Abstract
NK cells play an important role in innate immunity. A previous genome-wide association study demonstrated an association between a 17q12 allele (rs9916629C) and lower frequency of CD3−CD56+ NK cells in peripheral blood. We performed an analysis that not only replicates the original result of the genome-wide association study (p = 0.036) but also defines the specific cell subpopulations and functions that are modulated by the rs9916629 polymorphism in a cohort of 96 healthy adult subjects using targeted multiparameter flow cytometric profiling of NK cell phenotypes and functions. We found that rs9916629C is associated with alterations in specific NK cell subsets, including lower frequency of predominantly cytotoxic CD56dim NK cells (p = 0.011), higher frequency of predominantly regulatory CD56bright NK cells (p = 0.019), and a higher proportion of NK cells expressing the inhibitory NKG2A receptor (p = 0.0002). Functionally, rs9916629C is associated with decreased secretion of macrophage inflammatory protein-1β by NK cells in the context of Ab-dependent cell-mediated cytotoxicity (p = 0.039) and increased degranulation in response to MHC class I-deficient B cells (p = 0.017). Transcriptional profiling of NK cells suggests that rs9916629 influences the expression of transcription factors such as TBX21, which has a role in NK cell differentiation, offering a possible mechanism for the phenotypic and functional differences between the different alleles. The rs9916629C allele therefore has a validated effect on the proportion of NK cells in peripheral blood and skews NK cells toward a specific phenotypic and functional profile, potentially influencing the impact that these innate immune cells have on infection and autoimmunity.
Natural killer cells represent a key component of the innate immune system and are able to rapidly eliminate tumor cells or virally infected cells without prior Ag sensitization (1-4). NK cells are broadly defined as a lymphocyte subset expressing CD56 and/or CD16 on their surface in the absence of T or B cell lineage markers. The level of CD56 surface expression further distinguishes two functionally distinct subsets of NK cells: 1) a smaller subset of CD56bright cells that exhibit a more immunoregulatory profile that primarily involves secretion of cytokines (~10% of NK cells in the blood), and 2) the major subset of CD56dim cells that exert mostly cytotoxic functions (~90% of NK cells in the blood) (5, 6). NK cells represent ~5–15% of total circulating lymphocytes, but the proportion of NK cells varies across human populations and this variation is in part heritable (7, 8), potentially conferring a differential capacity to fight infection or malignancy.
In addition to their role in antimicrobial, antiviral, and antitumor immunity, NK cells have recently also been implicated in inflammatory and autoimmune disorders such as type I diabetes (9, 10) and multiple sclerosis (11, 12). Genetic variations that influence the proportion of NK cells may also contribute to the clinical outcomes in these diseases. We previously reported that multiple sclerosis patients display a reduced frequency of the CD3−CD4− CD8lowCD56+ subset of NK cells (13). Interestingly, certain disease-modifying therapies being tested in multiple sclerosis influence NK cell subset distribution and function; for example, daclizumab (an anti–IL-2Rα mAb) induces the expansion and activation of regulatory CD3−CD56bright NK cells (14, 15). Thus, an enhanced understanding of the genetic factors influencing variation in NK subset phenotype and NK cell function will be important for clinical translation of certain molecules: genotypes that are associated with NK cell frequency, subset distribution, and specific functions might provide a useful tool to predict risk and outcome in certain diseases and serve as potential therapeutic targets.
Beyond the basic distinction of regulatory and cytotoxic NK cells, these innate immune effector cells can be segregated into a vast repertoire of subsets based on their cell receptor profiles, each with a unique capacity to recognize infected or malignant target cells (16). The major classes of receptors include the killer Igrelated receptors (KIRs) that bind to MHC molecules, the c-type lectins (NKG2) that bind to stress inducible molecules such as MICA/B and ULBP, the natural cytotoxicity receptors that bind to viral hemagglutinins, as well as the FcγRIIIa receptor (CD16) that binds to the Fc region of IgG Abs (16). NK cell clones, either constitutively or upon activation, express a wide range of additional receptors that can further modulate target cell recognition.
A recent genome-wide association study in healthy subjects identified two single nucleotide polymorphisms (SNPs) on chromosome 17q12 (rs1838149 and rs9916629; r2 = 1.0 in HapMap 2 CEU subjects, Utah residents with ancestry from northern and western Europe) that are associated with the frequency of CD3− CD56+ NK cells (7). However, this study only addressed how these polymorphisms affected the overall frequency of NK cells without investigating whether specific NK cell subsets or functions were modulated preferentially. Given the complexity of NK cell population structure and function, we were thus interested in not only replicating this result but also defining whether this locus is associated with alterations in specific NK cell subpopulations or functions that could predict differential disease outcomes. In this study, multiparameter flow cytometric phenotyping was coupled with functional profiling of NK cells and gene expression analysis of purified NK cell populations of individuals carrying the different variants at this locus. We report a replication of the original association and a novel observation that the candidate SNP (rs9916629) is associated with an altered NK phenotypic and functional profile.
Materials and Methods
Study subjects and genotyping of the candidate gene
The Institutional Review Board of Partners Healthcare approved the study. Subjects gave written informed consent for their DNA analysis and immune cell profiling. To study the association between the candidate SNP and NK cell phenotype and function, we drew subjects from a living biobank of >1600 healthy adult blood donors between 18 and 50 y age who were recruited from the Boston area as part of the Brigham and Women’s Hospital PhenoGenetic Project, which is an ongoing effort to understand how genetic variations affect the immune system and influence the risk of inflammatory diseases (see Supplemental Table I for demographic details). The candidate SNP (rs9916629) was genotyped using the iPLEX Sequenom MassARRAY platform (genotype call rate > 95%, Hardy–Weinberg Equilibrium p > 0.001). Genotype frequencies in subjects are described below: 8.3% with the CC genotype, 44.8% with the CT genotype, and 46.9% with the TT genotype.
Multiparameter flow cytometric phenotypic profiling of NK cells
We conducted a comprehensive phenotypic profiling of NK cells using a multiparameter flow cytometric approach. All blood samples were collected between 8:00 am and 12:00 pm in the Center for Clinical Investigation at the Brigham and Women’s Hospital to minimize circadian fluctuations in lymphocyte counts and subset frequencies. PBMCs were isolated from fresh whole blood of each subject within 4 h after venipuncture by Ficoll-Hypaque density gradient centrifugation (Sigma-Aldrich, St. Louis, MO), and 106 PBMCs were incubated for 15 min with different fluorescently labeled Abs. The following Abs were used: CD3 Pacific Blue, CD16 allophycocyanin-Cy7 or Alexa Fluor 700, CD56 PE-Cy7 or Alexa Fluor 700, CD161 (KLRB1) PE-CY5, CD94 (KLRD1) FITC, CD314 (NKG2D) allophycocyanin, CD335 (NKp46) PE, CD337 (NKp30) Alexa Fluor 647, CD336 (NKp44) PE, perforin FITC (all from BD Bioscience, San Jose, CA); and CD159A (NKG2A) PE (Beckman Coulter). For intracellular staining of perforin, cells were then permeabilized using Fix/Perm solution (BD Biosciences) followed by incubation with the respective Ab for 30 min. Data were acquired on a multiparametric LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software v9.3 (Tree Star, Ashland, OR). NK cells were defined as CD3− lymphocytes expressing CD56 and/or CD16. NK cells were defined as CD3−CD56+/−CD16+/−. Furthermore, CD56bright NK cells were defined as CD3−CD56+CD16−. CD56dim cells were defined as CD3− CD56+CD16+(high or low). Frequency of all cells expressing the tested markers was assessed for all NK cells and for the CD56bright and CD56dim subsets separately. Of note, cytometric data generation and manual parameter extraction were performed by an investigator who was blinded to each subject’s genotype.
Functional NK cell assay using flow cytometry
The capacity of NK cells to secrete cytokines and degranulate was examined following stimulation of PBMCs with different NK target cell lines, each at a 10:1 E:T ratio: 1) Ab-dependent cell-mediated cytotoxicity (p815 cells; a mouse leukemic cell line [American Type Culture Collection], preincubated for 1 h with 1 mg/ml p815-specific Ab [Abcam, Cambridge, MA]), 2) MHC-devoid target cell line (K562 cells; American Type Culture Collection), and 3) MHC class I-deficient B cell line (721.221 cells; American Type Culture Collection). Treatment with PMA (2.5 μg/ml) and ionomycin (1 μg/ml) was used as positive control (Sigma-Aldrich), and incubation of PBMCs with media alone (RPMI 1640 medium with 10% FCS) or PBMCs with p815 cells without the coating Ab was used as the negative control for the respective experiments. Brefeldin A (0.5 μg/ml; Sigma-Aldrich), 0.3 μg/ml monensin (GolgiStop; BD Biosciences), and anti–CD107a-PE-Cy5 Ab (a surrogate marker of cell degranulation and cytotoxicity; BD Biosciences) were added directly to the stimulation conditions and cells were incubated at 37°C in 5% CO2 for 6 h. Following this period of coculture, cells were washed and stained with the immunophenotype markers (CD3, CD16, CD56) as described above. Cells were then washed, fixed, and permeabilized using Fix/Perm solutions (BD Biosciences) according to the manufacturer’s instructions. Intracellular cytokine staining for IFN-γ and macrophage inflammatory protein (MIP)-1β (also known as CCL4) was performed using anti–IFN-γ-FITC and anti–MIP-1β Abs (both BD Biosciences). Data were acquired on a BD LSRII flow cytometer and analyzed using FlowJo software v9.3.
RNA expression and expression quantitative trait locus analysis
From 20 subjects with genotype data at rs9916629, RNA expression profiles in purified NK cell populations were generated using the Illumina Bead-Array platform (Illumina, San Diego, CA). Briefly, NK cells were purified using the RosetteSep separation kit (Stemcell Technologies, Vancouver, BC, Canada) on whole blood following cell lysis. RNA was extracted using an RNA isolation kit (Qiagen, Valencia, CA). The Illumina platform reported average signal, bead SE, total number of beads detected per probe, and detection p value. Our samples contained expression data from 48,760 probes that met the Illumina threshold for significant detection (detection p value cutoff of 0.01). These probes cover a total of 37,780 transcripts. After removing probes without expression in any sample, expression data for 14,431 probes remained. With C as the reference allele, we performed linear regression using an additive model for each probe against the genotype at rs9916629.
Statistical analysis
We used linear regression models to explore the correlation between the candidate SNP (rs9916629) and the quantitative traits of NK profile obtained in the flow cytometric assessment. To meet the assumptions of our linear regression models, and to limit the impact of outliers, we assessed quantitative NK cell frequencies for normality and performed a square root or natural log transformation where appropriate. Because the magnitude of the β estimate from the linear model is not comparable between transformed and untransformed values, results are presented in terms of the p value and direction of effect. Age and sex were used as covariates in multivariate analyses. For frequencies of NK cells that express cytokines or degranulate following stimulation with the different target cells, the model was further adjusted for background cytokine secretion or degranulation (stimulation with media alone or p815 cells without Ab, respectively). When a statistically significant association was found using the primary additive model for the candidate SNP, we next explored whether this effect was dominant or recessive using linear regression and based on the observed median frequencies in each genotype group. Statistical analyses were performed using PLINK software, version 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (17) and SAS, version 9.2 (SAS Institute, Cary, NC).
Results
A previous study found that a polymorphism at the 17q12 locus was associated with the frequency of the CD3−CD56+ NK cells in circulation (7), but whether this SNP affects all NK cells or specific NK cell populations was not examined. In this study, we sought to dissect the impact of the 17q12 locus on specific NK cell subpopulations and their functions. To address these questions, we leveraged a living biobank of >1600 healthy adult blood donors between 18 and 50 years of age (the Brigham and Women’s Hospital PhenoGenetic Project) and captured a detailed profiling of NK cell immunophenotypes and functional capacity from 96 randomly selected subjects (see Supplemental Table I for demographic details). Outcome measures included NK cell subpopulation frequency and functional response to different activation pathways. In our data processing pipeline, we comprehensively assessed the frequency of nine specific cell surface markers on NK cells using multicolor flow cytometry. Additionally, NK cell function was assessed in response to three different stimuli using intracellular cytokine staining for IFN-γ and MIP-1β, and NK cell degranulation, which serves as a surrogate marker for NK cell-mediated cytotoxicity (18). The frequency of NK cell subsets was expressed as a percentage of CD3−CD56+ bulk NK cells, CD56bright, or CD56dim cells. We performed association analyses between the rs9916629 variant and the quantitative traits derived from NK cell cytometric profiles to define the impact of this SNP on NK cells.
In our primary analysis, we set out to replicate the previously published association between the rs9916629C allele and reduced NK cell frequency in the peripheral circulation. We replicated the finding by Ferreira et al. (7) in our cohort of 96 healthy adult subjects: rs9916629C is associated with a lower frequency of CD3−CD56+ NK cells (p = 0.015 unadjusted; p = 0.036 after adjusting for age and sex, which independently influence the frequency of NK cells; median NK cell frequency [quartile 1–quartile 3] for each genotype as below: CC = 5.86% [4.23–9.63%], CT = 5.71% [4.39–9.02%], TT = 8.15% [5.54–13.4%]). There is no statistically significant association between rs9916629C and the absolute counts of total NK cells. Given that NK cells can be further divided into a minor regulatory CD56bright or the major cytotoxic CD56dim NK cell subpopulations in the blood, we next examined whether the rs9916629C allele was associated with any specific alteration in either of these two NK cell subpopulations. Interestingly, we found that rs9916629C was associated with a significant perturbation of NK cell subpopulations, marked by an increase in the frequency of CD56bright NK cells (p = 0.019) and a reduction in the overall frequency of CD56dim NK cells (p = 0.0069) (Fig. 1, Table I). Moreover, after adjusting for age and sex in a multivariate analysis, the rs9916629C association with elevated CD56bright NK cells (p = 0.019) and with reduced CD56dim NK cells (p = 0.011) persisted (Table I). When the analysis was restricted to a subset of subjects who are self-reported non-Hispanic whites (72% of the subjects), we observed the same association between rs9916629C and the primary outcomes (CD56bright, p = 0.029; CD56dim, p = 0.036), and the association in this subset of subjects trended toward statistical significance after adjusting for age and sex (CD56bright, p = 0.052; CD56dim, p = 0.065). Subjects of other ancestries were too few in number to analyze separately. Overall, the rs9916629C allele is thus associated with NK cell subset redistribution that includes a higher frequency of the regulatory CD56bright NK cells in addition to a general decrease in the total frequency of circulating NK cells.
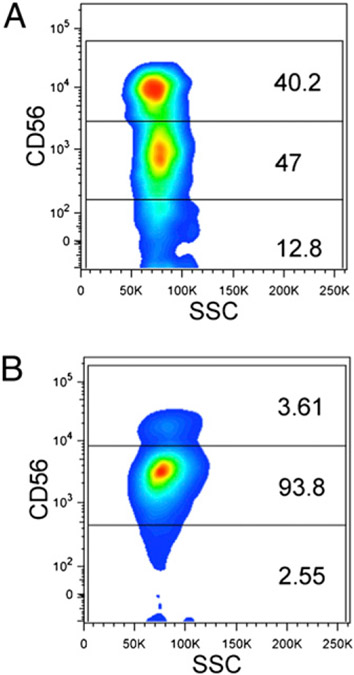
FIGURE 1.
Representative flow cytometry data for the primary outcome from two subjects. (A) Homozygote for the reference allele C; (B) homozygote for the T allele at the rs9916629 SNP.
Table I.
Candidate SNP (rs9916629) and primary outcome of NK cytometric profile in the healthy cohort
| Unadjusted |
Multivariatea |
|||
|---|---|---|---|---|
| NK Cells | Direction of Effect |
p | Direction of Effect |
p |
| CD56bright | ↑ | 0.019 | ↑ | 0.019 |
| CD56dim | ↓ | 0.0069 | ↓ | 0.011 |
Genotype and quantitative trait association was analyzed using PLINK with the C allele of rs9916629 as the reference allele. Direction of effect is relative to the C allele and is determined from the beta value.
In the multivariate analysis, the association between rs9916629 and the phenotype was adjusted for age and sex as covariates.
Multiparameter flow cytometric phenotypic profiling of NK cells
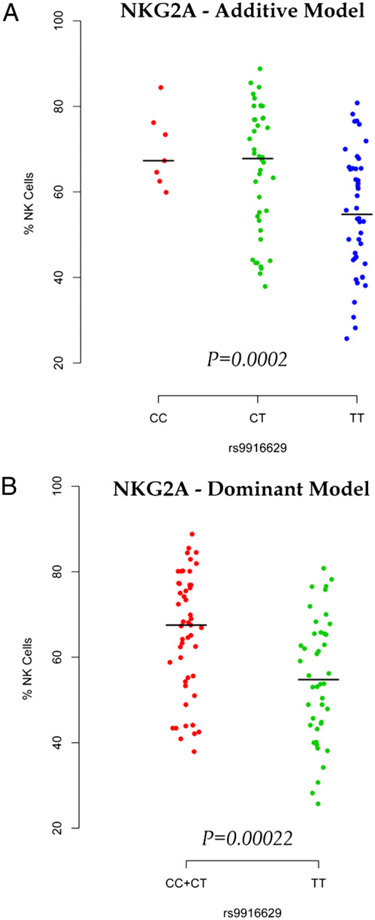
To further define whether the rs9916629C allele modulated the frequency of specific NK cell subpopulations, we next used targeted immunophenotypic profiling to extend our understanding of the impact of the SNP on the frequency of NK cells expressing particular NK cell receptors. Interestingly, we found an association between rs9916629C and the frequency of NK cells expressing the inhibitory NK cell receptor NKG2A (or CD159A) (Table II). Using our default additive model, the frequency of NK cells expressing the inhibitory NKG2A receptor increases for each additional rs9916629C allele (p = 0.00046 unadjusted; p = 0.0002 after adjusting for age and sex) (Fig. 2A), meeting a Bonferroni-corrected p value threshold for multiple comparisons of 0.0056 (given that one SNP was tested with nine markers of NK cell subpopulations). Because rs9916629CT and rs9916629CC subjects had similar median NKG2A frequencies (67.8 and 67.3%, respectively), we also tested the association between rs9916629 and NKG2A using a dominant model (rs9916629CT/CC versus rs9916629TT) and observed a similar result (Fig. 2B, Supplemental Table II) as that seen in the additive model for this SNP. Thus, we cannot distinguish whether the effect of rs9916629C is additive or dominant at this time.
Table II.
Candidate SNP (rs9916629) and secondary outcome of NK cell subpopulations in the healthy cohort
| Variable | Median Percentage of Expression (Quartile 1–Quartile 3) |
Unadjusted |
Multivariatea |
||||
|---|---|---|---|---|---|---|---|
| CC (n = 8) | CT (n = 43) | TT (n = 45) | Direction of Effect |
p | Direction of Effect |
p | |
| CD161/NK | 71.8 (49.9–76.6) | 72.7 (61.8–80.4) | 58.75 (47.85–72.85) | ↑ | 0.026 | ↑ | 0.038 |
| CD94/NK | 68.4 (58.6–74.4) | 64.4 (48.8–71.6) | 57.4 (45.25–67.6) | ↑ | 0.064 | ↑ | 0.030 |
| KIR/NK | 30.8 (25.75–36.4) | 28.8 (23.5–41) | 33.9 (24.7–40.5) | ↓ | 0.81 | ↑ | 0.97 |
| NKG2A/NK | 67.3 (62.5–76.2) | 67.8 (53.3–77.2) | 54.75 (44.3–65.5) | ↑ | 0.00046 | ↑ | 0.00020 |
| NKG2D/NK | 88.1 (80.7–96.6) | 85.8 (81.8–92.1) | 87.55 (80.55–91.45) | ↑ | 0.50 | ↑ | 0.66 |
| NKp30/NK | 70.7 (70.7–70.7) | 87.45 (82.7–92.2) | 87.05 (84.2–94) | ↓ | 0.11 | ↓ | 0.299 |
| NKp44/NK | 4.09 (2.59–6.19) | 3.7 (3.19–5.46) | 3.56 (2.89–5.04) | ↑ | 0.41 | ↑ | 0.64 |
| NKp46/NK | 94.6 (91.4–95.6) | 91.6 (86.9–95.2) | 90.8 (83.7–94.4) | ↑ | 0.068 | ↑ | 0.076 |
| Perforin/NK | 71.25 (64.85–73.75) | 77.9 (70.4–83.1) | 81.3 (74.3–86.7) | ↓ | 0.11 | ↓ | 0.11 |
| CD161/CD56bright | 47.3 (39.2–72.1) | 62.65 (44.8–69.9) | 50.65 (39–63.65) | ↑ | 0.086 | ↑ | 0.10 |
| CD94/CD56bright | 97.3 (93.1–99.4) | 96.15 (91.5–98.7) | 97.7 (95.3–98.75) | ↓ | 0.094 | ↓ | 0.14 |
| KIR/CD56bright | 5.94 (2.33–9.56) | 5.33 (2.68–8.65) | 4.42 (3.05–6.05) | ↑ | 0.61 | ↑ | 0.60 |
| NKG2A/CD56bright | 93.5 (91.2–97.7) | 92.7 (86.5–95.6) | 93.05 (89.4–95.4) | ↓ | 0.62 | ↓ | 0.74 |
| NKG2D/CD56bright | 86.9 (80.9–95.5) | 83.7 (76.1–91.1) | 86.5 (78.3–92.5) | ↓ | 0.80 | ↓ | 0.74 |
| NKp30/CD56bright | 97.5 (97.5–97.5) | 96.35 (93–99.7) | 95.9 (95.8–96.6) | ↑ | 0.55 | ↓ | 0.66 |
| NKp44/CD56bright | 15.9 (8.47–19.4) | 16.3 (10.8–20.7) | 18.1 (12.2–22.2) | ↓ | 0.32 | ↓ | 0.26 |
| NKp46/CD56bright | 98.7 (97.3–98.9) | 98.6 (96–99.1) | 99 (98.2–99.5) | ↓ | 0.079 | ↓ | 0.095 |
| Perforin/CD56bright | 6.61 (3.71–11.13) | 7.38 (4.21–10.9) | 6.53 (4.18–13.5) | ↑ | 0.51 | ↑ | 0.60 |
| CD161/CD56dim | 77.2 (50.5–87.9) | 76.15 (62.8–83.3) | 61.05 (49.1–76.6) | ↑ | 0.024 | ↑ | 0.043 |
| CD94/CD56dim | 66.2 (47.6–73) | 61.2 (46.6–70.9) | 56.4 (41.5–71.2) | ↑ | 0.22 | ↑ | 0.16 |
| KIR/CD56dim | 37 (31.7–42.75) | 35.3 (26.4–45.3) | 36.5 (26.8–45.1) | ↑ | 0.89 | ↑ | 0.73 |
| NKG2A/CD56dim | 65.5 (61.8–75.6) | 66.05 (51.2–79.2) | 53.85 (38.3–66.15) | ↑ | 0.0024 | ↑ | 0.0017 |
| NKG2D/CD56dim | 88.9 (82.1–97.1) | 88.25 (83.7–93.1) | 87.65 (80.35–92.3) | ↑ | 0.39 | ↑ | 0.51 |
| NKp30/CD56dim | 63.9 (63.9–63.9) | 84.45 (76.5–92.4) | 84.3 (80.8–93.2) | ↓ | 0.12 | ↓ | 0.35 |
| NKp44/CD56dim | 2.46 (2.1–3.97) | 2.31 (1.92–3.09) | 2.36 (2.03–3.19) | ↓ | 0.66 | ↓ | 0.48 |
| NKp46/CD56dim | 95.3 (91.3–95.9) | 93.3 (87.1–96.3) | 91.2 (83.4–94.7) | ↑ | 0.068 | ↑ | 0.085 |
| Perforin/CD56dim | 82.4 (77.7–84.45) | 87.8 (82.6–90.6) | 89.5 (85.6–92.6) | ↓ | 0.25 | ↓ | 0.21 |
Genotype and quantitative trait association were analyzed using PLINK with the C allele of rs9916629 as the reference allele. Direction of effect is relative to the C allele and is determined from the beta value. Boldface indicates a p value meeting a Bonferroni-corrected threshold for multiple comparisons. Alternative marker names: CD94 (KLRD1), CD159A (NKG2A), CD161 (KLRB1, NKR-P1A), CD314 (NKG2D), CD335 (NKp46), CD336 (NKp44).
In the multivariate analysis, association between rs9916629 and the phenotype was adjusted for age and sex as covariates.
FIGURE 2.
The effect of rs9916629C on the frequency of NKG2A-expressing NK subpopulation using additive (A) and dominant model (B). We compare the homozygotes for the alternative allele (TT) to the combined heterozygotes (CT) and homozygotes (CC) for the reference C allele. The reference allele frequency is 0.31.
Whereas NKG2A is expressed on both CD56bright and CD56dim NK cells, it is expressed with a higher frequency and at higher intensity on CD56bright NK cells than on CD56dim NK cells. We next assessed whether rs9916629C influences the overall NKG2A expression or expression of this inhibitory NK cell receptor on one of the two major NK cell subsets. We found that rs9916629C was associated with a higher frequency of CD56dim NK cells expressing NKG2A (p = 0.0024 unadjusted; p = 0.0017 after adjusting for age and sex) but did not influence the frequency of NKG2A-expressing CD56bright NK cells (Supplemental Table II). Of note, we also found a suggestive association between rs9916629C and a higher frequency of bulk NK cells expressing CD161 (p = 0.025 unadjusted; p = 0.038 after adjusting for age and sex) and a higher frequency of bulk NK cells expressing CD94 (p = 0.064 unadjusted; p = 0.030 after adjusting for age and sex). Thus, rs9916629C is specifically associated with an enrichment of NKG2A+CD56dim NK cells that also express higher levels of the NKG2A-heterodimeric partner CD94 and the inhibitory NK cell receptor CD161 in the peripheral blood.
Functional NK cell assay using flow cytometry
Because of the observed association between rs9916629C and the inhibitory NKG2A receptor, we next explored whether the rs9916629C variant influences the functional capacity of NK cells. To do so, we incubated PBMCs from each subject with PMA/ionomycin or one of the three tumor target cell lines, each stimulating a distinct NK activation pathway: 1) Ab-dependent cell-mediated cytotoxicity (ADCC) using Ab-coated p815 cells; 2) NKG2D-mediated activation using the MHC-devoid target cells line, K562 cells, in the absence of the inhibitory signal through interaction between MHC class I and KIR; or 3) natural cytotoxicity receptor (including NKp30 and NKp46)-mediated activation using an MHC class I-deficient B cell line, 721.221 cells, in the absence of MHC/KIR inhibition. For each stimulation condition, three functional properties of NK cells were interrogated: 1) degranulation, using surface CD107a as a surrogate marker of cytotoxicity (18); 2) secretion of IFN-γ, which has antiviral and proinflammatory properties; and 3) secretion of MIP-1β, a chemoattractant (Table III). Because three outcome measures were assessed for each of the four stimulation conditions (a total of 12 comparisons), we implemented a Bonferroni-corrected threshold of significance of p = 0.0042 in our primary analysis, which focused on bulk NK cells. We found that rs9916629 influenced the functional capacity of NK cells. Specifically, the rs9916629C allele was associated with increased expression of CD107a on the surface of NK cells following PMA/ionomycin stimulation (p = 0.0014 after adjusting for background staining; p = 0.0011 after adjusting for background, age, and sex) (Table III). This enhanced detection of a degranulation marker with rs9916629C was seen in both CD56bright and CD56dim NK cells, suggesting that both subsets of NK cells are more functionally active following maximal stimulation in the presence of this polymorphism. Similarly, there was a suggestive association between rs9916629C and enhanced degranulation in bulk NK cells as well as in CD56bright and CD56dim NK cells following stimulation with 721.221 cells (bulk, p = 0.017; CD56bright, p = 0.023; CD56dim, p = 0.018; after adjusting for background staining, age, and sex) (Table III), suggesting heightened functional activation in response to MHC class I-deficient B cells. Furthermore, we observed suggestive evidence of association between rs9916629C and reduced MIP-1β secretion by bulk NK cells following activation with Ab-coated target cells, which requires binding of the constant part of an Ab to FcγRIII (CD16) on the surface of NK cells to induce ADCC by NK cell activation (p = 0.016 after adjusting for background; p = 0.039 after adjusting for background, age, and sex) and to a lesser extent following stimulation through the NKG2D activation pathway with an MHC-devoid target cell line K562 (p = 0.039 after adjusting for background; p = 0.10 after adjusting for background, age, and sex). Taken together, rs9916629C was associated with a number of specific alterations, suggesting enhanced NK cell degranulation and reduced cytokine secretion in the presence of this allele.
Table III.
Candidate SNP (rs9916629) and secondary outcome of NK cell functional response following stimulation
| Condition | Variable | Median Percentage of Expression (Quartile 1–Quartile 3) |
Unadjusted |
Multivariatea |
||||
|---|---|---|---|---|---|---|---|---|
| CC (n = 8) | CT (n = 43) | TT (n = 45) | Direction of Effect |
p | Direction of Effect |
p | ||
| 721.221 | CD107a/NK | 11.18 (9.02–14.9) | 9.72 (7–14.9) | 8.04 (5.7–10.4) | ↑ | 0.021 | ↑ | 0.017 |
| 721.221 | IFN-γ/NK | 3.76 (2.6–7.7) | 3.2 (1.62–5.07) | 3.38 (2.19–4.89) | ↑ | 0.76 | ↑ | 0.82 |
| 721.221 | MIP-1β/NK | 30.8 (28.7–41.8) | 31.1 (22.5–42.3) | 37.15 (31.25–41.95) | ↓ | 0.062 | ↓ | 0.11 |
| 721.221 | CD107a/CD56bright | 10.12 (8.3–15.7) | 9.83 (5.64–14.3) | 6.5 (4.45–10.6) | ↑ | 0.02 | ↑ | 0.023 |
| 721.221 | IFN-γ/CD56bright | 1.44 (0.87–3.14) | 1.32 (0.71–2.66) | 1.5 (1–2.62) | ↓ | 0.89 | ↓ | 0.84 |
| 721.221 | MIP-1β/CD56bright | 15.7 (9.26–27.7) | 15.7 (12.25–22.55) | 15.6 (11.2–25.25) | ↓ | 0.63 | ↓ | 0.77 |
| 721.221 | CD107a/CD56dim | 12.2 (7.95–14.6) | 9.72 (6.97–14.5) | 7.68 (5.47–9.92) | ↑ | 0.021 | ↑ | 0.018 |
| 721.221 | IFN-γ/CD56dim | 4.8 (2.98–8.82) | 3.39 (1.88–5.39) | 3.59 (2.29–5.44) | ↑ | 0.58 | ↑ | 0.63 |
| 721.221 | MIP-1β/CD56dim | 38.8 (32.4–47.2) | 38.2 (28.6–46.9) | 41.6 (34.1–48.35) | ↓ | 0.18 | ↓ | 0.24 |
| K562 | CD107a/NK | 25.55 (23.15–34.65) | 26.2 (20.9–33.9) | 25.1 (16.6–30.7) | ↑ | 0.092 | ↑ | 0.06 |
| K562 | IFN-γ/NK | 5.31 (2.42–8.3) | 3.83 (2.84–6.02) | 4.34 (2.65–7.02) | ↑ | 0.77 | ↑ | 0.72 |
| K562 | MIP-1β/NK | 44.4 (40.5–48.9) | 43.1 (35.1–51.3) | 47.3 (39.85–56.25) | ↓ | 0.039 | ↓ | 0.10 |
| K562 | CD107a/CD56bright | 22.65 (15.35–28.15) | 20 (12–27.7) | 16.8 (13.3–23.6) | ↑ | 0.18 | ↑ | 0.17 |
| K562 | IFN-γ/CD56bright | 1.21 (0.75–2.68) | 1.53 (0.84–2.04) | 1.49 (0.89–2.11) | ↓ | 0.92 | ↑ | 0.92 |
| K562 | MIP-1β/CD56bright | 16.6 (8.44–23.5) | 18.5 (14.5–22.3) | 18.75 (11.85–23.9) | ↓ | 0.48 | ↓ | 0.69 |
| K562 | CD107a/CD56dim | 26.7 (25.15–40.7) | 27.7 (21.6–35) | 26 (17.5–31) | ↑ | 0.081 | ↑ | 0.056 |
| K562 | IFN-γ/CD56dim | 6.1 (3.27–10.11) | 4.48 (3.35–6.49) | 5.03 (2.86–7.57) | ↑ | 0.45 | ↑ | 0.41 |
| K562 | MIP-1β/CD56dim | 51.7 (45.7–59.9) | 53 (41.9–57.2) | 52.3 (42.35–62.6) | ↓ | 0.31 | ↓ | 0.53 |
| 815AB | CD107a/NK | 37.1 (25.5–42.65) | 36.5 (30.4–42.9) | 33.1 (24.9–42.1) | ↑ | 0.64 | ↑ | 0.48 |
| 815AB | IFN-γ/NK | 7.09 (5.65–10.54) | 5.5 (3.61–7.88) | 7.73 (4.21–10.84) | ↓ | 0.44 | ↓ | 0.57 |
| 815AB | MIP-1β/NK | 67.6 (59.1–78.6) | 72.4 (60–79.9) | 80.8 (72.95–85.4) | ↓ | 0.016 | ↓ | 0.039 |
| 815AB | CD107a/CD56bright | 18.6 (10.16–27.65) | 18.3 (11.7–25.7) | 15 (9.27–20.6) | ↑ | 0.5 | ↑ | 0.48 |
| 815AB | IFN-γ/CD56bright | 1.37 (1.24–2.62) | 1.34 (0.66–2.11) | 1.61 (0.86–2.15) | ↑ | 0.82 | ↑ | 0.84 |
| 815AB | MIP-1β/CD56bright | 26.2 (10.7–29.2) | 19.5 (13.4–26.1) | 20.55 (14.8–28) | ↓ | 0.49 | ↓ | 0.67 |
| 815AB | CD107a/CD56dim | 42.55 (30.25–46.4) | 38.6 (32.4–45.1) | 33.7 (25.8–44.1) | ↑ | 0.44 | ↑ | 0.34 |
| 815AB | IFN-γ/CD56dim | 9.8 (6.61–14.4) | 7.22 (5.41–10.1) | 9.41 (5.19–12.3) | ↑ | 0.89 | ↑ | 0.82 |
| 815AB | MIP-1β/CD56dim | 77.4 (72–90.7) | 84.7 (76.5–87.8) | 86.75 (79.85–90.15) | ↓ | 0.19 | ↓ | 0.3 |
| PMA | CD107a/NK | 33.45 (13.7–48.35) | 13.9 (8.39–28.3) | 10.3 (5.88–15.3) | ↑ | 0.0014 | ↑ | 0.0011 |
| PMA | IFN-γ/NK | 23.45 (15.8–38.9) | 9.95 (5.11–18.1) | 9.59 (3.41–18.7) | ↑ | 0.031 | ↑ | 0.054 |
| PMA | MIP-1β/NK | 75.1 (64.3–81.7) | 55.6 (47–65.2) | 58.85 (34.65–73.9) | ↑ | 0.38 | ↑ | 0.44 |
| PMA | CD107a/CD56bright | 59.35 (30.35–75.95) | 36.5 (20.9–61.3) | 24.5 (15.4–42.3) | ↑ | 0.0055 | ↑ | 0.007 |
| PMA | IFN-γ/CD56bright | 26.15 (21.35–52.35) | 20.2 (9.15–29.5) | 16.3 (10.8–26.4) | ↑ | 0.04 | ↑ | 0.056 |
| PMA | MIP-1β/CD56bright | 82.9 (80.6–93.6) | 72.7 (63–81.8) | 74.05 (60.5–85) | ↑ | 0.46 | ↑ | 0.4 |
| PMA | CD107a/CD56dim | 19.6 (8.46–35.4) | 9.85 (4.63–16.7) | 5.72 (3.78–10.7) | ↑ | 0.0096 | ↑ | 0.0098 |
| PMA | IFN-γ/CD56dim | 23.9 (11.65–30.7) | 7.99 (2.8–15.4) | 7.81 (2.42–18.4) | ↑ | 0.1 | ↑ | 0.17 |
| PMA | MIP-1β/CD56dim | 69.4 (60.5–80.3) | 61.5 (43.3–72.5) | 62.55 (29.25–77.05) | ↑ | 0.27 | ↑ | 0.34 |
Genotype and quantitative trait association were analyzed using PLINK with the C allele of rs9916629 as the reference allele. Direction of effect is relative to the C allele and is determined from the beta value. In the multivariate analysis, association between rs9916629 and the phenotype was further adjusted for age and sex as covariates. Boldface indicates a p value meeting a Bonferroni-corrected threshold for multiple comparisons. 721.221, HLA class I-deficient B cell line; 815AB, p815 Ab (a mouse leukemic cell line coated with specific Ab; Ab-dependent cell-mediated cytotoxicity); CD107a, marker of degranulation; IFN-γ, IFN-γ–secreting NK cells; K562, MHC-devoid target cell line; MIP-1β, macrophage inflammatory protein-1β–secreting NK; PMA, phorbol 12-myristate 13-acetate and ionomycin (positive control).
Outcomes were adjusted for the appropriate background (media alone or uncoated p815 cell line).
RNA expression and expression quantitative trait locus analysis
To investigate potential mechanisms by which rs9916629C might alter NK cell subset distribution and function, we examined the RNA expression profile of bulk CD3−CD56+ NK cells in a subset of subjects (n = 20; 21% of total study population). Performing an expression quantitative trait locus analysis, we first assessed genes in the 17q12 locus, since such genes are the most likely to be directly influenced by genetic variation in their vicinity. Because rs9916629 is located near SLFN13, a member of the family of Schlafen genes that are all found in a cluster at 17q12, we assessed its effect on the level of expression of all SLFN genes on which data were available: the level of expression of probe sets interrogating SLFN1, 5, 11, 12, 12L, and 13 were not associated with rs9916629. On an exploratory basis, we assessed our RNA data for an effect of rs9916629 on all other interrogated transcripts, and we observed changes in the expression levels of 126 genes within the purified NK cell population that were suggestively associated with rs9916629C (p < 0.01; Supplemental Table III). Given our small sample size, no single transcript had significant evidence of association after correcting for the testing of multiple hypotheses. Among the suggested associations, interestingly, we found that rs9916629C was associated with increased expression of TBX21 (p = 0.0037), which encodes the T-box transcription factor 21 (also known as T-BET) (Supplemental Fig. 1) previously implicated in the development and differentiation of NK cells (19). When applying the Ingenuity pathway analysis tools (Ingenuity Systems, http://www.ingenuity.com) to identify the networks of genes whose expressions are coordinately influenced by rs9916629C, we observed, in this unsupervised analysis, that the best scoring model (score of 20) contained TBX21 as well as 12 other genes, suggesting that increased TBX21 expression may be part of a transcriptional program influenced by rs9916629C.
Discussion
NK cells are an important effector component of the innate immune system that play a central role in eliminating infected or malignantly transformed cells as well as in qualitatively modulating adaptive immunity. Thus, identifying the genetic factors influencing NK cell frequency, phenotype, or function may lead to improved prediction models for disease susceptibility or clinical outcomes and discovery of potential therapies that specifically target the function of NK cells. Following the recent report of an SNP (rs9916629) linked to reduced NK cell frequency (7), we sought to replicate this finding and to examine the previously unaddressed questions of whether genetic variation at this locus affects particular NK cell subpopulations and functional profiles. Informed by the functionally distinct NK cell subsets distinguished based on the intensity of CD56 staining, CD56bright and CD56dim, our analytic approach uncovers a more complex effect of the 17q12 locus on NK cells and refines the original association within a much smaller subject sample size (n = 96) than the original study (n = 2538), which used a coarser CD3−CD56+ definition for NK cells (7). Given their distinct functions, the failure to distinguish the CD56+ NK cell subsets may, as in this case, result in averaging opposing effects in different sub-populations and dilution of an association with the NK cell phenotype. Because age affects NK cells, it is also important that our study extends previous findings to an adult population since the subjects in the cohort studied by Ferreira et al. (7) were pre-dominantly adolescents.
The detailed profiling of NK cell immunophenotypes and functional responses that we have generated provides additional insights into how the rs9916629C variant influences NK cell frequency and response. Overall, several observations support the conclusion that individuals bearing one or two copies of the rs9916629C allele exhibit an altered peripheral NK cell profile when compared with individuals lacking the rs9916629C allele. First, rs9916629C is associated with an intriguing redistribution of NK cell subsets with a reduction in the CD56dim NK cells and an expansion of the CD56bright NK cells. These immmunoregulatory CD56bright NK cells secrete copious amounts of cytokines and chemokines that are essential in initiating the early inflammatory response. Second, this allele is associated with the preferential accumulation of NK cells expressing the inhibitory c-type lectin NKG2A and its heterodimeric partner, CD94. Third, rs9916629C shows a robust correlation with enhanced degranulation (CD107a) by bulk NK cells as well as by CD56bright and CD56dim cells following maximum stimulation in response to PMA/ionomycin and following stimulation with an MHC class I-deficient B cell line. Finally, this allele is associated with decreased secretion of the chemoattractant MIP-1β by bulk NK cells following FcγRIIIa activation in the context of ADCC, and possibly following exposure to an MHC-devoid target cell line. These results clearly demonstrate that genetic variation at the 17q12 locus has pleiotropic effects on both the phenotype and function of NK cells.
Note that because this variant emerged from a genome-wide association scan, it is probably not the causal variant within this locus. Fine mapping with interrogation of all genetic variants in the region will be necessary to identify the causal variant. In the interim, rs9916629 is a good surrogate marker that captures the effect of the 17q12 locus, which may be dominant or additive, as our current data fit either model equally well. Although the impact of rs9916629 on the distribution of NK cell surface phenotypes is more clearly appreciated, the functional impact of the SNP is not as clear in part due to the heterogeneity in NK cell activity from subject to subject. Functional heterogeneity is linked to differences in KIR/HLA backgrounds that alter the functional licensing of NK cells, resulting in different levels of activity against generic target cell lines. Only extremely large cohort studies could normalize for the impact of the extreme genetic variability within the KIR and HLA loci. In our relatively small cohort, the rs9916629C was associated with alterations in NK cell responsiveness to PMA, more readily than MHC-devoid target cell lines, strongly suggesting that CD56bright, rather than CD56dim, NK cells are functionally modulated by this genetic variant.
Although NKG2A serves as the dominant self-sensing inhibitory signal on CD56bright NK cells, it is also expressed on the CD56dim NK cells. Interestingly, high expression level of NKG2A has been suggested as a marker of terminal differentiation in NK cells, CD56brightNKG2A+KIR− NK cells are thought of as precursors of CD56dim cells (20), and expression of NKG2A varies widely in the setting of different viral infections and likely results in altered NK cell functional activity (21-23). Moreover, the predominant self-ligand for NKG2A is the MHC class I Ag E (HLA-E), a relatively nonpolymorphic variant of the major histocompatibility genes, which presents the leader peptide of other MHC class I genes (24). Whereas HLA-E is typically expressed at low levels, inflammation due to infection or malignancy has been shown to alter the expression of this NKG2A ligand, and this interaction between NKG2A and HLA-E likely plays an important role in providing inhibitory signals to peripheral NK cells. Furthermore, there is evidence that the NKG2A/HLA-E interaction may also play a critical role during NK cell development, where NKG2A is expressed early in the developmental pathway of this lymphocytic cell subset (20, 25). Given that our study involved detailed characterization of NK cells but not their targets, we did not examine the expression of HLA-E to determine whether increased ligand expression may increase NKG2A expression or NK cell expansion. We did not observe any obvious change in HLA-E expression in the transcriptional analysis of purified NK cell populations in individuals that possess the rs9916629C allele (data not shown). It is plausible that changes in ligand expression on other more relevant cells (e.g., stromal cells, APCs) may more profoundly impact NK cell development and warrant further investigation to define the specific mechanism by which the SNP may alter NK cell development. Taken together, the observations that rs9916629 is associated with not only a shift toward the CD56bright NK subset, which typically expresses more NKG2A, but also with an overall increased expression of this marker on CD56dim NK cells as well as an accompanying enhancement of NK cell degranulation (a hallmark effector function of CD56dim NK cells) potentially reveal a previously unappreciated role of this inhibitory receptor in NK cell licensing (16) or education (4, 26). The functional activity of NK cells is determined early in NK cell development through the interaction between self-ligands and inhibitory NK cell receptors such as KIRs and NKG2A. It is plausible that rs9916629C, by affecting NKG2A expression, may also influence NK cell licensing, as we observed enhanced NK cell degranulation and elevated NKG2A expression that are associated with this SNP.
To gain further insights into the role of this SNP in modulating NK cell development, we performed an analysis of RNA expression in a subset of subjects. Among these individuals, we did not find an effect on nearby genes of the SLFN family but noticed suggestive associations between the rs9916629C allele and many different transcripts, which include increased TBX21 RNA expression with rs9916629C. TBX21 has been implicated in autoimmune diseases, including experimental autoimmune encephalomyelitis (27, 28). Intriguingly, TBX21 is also a master regulator of commitment to the Th1 lineage, is expressed at the T/NK cell lymphocyte progenitor phase, and regulates the expression of IFN-γ in Th1-derived cells and NK cells (19, 29-31). NK cell differentiation is dramatically reduced in mice lacking the murine TBX21 homolog (19), demonstrating the essential role of this transcription factor in NK cell development. Furthermore, the observation that TBX21 regulates the expression of sphingosine 1-phosphate receptor 5 on NK cells, which is essential for NK cell egress from the lymph nodes, suggests that TBX21 may also influence the distribution of NK cells in blood (32). We therefore speculate that elevated TBX21 expression, in the presence of rs9916629C, may affect NK cell differentiation. This is supported by our observation that the rs9916629C allele was associated with higher TBX21 RNA expression in human NK cells as well as a lower frequency of CD56dim NK cells and higher frequency of CD56bright NK cells in the peripheral circulation. Thus, rs9916629C may alter NK cell frequency and phenotype by influencing the expression of early transcription factors required for NK cell development. This interesting hypothesis will require further investigation to be rigorously tested and elaborated.
Although we have provided possible explanations for the association between the candidate SNP and the observed NK phenotype, the exact mechanism by which the 17q12 locus affects TBX21 expression and NK cell biology remains unclear. Given the large distance between TBX21 and rs9916629 (12 Mb), it is unlikely that the rs9916629 variant has a direct effect on TBX21 expression. The influence on TBX21 expression is likely mediated by an indirect effect of the chromosomal region containing rs9916629. Interestingly, the rs9916629 SNP is found between two members of the Schlafen family of genes (SLFN12L and SLFN13) (University of California at Santa Cruz Genome Browser, http://genome.ucsc.edu) (33). Although little is known about these genes, other members of this gene family, which are located nearby, have been implicated as negative regulators of lymphocyte and thymocyte proliferation (34). Furthermore, SLFN2 and SLFN5 are involved in responses to type I IFN stimulation (35, 36), which plays an important role in NK cell activation. Thus, it is plausible that rs9916629 influences the function of a specific Schlafen family gene that may regulate the expression of TBX21, which may in turn regulate NK cell differentiation and maturation. Fine mapping of the 17q12 locus will provide insights into the identity of the gene(s) that may mediate the association between rs9916629 and NK cell phenotypes and functions.
In conclusion, we validate and refine the association of the 17q12 locus with NK cell frequency by demonstrating that an SNP in this locus is associated with the frequency of two major subsets of NK cells: the rs9916629C allele is associated with a larger proportion of regulatory CD56bright and a smaller proportion of cytotoxic CD56dim cells. The shift toward the CD56bright NK cell profile occurs in conjunction with immunophenotypic and functional alterations that suggest a possible mechanism by which the 17q12 locus influences the differentiation and function of NK cells: it may alter the function of members of the Schlafen gene family with repercussions on transcriptional programs that include the NK cell fate-determining gene TBX21.
Supplementary Material
Acknowledgments
We thank participants in the Brigham and Women’s Hospital PhenoGenetic Project. We also acknowledge Drs. David Goldstein and Kevin Shianna’s laboratory for generating RNA expression data from NK cells.
This work was partially supported by a Ragon Institute pilot study grant (to P.L.D.J. and G.A.). Z.X. is a recipient of the Clinician Scientist Development award from the National Multiple Sclerosis Society and the American Academy of Neurology and is a fellow in the Clinical Investigator Training Program (Beth Israel Deaconess Medical Center–Harvard Medical School, in collaboration with Pfizer, Inc. and Merck & Co.). P.L.D.J. is a Harry Weaver Neuroscience Scholar of the National Multiple Sclerosis Society. C.T.B. is partially supported by a grant from the University of Basel, Basel, Switzerland.
Abbreviations used in this article:
- ADCC
Ab-dependent cell-mediated cytotoxicity
- KIR
killer Ig-related receptor
- MIP
macrophage inflammatory protein
- SNP
single nucleotide polymorphism
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Berger CT, and Alter G. 2011. Natural killer cells in spontaneous control of HIV infection. Curr. Opin. HIV AIDS 6: 208–213. [DOI] [PubMed] [Google Scholar]
- 2.Cerwenka A, and Lanier LL. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol 1: 41–49. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, et al. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. USA 105: 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, and Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331: 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper MA, Fehniger TA, and Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22: 633–640. [DOI] [PubMed] [Google Scholar]
- 6.Caligiuri MA 2008. Human natural killer cells. Blood 112: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira MA, Mangino M, Brumme CJ, Zhao ZZ, Medland SE, Wright MJ, Nyholt DR, Gordon S, Campbell M, McEvoy BP, et al. ; and International HIV Controllers Study. 2010. Quantitative trait loci for CD4: CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am. J. Hum. Genet 86: 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall MA, Ahmadi KR, Norman P, Snieder H, MacGregor AJ, Vaughan RW, Spector TD, and Lanchbury JS. 2000. Genetic influence on peripheral blood T lymphocyte levels. Genes Immun. 1: 423–427. [DOI] [PubMed] [Google Scholar]
- 9.Grieco FA, Vendrame F, Spagnuolo I, and Dotta F. 2011. Innate immunity and the pathogenesis of type 1 diabetes. Semin. Immunopathol 33: 57–66. [DOI] [PubMed] [Google Scholar]
- 10.Rodacki M, Milech A, and de Oliveira JE. 2006. NK cells and type 1 diabetes. Clin. Dev. Immunol 13: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi R, Laroni A, and Weiner HL. 2010. Role of the innate immune system in the pathogenesis of multiple sclerosis. J. Neuroimmunol 221: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schleinitz N, Vély F, Harlé JR, and Vivier E. 2010. Natural killer cells in human autoimmune diseases. Immunology 131: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Jager PL, Rossin E, Pyne S, Tamayo P, Ottoboni L, Viglietta V, Weiner M, Soler D, Izmailova E, Faron-Yowe L, et al. 2008. Cytometric profiling in multiple sclerosis uncovers patient population structure and a reduction of CD8low cells. Brain 131: 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA, McFarland H, Henkart PA, and Martin R. 2006. Regulatory CD56bright natural killer cells mediate immunomodulatory effects of IL-2Rα-targeted therapy (daclizumab) in multiple sclerosis. Proc. Natl. Acad. Sci. USA 103: 5941–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin JF, Perry JS, Jakhete NR, Wang X, and Bielekova B. 2010. An IL-2 paradox: blocking CD25 on T cells induces IL-2-driven activation of CD56bright NK cells. J. Immunol 185: 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanier LL 2005. NK cell recognition. Annu. Rev. Immunol 23: 225–274. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, and Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alter G, Malenfant JM, and Altfeld M. 2004. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294: 15–22. [DOI] [PubMed] [Google Scholar]
- 19.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, and Glimcher LH. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity 20: 477–494. [DOI] [PubMed] [Google Scholar]
- 20.Béziat V, Descours B, Parizot C, Debré P, and Vieillard V. 2010. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE 5: e11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison RJ, Ettorre A, Little AM, and Khakoo SI. 2010. Association of NKG2A with treatment for chronic hepatitis C virus infection. Clin. Exp. Immunol 161: 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tjwa ET, van Oord GW, Hegmans JP, Janssen HL, and Woltman AM. 2011. Viral load reduction improves activation and function of natural killer cells in patients with chronic hepatitis B. J. Hepatol 54: 209–218. [DOI] [PubMed] [Google Scholar]
- 23.Zhang R, Xu J, Hong K, Yuan L, Peng H, Tang H, Ma P, Zhang Y, Xing H, Ruan Y, and Shao Y. 2007. Increased NKG2A found in cytotoxic natural killer subset in HIV-1 patients with advanced clinical status. AIDS 21(Suppl 8): S9–S17. [DOI] [PubMed] [Google Scholar]
- 24.Braud VM, Allan DS, O’Callaghan CA, Söderström K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391: 795–799. [DOI] [PubMed] [Google Scholar]
- 25.Sivakumar PV, Gunturi A, Salcedo M, Schatzle JD, Lai WC, Kurepa Z, Pitcher L, Seaman MS, Lemonnier FA, Bennett M, et al. 1999. Cutting edge: expression of functional CD94/NKG2A inhibitory receptors on fetal NK1.1+Ly-49− cells: a possible mechanism of tolerance during NK cell development. J. Immunol 162: 6976–6980. [PubMed] [Google Scholar]
- 26.Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25: 331–342. [DOI] [PubMed] [Google Scholar]
- 27.Peng SL 2006. The T-box transcription factor T-bet in immunity and auto-immunity. Cell. Mol. Immunol 3: 87–95. [PubMed] [Google Scholar]
- 28.Nath N, Prasad R, Giri S, Singh AK, and Singh I. 2006. T-bet is essential for the progression of experimental autoimmune encephalomyelitis. Immunology 118: 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SA, and Weinmann AS. 2010. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol. Rev 238: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, and Glimcher LH. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100: 655–669. [DOI] [PubMed] [Google Scholar]
- 31.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, and Glimcher LH. 2002. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295: 338–342. [DOI] [PubMed] [Google Scholar]
- 32.Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, Xu Y, Roots CM, Beilke JN, Banerjee A, et al. 2009. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J. Exp. Med 206: 2469–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz DA, Katayama CD, and Hedrick SM. 1998. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9: 657–668. [DOI] [PubMed] [Google Scholar]
- 35.Katsoulidis E, Carayol N, Woodard J, Konieczna I, Majchrzak-Kita B, Jordan A, Sassano A, Eklund EA, Fish EN, and Platanias LC. 2009. Role of Schlafen 2 (SLFN2) in the generation of interferon a-induced growth inhibitory responses. J. Biol. Chem 284: 25051–25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N, Sawicki KT, Munshi HG, and Platanias LC. 2010. Role of interferon alpha (IFNα)-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J. Biol. Chem 285: 40333–40341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.