Abstract
Background and purpose:
Artificial intelligence advances have stimulated a new generation of autosegmentation, however clinical evaluations of these algorithms are lacking. This study assesses the clinical utility of deep learning-based autosegmentation for MR-based prostate radiotherapy planning.
Materials and methods:
Data was collected prospectively for patients undergoing prostate-only radiation at our institution from June to December 2019. Geometric indices (volumetric Dice-Sørensen Coefficient, VDSC; surface Dice-Sørensen Coefficient, SDSC; added path length, APL) compared automated to final contours. Physicians reported contouring time and rated autocontours on 3-point protocol deviation scales. Descriptive statistics and univariable analyses evaluated relationships between the aforementioned metrics.
Results:
Among 173 patients, 85% received SBRT. The CTV was available for 167 (97%) with median VDSC, SDSC, and APL for CTV (prostate and SV) 0.89 (IQR 0.83–0.95), 0.91 (IQR 0.75–0.96), and 1801 mm (IQR 1140–2703), respectively. Physicians completed surveys for 43/55 patients (RR 78%). 33% of autocontours (14/43) required major “clinically significant” edits. Physicians spent a median of 28 min contouring (IQR 20–30), representing a 12-minute (30%) time savings compared to historic controls (median 40, IQR 25–68, n = 21, p < 0.01). Geometric indices correlated weakly with contouring time, and had no relationship with quality scores.
Conclusion:
Deep learning-based autosegmentation was implemented successfully and improved efficiency. Major “clinically significant” edits are uncommon and do not correlate with geometric indices. APL was supported as a clinically meaningful quantitative metric. Efforts are needed to educate and generate consensus among physicians, and develop mechanisms to flag cases for quality assurance.
Keywords: Radiation oncology, Deep learning, Radiologic technology, Program evaluation, Prostatic neoplasms
Contour delineation is an integral part of radiation treatment planning in the modern era. Both intra- and inter-observer variation are common [1–7], with some evidence suggesting that specialization or expertise could influence contour quality [8] and subsequent patient outcomes [9]. With increasingly complex treatments, contouring has also become a time-consuming process that can limit clinic workflow efficiency [10]. Previous work by our group suggests that radiation oncologists in the United States spend a median of 6 hours per week contouring, despite frequent assistance from dosimetrists and trainees (data in submission), which is comparable to the 5 hours per week reported in a similar survey among radiation oncologists in New Zealand and Australia [11].
Autosegmentation aims to address these challenges by reducing both time spent manually contouring and contour variation. To date, atlas- and model-based methods have demonstrated time savings of up to 30% when automating normal tissues, or organs at risk (OAR) [12,13]. Recent advances in artificial intelligence have spurred a new generation of autosegmentation tools based on deep learning. In many cases, deep learning appears to have outperformed older methods in both accuracy and efficiency, and have been touted to match human performance [14–16]. Recent studies show that deep learning-based algorithms used for prostate autosegmentation specifically are able to perform at a level comparable to expert inter-observer variability based on geometric indices, with one study showing time savings of 12 min (46%) among a small cohort (n = 36) [17–19]. Geometric indices are most commonly reported but tend to correlate poorly with physician quality ratings [20]. Additionally, clinical utility and efficiency gains in routine practice have not been demonstrated, particularly for target volumes.
This study therefore aims to comprehensively evaluate the implementation of an in-house developed, MRI-based deep learning autosegmentation algorithm for both OARs and target volumes in short-course prostate radiation [21]. Primary endpoints include physician-reported time spent contouring, physician-assessed quality scores, and standard geometric indices. Secondarily we investigated the correlations between physician and geometric contour assessments, and evaluate changes in the magnitude of physician edits throughout the study period. We hypothesized that physician feedback would not correlate well with objective geometric measures, and that the magnitude of physician edits would decrease over the study period due to a learned reliance on the algorithm, a behavioral phenomenon known as automation bias [22].
Materials and methods
Study design
This is an observational quality improvement study of the clinical implementation of an in-house deep learning-based automated contouring system designed to generate both OARs and target volumes for short-course prostate radiation. Clinical data including physician feedback was collected prospectively as part of routine care and approved by the Institutional Review Board at XXX for retrospective analysis.
Patient cohort and simulation
The study cohort included consecutive patients with prostate cancer treated from June-December 2019 with prostate-only radiation at our institution. All patients underwent implantation of three intraprostatic fiducials and a hydrogel rectal spacer approximately 1 week before undergoing simulation in our institution’s MR-only workflow. Patients were simulated supine with 3 T MRI with a full bladder (1–2 cups of water 30–45 min prior) and empty rectums (enema administered 2–3 h prior). MR images were acquired for contouring (T2w axial, voxel size 0.5 mm × 0.5 mm × 3 mm), synthetic-CT generation (3D FFE-based) and fiducial identification (3D bFFE-based). Acquisition time was 25 min and synthetic-CT was generated at the console using the commercial software, MRCAT [23]. Further details about the simulation method used have been previously described [21,24].
Autosegmentation algorithm and contouring process
A detailed explanation of the autosegmentation algorithm development can be found in earlier publications [21]. In brief, an expert-delineated cohort of 50 MRI images obtained via clinical routine on one dedicated MR scanner at our institution was selected to use as a training dataset. Delineations included the clinical target volume (CTV, which included prostate and entire seminal vesicles combined, per institutional standard) and relevant OARs (namely: rectum including anal canal, penile bulb, and bladder). For treatment planning, a uniform PTV expansion of 3 mm is added to the CTV without further edits. The deep learning architecture used for autosegmentation was the publicly available DeepLabV3 + developed initially by Chen et al. [25–28] for use in general purpose 2D computer vision tasks. A transfer learning approach was used to train the model, which leveraged generic convolutional filters generated from training on millions of natural images available in computer vision challenges like Microsoft Common Objects in Context (COCO) and PASCAL Visual Object Classes (VOC). In order to apply this technique, pre-processing of the T2w scans to multiple 8-bit three channel (false color) images was done. This involved converting each axial image into a set of 3 images, where each image received a separate preprocessing consisting of either a simple down-sample from 16-bit to 8-bit (1st channel), additional image inversion (2nd channel) or apply contrast limited adaptive histogram equalization (3rd channel).
Prior to physician review, contours were generated automatically on the T2w axial scan then were subsequently mapped onto registered synthetic-CT simulation scans using the implanted fiducials as reference. A standard post-processing algorithm of the contours was applied through vendor software (MIM Software, Inc) to remove potential stray pixels and smoothen contours. The treating physician then reviewed and edited these contours as needed prior to approving for subsequent treatment planning.
Geometric contour assessment
DICOM RT structure sets containing the CTV, rectum, penile bulb, and bladder were automatically saved both at baseline (before any manual edits) and at plan approval (after physician edits as well as dosimetrist/planner edits required to proceed with treatment planning). A quantitative comparison using three geometric measures was performed on each structure to assess the similarity of the automated contours to the approved contours in the clinical setting. Additionally, a composite of all contours (average of CTV + each OAR) was calculated in order to assess the correlation of these measures with the quality score and time spent contouring recorded by the treating physician on the entire case (further described below). We included the most commonly referenced volumetric Dice-Sørensen Coefficient (VDSC), as well as surface Dice-Sørensen Coefficient (SDSC) which compares the relative contour surface overlap above a clinically determined tolerance parameter, τ, which has potentially improved correlation with time savings compared to VDSC [16,29,30]. More specifically, VDSC is represented by the volume overlap of two structures V1 and V2 shown in equation 1.
SDSC with tolerance τ is defined as the summation of the intersection of each surface (S1 and S2) with respect to its expanded boundary surface with size τ (B1 and B2) divided by the total surface area of both, as shown in equation 2.
A τ value of 3 mm was set for this study given the resolution of the input MR axial T2-weighted images have similar voxel size.
Added Path Length (APL) was collected as a third metric based on work by Vaassen et al. [31] showing that this distance measurement correlates best with absolute time spent editing contours. APL is an absolute distance measurement of amount of surface adjusted from the original (autosegmented) contour, summed over all slices. Since it is not relative to the size/shape of the contour in question, it directly represents the amount of editing made, which makes it distinct from metrics like VDSC and SDSC.
Physician feedback and contour assessment
A short questionnaire was integrated into the clinical workflow during the first 2 months to assess the physician-perceived quality of automated contours. Physicians provided a global quality rating on a 3-point scale similar to that previously reported from clinical trial quality assurance for defining protocol deviations [32], with “1” indicating automated contours were acceptable without edits, “2” indicating the need for minor edits, and “3” suggesting that major, clinically significant edits were needed. Self-reported time spent contouring and optional free-text comments about the autocontours were additionally collected to provide further insight into physicians’ experience and perceptions of the clinical utility of the autosegmented contours.
Localizing frequent physician edits
To further characterize physician edits, three-dimensional (3D) heat maps were generated to visualize the specific locations of the most-edited regions for the CTV, rectum, and bladder. All initial and final (edited) contour masks were first deformably registered using the open-source software package SimpleITK [29]. By counting the number of pixels with vector magnitudes exceeding 0.5 mm (one-pixel length) within each deformation map, and then summing all voxels across patient datasets, the resultant heat maps highlight aggregate regions of frequent contour adjustment. All patients were aligned using the centroid of each contour and visualization is done by mapping this heat map on a single representative MR scan.
Statistical analysis
Descriptive statistics were calculated for all collected metrics. Spearman’s rank correlation coefficients between the three quantitative metrics and time spent contouring were generated in order to determine which metrics correspond best with physician effort. Further correlations were calculated in relation to the magnitude of physician edits over time. Due to the binary nature of available physician-reported quality scores (no “1 = acceptable, no edits”) Wilcoxon rank sum tests were used to assess the relationship between geometric indices and physician quality scores for automated contours. Two-sided p-values with adjusted alpha levels less than 0.006 were applied to all statistical tests, in order to account for multiple comparisons (Bonferroni correction). All computations were performed and generated using Rv3.6 (R Core Team, 2019, Vienna, Austria).
Results
A total of 173 patients with intact prostate cancer undergoing radiation to the prostate only were eligible for inclusion, as seen in Fig. A.1 in Appendix A. Treatment was provided by 18 physicians across 7 campuses, with each physician treating a median of 8 patients on study (IQR 6–13) during the 6-month study period. 72% of patients (n = 124) were treated with SBRT alone, 15% (n = 26) were treated with SBRT after brachytherapy, and 13% (n = 23) received moderately hypofractionated radiotherapy.
For the 167 cases for which complete CTV data was available, the median SDSC and VDSC for CTV final vs. initial automated contours was 0.91 (IQR 0.75–0.96) and 0.89 (IQR 0.83–0.95), respectively. When taking OAR contours into account to calculate a composite geometric index, the aforementioned coefficients increased to 0.95 (IQR 0.88–0.98) and 0.94 (IQR 0.90–0.97). Data distribution for these two metrics is displayed with histograms in Fig. A.2 in Appendix A. CTV contours had a median APL of 1801 mm (IQR 1140–2703), which increased to 3062 mm (IQR 2011–4837) with the addition of OARs. This is illustrated in Fig. 1, which shows the APL distribution for each volume of interest as overlaid kernel density plots. Detailed breakdown of all three metrics across anatomic sites is shown in Table 1.
Fig. 1.
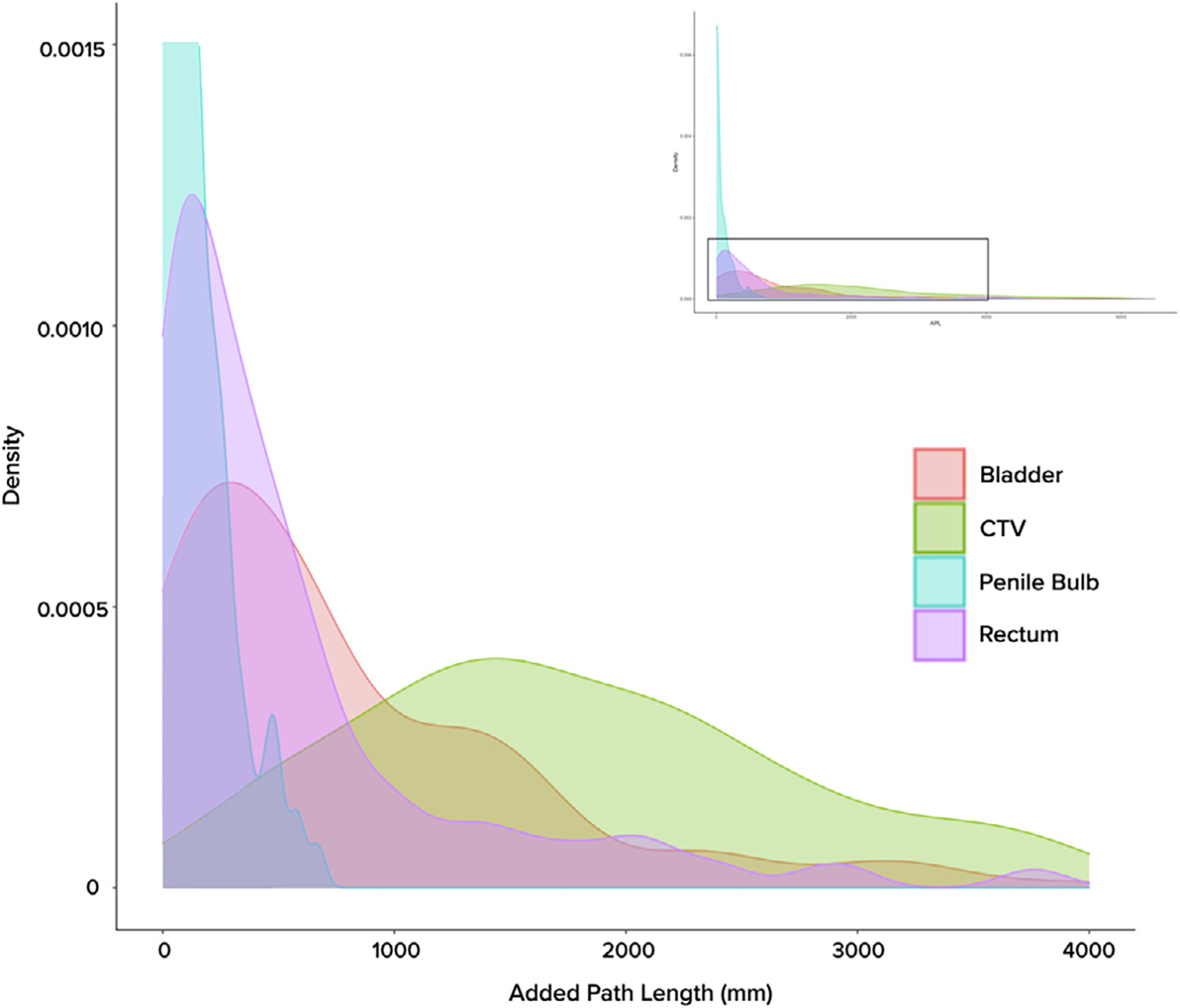
Distribution of Added Path Length (APL) across volumes of interest, as represented by kernel density plots. As histogram variants, kernel density plots allow for smoother visualization of data spread across a continuous interval. Full graphical representation of the curves can be seen in the upper right inlay, with the magnified area indicated with the viewfinder.
Table 1.
Comparison of geometric evaluation metrics across volumes of interest.
| Median (IQR) | Surface DSC* | Volumetric DSC* | Added Path Length [mm]* |
|---|---|---|---|
|
| |||
| Prostate & Seminal Vesicles (CTV; n = 167) | 0.91 (0.75–0.96) | 0.89 (0.83–0.95) | 1801 (1140–2703) |
| Bladder (n = 173) | 0.99 (0.96–1.00) | 0.99 (0.98–1.00) | 577 (203–1280) |
| Penile Bulb (n = 168) | 1.00 (0.95–1.00) | 0.97 (0.85–1.00) | 17 (2–136) |
| Rectum (n = 172) | 0.99 (0.93–1.00) | 0.97 (0.94–0.99) | 348 (90–681) |
| Composite (CTV + OARs; n = 163) | 0.95 (0.88–0.98) | 0.94 (0.90–0.97) | 3062 (2011–4837) |
Abbreviations: DSC, Dice-Sørensen Coefficient; CTV, clinical target volume; OAR, organ at risk; IQR, interquartile range.
Comparing final vs. automated contour datasets.
Physician rating of contour quality was available for 43 patients treated in the first 2 months (early pilot phase) of the evaluation period, with an overall response rate of 78% (43/55). No difference was observed between the non-responder and responder cohorts when assessing VDSC on univariate analysis. Physicians reported a median of 28 min spent contouring (IQR 20–30), reflecting a 12-minute (30%) reduction in time compared to historic controls collected immediately prior to algorithm implementation (median 40 min, IQR 25–68, n = 21, p < 0.01). As shown in Fig. 2, 65% of automated contour sets (n = 28) received a quality score of 2 (accept with minor edits) from the treating clinician, while 35% (n = 15) required major, clinically significant edits (score of 3). No automated contour sets were deemed acceptable without edits (quality score of 1).
Fig. 2.
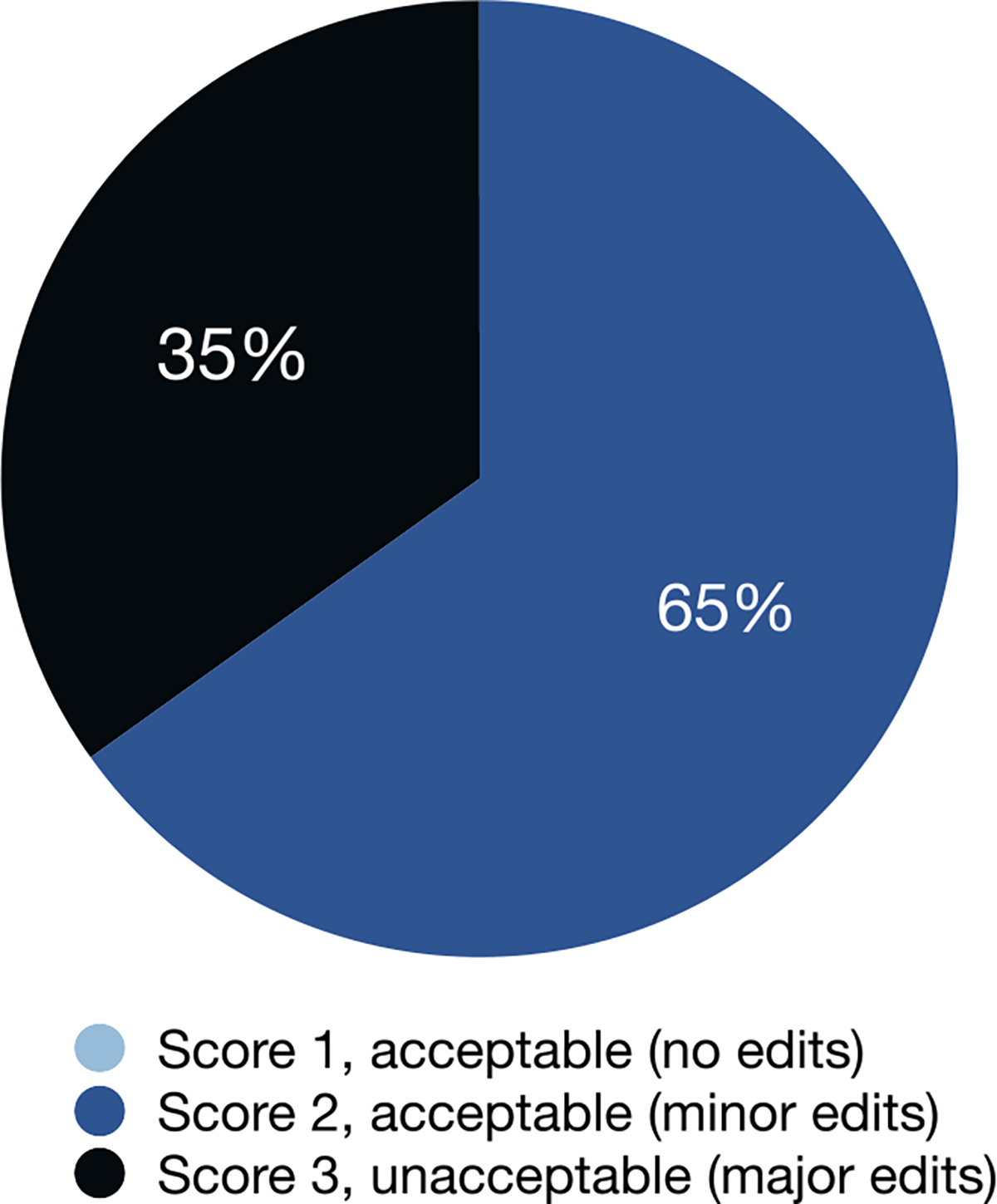
Physician scoring of automated contours (n = 43).
Correlation coefficients between each geometric index and time spent contouring were calculated, as shown in Fig. 3. Almost all correlations were statistically significant (rejecting the null hypothesis of no correlation) but the correlation was only weak-moderate (0.2–0.5). Geometric indices were not significantly different between those scored “2 = minor edits” and “3 = major edits” (all p > 0.3). Over time, there was no change in the magnitude of edits based on any geometric indices (all p > 0.1).
Fig. 3.
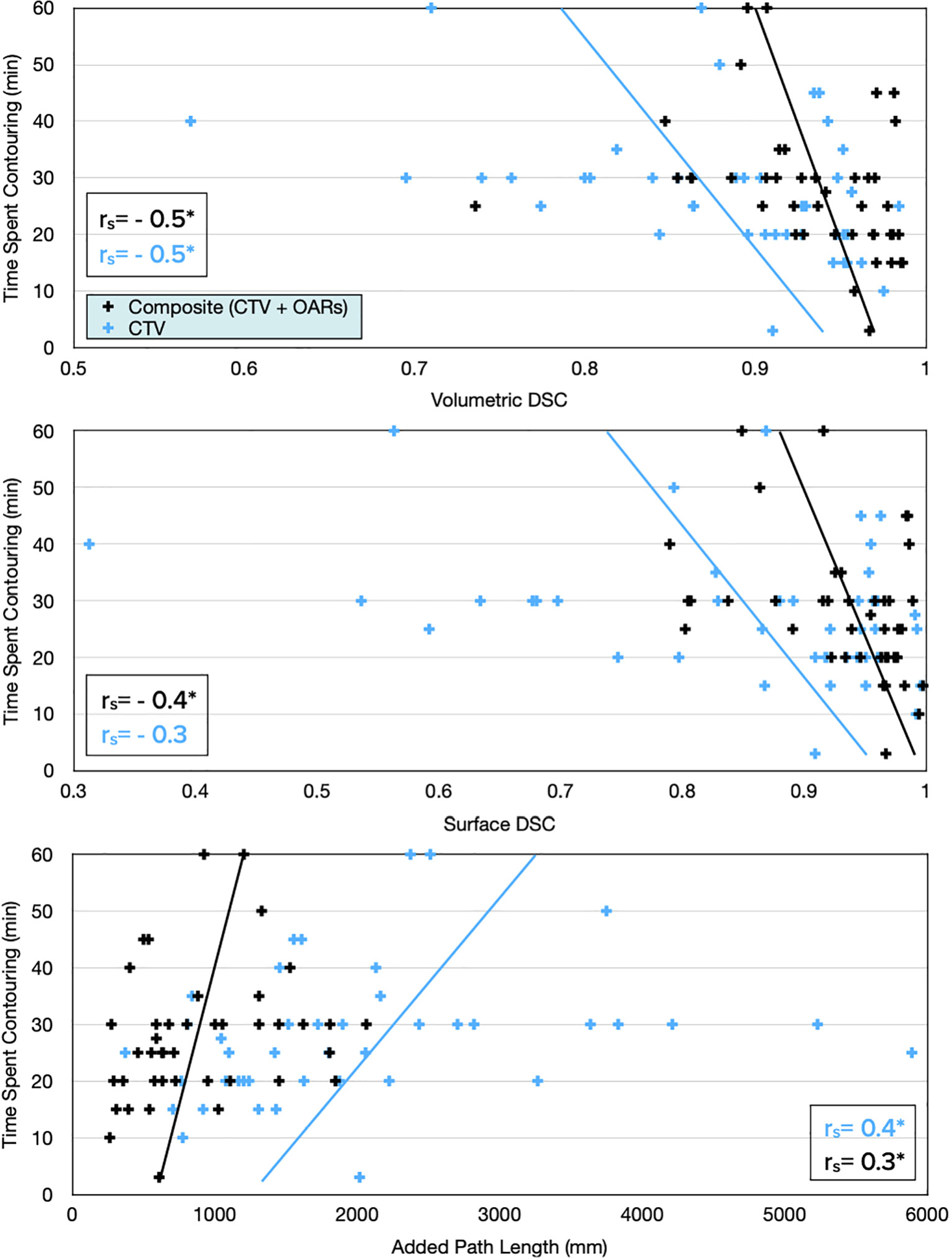
All geometric measures correlated with physician-reported time spent contouring. Statistically significant correlation coefficients (rs) indicated with an asterisk (*).
Common automated contour errors were recorded from treating physician survey comments and categorized by anatomic location. Of the 25 optional free-text comments collected from the physician survey, 88% (n = 22) referenced CTV contours while 72% (n = 18) referenced OARs. Specific CTV comments specified the prostate apex and SV, while notable OARs were bladder and rectum each with 7 comments. The overall sentiments of comments were coded as “negative,” “positive,” or “mixed” following qualitative analysis by a single reviewer. 64% (n = 14) of CTV-related comments were negative, 23% (n = 5) were positive, and the remaining 14% (n = 3) were mixed or had a neutral tone. In contrast, a higher proportion of OAR-related comments were positive (44%, n = 8), with only 28% (n = 5) of comments coded as negative and the remaining 28% (n = 5) deemed mixed.
Three-dimensional heat maps characterizing aggregate physician edits across all patient datasets can be seen in Fig. 4. Highlighted areas, corresponding to a structure’s most edited region, included the prostate apex, the rectosigmoid junction, and several structure interfaces (e.g. CTV-rectum, bowel-bladder, CTV-bladder).
Fig. 4.
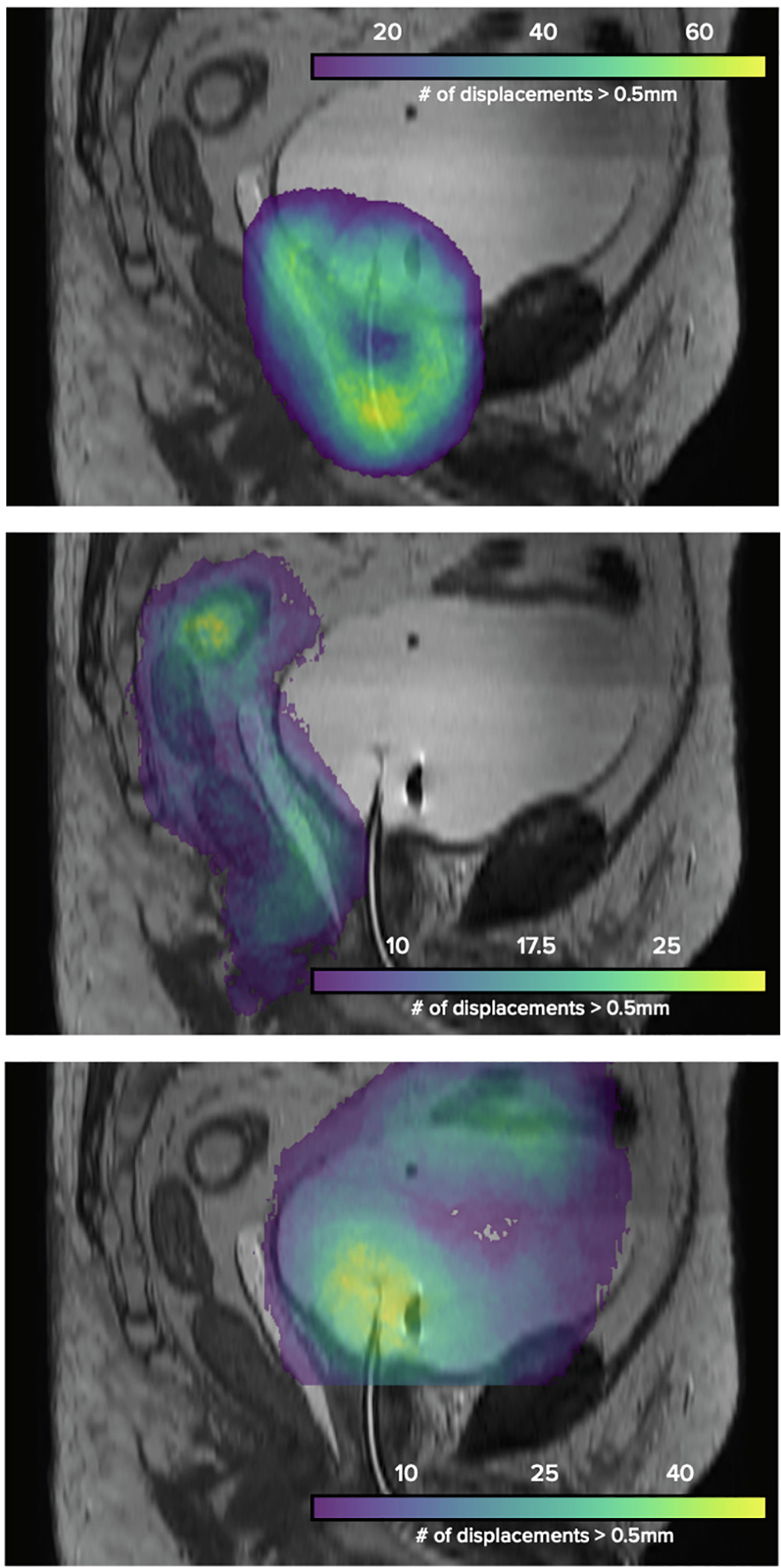
Regions of frequent physician edits across all patient datasets, with magnitude of displacements represented using 3D heatmaps.
Discussion
In this largest-to-date study of clinical implementation and physician assessment of deep learning-based autosegmented contours for prostate-only radiation, we found a high utility of both OARs and CTV with 65% of cases requiring no more than minor edits, and a resultant median time savings of 12 min (30% of total time spent contouring) for physicians. The high geometric similarity between initial and final contours, and subsequent efficiency benefits provided by this deep learning-based algorithm are consistent with prior research [16,17]. We confirm limitations of geometric indices in determining the subset of cases requiring major (clinically significant) edits, highlight the potential utility of aggregating edits onto a 3D heat map to localize common regions requiring edits, and reinforce the importance of physician engagement in improving the clinical utility of autosegmentation.
From both geometric indices and physician feedback, we conclude that edits predominantly occurred to the CTV contour. This is consistent with prior studies examining autosegmentation accuracy [18], and is likely in part reflective of the critical importance of accurate CTV delineation in the context of prostate SBRT. Furthermore, aggregated edits correspond to physician comments highlighting specifically the prostate apex and seminal vesicles, which are areas previously demonstrated to generate high inter-observer contouring variation [4]. Importantly, limited edits were needed at the prostate-rectum interface, which is often considered at highest risk for contributing to radiation toxicity, and of increasing concern with hypofractionated radiation [33]. This may be a result of using a spacer as standard of care for patients included in this study. This is further supported by higher geometric similarity of initial and final rectal contours in our model compared to that of a recently published MR-based deep learning autosegmentation algorithm for prostate OARs (VDSC 0.97 vs 0.88, respectively) [34]. In combination, these data suggest progress toward clinically useful models for autosegmentation of both OARs and target volumes when using MR-based deep learning techniques.
Interestingly, we did not see a reduction in the magnitude of physician contour edits over time, despite prior evidence that secular changes can occur when humans become accustomed to automation through a phenomenon called automation bias, which can have both positive (reducing inter-observer variation) and negative consequences (regressing to a potentially faulty standard) [22].
Immediately following the study period, we sought to further improve the model by integrating the final edited contours (primarily CTV), and were surprised to find minimal improvement in model performance. This appears to reflect the challenge created by inter-observer variability. We hypothesize that model improvement will require active engagement from physicians to generate consensus and concerted educational efforts to limit clinically insignificant edits. Meanwhile, our physicists and computer scientists can refine irregularities, focus on performance in areas identified by the heatmap to have greatest difference between initial and final contours, and generate dashboards to support rapid review and feedback for physicians (Fig. A.3 in Appendix A). Despite current algorithm limitations, no physicians requested a return to prior manual-only methods.
Finally, this study aimed to determine the utility of quantitative metrics (VDSC, SDSC and APL) in assessing deep learning algorithms in clinical practice. VDSC and APL correlated with physician reported time savings, although weakly [35]. APL does appear to provide more relevant and complete information than VDSC about contour edits across the patient cohort analyzed, given emphasis on absolute length of edits. Of the OARs, the penile bulb and rectum were less edited than the bladder. Combined with the localization heat map, this allows for a comprehensive understanding of the magnitude and position of potential model deficiencies from which to improve. These conclusions would be difficult to make using VDSC alone, given the relatively similar scores for OARs and perceived high score for CTV.
There are several limitations to the current study. First, physician assessments were limited (n = 43) due to declining response rates at the end of 2 months, in order to reduce bias in the sample for analysis. This limits power of testing to correlate physician-reported quality to geometric indices. Additionally, power in this study is limited by the 3-point scale compared to a continuous scale, as is available with geometric indices. Nonetheless, due to the importance of assessing clinical relevance associated with physician ratings, consideration of a 5-point scale has been previously proposed and we would agree with that approach [36]. The quality scores are furthermore an aggregate for the case to simplify the survey and facilitate physician participation, though that further limits the ability to correlate quality ratings to geometric indices. Nonetheless, from the available data, quality scores appear to primarily reflect the CTV, due in large part to the high accuracy of the OAR autocontours, giving us confidence with this approach to evaluation. The lack of a true gold standard for prostate contour delineation is another inherent limitation to the study, with inter-observer variability making it difficult to truly assess the performance of the algorithm. And finally, during the study period, the institutional standard was to contour the entire seminal vesicle, thereby potentially under-reporting required physician edits in the setting of a risk-based approach. Optimal autosegmentation algorithms would account for clinical risk factors in addition to imaging characteristics.
In conclusion, a deep learning autosegmentation tool, developed using clinical institutional data, was successfully implemented for MR-based planning for intact prostate cancer with most (65%) of patients requiring no more than minor edits, and resulting in physician a median time savings of 12 min. Further time savings may be limited by human factors such as inter-observer variability. We describe a framework for clinical evaluation and physician engagement in clinical autosegmentation implementation. To fully realize the benefits of deep learning autosegmentation, greater contouring consensus and ongoing education will be required.
Supplementary Material
Acknowledgement
We acknowledge the expertise of computer scientist Harini Verraraghavan, PhD for input on autosegmentation algorithm development, as well as all radiation oncologists at Memorial Sloan Kettering treating prostate cancer that contributed feedback regarding algorithm performance.
Funding statement
This work is supported by an MSK Core Grant (P30 CA008748). Additional funding provided by the Radiologic Society of North American (RSNA) (EI1902, E.F.G), Agency for Healthcare Research and Quality (AHRQ) (R18 HS026881, E.F.G.), National Cancer Institute (K08 CA252640, E.F.G), Varian (J.D.), Elekta (J.D.), and Breast Cancer Research Foundation (BCRF) (J.D.). The funders/sponsors had no role in the design and conduct of this study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Abbreviations:
- VDSC
volumetric Dice-Sørensen coefficient
- SDSC
surface Dice-Sørensen coefficient
- APL
added path length
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Disclosures
Erin Gillespie is a co-founder of the educational website eContour.org. Joseph Deasy is a co-founder and shareholder of PAIGE. AI. Michael Zelefsky serves as a consultant for Boston Scientific.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.radonc.2021.02.040.
References
- [1].Bekelman JE, Wolden S, Lee N. Head-and-neck target delineation among radiation oncology residents after a teaching intervention: a prospective, blinded pilot study. Int J Radiat Oncol Biol Phys 2009;73:416–23. [DOI] [PubMed] [Google Scholar]
- [2].Bhardwaj AK, Kehwar TS, Chakarvarti SK, Sastri GJ, Oinam AS, Pradeep G, et al. Variations in inter-observer contouring and its impact on dosimetric and radiobiological parameters for intensity-modulated radiotherapy planning in treatment of localised prostate cancer. J Radiother Pract 2008;7:77–88. [Google Scholar]
- [3].Brouwer CL, Steenbakkers RJ, van den Heuvel E, Duppen JC, Navran A, Bijl HP, et al. 3D variation in delineation of head and neck organs at risk. Radiat Oncol 2012;7. 10.1186/1748-717X-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fiorino C, Reni M, Bolognesi A, Cattaneo GM, Calandrino R. Intra- and inter-observer variability in contouring prostate and seminal vesicles: implications for conformal treatment planning. Radiother Oncol 1998;47:285–92. [DOI] [PubMed] [Google Scholar]
- [5].Nakamura K, Shioyama Y, Tokumaru S, Hayashi N, Oya N, Hiraki Y, et al. Variation of clinical target volume definition among Japanese radiation oncologists in external beam radiotherapy for prostate cancer. Jpn J Clin Oncol 2008;38:275–80. [DOI] [PubMed] [Google Scholar]
- [6].Petric P, Dimopoulos J, Kirisits C, Berger D, Hudej R, Pötter R. Inter- and intraobserver variation in HR-CTV contouring: intercomparison of transverse and paratransverse image orientation in 3D-MRI assisted cervix cancer brachytherapy. Radiother Oncol 2008;89:164–71. [DOI] [PubMed] [Google Scholar]
- [7].Steenbakkers RJ, Duppen JC, Fitton I, Deurloo KE, Zijp L, Uitterhoeve AL, et al. Observer variation in target volume delineation of lung cancer related to radiation oncologist-computer interaction: a ‘Big Brother’ evaluation. Radiother Oncol 2005;77:182–90. [DOI] [PubMed] [Google Scholar]
- [8].Boero IJ, Paravati AJ, Xu B, Cohen EEW, Mell LK, Le Q-T, et al. Importance of radiation oncologist experience among patients with head-and-neck cancer treated with intensity-modulated radiation therapy. J Clin Oncol 2016;34:684–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fairchild A, Straube W, Laurie F, Followill D. Does quality of radiation therapy predict outcomes of multicenter cooperative group trials? A literature review. Int J Radiat Oncol Biol Phys 2013;87:246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Austin-Seymour M, Chen GTY, Rosenman J, Michalski J, Lindsley K, Goitein M. Tumor and target delineation: current research and future challenges. Int J Radiat Oncol Biol Phys 1995;33:1041–52. [DOI] [PubMed] [Google Scholar]
- [11].Leung J, Forstner D, Chee R, James M, Que E, Begum S. Faculty of Radiation Oncology 2018 workforce census. J Med Imaging Radiat Oncol 2019;63:852–61. [DOI] [PubMed] [Google Scholar]
- [12].Walker GV, Awan M, Tao R, Koay EJ, Boehling NS, Grant JD, et al. Prospective randomized double-blind study of atlas-based organ-at-risk autosegmentation-assisted radiation planning in head and neck cancer. Radiother Oncol 2014;112:321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang J, Amini A, Williamson R, Zhang L, Zhang Y, Komaki R, et al. Automatic contouring of brachial plexus using a multi-atlas approach for lung cancer radiotherapy. Pract Radiat Oncol 2013;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ahn SH, Yeo AU, Kim KH, Kim C, Goh Y, Cho S, et al. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat Oncol 2019;14. 10.1186/s13014-019-1392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol 2018;126:312–7. [DOI] [PubMed] [Google Scholar]
- [16].Nikolov S, Blackwell S, Mendes R, De Fauw J, Meyer C, Hughes C, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. ArXiv. 2018:1809.04430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kiljunen T, Akram S, Niemelä J, Löyttyniemi E, Seppälä J, Heikkilä J, et al. A deep learning-based automated CT segmentation of prostate cancer anatomy for radiation therapy planning-A retrospective multicenter study. Diagnostics (Basel) 2020;10:959. 10.3390/diagnostics10110959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wong J, Fong A, McVicar N, Smith S, Giambattista J, Wells D, et al. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020;144:152–8. [DOI] [PubMed] [Google Scholar]
- [19].Zabel WJ, Conway JL, Gladwish A, Skliarenko J, Didiodato G, Goorts-Matthews L, et al. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract Radiat Oncol 2021;11:e80–9. [DOI] [PubMed] [Google Scholar]
- [20].Duke SL, Tan L-T, Jensen NBK, Rumpold T, De Leeuw AAC, Kirisits C, et al. Implementing an online radiotherapy quality assurance programme with supporting continuous medical education - report from the EMBRACE-II evaluation of cervix cancer IMRT contouring. Radiother Oncol 2020;147:22–9. [DOI] [PubMed] [Google Scholar]
- [21].Elguindi S, Zelefsky MJ, Jiang J, Veeraraghavan H, Deasy JO, Hunt MA, et al. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys Imaging Radiat Oncol 2019;12:80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Philpotts LE. Can computer-aided detection be detrimental to mammographic interpretation?. Radiology 2009;253:17–22. [DOI] [PubMed] [Google Scholar]
- [23].Köhler MV, Van Grootel M, Hoogeveen R, Kemppainen R, Renisch S. MR-only simulation for radiotherapy planning. Philips White Paper 2015. [Google Scholar]
- [24].Tyagi N, Fontenla S, Zelefsky M, Chong-Ton M, Ostergren K, Shah N, et al. Clinical workflow for MR-only simulation and planning in prostate. Radiat Oncol 2017;12. 10.1186/s13014-017-0854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen L-CC, Zhu M, Papandreou Y, Zoph G, Schroff G, Adam F, et al. , Searching for efficient multi-scale architectures for dense image prediction. Appl Phys Lett 2018;99. [Google Scholar]
- [26].Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018;40:834–48. [DOI] [PubMed] [Google Scholar]
- [27].Chen L-CP, Schroff F, Adam H. Rethinking atrous convolution for semantic image segementation arXiv. 2017. [Google Scholar]
- [28].Zhu YH. An adaptive histogram equalization algorithm on the image gray level mapping. Physics Procedia 2012;25:601–8. [Google Scholar]
- [29].Gooding MJ, Smith AJ, Tariq M, Aljabar P, Peressutti D, Stoep J, et al. Comparative evaluation of autocontouring in clinical practice: A practical method using the Turing test. Med Phys 2018;45:5105–15. [DOI] [PubMed] [Google Scholar]
- [30].Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vaassen F, Hazelaar C, Vaniqui A, Gooding M, van der Heyden B, Canters R, et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys Imaging Radiat Oncol 2020;13:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Francolini G, Thomsen MS, Yates ES, Kirkove C, Jensen I, Blix ES, et al. Quality assessment of delineation and dose planning of early breast cancer patients included in the randomized Skagen Trial 1. Radiother Oncol 2017;123:282–7. [DOI] [PubMed] [Google Scholar]
- [33].Morgan SC, Hoffman K, Loblaw DA, Buyyounouski MK, Patton C, Barocas D, et al. Hypofractionated radiation therapy for localized prostate cancer: executive summary of an ASTRO, ASCO, and AUA evidence-based guideline. Pract Radiat Oncol 2018;8:354–60. [DOI] [PubMed] [Google Scholar]
- [34].Savenije MHF, Maspero M, Sikkes GG, van der Voort van Zyp JRN, AN TJK, Bol GH, et al. Clinical implementation of MRI-based organs-at-risk autosegmentation with convolutional networks for prostate radiotherapy. Radiat Oncol. 2020;15:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kiser K, Barman A, Stieb S, Fuller CD, Giancardo L. Novel autosegmentation spatial similarity metrics capture the time required to correct segmentations better than traiditional metrics in a thoracic cavity segmentation workflow. medRxiv 2020. 10.1101/2020.05.14.20102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ghooi RB, Bhosale N, Wadhwani R, Divate P, Divate U. Assessment and classification of protocol deviations. Perspect Clin Res 2016;7:132–6. 10.4103/2229-3485.184817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


