2301417, AB_882455, AB_10615604, AB_11143446, AB_2167511, AB_2313614, AB_1608077, AB_231491
Introduction:
There are thousands of neuronal types in the central nervous system (CNS) that have different morphologies, intrinsic properties, and connectivity patterns (Luo et al., 2008; Masland, 2012; Nelson et al., 2006). Derived from a common progenitor pool, each neuronal type uses a combination of transcriptional regulation and environmental cues to express a set of genes critical for inclusion into specific circuits. A description of the mechanisms that generate neuronal diversity not only provides insight into circuit formation and function, but also leads to potential gene-based strategies for the regeneration of specific cell types when damaged due to injury or disease.
In the mouse visual system, the retina transforms the visual scene into more than 20 different channels of information, each mediated by a unique type of retinal ganglion cell (RGC) (Baden et al., 2016; Masland, 2012). All RGC types send axonal projections to specific subcortical retinal recipient areas that are responsible for executing reflexive and planned behaviors (Dhande and Huberman, 2014). For example, OFF and ON type RGCs fire action potentials in response to changes in luminance; other RGC types respond to movement in a specific direction regardless of luminance changes (ON-OFF direction-selective (DS) RGCs). Most OFF, ON, and ON-OFF DS RGCs send axons to the dorsal lateral geniculate nucleus (dLGN) and superior colliculus (SC), which are known as image-forming areas because of their roles in conscious or reflexive movements. Conversely, the axons of intrinsically photosensitive RGCs (ipRGCs) and ON DS RGCs project to non-image-forming nuclei that regulate eye movements, image stabilization (Dhande et al., 2013), pupil constriction (Chen et al., 2011), and day-night activity cycles (Cosenza and Moore, 1984; Harrington, 1997; Hattar et al., 2002) (Morin et al., 2003). How do these different RGC types emerge? The relative contributions of environment and genetics to specifying RGC types remain largely unknown.
Two lines of evidence suggest that RGC type specification is primarily under genetic, rather than environmental control: 1) much of the morphological specificity of RGC types is formed before eye opening, and 2) RGC type-specific morphology and physiology do not dramatically change if retinal activity is perturbed (Anishchenko et al., 2010; Elstrott et al., 2008; Kerschensteiner et al., 2008; Yamagata and Sanes, 1995). Therefore, a favored working hypothesis used in other neural systems (Tanabe and Jessell, 1996) is that different RGC types express specific sets of transcription factors, which in turn control genes important for type-specific features. We currently lack a good understanding of the control mechanisms that direct each RGC type to express its complement of transcription factors. Neuronal differentiation studies in the mammalian cortex and retina (Guo et al., 2013; Cepko, 2014; Franco et al., 2012) and in Drosophila (Pearson and Doe, 2004) have led to several models. Dedicated RGC progenitors could each give rise to a specific RGC type; alternatively, multipotent progenitors could generate different RGCs during a series of temporal phases (Kang and Reichert, 2015). Another possibility is that RGC types are generated stochastically after differentiation; in this case, feedback mechanisms would ensure that an appropriate number of each type is generated.
To begin answering these questions, we first sought to identify transcription factors that are expressed in non-overlapping classes in the developing or adult mouse retina. We previously showed that the basic helix-loop-helix transcription factor Islet-2 (Isl2) is expressed in a set of RGCs that selectively project to the contralateral image-forming retinal recipient areas (SC and LGN) and include ON and OFF alpha-RGCs (indicated by expression of SMI-32 antigen) but not ON-OFF DS RGCs (Triplett et al., 2014). On the other hand, the T-Box transcription factor Tbr2 is expressed in non-overlapping RGC types that project to non-image-forming areas of the brain including the pretectum, suprachiasmatic nucleus (SCN), and ventral lateral geniculate nucleus (vLGN), and include all melanopsin-expressing ipRGCs (Mao et al., 2014; Sweeney et al., 2014).
Here we show that the homeodomain-containing special AT-rich binding proteins 1 and 2 (Satb1 and Satb2) are expressed in overlapping populations of RGCs that show very little overlap with Isl2-expressing or Tbr2-expressing RGCs, either in early postnatal or adult mouse retina. We characterize Satb1 and Satb2 expressing RGCs in two ways. One is by using transgenic BAC lines that express GFP in defined RGC types to see if Satb1 and/or Satb2 are expressed in ON-OFF DS RGCs; the other is by looking at marker co-expression. We also analyze these three RGC classes (Isl2, Tbr2, and Satb1/Satb2) in terms of gene expression onset, progenitor vs. postmitotic RGC expression, and birthdate.
Materials and Methods:
Mouse lines:
Most experiments were performed on Isl2-GFP transgenic reporter mice, genotyped by PCR for GFP (Triplett et al., 2014) or using a GFP-exciting flashlight and detecting filter glasses to visualize expression in newborn pups (P0-P4) (BlueStar model Bls2). C57Bl/6 mice (Charles River) were used for experiments that did not require visualization of Isl2. CB2-GFP, DRD4-GFP, THRH-GFP and Cdh3-GFP mice were genotyped as described previously (Huberman et al., 2008; Huberman et al., 2009; Rivlin-Etzion et al., 2011; Osterhout et al., 2011).
Both male and female mice were used for these experiments, with no differences noted between sexes. Approximately 50 Isl2-GFP, 10 C57 Bl/6, and 20 mice from GFP-labeled lines were used in these experiments. All procedures were performed in accordance with the UCSC Institutional Animal Care and Use Committee.
EdU-based cellular birth dating:
EdU is a thymidine analog that integrates into the DNA of dividing cells during replication (Salic and Mitchison, 2008). Pregnant dams were injected intraperitoneally with EdU (Invitrogen/Thermo Fisher) dissolved in PBS (136.9 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.4 mM K2PO4) at 50 mg/kg bodyweight. To detect EdU incorporation, we first performed click-chemistry aided staining (Invitrogen/Thermo Fisher) before immunohistochemical detection of other antigens.
Immunohistochemistry:
Postnatal mice were sacrificed and intracardially perfused with ice-cold PBS followed by ice-cold paraformaldehyde (PFA) (pH 7.4, 4% in PBS). Eyes were dissected out and fixed in 4% PFA in PBS for either 30 minutes at room temperature or overnight at 4°C. The retinas were isolated, placed in 30% sucrose in PBS overnight, embedded in Tissue-Tek OCT (Sakura Finetek, USA, Torrance, CA, USA), cryosectioned at 12-25 μm on a CM1520 Cryostat (Leica Microsystems, Buffalo Grove, IL, USA) maintained at −20 to −25°C, and collected on Superfrost Plus glass slides (Thermo Fisher). In some cases, we first performed click-chemistry aided EdU staining before immunohistochemical detection of antigens (Invitrogen/Thermo Fisher). All immunostained sections were blocked for 1h in 5% heat inactivated serum (either horse, goat or donkey) in PBS + 0.1% Triton-X100. Primary and secondary antibodies were diluted in this blocking medium, and multiple washes were done with PBS. DAPI (Sigma D8417) was included in a penultimate PBS rinse at a dilution of 1:2000 to visualize nuclei. The following primary antibodies were used:
| Antigen | Immunogen | Manufacturer, Cat #, RRID, species raised in, clonality |
Concentration |
|---|---|---|---|
| Satb2 | Synthetic peptide conjugated to KLH derived from within residues 700 to the C-terminus of mouse SatbSatb2. | Abcam ab34735, AB_2301417, rabbit, polyclonal | 1:1000 |
| Satb2 | Recombinant fragment corresponding to human Satb2 (C terminal) | Abcam ab51502, AB_882455, mouse, monoclonal | 1:1000 |
| Tbr2 | KLH-conjugated linear peptide corresponding to mouse Tbr2 | Millipore AB15894, AB_10615604, chicken, polyclonal | 1:1000 |
| Satb1 | KLH-conjugated linear peptide correcponding to the N terminus corresponding to human Satb1 | Santa Cruz biotech SC-5989 goat, polyclonal | 1:1000 |
| Tbr2 (Eomes) | Linear peptide corresponding to mouse Tbr2 (KLH) | LifeSpan Bio LS-B6801, AB_11143446, chicken, polyclonal | 1:1000 |
| Tbr2 (Eomes) | KLH-conjugated peptide from residue 650 to the C-terminus of mouse Tbr2/Eomes | Abcam ab23345, AB_778267, rabbit, polyclonal | 1:1000 |
| Brn3a | Amino acids 108-138 (C-20) of human Brn3a | SCBT sc31984, AB_2167511, goat, polyclonal | 1:1000 |
| CART | Nucleotides 61-102 of CART (C4 peptide) | Phoenix Pharmaceuticals 55-102, AB_2313614, rabbit, polyclonal | 1:1000 |
| Melanopsin (Opn4) | 15 most N-terminal amino acids of mouse melanopsin extracellular domain | Advanced Targeting Systems AB-N38, AB_1608077, rabbit, polyclonal | 1:1000 |
| SMI32 | Homogenized hypothalamus from Fischer 344 rats | Covance SMI-32P, AB_2314912, mouse, monoclonal | 1:2500 |
Antibody characterization:
Anti-Satb2 (Abcam ab34735):
In ES cells, Satb2 siRNA eliminates Satb2 as measured with this antibody, but not Satb1 (Agrelo et al., 2009).
Anti-Satb2 (Abcam ab51502):
While advertised as a Satb2 antibody, we learned that this antibody recognizes Satb1 and Satb2 using cortical tissue derived from Satb2 knock out mice (Professor Bin Chen, UCSC). It labels more cells in the RGC layer than the Satb2 antibody described above (Abcam ab34735). When used for co-staining, all of the cells labeled by the ab34735 were also labeled by ab51502, but conversely some cells were only labeled by ab51502. Subsequent staining combining ab51502 with an antibody against Satb1 (SCBT sc-5989) found that all cells labeled by the Satb1 antibody were also labeled by ab51502. From this, we concluded that ab51502 recognizes both Satb1 and Satb2 antigens.
Anti-Satb1 (Santa Cruz biotech SC-5989):
In Th4 cells, Satb1 siRNA reduces Satb1 protein as measured with this antibody (Ahlfors et al., 2010).
Anti-Tbr2 (Millipore AB15894):
As expected, this antibody labels cells in the subventricular zone in embryonic mouse brain (Vasistha et al., 2015); in the retina, it displays a pattern identical to that which we previously reported using two another antibody other antibodies against Tbr2 including LS-B6801 and Abcam ab23345 (Sweeney et al., 2014).
Anti-Tbr2 (Bio LS-B6801 and Abcam ab23345):
The expression pattern of these antibodies corresponds to previous reports and conditional knockout of Tbr2 leads to a loss of signal in the retina (Mao et al., 2014; Sweeney et al., 2014).
Anti-Brn3a (SCBT sc31984):
In the mouse retina the staining pattern matches reports showing RGC localization in rat and mouse (Ogata et al., 2012) retina with the same antibody (Sweeney et al., 2014), and a different anti-Brn3a antibody (Wang et al., 2013).
Anti-CART (55-102):
While there is normally staining in rat pelvic ganglia, this staining disappears after antibody is preabsorbed with the CART peptide (Dun et al., 2000) . In mouse retina, staining matches previously reported expression patterns (Kay et al., 2011; Sweeney et al., 2014) . In retinal cryosections, CART is expressed by a subset of RGCs and is subcellularly localized to “hat-like” Golgi regions apical to the nuclei.
Anti-Melanopsin (AB-N38):
In melanopsin knockout mice , this antibody showed no reactivity (Panda et al., 2002) (Berson et al., 2010) . In mouse retina it matches previously reported expression patterns (Berson et al., 2010).
Anti-SMI32 (SMI-32P):
The expression pattern for this antibody in mouse retina corresponds with previous reports (Straznicky et al., 1992).
Microscopy:
Sections were imaged using a Leica DM5500 B Widefield Microscope, with either a 10x/0.3 or 20x/0.5 objective and Leica filter cubes optimized for DAPI, GFP, Cy3 and Cy5, or on an Olympus BX51 epifluorescent microscope (Olympus, Center Valley, PA, USA) equipped with QImaging Retiga EXi digital camera (QImaging, Surrey, BC, Canada).
Data analysis:
Cells were manually counted with ImageJ. DAPI and lamination patterns were used to restrict analysis to the RGC layer where applicable. GraphPad’s Prism software program was used to calculate and graph means, standard deviations and standard errors, following cell counts. For some figures, brightness and contrast were changed in ImageJ to standardize the appearance of cells and enhance figure legibility.
Results:
Postnatal RGCs express either Tbr2, Isl2, and one or both of Satb1 and Satb2 with very little overlap.
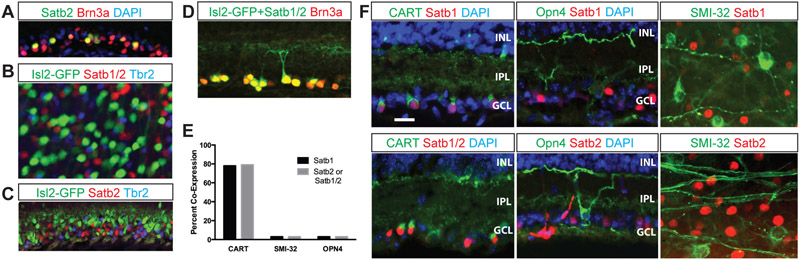
To identify transcriptional regulators involved in RGC type specification, we performed an immunohistochemistry screen to search for transcription factors that are expressed in subsets of RGCs in the developing or adult mouse retina. We conducted this screen in an Isl2-GFP BAC transgenic line; we have previously characterized and demonstrated that this line faithfully reports endogenous Isl2 expression in RGCs (Triplett et al., 2014). We initially found that the transcription factor Satb2 is expressed in a subset of non-Isl2-GFP RGCs that also co-express the transcription factor Brn3a in the adult retina (Fig. 1A). Brn3a is a transcription factor important for RGC development and is expressed in approximately 80% of RGCs (Badea et al., 2009; Quina et al., 2005; Sweeney et al., 2014). Remarkably, when immunostaining with an antibody that recognizes Satb2 and its homologue Satb1, we found that Satb1/2-expressing RGCs have very little overlap with Isl2-expressing or Tbr2-expressing RGCs in the mature retina (Fig. 1B, Table 1). This non-overlapping pattern of expression for Isl2, Satb2, and Tbr2 is already present at P0 (Fig. 1C).
Figure 1. Isl2, Satb1, Satb2, and Tbr2 mark distinct RGC classes.
20 μm cryosections through adult retinas (unless otherwise indicated) were immunostained with the antibodies indicated; Isl2-GFP is native GFP from the transgenic line used. Satb1/2 refers to the use of Abcam ab51502 antibody that recognizes both Satb1 and Satb2. The color of the text matches the color of the fluorescent label or the nuclear DAPI staining. A: Section through adult ganglion cell layer of the retina showing Satb2-expressing RGCs also express Brn3a. B: Flat-mounted adult retina shows that Isl2, Satb1/2 and Tbr2 are expressed in largely non-overlapping populations of RGCs. C: 20 μm P0 cryosection stained with Isl2, Satb2 and Tbr2 show largely non-overlapping expression. D: Nearly all Brn3a-expressing cells in the RGC layer co-express either Isl2 or Satb1/2 (98%; 360 Brn3a-expressing cells counted) E: Quantification of staining data from F for either Satb2 staining (Abcam 34735), Satb1 staining (SC SC-5989) or Satb1/2 staining (Abcam 51502). F: Adult cryosections (CART, Opn4) or retinal flat mount (SMI-32) show that very few SMI-32-expressing or Opn4-expressing RGCs express Satb1 or Satb2 (SMI-32: 0% of Satb1-expressing, 103 Satb1-expressing cells counted; 0% of Satb2-expressing, 118 Satb2-expressing cells counted; Opn4: 0% of Satb1-expressing, 102 Satb1-expressing cells counted; 0% of Satb1/2, 97 Satb2-expressing cells counted), but a large percentage (79% of Satb1, 130 Satb1-expressing cells counted; 79% of Satb1/2, 92 Satb1/2-expressing cells counted) of CART-expressing RGCs also express Satb1/2 (and Satb2). IPL, inner plexiform layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar in F applies to all panels, 20 μm.
Table 1.
Isl2, Satb2, and Tbr2 show low levels of co-expression in RGCs. Frequencies of RGCs expressing Isl2, Satb2, or Tbr2 were calculated using 20 μm cryosections from P10 mouse retina. N=5 animals/condition, 7 images quantified/animal. In total, 2631 cells were counted. Cell counts were summed across images for each animal; averages and S.D.s were calculated across animals.
| RGC type | Percentage ± S.D. |
|---|---|
| Isl2+ | 41% ± 3 |
| Satb2+ | 22% ± 4 |
| Tbr2+ | 28% ± 6 |
| Isl2+/Satb2+ | 3% ± 1 |
| Satb2+/Tbr2+ | 2% ± 1 |
| Isl2+/Tbr2+ | 3% ± 1 |
| Isl2+/Satb2+/Tbr2+ | 0.3% ± 0.3 |
Lacking a pan-RGC marker, we were only indirectly able to ask whether each RGC expresses at least one of the three transcription factors. Brn3a is expressed in 80% of adult RGCs but is not expressed in RGCs that express Tbr2, which comprise about 20% of total RGCs (Badea et al., 2009; Quina et al., 2005; Sweeney et al., 2014). In the adult retina we find that 98% of RGCs that express Brn3a also express either Isl2 or Satb1/2 (Fig 1D; n= 360 Brn3a-expressing cells), and very few Isl2-expressing or Satb1/2-expressing RGCs are Brn3a negative. Therefore, we can divide RGCs into three categories – Brn3a/Satb1/2-expressing, Brn3a/Isl2-expressing and Tbr2-expressing – and together these categories comprise nearly all RGCs; however, it is possible that a small percentage of RGCs do not express any of these transcription factors.
Satb1 and Satb2 are expressed in overlapping subsets of On-Off DS RGCs.
Based on the percentage of RGCs that express Satb1 or Satb2, we hypothesized that these were a collection of types that share common features rather than a single RGC type. To test this idea, we determined the degree of overlap between Satb1 and Satb2 expression and markers of defined RGC types. We find that 79% of Satb1- and Satb2-expressing RGCs also express the ON-OFF DS marker CART (although CART is not a perfect marker of DS RGCs) (Kay et al., 2011), and have very little co-expression with non-direction selective RGCs labeled with Smi32 and melanopsin (OPN4, Fig. 1E, F).
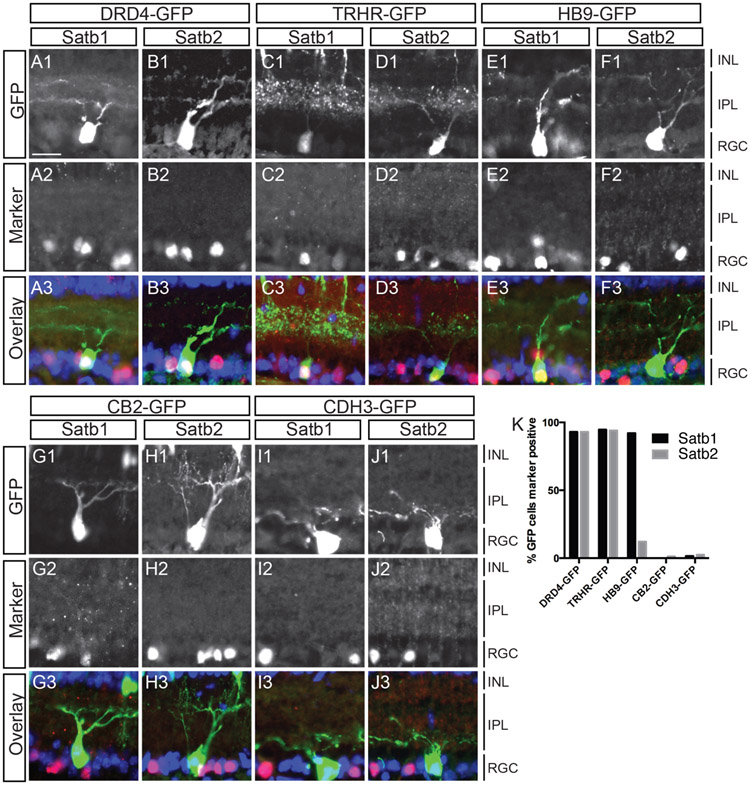
Consistent with the hypothesis that Satb1 and Satb2 are expressed in ON-OFF DS RGCs, we find that Satb1 and Satb2 are almost always expressed in the GFP-labeled posterior DS RGCs labeled by the DRD4-GFP (>90%) and TRHR-GFP (>90%) transgenic mouse lines (Huberman et al., 2009; Rivlin-Etzion et al., 2011, Fig. 2A-D). Satb1 (>90%) is also expressed in the HB9-GFP line that labels upward motion DS RGCs (Trenholm et al., 2011) while Satb2 is expressed in only 3% of HB9-GFP DS RGCs (Fig. 2E, F). We also find that neither of Satb1 or Satb2 are expressed in OFF-transient, CB2-GFP labeled RGCs (Huberman et al., 2008) (<5%), or in the non-image forming, Cdh3-GFP RGCs (<5%, Fig. 2G-J, (Osterhout et al., 2011).
Figure 2. Satb1 and Satb2 are expressed in GFP-labeled On-Off DS RGCs but not in other defined RGC types.
(A-J) Sections through P8 eyes from BAC transgenic GFP lines were immunostained with antibodies against Satb1 and Satb2. Anti-GFP antibodies were used to increase signal and sections were treated with DAPI (blue) to visualize cell nuclei. Nearly all DRD4-GFP RGCs express Satb1 (92%; n=57) and Satb2 (96%; n=52). TRHR-GFP RGCs also express both Satb1 (95%; n=114) and Satb2 (94%; n=50). HB9-GFP RGCs express Satb1 (92%; n=51) but not Satb2 (3%; n=78) (E-F). Very few CB2-GFP RGCs were labeled by Satb1 (1%; n=85) or Satb2 (12%; n=74) and similarly, very few Cdh3-GFP RGCs expressed Satb1 (1.5%; n=66) or Satb2 (1.1%; n=85)(G-J). (K) Graphical representation of A-J. ONL, Outer nuclear layer; IPL, inner plexiform layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar: 20 uM in A for panels A-J.
Isl2, Tbr2, and Satb2 are all expressed in post-mitotic RGCs but differ in their onset of expression.
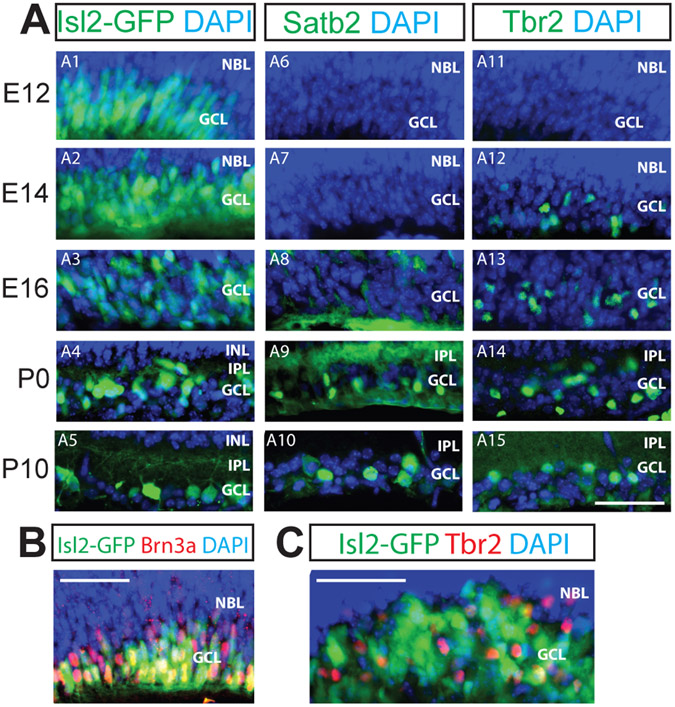
Within the retina, the different cell types – RGCs, amacrine cells, bipolar cells, horizontal cells, photoreceptors and Muller glia – are generated from retinal progenitor cells (RPCs) in successive yet overlapping temporal waves (Cepko et al., 1996). RGCs are among the earliest-born; they begin to appear at embryonic day 8 (E8) and are completely generated by E17, with birth rate peaking at E11 (Farah and Easter, 2005; Voinescu et al., 2009). To determine the onset of expression of these transcription factors, we examined the expression of Isl2, Satb2, and Tbr2 in the mouse retina on different days of retinal development (E12, E14, E16, P0 and P10). In agreement with previous reports (Pak et al., 2004), we detected widespread Isl2 in the ganglion cell layer of the retina at the earliest embryonic time points examined (Figs. 3A-A5), and saw 97% overlap with Brn3a-expressing RGCs at E14 (Fig. 3B). Isl2 gradually refined into its mature pattern by P0. This refinement correlates with the onset of expression of Satb2 (Fig. 3A6-A10) and Tbr2 (Fig. 3A11-3A15), consistent with (Mao et al., 2008). Even at E16, Isl2 and Tbr2 mark separate RGC populations (Fig. 3C).
Figure 3. Developmental onset of expression differs for Isl2, Satb2 and Tbr2.
Panels are labeled as in Figure 1. 20 μm (A, C) or 12 μm (B) retinal cryosections at the embryonic ages indicated on the left were immunostained with the antibodies indicated. A: Isl2 is widely expressed in the RGC layer at E12 and is gradually refined to its final expression pattern (A1-A5). Satb2 expression is first detected at E16 and becomes refined to the GCL at P0; Tbr2 expression is first detected at E14 (A11-A15). B: At E14, nearly all Brn3a-expressing RGCs also express Isl2 (97% of Brn3a-expressing cells; 347 Brn3a-expressing cells counted). C: At E16, few Tbr2-expressing RGCs colocalize with Isl2-expressing RGCs. NBL, neuroblast layer; IPL, inner plexiform layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bars, 50 μm.
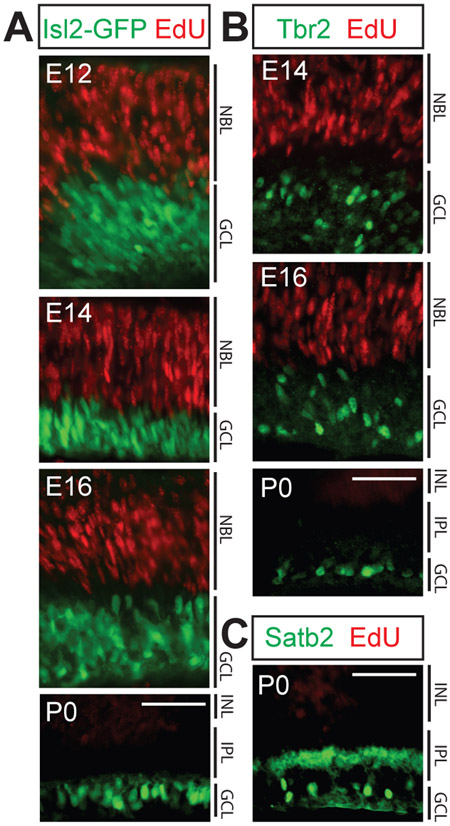
In some developmental systems, distinct types of progenitor cells give rise to progeny that share their marker expression profiles. A striking example of this is that of the Cdh6-expressing RPCs, which give rise to mainly Cdh6-expressing RGCs (and other neuronal cell types in the retina) (De la Huerta et al., 2012). We wanted to determine whether Isl2, Satb2, or Tbr2 expression is restricted to postmitotic RGCs or if these transcription factors can also be found in proliferative RPCs, which might give rise to a limited repertoire of RGCs. To do this, we labeled dividing cells with a pulse of EdU introduced by IP injection of timed-pregnant dams or neonatal mice, sacrificed the animals 4 hours after EdU injection, and stained the retinas to visualize EdU incorporation and transcription factor expression in retinal sections. Consistent with previous work that showed Isl2 and Tbr2 are not expressed in progenitors (Mao et al., 2008; Pak et al., 2004), we found no time point at which any of the three transcription factors were expressed in EdU labeled cells in the retina (even outside of the ganglion cell layer), or within the neuroblast layer (NBL) where the RPCs reside. This demonstrates that Isl2, Satb2, and Tbr2 are only expressed post-mitotically in RGCs (Figs. 4A-C).
Figure 4. Isl2, Satb2 and Tbr2 expression is limited to post-mitotic RGCs.
20 μm retinal cryosections from mice injected with EdU at the ages indicated and sacrificed 4 hours later were immunostained with the antibodies indicated, following EdU visualization. A: EdU labeled RPCs at E12, E14, E16, and P0 do not express Isl2. B: Tbr2 is not expressed by progenitor cells at E14, E16 or P0. C: Satb2 is not expressed by progenitor cells at P0. NBL, neuroblast layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. For all three transcription factors, no cellular colocalization with Edu label was observed: n=3-5 animals/age, 5-7 sections/animal examined. Scale bars, 50 μm.
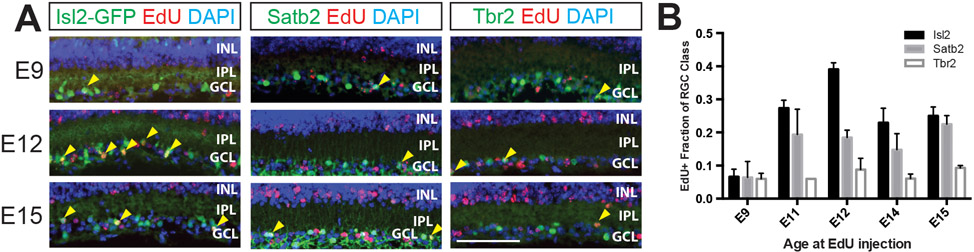
There is no specific temporal order in generating the Isl2/Satb2/Tbr2 RGC classes.
Different neuronal cell types are born in successive temporal waves in the mammalian cerebral cortex, spinal cord, and retina (Berry et al., 1964; Soula et al., 2001; Livesey and Cepko, 2001). Recently, it was shown that this is also true for some RGC types (Osterhout et al., 2014). To test if the RGC classes defined by Isl2, Satb2, and Tbr2 are born in a temporal order, we used EdU to label dividing cells at several embryonic time points from E9 to E15, spanning the window of RGC genesis. We then euthanized the pups at P10 and calculated the percentage of mature RGCs that express Isl2, Satb2, or Tbr2 and terminally differentiated on a particular day (EdU labeled). We found that RGCs born at each time point examined could express each transcription factor (Figs. 5A, B). Therefore, despite the fact they are first detectable at different developmental points (Fig. 3), we find no evidence that Isl2-, Satb2-, or Tbr2-expressing RGCs are biased to be born on a particular day of RGC genesis. However, we note that at each time point, we detected only low percentages of Tbr2-expressing RGCs being born. Since we normalized by the total number of Tbr2-expressing RGCs present in the retina at P10, this result is surprising and may mean that there is a sharp peak of Tbr2-expressing RGC birth that was missed in our time course.
Figure 5. Isl2-, Satb2- and Tbr2-expressing RGCs are born over similar, broad windows of RGC differentiation.
20 μm P10 retinal cryosections from mice injected with EdU at the ages indicated were immunostained with the antibodies indicated after EdU visualization. A: RGCs expressing Isl2, Satb2, and Tbr2 can be EdU labeled at E9, E12 and E15. Yellow arrowheads indicate cells co-expressing EdU (red) and transgenic GFP (Isl2-GFP) or the transcription factor stained for (Satb2, Tbr2) in each column (green). Scale bar, 50 μm. B: Quantification of EdU birthdating experiment for five ages. The Y-axis measures the fraction of mature RGCs of each class that is EdU labeled, following mitotic labeling on the embryonic day indicated. INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. N=3-5 animals, 5-6 images quantified per animal. Error bars represent S.E.M.
Discussion:
The retina divides the visual world into distinct channels of information; RGCs extend axons to relay this visual information to higher processing centers within the brain. RGCs can be subdivided into classes using a variety of measures including morphology, gene expression, connectivity, and characteristic functional response profiles. Our work shows that almost all RGCs can be divided into three classes based on their expression of Isl2, Satb1/Satb2, or Tbr2. Based on their onset of expression in post-mitotic RGCs, co-expression with RGC type specific markers, and function in other developmental systems, we hypothesize that these transcription factors mediate RGC type identity by segregating RGCs into three distinct functional classes: contrast-sensitive RGCs, On-Off direction-selective RGCs, and non-image-forming RGCs.
Isl2, Satb1/2, and Tbr2 subdivide RGCs into three molecular types.
Here we have shown that nearly all mature RGCs express one of the transcription factors Isl2, Satb1 and/or Satb2, or Tbr2 This hints that a precise transcriptional network mediates RGC type identity. Isl2 has been shown to play a role in specifying ipsilateral vs. contralateral axonal projections of ventral-temporal RGCs early in development (Pak et al., 2004). In the mature retina, Isl2 is restricted to a population of RGCs with characteristic dendritic stratification in layer S3 of the IPL, suggesting that Isl2-expressing RGCs share common functional properties (Triplett et al., 2014). Expression of Isl2 in known types of transient-OFF and OFF-alpha RGCs (marked by CB2-GFP and SMI-32, respectively) combined with lack of expression in posterior DS RGCs (marked by DRD4-GFP) provide evidence that Isl2-expressing RGCs are primarily contrast-sensitive and non-DS (Triplett et al., 2014). Tbr2 is expressed in several types of Brn3a-negative non-image-forming RGCs, including ipRGCs. In Tbr2-null mice, the Tbr2 cells either die or fail to develop; furthermore, the pupillary light reflex, mediated by non-image-forming RGCs, is greatly reduced (Mao et al., 2014; Sweeney et al., 2014).
While Isl2- and Tbr2- expressing RGCs have been previously described, very little is known about the RGC types that express Satb1 and Satb2. One study showed that they are expressed in a subset of Foxp2 expressing RGCs (Rousso et al., 2016), but did not do extensive analysis of the RGC types that express Satb1 and Satb2. Here we show that Satb1 and Satb2 are expressed in the posterior ON-OFF DS RGCs labeled in the Drd4- and TRHR-GFP lines, and Satb1 is expressed in the upward ON-OFF DS RGCs labeled in the HB9-GFP mouse line. Satb1 and Satb2 also show a large overlap of expression (79%) in RGCs with the putative ON-OFF DS marker CART. Taken together, these results suggest that Satb1 and Satb2 are expressed in ON-OFF DS RGCs. However, we cannot definitively conclude that Satb1 and/or Satb2 are expressed in all ON-OFF DS RGCs because we do not have a marker of the anterior or dorsally tuned ON-OFF DS RGCs and because some ON-OFF DS RGCs do not express CART (Dhande et al., 2013). Future functional analysis of Satb1- and Satb2-expressing RGCs will need to be performed to confirm our hypothesis.
No evidence for RGC type-specific progenitors
Isl2, Satb2 and Tbr2 are only found in postmitotic cells (Fig. 3), excluding a model in which RPCs express these transcription factors and give rise to class-restricted progeny as shown for Cdh6 expressing RPCs (De la Huerta et al., 2012). Birthdating experiments show that there is no strict temporal order of birth for these three RGC classes, thus discounting a temporal model for generating these three RGC classes. This is in agreement with previous work showing that three non-overlapping RGC types are born in a highly overlapped fashion (De la Huerta et al., 2012). However, another study showed that there are differences in birth timing for three other non-overlapping RGC types (Osterhout et al., 2014), including a comparatively early peak birth date for the RGC types labeled by the Cdh3-GFP mouse line, which make up ~9% of the Tbr2-expressing RGCs (Sweeney et al., 2014). Thus, some RGC types within the classes that we propose may show a temporal bias in birth date that is absent at the class level.
Taken together, these data suggest that cell identity emerges gradually in postmitotic RGCs. It could be that expression levels of certain genes activate mutual inhibition and/or positive feedback-like transcriptional programs that solidify cell identity. The transcription factors described here – Isl2, Satb1, Satb2, and Tbr2 – may be involved directly in such a process. Several pieces of evidence suggest that they play a role in generating RGC identity. First, their expression patterns are non-overlapping and together, they account for nearly all RGCs; this positions them as potential master regulators of RGC identity. Second, the internal division of these three classes into Brn3a-coexpressing (Satb1, Satb2-expressing or Isl2-expressing) and Brn3a non-expressing (Tbr2-expressing) is suggestive of their potential to influence the expression of other transcription factors. Alternatively, it may be that other genes upstream of these transcription factors are necessary for generating the RGC classes that we describe, and that Isl2, Satb1, Satb2, and Tbr2 are simply markers for these classes. In the future, gain and loss of function experiments will explore the role that these genes play in RGC development.
In glaucoma, a common neurodegenerative disease, patients become blind due to RGC death. Cell replacement therapy, i.e. injection of RGCs or RPCs into the glaucoma patient’s retina, is an attractive mode of treatment currently under study (Riazifar et al., 2014; Sluch et al., 2015). The success of this approach will depend on our collective ability to faithfully generate the endogenous RGC types, each with its own functional role. In other CNS circuits, reprogramming and transdifferentiation experiments have powerfully shown that transcription factor induction is sufficient to generate specific neuronal types (Pfisterer et al., 2011; Yang et al., 2011). The data presented here comprise one step toward the goal of faithful RGC recapitulation. Future work will examine which, if any, of Isl2, Satb1, Satb2, and Tbr2 – are necessary and/or sufficient to drive RGC identity.
Acknowledgements:
This study was funded by R01 E4014689, R01 EYO22117 (D.A.F); CIRM Postdoctoral Fellowships TG2-01157 (to K.N.J. and N.T.S.). CIRM Facilities Grant FA1-00617-1. We thank Jena Yamada for technical help and comments on the manuscript.
Footnotes
Conflict of interest statement: The authors state that they have no conflict of interest to declare.
References
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. (2009). SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell 16, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors H, Limaye A, Elo LL, Tuomela S, Burute M, Gottimukkala KV, Notani D, Rasool O, Galande S, and Lahesmaa R (2010). SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood 116, 1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishchenko A, Greschner M, Elstrott J, Sher A, Litke AM, Feller MB, and Chichilnisky EJ (2010). Receptive field mosaics of retinal ganglion cells are established without visual experience. J Neurophysiol 103, 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, and Nathans J (2009). Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61, 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Roman Roson M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M, Rogers AW, and Eayrs JT (1964). Pattern of Cell Migration during Cortical Histogenesis. Nature 203, 591–593. [DOI] [PubMed] [Google Scholar]
- Berson DM, Castrucci AM, and Provencio I (2010). Morphology and mosaics of melanopsin-expressing retinal ganglion cell types in mice. J Comp Neurol 518, 2405–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C (2014). Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci 15, 615–627. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, and Ezzeddine D (1996). Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A 93, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Badea TC, and Hattar S (2011). Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza RM, and Moore RY (1984). Afferent connections of the ventral lateral geniculate nucleus in the rat: an HRP study. Brain Res 310, 367–370. [DOI] [PubMed] [Google Scholar]
- De la Huerta I, Kim IJ, Voinescu PE, and Sanes JR (2012). Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci U S A 109, 17663–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, and Huberman AD (2013). Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J Neurosci 33, 17797–17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, and Huberman AD (2014). Retinal ganglion cell maps in the brain: implications for visual processing. Curr Opin Neurobiol 24, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun NJ, Dun SL, Wong PY, Yang J, and Chang J (2000). Cocaine- and amphetamine-regulated transcript peptide in the rat epididymis: an immunohistochemical and electrophysiological study. Biol Reprod 63, 1518–1524. [DOI] [PubMed] [Google Scholar]
- Elstrott J, Anishchenko A, Greschner M, Sher A, Litke AM, Chichilnisky EJ, and Feller MB (2008). Direction selectivity in the retina is established independent of visual experience and cholinergic retinal waves. Neuron 58, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, and Easter SS Jr. (2005). Cell birth and death in the mouse retinal ganglion cell layer. J Comp Neurol 489, 120–134. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, and Muller U (2012). Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 337, 746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JL, and Chen B (2013). Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron 80, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington ME (1997). The ventral lateral geniculate nucleus and the intergeniculate leaflet: interrelated structures in the visual and circadian systems. Neurosci Biobehav Rev 21, 705–727. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, and Yau KW (2002). Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, and Barres BA (2008). Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron 59, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, and Barres BA (2009). Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron 62, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KH, and Reichert H (2015). Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res 359, 33–45. [DOI] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, and Sanes JR (2011). Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci 31, 7753–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner D, Liu H, Cheng CW, Demas J, Cheng SH, Hui CC, Chow RL, and Wong RO (2008). Genetic control of circuit function: Vsx1 and Irx5 transcription factors regulate contrast adaptation in the mouse retina. J Neurosci 28, 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, and Cepko CL (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci 2, 109–118. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, and Svoboda K (2008). Genetic dissection of neural circuits. Neuron 57, 634–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, and Klein WH (2008). Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development 135, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Li H, Zhang Z, Kiyama T, Panda S, Hattar S, Ribelayga CP, Mills SL, and Wang SW (2014). T-box transcription regulator Tbr2 is essential for the formation and maintenance of Opn4/melanopsin-expressing intrinsically photosensitive retinal ganglion cells. J Neurosci 34, 13083–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH (2012). The neuronal organization of the retina. Neuron 76, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Blanchard JH, and Provencio I (2003). Retinal ganglion cell projections to the hamster suprachiasmatic nucleus, intergeniculate leaflet, and visual midbrain: bifurcation and melanopsin immunoreactivity. J Comp Neurol 465, 401–416. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Hempel C, and Sugino K (2006). Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol 16, 571–576. [DOI] [PubMed] [Google Scholar]
- Ogata G, Stradleigh TW, Partida GJ, and Ishida AT (2012). Dopamine and full-field illumination activate D1 and D2-D5-type receptors in adult rat retinal ganglion cells. J Comp Neurol 520, 4032–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, El-Danaf RN, Nguyen PL, and Huberman AD (2014). Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep 8, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu SW, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. (2011). Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 71, 632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak W, Hindges R, Lim YS, Pfaff SL, and O'Leary DD (2004). Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell 119, 567–578. [DOI] [PubMed] [Google Scholar]
- Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, and Kay SA (2002). Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298, 2213–2216. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, and Doe CQ (2004). Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol 20, 619–647. [DOI] [PubMed] [Google Scholar]
- Pfisterer U, Wood J, Nihlberg K, Hallgren O, Bjermer L, Westergren-Thorsson G, Lindvall O, and Parmar M (2011). Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle 10, 3311–3316. [DOI] [PubMed] [Google Scholar]
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O'Leary DD, Goulding M, and Turner EE (2005). Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci 25, 11595–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazifar H, Jia Y, Chen J, Lynch G, and Huang T (2014). Chemically induced specification of retinal ganglion cells from human embryonic and induced pluripotent stem cells. Stem Cells Transl Med 3, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Qiao M, Kagan RD, Yamagata M, Palmiter RD, and Sanes JR (2016). Two Pairs of ON and OFF Retinal Ganglion Cells Are Defined by Intersectional Patterns of Transcription Factor Expression. Cell Rep 15, 1930–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluch VM, Davis CH, Ranganathan V, Kerr JM, Krick K, Martin R, Berlinicke CA, Marsh-Armstrong N, Diamond JS, Mao HQ, et al. (2015). Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci Rep 5, 16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soula C, Danesin C, Kan P, Grob M, Poncet C, and Cochard P (2001). Distinct sites of origin of oligodendrocytes and somatic motoneurons in the chick spinal cord: oligodendrocytes arise from Nkx2.2-expressing progenitors by a Shh-dependent mechanism. Development 128, 1369–1379. [DOI] [PubMed] [Google Scholar]
- Straznicky C, Vickers JC, Gabriel R, and Costa M (1992). A neurofilament protein antibody selectively labels a large ganglion cell type in the human retina. Brain Res 582, 123–128. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, and Feldheim DA (2014). Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci 34, 5447–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, and Jessell TM (1996). Diversity and pattern in the developing spinal cord. Science 274, 1115–1123. [DOI] [PubMed] [Google Scholar]
- Trenholm S, Johnson K, Li X, Smith RG, and Awatramani GB (2011). Parallel mechanisms encode direction in the retina. Neuron 71, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JW, Wei W, Gonzalez C, Sweeney NT, Huberman AD, Feller MB, and Feldheim DA (2014). Dendritic and axonal targeting patterns of a genetically-specified class of retinal ganglion cells that participate in image-forming circuits. Neural Dev 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha NA, Garcia-Moreno F, Arora S, Cheung AF, Arnold SJ, Robertson EJ, and Molnar Z (2015). Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cereb Cortex 25, 3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, and Sanes JR (2009). Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol 517, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fox MA, and Povlishock JT (2013). Diffuse traumatic axonal injury in the optic nerve does not elicit retinal ganglion cell loss. J Neuropathol Exp Neurol 72, 768–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (1995). Target-independent diversification and target-specific projection of chemically defined retinal ganglion cell subsets. Development 121, 3763–3776. [DOI] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Sudhof TC, and Wernig M (2011). Induced neuronal cells: how to make and define a neuron. Cell Stem Cell 9, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]