Abstract
The so-called symbiotic region of the Bradyrhizobium japonicum chromosome (C. Kündig, H. Hennecke, and M. Göttfert, J. Bacteriol. 175:613–622, 1993) was screened for the presence of genes controlled by the nitrogen fixation regulatory protein NifA. Southern blots of restriction enzyme-digested cosmids that represent an ordered, overlapping library of the symbiotic region were competitively hybridized with in vitro-labeled RNA from anaerobically grown wild-type cells and an excess of RNA isolated either from anaerobically grown nifA and rpoN mutant cells or from aerobically grown wild-type cells. In addition to the previously characterized nif and fix gene clusters, we identified three new NifA-regulated genes that were named nrgA, nrgB, and nrgC (nrg stands for NifA-regulated gene). The latter two probably form an operon, nrgBC. The proteins encoded by nrgC and nrgA exhibited amino acid sequence similarity to bacterial hydroxylases and N-acetyltransferases, respectively. The product of nrgB showed no significant similarity to any protein with a database entry. Primer extension experiments and expression studies with translational lacZ fusions revealed the presence of a functional −24/−12-type promoter upstream of nrgA and nrgBC and proved the NifA- and RpoN (ς54)-dependent transcription of the respective genes. Null mutations introduced into nrgA and nrgBC resulted in mutant strains that exhibited wild-type-like symbiotic properties, including nitrogen fixation, when tested on soybean, cowpea, or mung bean host plants. Thus, the discovery of nrgA and nrgBC further emphasizes the previously suggested role of NifA as an activator of anaerobically induced genes other than the classical nitrogen fixation genes.
Nitrogen-fixing rhizobia belonging to any of the four genera Azorhizobium, Bradyrhizobium, Rhizobium, and Sinorhizobium are able to establish an endosymbiotic interaction with specific leguminous host plants. The transition from the free-living to the symbiotic life style is initiated by the exchange of specific signal molecules between compatible symbiotic partners. Eventually this leads to the formation of root nodules (or in some instances stem nodules) hosting the bacterial partner as an intracellular microsymbiont (for reviews, see references 10 and 57). The induction of a number of symbiotic genes, including those specifying the nitrogen fixation apparatus, is coordinated together with nodule development via the micro-oxic conditions prevailing in the central nodule tissue (16, 53). Perception and transduction of the low-oxygen signal are mediated by conserved regulatory proteins that are integrated into species-specific networks in different rhizobia (15, 16, 32).
Two oxygen-responsive regulatory cascades are present in the soybean symbiont Bradyrhizobium japonicum, the FixLJ-FixK2 cascade and the RegSR-NifA cascade. In response to low-oxygen conditions, the FixJ protein of the FixLJ two-component regulatory pair becomes phosphorylated and activates expression of the subordinate regulatory gene fixK2, which, in turn, controls a number of functions associated with microaerobic or anaerobic metabolism (42). The environmental signal for the two-component system of the second cascade, RegSR (5), is not yet known; however, the activity of the transcriptional activator NifA, whose expression is partially controlled by RegR, is directly affected by the oxygen status (16). Among the targets of NifA are eight nif genes that are directly involved in nitrogen fixation and also the fixRnifA operon, which is subject to NifA-dependent autoregulation under low-oxygen conditions (5, 15). Furthermore, NifA controls expression of the fixA and fixBCX genes, which are essential for symbiotic nitrogen fixation.
NifA activates gene expression in concert with RNA polymerase containing the specialized ς factor ς54, which enables the core polymerase to recognize −24/−12-type promoters. Notably, two highly conserved genes encoding ς54 (rpoN1 and rpoN2) are present in B. japonicum (36). Mutant analysis showed that their products can functionally replace each other with regard to their role in nitrogen fixation. NifA normally binds to upstream activator sequences (UAS) and interacts with the RNA polymerase holoenzyme via loop formation by the intervening DNA. DNA bending may be facilitated by the integration host factor bound to a site located between the UAS and the core promoter region. Transcription is initiated by productive interaction of the holoenzyme with NifA, catalyzing open complex formation in an ATP-dependent reaction (see reference 11 and references therein).
The key role of B. japonicum NifA in symbiotic nitrogen fixation is documented by the pleiotropic phenotype of nifA mutants. Such mutants not only fail to fix nitrogen but also elicit numerous small nodules whose necrotic interior is reminiscent of a hypersensitive response characteristic of noncompatible host-pathogen interactions (17, 34, 54). On the basis of this observation, we speculated that in the wild type, NifA may control as-yet-unknown bacterial genes involved in the suppression of a potential plant defense reaction and in the maintenance of a balanced host-symbiont interaction. In the search for such genes, we found two new NifA-dependent targets, namely, a chaperonin-encoding operon (groESL3) (18) and a promoter (ndp) which is not closely associated with an obvious gene (58). Yet, neither of the two is essential for symbiosis.
In the present work, we have applied competitive RNA-DNA hybridization to explore the global regulatory scope of NifA. Our analysis was focused on a genomic region of approximately 400 kb of the 8,700-kb B. japonicum chromosome, as it turned out that many symbiotic genes are clustered in this region (the symbiotic region) (38). We speculated, therefore, that additional NifA targets might be located there. Moreover, this region was represented in an ordered cosmid library that was available in our laboratory and whose nucleotide sequence is currently being determined (M. Göttfert, unpublished data). The NifA-dependent transcription pattern of the symbiotic region was analyzed by the competitive hybridization method (14, 45), which led to the identification of three novel NifA-regulated genes (termed nrg) of B. japonicum.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories, Inc., Gaithersburg, Md. |
| S17-1 | Smr SprhsdR (RP4-2 kan::Tn7 tet::Mu, integrated in the chromosome) | 52 |
| B. japonicum | ||
| 110spc4 | Spr wild type | 49 |
| A9 | Spr KmrnifA::aphII | 17 |
| N50-97 | Spr Kmr SmrrpoN1::aphII rpoN2::Ω | 36 |
| 8236 | Spr KmrnrgA::aphII (opposite orientation) | This work |
| 8620 | Spr KmrnrgBC::aphII | This work |
| 8233a | Spr TcrnrgA′-′lacZ chromosomally integrated | This work |
| 8626 | Spr Tcrorf110′-′lacZ chromosomally integrated | This work |
| 8627a | Spr TcrnrgB′-′lacZ chromosomally integrated | This work |
| 8628a | Spr TcrnrgC′-′lacZ chromosomally integrated | This work |
| Plasmids | ||
| pUC18 | Apr | 43 |
| pSUP202 | Apr Cmr TcroriT from RP4 | 52 |
| pSUP202pol4 | Tcr (pSUP202) part of polylinker from pBluescript II KS+ between EcoRI and PstI | 18 |
| pSUP480 | Tcr, ′lacZ part from pNM480 in pSUP202pol4 | H. M. Fischer, unpublished data |
| pSUP481 | Tcr, ′lacZ part from pNM481 in pSUP202pol4 | H. M. Fischer, unpublished data |
| pBSL14/15 | Apr Kmr | 2 |
| pRJ8227 | Apr (pUC18) B. japonicum 1.8-kb EcoRI fragment of cosmid 11; nrgA | This work |
| pRJ8610 | Apr (pUC18) B. japonicum 11.5-kb EcoRI fragment from cosmid 16; groESL3 | This work |
| pRJ8611 | Apr (pUC18) B. japonicum 4.1-kb EcoRI fragment from cosmid 16; nrgBC | This work |
| Cosmids | ||
| Lorist6 | Kmr, bacteriophage λ replicon | 21 |
| Cosmids 6–18 | Kmr, ordered library of the symbiotic region in B. japonicum (Sau3A fragments of B. japonicum genomic DNA in Lorist6) | 37 |
The same nrgA′-, nrgB′-, and nrgC′-′lacZ fusions were also integrated into the chromosome of B. japonicum A9 and N50-97. The resulting strains were given the same numbers preceded by A (A9 derivatives) or N (N50-97 derivatives); see Table 3.
Media and growth conditions.
Luria-Bertani medium (40) was used for growth of Escherichia coli cells and contained the following concentrations (micrograms per milliliter) of antibiotics for plasmid selection: ampicillin, 200; kanamycin, 30; and tetracycline, 10. Peptone-salts-yeast extract (PSY) medium (49) supplemented with 0.1% l-arabinose was used for routine aerobic cultures of B. japonicum, whereas yeast extract-mannitol (YEM) medium supplemented with 10 mM KNO3 (8) was used for anaerobic B. japonicum cultures and corresponding aerobic control cultures. Anaerobic cultures were kept under argon in rubber-stoppered serum bottles. Concentrations (micrograms per milliliter) of antibiotics for use in B. japonicum cultures were as follows: spectinomycin, 100; kanamycin, 100; streptomycin, 50; and tetracycline, 50 (solid media) or 25 (liquid media).
DNA work and sequence analysis.
Recombinant DNA work and Southern blotting were performed according to standard protocols (50). For homologous hybridizations, we used digoxigenin-labeled probes generated by PCR or by elongation of random hexanucleotides with the Klenow fragment of DNA polymerase (DIG DNA Labeling Kit; Roche Diagnostics, Rotkreuz, Switzerland). B. japonicum chromosomal DNA was isolated as described previously (26). For computer-assisted analyses of DNA and protein sequences, we used the software package (version 8) of the Genetics Computer Group of the University of Wisconsin, Madison), and the MAC program DNA-STRIDER, version 1.2. Homology searches were performed by using the National Center for Biotechnology Information BLAST network server (http://www.ncbi.nlm.nih.gov/BLAST/).
RNA extraction and labeling.
B. japonicum cells were grown anaerobically in 500-ml serum bottles filled with 400 ml of YEM medium for at least 4 to 7 days to an optical density (600 nm) of 0.2 to 0.4. Spectinomycin was the only antibiotic used in these cultures. After the cultures were cooled on ice-water, cells were harvested by centrifugation and washed with 0.9% (wt/vol) NaCl, and the cell pellets were immediately frozen in liquid nitrogen and stored at −80°C. For RNA isolation, 200 to 400 mg (wet weight) of cells was resuspended in 10 ml of cold 20 mM Na-acetate (pH 5.5)–1 mM EDTA plus 0.5% (wt/vol) (final concentration) sodium dodecyl sulfate (SDS), followed by extraction with 10 ml of prewarmed (65°C) acidic phenol (pH 5.5). The phenol extraction was repeated with 10 ml of phenol-CHCl3-isoamylalcohol (49.5:49.5:1), and RNA was ethanol precipitated. The samples were treated with RQ1 RNase-free DNase (5 U) (Promega, Madison, Wis.) for 15 min at 37°C to remove potential contaminating DNA. After an additional phenol extraction, the RNA was ethanol precipitated and dissolved in diethyl pyrocarbonate-treated H2O. RNA yields were approximately 100 μg per 100 mg (wet weight) of cells as determined by spectrophotometry. Radioactive end labeling of RNA was performed as described previously (14). RNA (15 to 22 μg) was partially hydrolyzed by incubation in NaOH (125 mM final concentration) for 25 min on ice and labeled with T4 polynucleotide kinase (20 U) (MBI Fermentas, Vilnius, Lithuania) and [γ-32P]ATP for 90 min at 37°C. Unincorporated label and low-molecular-weight RNA fragments were removed by gel filtration (Sephadex G-50; Amersham Pharmacia Biotech), and 5 × 107 to 10 × 107 cpm of labeled RNA was used for competitive hybridizations.
Competitive RNA-DNA hybridizations.
Competitive hybridizations were performed as described previously (14, 45) with minor modifications. At least 1 μg of each of 13 cosmids (numbered from 6 to 18) representing the symbiotic region of the B. japonicum chromosome was digested with EcoRI. The resulting DNA fragments were separated on 1% agarose gels and transferred by Southern blotting to Hybond-N nylon membranes (Amersham Pharmacia Biotech). Prehybridizations were performed in 30 ml of PHS solution (50 mM Tris-HCl [pH 7.4], 1 M NaCl, 1% SDS, 0.2% bovine serum albumin, 0.2% Ficoll 400, 0.2% polyvinylpyrollidone, 0.2% Na-pyrophosphate) at 65°C for 8 h with at least 130 to 200 μg of nonlabeled RNA isolated from anaerobically grown B. japonicum strain A9 or N50-97 or from aerobically grown wild-type cells. Subsequently, RNA isolated from anaerobically grown wild-type cells, end labeled as described above, was added to the prehybridization solution, and competitive hybridizations were performed at 65°C for 16 h. Membranes were washed three times for 30 min at 65°C in prewarmed 1× SSC (150 mM NaCl, 15 mM sodium citrate)–1% SDS (20 ml of solution per wash step) and finally for 15 min at room temperature in 20 ml of 0.2× SSC. Hybridizing bands were analyzed with a PhosphorImager (Molecular Dynamics) after exposure of the membranes for at least 24 h.
Transcript mapping.
The transcriptional start sites of nrgA and nrgBC were mapped with primer extension experiments. Two 30-mers were used as primers for nrgA mapping (oligonucleotide 8227-4, 5′-GTTTGCATTCGCACATTTGATATCCGACTC515-3′; oligonucleotide 8227-5, 5′-CCAAATTTTCTGTCTACCTGTCAGAGTTAC466-3′ [position numbers refer to those in the sequence deposited in the GenBank database]). A 28-mer (oligonucleotide 8611-1, 5′-CTCATACGTCGGACAAGCCGGGTCGAGC557-3′) and a 25-mer (oligonucleotide 8611-2, 5′-GGGCATGCGATGTCATGTCTTCTCC597-3′) were used for nrgBC mapping. RNA was isolated as described previously (3) from B. japonicum strains 110spc4 (wild type), A9 (nifA), and N50-97 (rpoN1/2) grown anaerobically for 5 days in YEM medium containing 10 mM KNO3 and, for a control, also from wild-type cells cultured aerobically for 3 days in the same medium. The latter cells had to be harvested by centrifugation at 14,000 × g because of the pronounced synthesis of extracellular slime. Approximately 5 μg of RNA and at least 100,000 cpm of radiolabeled primer (200 to 500 fmol) were used for each primer extension experiment, which was performed as described previously (3, 5). Extension products were purified by phenol extraction followed by ethanol precipitation before they were loaded on 6% denaturing polyacrylamide gels.
Construction of B. japonicum nrgA and nrgBC mutant strains.
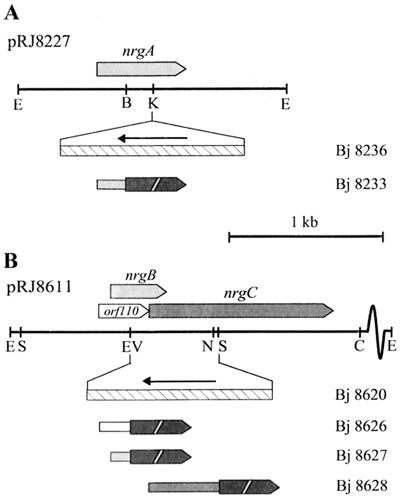
The nrgA gene was mutagenized by insertion of a 1.2-kb KpnI kanamycin resistance gene cassette (aphII), isolated from pBSL15, into its unique KpnI site (see Fig. 2A). The nrgBC genes were mutated by deleting a 0.56-kb EcoRV-NheI fragment, which was replaced by a NheI-SmaI aphII fragment from pBSL14 (see Fig. 2B). Appropriate DNA fragments containing the mutated nrgA or nrgBC genes were cloned into the vector pSUP202pol4 and mobilized into B. japonicum 110spc4 as described previously (26). Cointegrate-containing exconjugants (resulting from single crossover) were distinguished from true marker exchange mutants (resulting from double crossover) by the vector's tetracycline resistance. The correct genomic structures of all mutant strains were confirmed by Southern blot analysis of genomic DNAs.
FIG. 2.
Physical map of the EcoRI fragments carrying newly identified NifA-regulated genes. The orientation and location of nrgA on pRJ8227 (A) and of orf110 and nrgBC on pRJ8611 (B) are indicated. The structures of the nrgA and nrgBC mutations are shown along with the corresponding B. japonicum (Bj) strain numbers; hatched bars with horizontal arrows refer to inserted aphII (Kmr) cassettes and their orientation. Translational lacZ fusions to nrgA, orf110, nrgB, and nrgC are indicated by horizontal bars fused to black arrows (′lacZ) and are specified by the number of the strain harboring the respective chromosomally integrated fusion (Table 1). Relevant restrictions sites: B, BamHI; C, ClaI; E, EcoRI; EV, EcoRV; K, KpnI; N, NheI; S, SmaI.
Construction of chromosomally integrated nrgA′-, nrgB′-, and nrgC′-′lacZ fusions.
Translational lacZ fusions were constructed by making use of gene-internal restriction sites (see Fig. 2) and the mobilizable lacZ fusion vectors pSUP480 (for orf110 and nrgC) and pSUP481 (for nrgA and nrgB). The nrgA gene was fused at a BamHI site corresponding to Arg-70 in the predicted NrgA protein. The overlapping reading frames orf110 and nrgB were fused at their common EcoRV site corresponding to Asp-70 in the putative Orf110 protein and Ala-61 in the NrgB protein. The fusion to nrgC was constructed at a SmaI site corresponding to Pro-151 of the NrgC protein. The lacZ fusion constructs including appropriate portions of B. japonicum upstream DNA were conjugated into B. japonicum 100spc4 (wild type), A9 (nifA), and N50-97 (rpoN1/2). Those clones that contained the entire lacZ fusion plasmid integrated via single crossover at the homologous chromosomal position were selected by plating the exconjugants on tetracycline-containing plates. The genomic structures of all resulting strains were verified by Southern blot analysis.
β-Galactosidase assays.
β-Galactosidase activity assays were done as described previously (18).
Plant infection test.
The symbiotic phenotypes of the B. japonicum nrgA and nrgBC mutants were determined in infection tests using soybean [Glycine max (L.) Merr. cv. Williams], cowpea (Vigna unguiculata cv. Red Caloona), and mung bean (Vigna radiata) as host plants. The tests were performed as described previously (23, 26). Soybean seeds were kindly provided by P. M. Gresshoff (University of Queensland, Brisbane, Australia), whereas cowpea and mung bean seeds were kind gifts from W. D. Broughton (University of Geneva, Geneva, Switzerland).
Nucleotide sequence accessions numbers.
The nucleotide sequences of the B. japonicum nrgA and nrgBC genes have been deposited in the GenBank database under accession numbers AF190732 and AF190733, respectively.
RESULTS
Sections of the symbiotic region that are transcribed in a NifA-dependent manner.
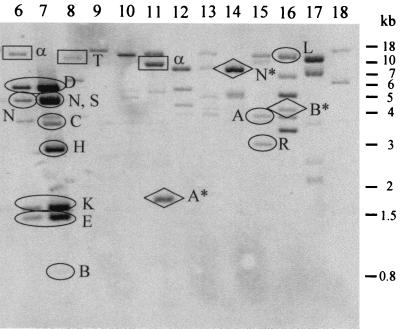
The competitive RNA-DNA hybridization technique described in Materials and Methods was used to probe the symbiotic region of B. japonicum for the presence of DNA segments that are transcribed under the control of NifA (Fig. 1). For comparison, we also performed analogous hybridization experiments with the rpoN1/2 mutant N50-97 and with competing RNAs from aerobically and anaerobically grown wild-type cells to detect regions whose transcription is dependent on RpoN or induced by anaerobiosis, respectively. The hybridization patterns were highly similar in all three experiments (data not shown).
FIG. 1.
Transcription pattern of the B. japonicum symbiotic region determined by competitive DNA-RNA hybridization. Radioactively labeled RNA from anaerobically grown wild-type cells was hybridized to membrane-bound, EcoRI-digested cosmid DNA in the presence of unlabeled competitor RNA isolated from anaerobically grown cells of nifA mutant A9 as described in Materials and Methods. Lane numbers refer to the respective cosmid numbers. Hybridizing EcoRI fragments carrying previously known NifA-dependent genes are marked with ellipses (class I genes [Table 2]), those with newly identified NifA-regulated genes are marked with diamonds (class II), and those with other, potentially NifA-dependent ORFs are marked with rectangles (class III). The hybridization signal of fragment B in lane 7 was observed only upon prolonged exposure times. Unmarked fragments were not further analyzed in this work. For explanation of letters, see Table 2.
The significance of the hybridization results was confirmed by the finding that many EcoRI fragments carrying previously characterized NifA- and RpoN-dependent nif and fix genes gave rise to strong hybridization signals. Examples include nif and fix genes of cluster I (15) present on cosmids 6 and 7 (Table 2). Similarly, the strong signal of the 11.5-kb EcoRI fragment of cosmid 16 could be assigned to the NifA-dependent groESL3 operon by subsequent hybridization experiments with appropriate subclones of this cosmid (data not shown).
TABLE 2.
Genes and ORFs located in NifA-dependently transcribed DNA regions identified by competitive hybridization as shown in Fig. 1
| Class | Location of hybridizing region
|
Hybridizing gene(s) or ORF(s) | Reference(s) or comments | |
|---|---|---|---|---|
| Cosmid(s)a | EcoRI fragment (kb)b | |||
| I (previously known NifA-dependent genes) | 6, 7 | D (5.5) | nifD′ | 30 |
| 6, 7 | K (1.6) | ′nifDK′ | 30, 55 | |
| 6, 7 | E (1.3) | ′nifKE′ | 1, 55 | |
| 6, 7 | N (4.8) | ′nifENX | 1 | |
| 7 | S (5.2) | nifS, nifB′ | 12 | |
| 7 | B (0.8) | ′nifBfrxA | 12, 13 | |
| 7 | H (2.8) | nifH | 19 | |
| 7 | C (4.1) | fixBC | 20 | |
| 15 | R (3.0) | fixRnifA′ | 56 | |
| 15 | A (3.8) | ′nifA, fixA | 24, 56 | |
| 16 | L (11.5) | groESL3 | 18 | |
| II (newly identified NifA-dependent genes) | 11 | A* (1.8) | nrgA | This work; NrgA homologous to N-acetyltransferases |
| 14 | N* (8.1) | hemN1 | HemN1 homologous to coproporphyrinogen III dehydrogenase | |
| 16 | B* (4.1) | nrgBC | This work; NrgC homologous to hydroxylases | |
| III (putative NifA-dependent ORFsc) | 6 | α (11) | orf355-1 | This work; product 99% identical to product of orf355 present on RSRjα9 (48) |
| 8 | T (10.8) | orf228′ | This work; product 75% identical to product of orf228 (transposase) present on B. japonicum IS1632 (accession no. AB003134) | |
| 11 | α (9) | orf355-2 | This work; product 99% identical to product of orf355 present on RSRjα9 (48) | |
Based on (partial) sequence analyses and further hybridization with suitable subclones of selected EcoRI fragments, we divided NifA-dependent genes and open reading frames (ORFs) into three classes (Table 2). Class I comprises 16 previously known NifA-dependent genes. Class II consists of four newly identified genes, nrgA, nrgB, nrgC, and hemN1. A detailed study of nrgA and nrgBC is reported here. The analysis of the hemN1 gene will be presented elsewhere; briefly summarized, it turned out that B. japonicum possesses two hemN-like genes and that the hemN1 gene identified in this work apparently encodes a nonfunctional protein whose synthesis is controlled only partially by NifA and predominantly by FixK2. The alternative hemN gene, hemN2, whose sequence was previously deposited in the GenBank database (accession number AJ002517.1) is located outside the symbiotic gene region. Class III (Table 2) includes three ORFs (orf355-1, orf355-2, and orf228) whose deduced products are highly homologous to the products of the previously reported B. japonicum orf355 (48) and orf228 (GenBank accession no. AB003134). orf228, which is located on the insertion sequence element IS1632 of B. japonicum, is believed to encode a transposase. The potential NifA-dependent transcription and function(s) of class III ORFs were not studied in greater detail (see also Discussion).
Identification of nrgA and nrgBC.
Our work then focused on the analysis of the hybridizing 1.8- and 4.1-kb EcoRI fragments of cosmids 11 and 16, which were subcloned into pUC18, resulting in plasmids pRJ8227 and pRJ8611, respectively (Fig. 2). Sequence analysis of pRJ8227 revealed an ORF, named nrgA, which specifies a predicted protein of 195 amino acids and a molecular mass of 21,466 Da. The hybridizing region on pRJ8611 was narrowed down to a 2.2-kb EcoRI-ClaI fragment, whose nucleotide sequence revealed three partially overlapping ORFs (orf110, nrgB, and nrgC). They encode putative proteins of 110 (orf110), 121 (nrgB), and 388 (nrgC) amino acids with molecular masses of 11,937, 14,004, and 41,013 Da, respectively. The nrgB and nrgC genes probably form an operon, since the two genes overlap by 20 codons and no obvious promoter was detected immediately upstream of nrgC. Moreover, nrgB and nrgC appeared to be coregulated as deduced from our studies with respective lacZ fusions (see below). While database searches revealed no entries with significant similarity to the products of orf110 and nrgB, the NrgA and NrgC proteins were found to be homologous to bacterial N-acetyltransferases and hydroxylases, respectively (see Discussion).
Transcriptional analysis of nrgA and nrgBC.
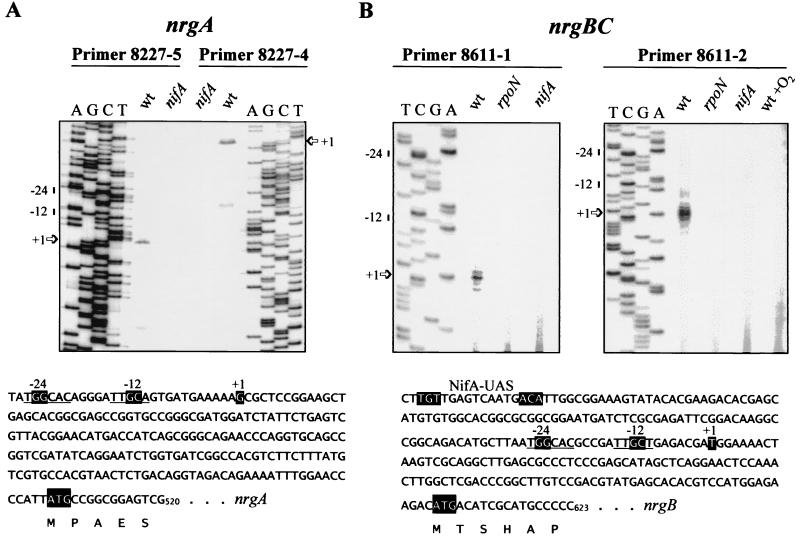
By inspection of DNA regions upstream of both the nrgA and orf110-nrgBC coding regions, we found DNA sequences that showed strong similarity to −24/−12-type promoters (nrgA, T287GGCAC-N5-TTGCA302 [Fig. 3A]; orf110-nrgBC, T474GGCAC-N5-TTGCT489 [Fig. 3B]). Moreover, a putative binding site for the transcriptional activator protein NifA was present at an appropriate distance of approximately 100 bp upstream of the presumptive nrgBC promoter (T364GT-N10-ACA379 [Fig. 3B]). No consensus NifA binding motif was found upstream of nrgA. Interestingly, a perfect copy of this motif is present in the 5′ coding region of nrgA (T530GT-N10-ACA545), yet its functional role at this unorthodox position is questionable.
FIG. 3.
Mapping of the transcription start sites of B. japonicum nrgA (A) and nrgBC (B) by primer extension. Total RNA was purified from anaerobically grown cells of wild-type B. japonicum (wt) or mutant strain A9 (nifA) or N50-97 (rpoN1/2) and used for primer extension experiments with the primers indicated, as described in Materials and Methods. An additional control experiment was performed with RNA isolated from aerobically grown wild-type cells and primer 8611-2 (wt +O2). The sequence ladders shown were generated with pRJ8227 (A) and pRJ8611 (B) plasmid DNA and the same primers used for the respective transcript mapping. The dominant transcription start sites for nrgA and nrgBC are marked by +1 and arrows. The nucleotide sequences of relevant DNA regions are shown at the bottom. A potential binding site for NifA (NifA-UAS), the −24/−12-type promoter core sequences, transcriptional start sites (+1), and putative translational start codons (ATG) are shown by white letters. Additional conserved nucleotides of −24/−12-type promoters are underlined. The numbering of nucleotide positions corresponds to that of the sequences deposited in the GenBank database.
The function of the putative promoters was confirmed by primer extension experiments using two different oligonucleotides for each of them (see Materials and Methods). Anaerobically grown cells of wild-type B. japonicum and of mutant strains A9 (nifA) and N50-97 (rpoN1/2) were used as sources for the isolation of template RNA. The results of these experiments are shown in Fig. 3. The 3′ end of the dominant elongation product obtained with the primers for nrgA and wild-type RNA corresponded to G314, which is located at the appropriate distance of 11 nucleotides downstream of the predicted −24/−12 promoter of nrgA (Fig. 3A). The minor elongation product ending at C301 is not associated with an obvious promoter and may have resulted from premature termination of the reverse transcription reaction. Regardless of the primer used, no primer extension product was obtained with RNA isolated from the nifA mutant A9. The results of primer extension experiments with orf110-nrgBC led to similar conclusions (Fig. 3B). The elongation products which were obtained with both primers and wild-type RNA, but not with RNA from the nifA or rpoN1/2 mutant, indicated the existence of a NifA- and RpoN-dependent transcript starting at T497, i.e., at a correct distance from the predicted promoter. Its dependence on the oxygen-labile NifA protein was further documented by the absence of an elongation product in the experiment with primer 8611-2 and RNA from aerobically grown wild-type cells (Fig. 3B, right panel).
Analysis of nrgA, orf110, and nrgBC expression with lacZ fusions.
Translational lacZ fusions to nrgA, nrgB, nrgC, and orf110 were constructed as described in Materials and Methods (see also Fig. 2) in order to quantitate expression from the −24/−12-type promoters identified above and also to test whether the ORFs and genes under investigation were translated. The fusions to nrgA, nrgB, and nrgC were integrated into the chromosome of wild-type B. japonicum and mutants A9 (nifA) and N50-97 (rpoN1/2). The orf110′-′lacZ fusion was introduced only into the wild type. Cells of all strains were grown under aerobic or anaerobic conditions, and β-galactosidase activity was determined (Table 3).
TABLE 3.
Expression of chromosomally integrated B. japonicum nrgA′-, nrgB′-, nrgC′-, and orf110′-′lacZ fusions in cells grown under different oxygen conditionsa
| Strain | Relevant genotype | β-Galactosidase activity (Miller Units)b
|
|
|---|---|---|---|
| Aerobicc | Anaerobicd | ||
| 8233 | nrgA′-′lacZ | 1.0 ± 0.8 | 188 ± 64 |
| A8233 | nrgA′-′lacZ nifA | 0.4 ± 0.0 | 2.5 ± 1.4 |
| N8233 | nrgA′-′lacZ rpoN1/2 | 0.5 ± 0.2 | 1.5 ± 1.0 |
| 8626 | orf110′-′lacZ | 0.3 ± 0.0 | 0.4 ± 0.6 |
| 8627 | nrgB′-′lacZ | 0.5 ± 0.0 | 117 ± 64 |
| A8627 | nrgB′-′lacZ nifA | 0.3 ± 0.0 | 0.3 ± 0.0 |
| N8627 | nrgB′-′lacZ rpoN1/2 | 0.3 ± 0.0 | 1.3 ± 1.0 |
| 8628 | nrgC′-′lacZ | 0.4 ± 0.0 | 161 ± 84 |
| A8628 | nrgC′-′lacZ nifA | 0.4 ± 0.0 | 1.2 ± 1.0 |
| N8628 | nrgC′-′lacZ rpoN1/2 | 0.3 ± 0.1 | 1.2 ± 0.8 |
The wild-type strain without an integrated lacZ fusion showed <1.5 Miller units under all growth conditions.
Numbers are mean vs ± standard errors from two (aerobic) or at least three (anaerobic; two independent experiments) cultures of individual strains, which were assayed in duplicate.
Aerobic cultures were grown for 3 days in PSY medium containing 100 μg of spectinomycin per ml.
Anaerobic cultures were grown for 6 days in YEM medium containing 10 mM KNO3 and 100 μg of spectinomycin per ml.
Regardless of the genetic background, no significant expression of any lacZ fusion was detected when cells were grown aerobically. By contrast, β-galactosidase activity derived from the fusions to nrgA, nrgB, and nrgC was drastically induced in the wild-type background under anaerobic conditions (∼190-, ∼340-, and ∼450-fold increases, respectively). No activities were detectable in the nifA or rpoN1/2 mutant strain grown under these conditions. No β-galactosidase activity was measurable also in strains harboring the orf110′-′lacZ fusion, from which we conclude that orf110 is not translated.
Symbiotic phenotypes of nrgA and nrgBC mutants.
The potential roles of nrgA and nrgBC in symbiotic nitrogen fixation were studied in infection tests with mutant strains 8236 (nrgA) and 8620 (nrgBC) (Fig. 2), using soybean, cowpea, and mung bean as the host plants. Nodulation and nitrogen fixation activity of 6 to 14 plants were evaluated 3 weeks after infection. The nitrogen fixation activities of both mutants did not differ significantly from that of the wild type on all three host plants tested (between 91 and 122% of wild-type Fix activity). The same result was found with regard to the size, the morphology, and the interior color of nodules, except for strain 8620, which elicited an increased number of nodules on cowpea (42 ± 12 and 21 ± 5 nodules for strain 8620 and the wild type, respectively). Thus, the products of nrgA and nrgBC are not essential for an effective B. japonicum-host plant symbiosis.
DISCUSSION
The DNA-RNA hybridization approach: advantages and drawbacks.
In the present study, we have applied competitive DNA-RNA hybridization to screen the symbiotic region of the B. japonicum chromosome for sections that are transcribed under the control of the oxygen-responsive regulator NifA. An original version of this method was based on differential DNA-cDNA hybridization of an ordered E. coli cosmid library (35), and it was applied to monitor alterations in the global transcription pattern in response to various external stimuli or regulatory mutations (7). Subsequently, modifications were made to this technique, including competitive DNA subtraction hybridization and DNA-RNA hybridization, and were used by several authors for the detection of differentially expressed, often symbiotic, genes in different rhizobia (6, 9, 14, 22, 45, 46). The success of our approach is documented by the identification of three new B. japonicum genes, nrgA, nrgB, and nrgC, which are associated with a ς54-dependent −24/−12-type promoter that is activated by NifA. With the two newly identified promoters, a total of 11 transcriptionally mapped NifA−/ς54-dependent promoters in the symbiotic region of B. japonicum are now known (15, 18, 38, and 58). A comparable number of 16 (putative) NifA-/ς54-dependent promoters were found in a transcriptional survey of the 536-kb symbiotic plasmid of Rhizobium sp. strain NGR234 (46).
Critical prerequisites for our successful screening included the availability of (i) an ordered cosmid library representing the symbiotic region, (ii) regulatory nifA and rpoN1/2 mutants used as a source for the competing RNA, and (iii) the emerging DNA sequence information originating from a partial genomic sequencing project (M. Göttfert, unpublished data). Probably we have not yet fully exploited the potential of the screening, as indicated by numerous hybridizing fragments that were not further characterized in this work. Additional candidate genes potentially controlled by NifA include, for example, nifV- and nifM-like genes whose products are involved in FeMo cofactor synthesis and activation of the nitrogenase Fe protein in free-living diazotrophs (reference 29 and references therein). Intriguingly, no rhizobial homologues of these genes have been identified so far.
Surprisingly, we have identified as many as approximately 35 specifically hybridizing EcoRI fragments within the approximately 400 kb of genomic B. japonicum DNA represented by the 13 cosmids. However, this rather large number does not reflect truly disparate NifA-dependent loci, because we could show in at least two cases that the repetitive element RSRjα was responsible for the observed hybridization signal (data not shown). Given the facts that the RSRjα elements are extremely well conserved and that several of them are located in the symbiotic region (25, 31), they have the potential to yield multiple hybridizing fragments. Hence, repetitive sequences which are transcribed in a NifA-dependent manner would render our approach less useful than initially anticipated.
All EcoRI fragments hybridizing in the experiment with competing RNA from the nifA mutant were also detected in the analogous experiment with competing RNA from the rpoN1/2 mutant (data not shown). This was to be expected in the light of the compulsory dependence on RpoN (ς54) of all NifA-activated promoters. The absence of a clear qualitative difference in the overall hybridization pattern in these two experiments indicated that the large majority of RpoN-dependent promoters in the symbiotic region are indeed activated by NifA and probably not by other activators working in concert with the ς54 RNA polymerase, such as NtrC. The only exception might be a gene present on the 10-kb EcoRI fragment of cosmid 13 that showed a strong differential hybridization with rpoN but not with nifA mutant RNA. Another notable aspect is that the hybridization pattern in the experiment with RNA from aerobically and anaerobically grown wild-type cells resembled very much that observed in both of the other hybridization experiments. We interpret this to mean that in the DNA region investigated, transcriptional activation in response to anaerobiosis is predominantly brought about by NifA and not by another oxygen-controlled regulator. This finding was not necessarily predictable, because at least one additional oxygen-responsive regulation system exists in B. japonicum, i.e., the FixLJ-FixK2 cascade (42). Interestingly, with hemN1 a gene was identified that apparently belongs to both the FixLJ-FixK2 and the RegSR-NifA regulons. At least one additional member of the FixLJ-FixK2 regulon namely, rpoN1 (38), had been previously mapped to the symbiotic region, so in theory this gene ought to have been detected in our hybridization experiment. Reasons why this was not the case could be a weak expression of rpoN1, an interference with NifA- and RpoN-dependent genes on the same EcoRI fragment, or a cross-hybridization with the highly similar, constitutively synthesized rpoN2 mRNA.
What is the role of the new Nrg proteins?
The question regarding the potential function of the newly identified genes nrgA, nrgB, and nrgC was addressed by phenotypic analyses of appropriate mutants and by database homology searches. The results from the plant infection tests clearly indicated that, under the applied laboratory conditions, none of these genes is essential for nitrogen fixation in symbiosis with the three hosts tested. Notably, nodulation of cowpea by the nrgBC null mutant 8620 seems to be slightly disturbed as indicated by the elevated number of nodules. Principally, we cannot rule out that additional subtle effects of the nrg mutations might be detected under more competitive field conditions. It was shown recently that disruption of the Sinorhizobium meliloti phbC gene, encoding poly-β-hydroxybutyrate synthase, resulted in a mutant that was outcompeted by the wild type in a mixed infection test, even though its ability to fix nitrogen was not affected (59). Alternatively, one could hypothesize that the function of the nrgA or nrgBC gene products in the respective mutants is replaced by potential homologues. However, Southern blot hybridization experiments performed under low-stringency conditions with suitable nrg probes were not indicative of this possibility (data not shown).
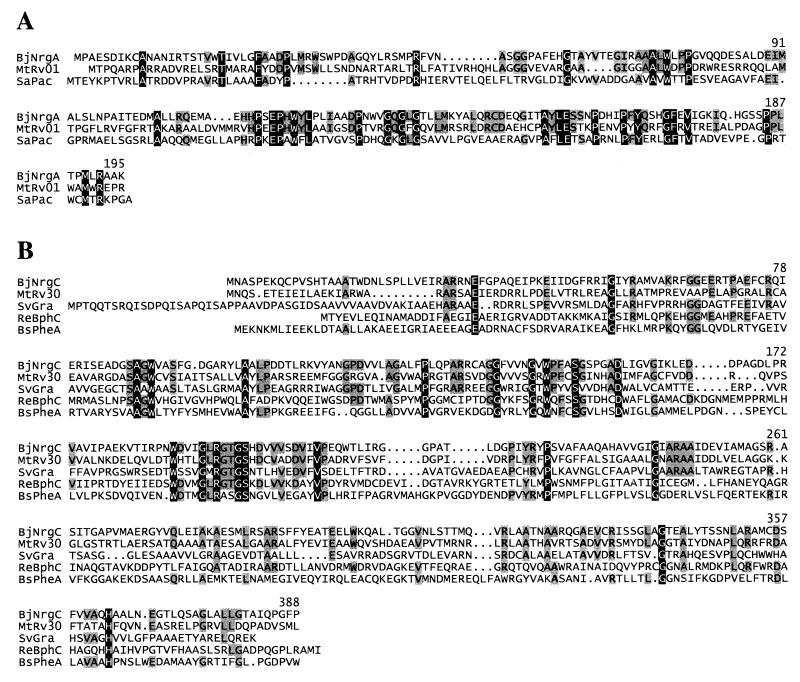
Searches with NrgA revealed that this predicted protein (195 amino acids) is similar to a number of N-acetyltransferases among which similarity to a puromycin N-acetyltransferase of Streptomyces anulatus was greatest (195 amino acids; 23% identity and 47% similarity [Fig. 4A]). Maximal similarity was found with a hypothetical protein of unknown function in Mycobacterium tuberculosis (201 amino acids; 33% identity and 53% similarity). Unfortunately, it was not possible to test the potential involvement of NrgA in puromycin resistance due to the intrinsic resistance of B. japonicum against this antibiotic (100 μg/ml). At any rate, it seems not compelling that NrgA is equivalent to a puromycin N-acetyltransferase, because the general similarity to several different N-acetyltransferases, particularly in the C-terminal half of the protein, allows for many possible substrates that could be envisaged for N acetylation by NrgA.
FIG. 4.
Amino acid sequence alignments of the predicted B. japonicum NrgA and NrgC proteins to their best homologues. (A) Alignment of NrgA (BjNrgA) to M. tuberculosis hypothetical protein Rv0133 (MtRv01; accession no. CAB07039) and to S. anulatus puromycin N-acetyltransferase (SaPac; accession no. P13249). (B) Alignment of NrgC (BjNrgC) to M. tuberculosis hypothetical protein Rv3094c (MtRv30; accession no. CAB08386), a putative S. violaceoruber hydroxylase (SvGra; accession no. CAA09642), an R. erythropolis hydroxylase (ReBphC; accession no. BAA25602), and a B. stearothermophilus phenol hydroxylase (BsPheA; accession no. AAA85688). Identical amino acids are emphasized by white letters; those amino acids of the Nrg proteins that are identical in at least one homologue of NrgA and two homologues of NrgC are highlighted in gray. Numbering refers to amino acid positions in the NrgA (A) and NrgC (B) proteins.
The search with NrgC (388 amino acids) identified hydroxylases from different bacteria as the homologous proteins (Fig. 4B). Examples include a putative hydroxylase of Streptomyces violaceoruber (400 amino acids; 26% identity and 49% similarity), a hydroxylase of Rhodococcus erythropolis (393 amino acids; 24% identity and 49% similarity), and a phenol hydroxylase of Bacillus stearothermophilus (400 amino acids; 23% identity and 46% similarity). Some of these proteins were shown to have indole oxidation activity in a colorimetric plate test (27, 33). When we applied this test to microaerobically grown B. japonicum wild-type and nrgBC mutant cells, no indole oxidation activity was detectable in either strain. Thus, the function of NrgC and the substrate that might be hydroxylated by NrgC remain obscure. Interestingly, as in the case of NrgA, the most similar protein to NrgC was a hypothetical, functionally undefined protein from M. tuberculosis (376 amino acids; 34% identity and 56% similarity).
It is interesting that by and large, the products of both nrgA and nrgC display similarity to enzymes that modify potentially toxic compounds. It is known that during the early stages of the symbiotic rhizobium-legume interaction, the host plant induces the synthesis of low-molecular-weight, phenolic compounds (e.g., phytoalexins) which are synthesized in response to pathogenic interactions and have antimicrobial activity (51; for reviews, see references 4 and 47). Moreover, it was shown previously that glyceollin, the phytoalexin of soybean, is present at elevated levels in nodules elicited by a B. japonicum nifA mutant (44). Therefore, it appears attractive to speculate that the NrgA and/or NrgC proteins contribute to overcoming the plant defense response. However, even if this is the case, the results from our plant infection tests imply that such a hypothetical function cannot be essential for the formation of a productive symbiotic interaction.
NifA, a global anaerobic regulator rather than a nitrogen fixation-specific regulator in rhizobia.
The identification of nrgA and nrgBC corroborates our earlier notion that NifA control is not restricted to genes directly concerned with nitrogen fixation (15). Other examples include the groESL3 operon and the glnII gene of B. japonicum (18, 39), rhizopine biosynthetic genes (mos) of S. meliloti (41), and the tyrosinase (polyphenol oxidase) structural gene melA of Rhizobium etli (28). Thus, NifA in rhizobia is a general regulator controlling microaerobically induced functions which may or may not be related to symbiosis. This implies that additional targets for NifA control might well be located outside the symbiotic region of the B. japonicum chromosome. Their identification could now be attempted by applying the method used in this study to a cosmid library representing the entire genome.
ACKNOWLEDGMENTS
We are grateful to Rémy Fellay for helpful experimental advice. Franziska Biellmann, Roger Frei, and Michael Spring are acknowledged for excellent technical assistance.
This work was supported by a grant from the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Aguilar O M, Taormino J, Thöny B, Ramseier T, Hennecke H, Szalay A A. The nifEN genes participating in FeMo cofactor biosynthesis and genes encoding dinitrogenase are part of the same operon in Bradyrhizobium species. Mol Gen Genet. 1990;224:413–420. doi: 10.1007/BF00262436. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–54. [PubMed] [Google Scholar]
- 3.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 4.Baron C, Zambryski P C. The plant response in pathogenesis, symbiosis, and wounding: variations on a common theme? Annu Rev Genet. 1995;29:107–129. doi: 10.1146/annurev.ge.29.120195.000543. [DOI] [PubMed] [Google Scholar]
- 5.Bauer E, Kaspar T, Fischer H M, Hennecke H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J Bacteriol. 1998;180:3853–3863. doi: 10.1128/jb.180.15.3853-3863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhagwat A A, Keister D L. Identification and cloning of Bradyrhizobium japonicum genes expressed strain-selectively in soil and rhizosphere. Appl Environ Microbiol. 1992;58:1490–1495. doi: 10.1128/aem.58.5.1490-1495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang S E, Daniels D L, Blattner F R. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175:2026–2036. doi: 10.1128/jb.175.7.2026-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel R M, Appleby C A. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P450, other haemoproteins, nitrate and nitrite reductases. Biochim Biophys Acta. 1972;275:347–354. doi: 10.1016/0005-2728(72)90215-0. [DOI] [PubMed] [Google Scholar]
- 9.David M, Domergue O, Pognonec P, Kahn D. Transcription patterns of Rhizobium meliloti symbiotic plasmid pSym: identification of nifA-independent fix genes. J Bacteriol. 1987;169:2239–2244. doi: 10.1128/jb.169.5.2239-2244.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dénarié J, Debellé F, Promé J C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 11.Dixon R. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in γ-Proteobacteria. Arch Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 12.Ebeling S, Hahn M, Fischer H M, Hennecke H. Identification of nifE-, nifN- and nifS-like genes in Bradyrhizobium japonicum. Mol Gen Genet. 1987;207:503–508. [Google Scholar]
- 13.Ebeling S, Noti J D, Hennecke H. Identification of a new Bradyrhizobium japonicum gene (frxA) encoding a ferredoxin-like protein. J Bacteriol. 1988;170:1999–2001. doi: 10.1128/jb.170.4.1999-2001.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellay R, Perret X, Viprey V, Broughton W J, Brenner S. Organisation of host-inducible transcripts on the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1995;16:657–667. doi: 10.1111/j.1365-2958.1995.tb02428.x. [DOI] [PubMed] [Google Scholar]
- 15.Fischer H M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer H M. Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol. 1996;4:317–320. doi: 10.1016/0966-842x(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 17.Fischer H M, Alvarez-Morales A, Hennecke H. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 1986;5:1165–1173. doi: 10.1002/j.1460-2075.1986.tb04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer H M, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 1993;12:2901–2912. doi: 10.1002/j.1460-2075.1993.tb05952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuhrmann M, Hennecke H. Rhizobium japonicum nitrogenase Fe protein gene (nifH) J Bacteriol. 1984;158:1005–1011. doi: 10.1128/jb.158.3.1005-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuhrmann M, Fischer H M, Hennecke H. Mapping of Rhizobium japonicum nifB-, fixBC-, and fixA-like genes and identification of the fixA promoter. Mol Gen Genet. 1985;199:315–322. [Google Scholar]
- 21.Gibson T J, Rosenthal A, Waterston R H. Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene. 1987;53:283–286. doi: 10.1016/0378-1119(87)90017-5. [DOI] [PubMed] [Google Scholar]
- 22.Girard L, Valderrama B, Palacios R, Romero D, Dávila G. Transcriptional activity of the symbiotic plasmid of Rhizobium etli is affected by different environmental conditions. Microbiology. 1996;142:2847–2856. [Google Scholar]
- 23.Göttfert M, Hitz S, Hennecke H. Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol Plant-Microbe Interact. 1990;3:308–316. doi: 10.1094/mpmi-3-308. [DOI] [PubMed] [Google Scholar]
- 24.Gubler M, Hennecke H. FixA, B and C genes are essential for symbiotic and free-living, microaerobic nitrogen fixation. FEBS Lett. 1986;200:186–192. [Google Scholar]
- 25.Hahn M. Genomstruktur von Bradyrhizobium japonicum: gemeinsames Vorkommen von repetitiven Sequenzen und Genen für die Wurzelknöllchensymbiose. Ph.D. thesis. Zürich, Switzerland: Eidgenössische Technische Hochschule; 1986. [Google Scholar]
- 26.Hahn M, Hennecke H. Localized mutagenesis in Rhizobium japonicum. Mol Gen Genet. 1984;193:46–52. [Google Scholar]
- 27.Hart S, Kirby R, Woods D R. Structure of a Rhodococcus gene encoding pigment product in Escherichia coli. J Gen Microbiol. 1990;136:1357–1363. doi: 10.1099/00221287-136-7-1357. [DOI] [PubMed] [Google Scholar]
- 28.Hawkins F K L, Johnston A W B. Transcription of a Rhizobium leguminosarum biovar phaseoli gene needed for melanin synthesis is activated by nifA of Rhizobium and Klebsiella pneumoniae. Mol Microbiol. 1988;2:331–337. doi: 10.1111/j.1365-2958.1988.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 29.Howard J B, Rees D C. Nitrogenase: a nucleotide-dependent molecular switch. Annu Rev Biochem. 1994;63:235–264. doi: 10.1146/annurev.bi.63.070194.001315. [DOI] [PubMed] [Google Scholar]
- 30.Kaluza K, Hennecke H. Fine structure analysis of the nifDK operon encoding the α and β subunits of dinitrogenase from Rhizobium japonicum. Mol Gen Genet. 1984;196:35–42. [Google Scholar]
- 31.Kaluza K, Hahn M, Hennecke H. Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol. 1985;162:535–542. doi: 10.1128/jb.162.2.535-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaminski P A, Batut J, Boistard P. A survey of symbiotic nitrogen fixation by rhizobia. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 431–460. [Google Scholar]
- 33.Kim I C, Oriel P J. Characterization of the Bacillus stearothermophilus BR219 phenol hydroxylase gene. Appl Environ Microbiol. 1995;61:1252–1256. doi: 10.1128/aem.61.4.1252-1256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klement Z. Hypersensitivity. In: Mount M S, Lacy G H, editors. Phytopathogenic prokaryotes. New York, N.Y: Academic Press Inc.; 1982. pp. 149–177. [Google Scholar]
- 35.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 36.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kündig C. Untersuchungen zur Genomstruktur und Charakterisierung des einzigen rRNA-Operons von Bradyrhizobium japonicum. Ph.D. thesis. Zürich, Switzerland: Eidgenössische Technische Hochschule; 1994. [Google Scholar]
- 38.Kündig C, Hennecke H, Göttfert M. Correlated physical and genetic map of the Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin G B, Chapman K A, Chelm B K. Role of the Bradyrhizobium japonicum ntrC gene product in differential regulation of the glutamine synthetase II gene (glnII) J Bacteriol. 1988;170:5452–5459. doi: 10.1128/jb.170.12.5452-5459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 41.Murphy P J, Heycke N, Trenz S T, Ratet P, de Bruijn F J, Schell J. Synthesis of an opine-like compound, a rhizopine, in alfalfa nodules is symbiotically regulated. Proc Natl Acad Sci USA. 1988;85:9133–9137. doi: 10.1073/pnas.85.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nellen-Anthamatten D, Rossi P, Preisig O, Kullik I, Babst M, Fischer H M, Hennecke H. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol. 1998;180:5251–5255. doi: 10.1128/jb.180.19.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligonucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 44.Parniske M, Fischer H M, Hennecke H, Werner D. Accumulation of the phytoalexin glyceollin I in soybean nodules infected by a Bradyrhizobium japonicum nifA mutant. Z Naturforsch. 1991;46:318–320. [Google Scholar]
- 45.Perret X, Fellay R, Bjourson A J, Cooper J E, Brenner S, Broughton W J. Subtraction hybridisation and shot-gun sequencing: a new approach to identify symbiotic loci. Nucleic Acids Res. 1994;22:1335–1341. doi: 10.1093/nar/22.8.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perret X, Freiberg C, Rosenthal A, Broughton W J, Fellay R. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1999;32:415–425. doi: 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- 47.Phillips D A, Kapulnik Y. Plant isoflavonoids, pathogens and symbionts. Trends Microbiol. 1995;3:58–64. doi: 10.1016/s0966-842x(00)88876-9. [DOI] [PubMed] [Google Scholar]
- 48.Ramseier T M, Göttfert M. Codon usage and G + C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156:270–276. doi: 10.1007/BF00262997. [DOI] [PubMed] [Google Scholar]
- 49.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Schmidt P E, Parniske M, Werner D. Production of the phytoalexin glyceollin-I by soybean roots in response to symbiotic and pathogenic infection. Bot Acta. 1992;105:18–25. [Google Scholar]
- 52.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulation of Gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Heidelberg, Germany: Springer Verlag; 1983. pp. 98–106. [Google Scholar]
- 53.Soupène E, Foussard M, Boistard P, Truchet G, Batut J. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc Natl Acad Sci USA. 1995;92:3759–3763. doi: 10.1073/pnas.92.9.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studer D, Gloudemans T, Franssen H J, Fischer H M, Bisseling T, Hennecke H. Involvement of the bacterial nitrogen fixation regulatory gene (nifA) in control of nodule-specific host-plant gene expression. Eur J Cell Biol. 1987;45:177–184. [Google Scholar]
- 55.Thöny B, Kaluza K, Hennecke H. Structural and functional homology between the α and β subunits of the nitrogenase MoFe protein as revealed by sequencing the Rhizobium japonicum nifK gene. Mol Gen Genet. 1985;198:441–448. [Google Scholar]
- 56.Thöny B, Fischer H M, Anthamatten D, Bruderer T, Hennecke H. The symbiotic nitrogen fixation regulatory operon (fixRnifA) of Bradyrhizobium japonicum is expressed aerobically and is subject to a novel, nifA-independent type of activation. Nucleic Acids Res. 1987;15:8479–8499. doi: 10.1093/nar/15.20.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weidenhaupt M, Fischer H M, Acuña G, Sanjuan J, Hennecke H. Use of a promoter-probe vector system in the cloning of a new NifA-dependent promoter (ndp) from Bradyrhizobium japonicum. Gene. 1993;129:33–40. doi: 10.1016/0378-1119(93)90693-w. [DOI] [PubMed] [Google Scholar]
- 59.Willis L B, Walker G C. The phbC (poly-β-hydroxybutyrate synthase) gene of Rhizobium (Sinorhizobium) meliloti and characterization of phbC mutants. Can J Microbiol. 1998;44:554–564. doi: 10.1139/w98-033. [DOI] [PubMed] [Google Scholar]