Abstract
Developing highly accurate and simple approaches to rapidly identify and isolate SARS-CoV-2 infected patients is important for the control of the COVID-19 pandemic. We, herein, reported the performance of a Cas12a-assisted RTF-EXPAR strategy for the identification of SARS-CoV-2 RNA. This assay combined the advantages of RTF-EXPAR with CRISPR-Cas12a can detect SARS-CoV-2 within 40 min, requiring only isothermal control. Particularly, the simultaneous use of EXPAR amplification and CRISPR improved the detection sensitivity, thereby realizing ultrasensitive SARS-CoV-2 RNA detection with a detection limit of 3.77 aM (∼2 copies/μL) in an end-point fluorescence read-out fashion, and at 4.81 aM (∼3 copies/μL) level via a smartphone-assisted analysis system (RGB analysis). Moreover, Cas12a increases the specificity by intrinsic sequence-specific template recognition. Overall, this method is fast, sensitive, and accurate, needing minimal equipment, which holds great promise to meet the requirements of point-of-care molecular detection of SARS-CoV-2.
Keywords: SARS-CoV-2 RNA, CRISPR-Cas12a, RTF-EXPAR, Isothermy, End-point fluorescence
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus, has caused the pandemic of coronavirus disease 2019 (COVID-19), threatening all aspects of human life (Anderson et al., 2021; Gorbalenya et al., 2020; Hanna et al., 2020; Matheson and Lehner, 2020). Mass screening of symptomatic and asymptomatic viral infections may allow for rapid intervention, and is helpful to slow down the fast spread of SARS-CoV-2 (Derakhshan et al., 2021; Kevadiya et al., 2021; Service, 2020).
Nucleic-acid detection is the most robust and accurate method for pathogen diagnostic, and PCR-based nucleic acid assay is considered the gold standard of COVID-19 confirmation (Broughton et al., 2020; Corman et al., 2020). However, PCR-based approaches need specialized equipment, expensive reagents, trained personnel, and a long turnaround time (>2 h) (Wang et al., 2022), which has been limited in resource-limited areas (Corman et al., 2020; Lee et al., 2021; Udugama et al., 2020). Therefore, developing rapid, portable, and easy-to-deploy point-of-care (POC) molecular diagnostics is a pressing need for frequent testing in lacking vaccines and effective therapeutics for COVID-19 (Derakhshan et al., 2021; Pokhrel et al., 2020; Song et al., 2021).
Compared with PCR methods, isothermal amplification has the advantages of short reaction time and no need for qualified personnel or sophisticated equipment, and is a suitable candidate for POC application (Glökler et al., 2021; Li and Macdonald, 2015). The common isothermal methods include loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) (Notomi et al., 2000; Piepenburg et al., 2006). They have been established for SARS-CoV-2 diagnosis outside of complex laboratories and can produce rapid results (<1 h) (Dao Thi et al., 2020; Feng et al., 2021; Pang et al., 2020; Xia and Chen, 2020). Another typical test in this category is Abbott's ID NOW COVID-19 which can obtain positive results within 5 min and negative results in 13 min based on a nicking endonuclease isothermal amplification reaction (NEAR) (Tauh et al., 2021; Tu et al., 2021). Notably, Carter et al. recently described a new isothermal method (called RTF-EXPAR) by combining exponential amplification reaction (EXPAR) with reverse transcription-free (RTF) step to convert RNA to DNA to achieve indirectly detection of the Orf1ab gene, allowing complete test within 10 min (Carter et al., 2021). Despite great progress, these isothermal amplification diagnostics often suffer from problems associated with nonspecific DNA amplification, followed by high rates of false-positive diagnosis (Chen et al., 2021b; Feng et al., 2021; Reid et al., 2018; Schneider et al., 2019; Tomita et al., 2008). Moreover, they are much less sensitive than RT-qPCR (Glökler et al., 2021). In addition, although these isothermal methods can avoid complicated thermal cycling, some of them still require complex readout instruments that are challenging for personal use (De Felice et al., 2022).
Consequently, many recent research efforts are focused on overcoming these drawbacks. One of the most effective and attractive strategies is to use RNA-guided CRISPR-Cas detection modalities following the isothermal amplification step (Li et al., 2019b; Wu et al., 2021). CRISPR-Cas system, a form of the prokaryotic adaptive immune system, has emerged as a powerful, easy-to-use, and field-deployable diagnostic technology for nucleic acid detection, in addition to successful gene editing (Liu et al., 2021; van Dongen et al., 2020; Wei et al., 2018; Zhao et al., 2021). A crucial feature of CRISPR-Cas processes is that their recognition interactions highly depend on the sequence of the template (Li et al., 2019b). Such sequence-specific recognition enabled the CRISPR-Cas to correct the possible Error-amplification. Besides, upon binding of targets, some Cas proteins, such as Cas12 and Cas13, unleash trans-cleavage (also called collateral cleavage) to degrade non-specific single-stranded nucleic acids, endowing Cas-based methods with improved sensitivity (Chen et al., 2018; Cox et al., 2017; Gootenberg et al., 2018; Li et al., 2018). In addition, such CRISPR-based tools could provide an easy-to-use readout using minimal instrumentation (Chen et al., 2021a; de Puig et al., 2021; Hang et al., 2022; Liang et al., 2019). Both RT-LAMP and RPA have been combined with CRISPR-Cas for clinical detection of SARS-CoV-2 (i.e. HOLMESv2, SHERLOCK, and DETECTR) (Chen et al., 2018; Gootenberg et al., 2017; Li et al., 2019a). The simplicity of these methods makes them attractive for POC testing (Chen et al., 2020; Zhou et al., 2022). However, they still require a total reaction time of approximately 1 h. Newly developed RT-EXPAR is ultrarapid (Carter et al., 2021), but thus far has not been found use in combination with CRISPR for accurate, sensitive, and simple SARS-CoV-2 detection.
Here, we show that RTF-EXPAR can be combined with CRISPR-Cas12a (named Cas12a-assisted RTF-EXPAR) for diagnostic of SARS-CoV-2 in 40 min using end-point fluorescence signal output, with detection limit as low as 3.77 aM (∼2 copies/μL). To further simplify the assay and provide a user-friendly diagnostic platform, we use the smartphone in combination with a mobile app for fluorescence results readout that allows detection of virus RNA down to 4.81 aM (∼3 copies/μL). This method needs minimal equipment, and is fast, sensitive, and accurate, expecting to meet the needs of POC detection of virus RNA.
2. Experimental section
2.1. Reagents and materials
Bst DNA polymerase (Large Fragment) derived from Bacillus stearothermophilus and 10 × thermoPol buffer were obtained from Nanjing Vazyme Biotech Co., Ltd. (Nanjing, China). Nt.BstNBI, BstNI, and 10 × NEBuffer 3.1 were purchased from New England Biolabs (NEB). LbCas12a (cpf1) was provided by Tolo Biotech Co., Ltd (Shanghai, China). Diethyl pyrocarbonate (DEPC)-treated water, deoxynucleotide triphosphates (dNTPs), 4S GelRed, DNA Marker, and all the DNA oligonucleotides were ordered from Sangon Biotechnology Co., Ltd. (Shanghai, China). SARS-CoV-2 RNA transcribed in vitro standard material was provided by National Institute of Metrology, China, while the gene composition and quantitative concentration were shown in Table S3. SARS-CoV-2 RNA, MERS-CoV RNA, SARS-CoV RNA, and crRNA were synthesized by Gene Pharma (Shanghai, China) and purified by HPLC. The corresponding sequences of the above-mentioned oligonucleotides were displayed in supporting information (SI, Tables S1 and S2).
2.2. Apparatus and instruments
Polyacrylamide gel electrophoresis (PAGE) was carried out on an electrophoresis analyzer (DYY-6C, Beijing, China) and gel images were performed on an imaging system (Tanon 5200Multi, Shanghai, China). In the benchtop detection, fluorescence signals were recorded on a multifunctional microplate reader (Synergy H4, BioTek, USA). In the smartphone-assisted detection, the samples were irradiated with a 302 nm UV lamp in a UV transmission reflectance analyzer (ZF-90, Shanghai, China), and photographed and analyzed with iPhone 12.
2.3. Optimization of crRNAs
The detailed process of activity comparison of crRNAs was provided in SI.
2.4. RTF reaction
RTF reaction is the first step in the overall assay, which converts viral RNA into short DNA via selective cleavage of DNA in RNA/DNA heteroduplexes by BstNI. For reaction, a total volume of 10 μL mixture of RTF reaction reagents was incubated at 50 °C for 5 min. This mixture was composed of SARS-CoV-2 RNA (4 μL, different concentrations), convertor DNA (2 μL, 10 nM), DEPC-treated water (2.2 μL), NEBuffer 3.1 (1 μL, 10 × ) and BstNI (0.8 μL, 10 U/μL).
2.5. EXPAR amplification
The product of RTF was used as the primer to trigger the EXPAR amplification, and 8 μL product was transferred into a test tube before the next operation. The preparation of the other EXPAR reagents (Parts A and B) was performed separately on ice. Part A was composed of template X′-X′-Y′ (2 μL, 100 nM), dNTPs (2 μL, 10 mM), thermoPol buffer (1 μL, 10 × ), MgSO4 (1.2 μL, 100 mM), and DEPC-treated water (0.8 μL). Part B consisted of NEBuffer 3.1 (0.2 μL, 10 × ), DEPC-treated water (3.8 μL), Nt.BstNBI (0.4 μL, 10 U/μL) and Bst DNA polymerase (0.6 μL, 1 U/μL). EXPAR amplification was conducted by immediately adding Parts A and B to the primer-containing test tube and incubating at 50 °C for 4 min, then stopped by heat treatment (95 °C for 10 min). The viability of RTF reaction and EXPAR amplification was demonstrated by PAGE, and the detailed procedure was placed in SI.
2.6. Cas12a-mediated assay
CRISPR-Cas12a cleavage was performed in the 30 μL reaction solution including 50 nM Cas12a-crRNA complex, 0.3 μM reporter DNA, 1 × TOLO Buffer 3, and 18 μL of RTF-EXPAR product. The above solution was allowed to incubate at 45 °C for 20 min. Subsequently, the volume was replenished to 110 μL with DEPC-treated deionized water, the final product was transferred to a multifunctional microplate reader for end-point fluorescence analysis. In addition, for simple visualization, the completed Cas12a-assisted RTF-EXPAR reaction tubes were irradiated with UV light (302 nm), followed by the fluorescence signal captured with a smartphone camera, and finally analyzed by app (Palette Cam). The complete operating procedure of this app (Fig. S5) and its specific information were presented in SI.
3. Results and discussion
3.1. Detection mechanism
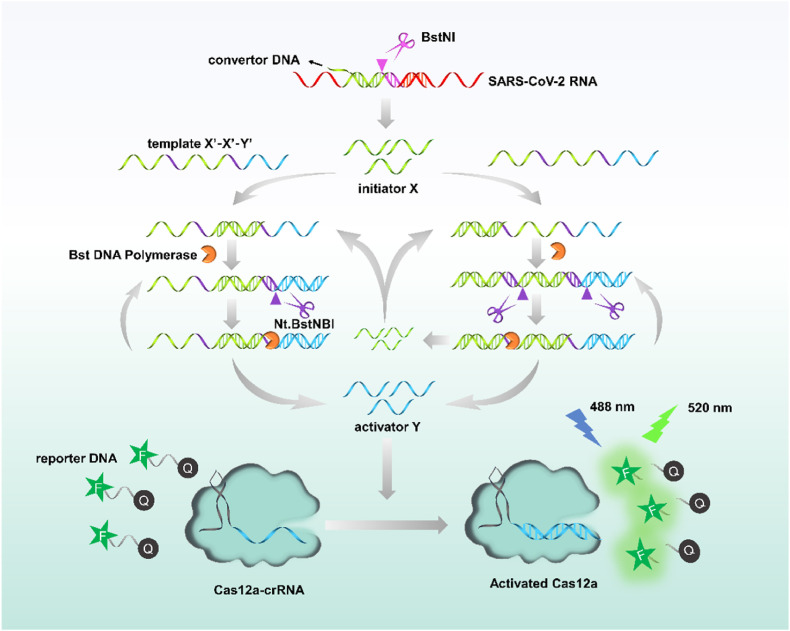
As shown in Scheme 1 , the procedures of the assay consisted of three steps: RTF transformation, EXPAR amplification, and CRISPR-Cas12a signal output. In this proof-of-principle work, an RTF convertor DNA was elaborately designed with the capability of partially hybridizing with the Orf1ab gene of SARS-CoV-2 RNA to form RNA/DNA heteroduplexes, which contains an endonuclease recognition site of BstNI (Table S4); an EXPAR amplification template oligonucleotide was designed including two repeat X′ sequences and one Y′ sequence, where X′ is complementary to initiator X, Y′ serves as the bridge for exponential amplification and CRISPR-Cas12a signal readout. In detail, once convertor DNA binds to the target RNA, it will be cut by BstNI, thus a short DNA (X) was released. This short X will hybridize with the template X′-X′-Y′ and experience a chain extension along the template with the assistance of Bst polymerase with strand displacement activity, generating a double-stranded DNA (dsDNA) containing two nicking sites (Table S4). The Nt.BstNBI thus cut this dsDNA in these nicking sites, producing a new X sequence and a new Y sequence. Newly formed X can be utilized as a primer to initiate new cycles of amplification reactions. The hybridization of X with X′-X′-Y′, under the collaboration of Bst polymerase and Nt.BstNBI, causes the repeated production of X and Y in this way, and finally amplifies plentiful X and Y to realize exponential amplification. Thereafter, generated activator Y binds to Cas12a-crRNA complexes and activates the trans-cleavage capability of Cas12a on random single-stranded DNA (ssDNA). Ultimately, the activated Cas12a cleaves the reporter DNA (FAM-TTATT-BHQ1) in a high-speed manner, thereby restoring the quenched fluorescence and producing a fluorescence signal. As a result, the target RNA can be quantified by monitoring the fluorescence signal.
Scheme 1.
Reaction mechanism of SARS-CoV-2 RNA detection based upon Cas12a-assisted RTF-EXPAR reaction.
Notably, EXPAR-based assays are prone to false-positive results due to non-specific amplification. In our strategy, in addition to introducing phosphate groups at the ends of the convertor and template DNAs, the specific recognition RTF-EXPAR product by CRISPR-Cas12a can effectively reduce false-positive signal resulting from non-specific amplification.
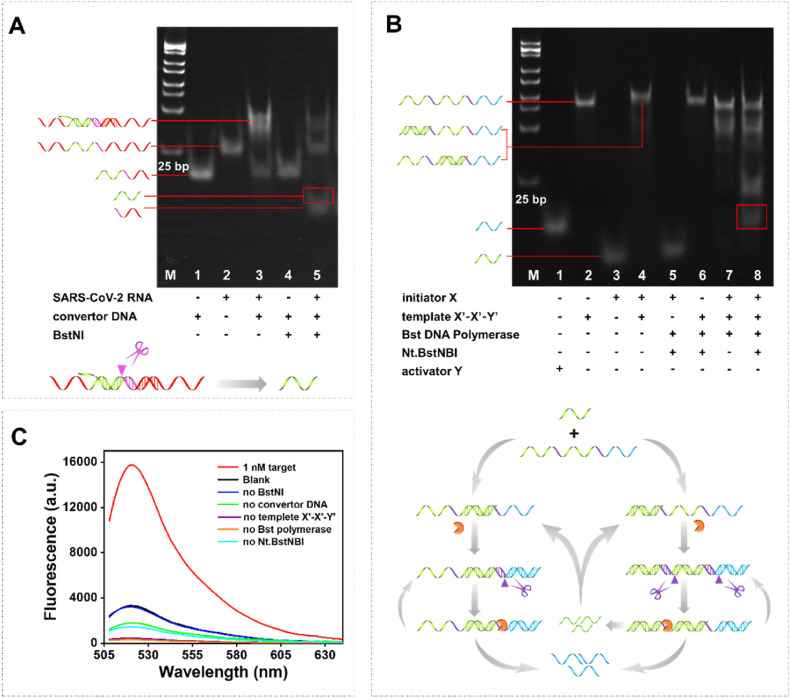
3.2. Viability analysis
To investigate the viability of the proposed SARS-CoV-2 RNA detection strategy, both PAGE and end-point fluorescence assay were adopted. We first validated the restriction enzyme BstNI-induced release of initiator X. As shown in Fig. 1 A, lanes 1 and 2 were the monomer of convertor DNA and target SARS-CoV-2 RNA, respectively. A band with slower migration appeared when incubated the monomer of convertor DNA and target SARS-CoV-2 RNA for an indicated period of time (lane 3), demonstrating the generation of RNA/DNA heteroduplex. As expected, in the presence of the target RNA, a new band corresponding to initiator X generated in lane 5 due to BstNI's unique capability that can cleavage DNA at a specified location within RNA/DNA heteroduplex. On the contrary, BstNI was unable to cleave the monomer of convertor DNA and no X generated (lane 4). Subsequently, the generation of Y sequence induced by EXPAR was investigated and the corresponding results were depicted in Fig. 1B. The monomer of activator Y, template X′-X′-Y′, and initiator X could be seen in lanes 1, 2, and 3, while duplex formed by initiator X and template X′-X′-Y′ (with the molar ratio of 1:1) could be respectively observed in lane 4. As expected, a new band corresponding to activator Y appeared obviously in lane 8, which was attributed to the function of the nicking endonuclease Nt.BstNBI and Bst DNA polymerase. However, no new band of activator Y generated upon the absence of template X′-X′-Y′ (lane 5), initiator X (lane 6), or nicking endonuclease Nt.BstNBI (lane 7).
Fig. 1.
Viability analysis. (A) PAGE analysis of the RTF feasibility. (B) PAGE analysis of the EXPAR feasibility. (C) End-point fluorescence assay of the SARS-CoV-2 RNA detection strategy. The target product band was circled in the red box.
To further demonstrate the feasibility of our assay, we verified it by means of end-point fluorescence analysis. The results were presented in Fig. 1C. In the presence of the target RNA, a strong fluorescence signal was observed. On the contrary, the absence of any of the components led to a low fluorescence signal. These results together with the PAGE analysis demonstrated our assay is a feasible strategy for SARS-CoV-2 RNA detection.
3.3. Optimization of experimental conditions
The crRNA sequence affects the trans-cleavage activity of CRISPR-Cas12a (Creutzburg et al., 2020), therefore we first optimized the sequence of crRNA. According to the crRNA design principle (Chen et al., 2018; Dai et al., 2019), we designed five crRNAs (Table S1) and explored them. As shown in Fig. S1, the trans-cleavage activities of CRISPR-Cas12a guided by different crRNAs were distinguishing, which was consistent with previous reports. To obtain efficient trans-cleavage efficiency, crRNA 2 with the highest activity was selected.
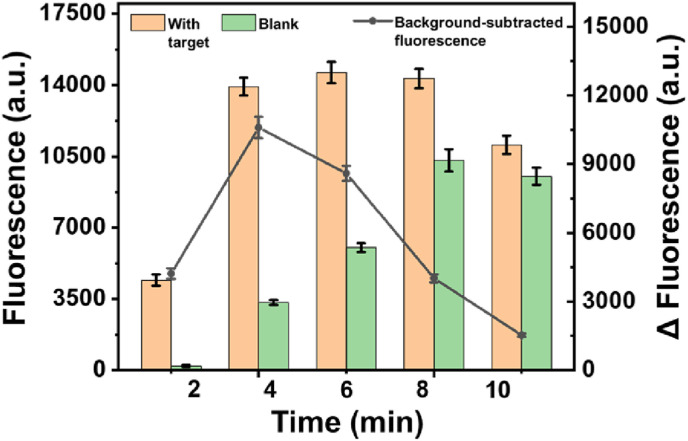
We then optimized the amounts of BstNI endonuclease, polymerase, Nt.BstNBI nicking enzyme and EXPAR reaction time those factors related to the performance of the amplification. As seen in Fig. S2, 0.8 U/μL of BstNI endonuclease, 0.03 U/μL of polymerase, 0.2 U/μL of Nt.BstNBI nicking enzyme had the best performance, respectively, according to the end-point background-subtracted fluorescence intensities (Δ FL). It was noted that the background fluorescence of the assay increased significantly upon the amplification time since that more reaction time will lead to more non-specific amplification products. This limitation could be overcome by careful optimization of the amplification time. The results were shown in Fig. 2 , an observed background-subtracted fluorescence intensity enabled clear detection of the 100 fM target at 4 min EXPAR time, which was thus selected for the following experiments.
Fig. 2.
Optimization of experimental condition. Amplification time of EXPAR. The concentration of SARS-CoV-2 RNA: 100 fM. Error bar means SD (n = 3).
We finally optimized the factors related to CRISPR-Cas12a. The reaction temperature makes a significant impact on the cleavage activity of Cas12a, therefore was explored. Compared with conventional reaction temperature (37 °C), a higher temperature setting at 45 °C exhibited a better cleavage activity on reporter DNA, which was chosen as an appropriate cleavage temperature (Fig. S3A). The concentration of Cas12a is highly correlated with the efficiency of the Cas12a-assisted amplification procedure and was examined. As seen in Fig. S3B, as the concentration of Cas12a increases, the signal experienced a sharp ascent trend before 50 nM, followed by a slight increase. Therefore, 50 nM was chosen as an applicable concentration. Subsequently, the cleavage time was optimized in the range from 10 min to 30 min. Fig. S3C showed that the signal kept in a steady state after the reaction time prolonged to 20 min, and 20 min was fixed as an appropriate condition.
3.4. Analytical performance
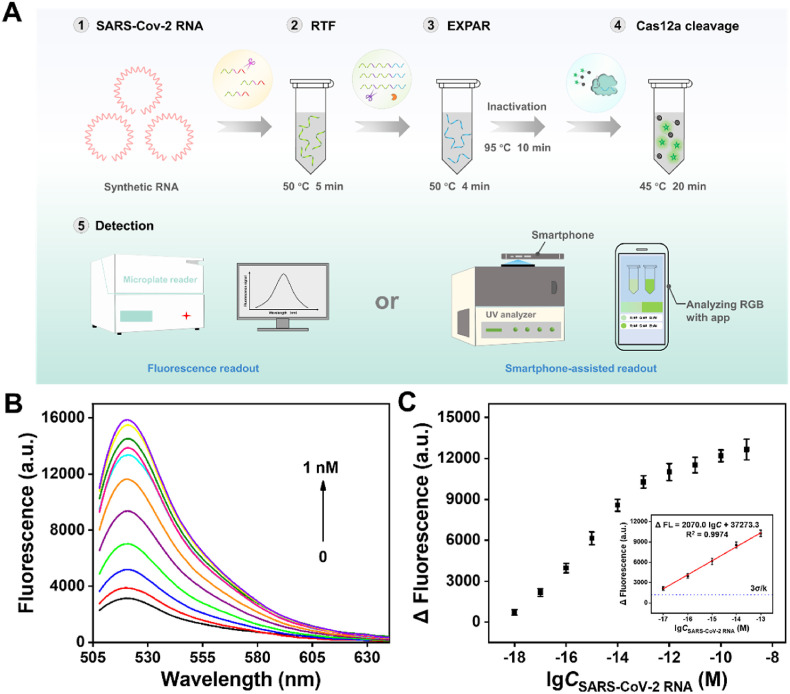
After the establishment of the optimized conditions, the detection performance of the Cas12a-assisted RTF-EXPAR strategy was evaluated. The overall operation of the assay for the detection of SARS-CoV-2 RNA was displayed in Fig. 3 A. As seen in Fig. 3B, the end-point fluorescence was increased in a target SARS-CoV-2 RNA concentration-dependent manner. A linear relationship (R2 = 0.9974) was obtained over the range from 10 aM to 100 fM (Fig. 3C). The corresponding linear regression equation was determined as Δ FL = 2070.0 lgC +37273.3. Based on 3σ/k (where σ is the standard deviation of the blank sample and k is the slope of the calibration curve), the detection limit can be calculated as low as 3.77 aM (∼2 copies/μL), which is superior to most reported SARS-CoV-2 detection methods (Table S5). The fly in the ointment is that the detection time is longer than the work of Carter et al. (2021).
Fig. 3.
Analytical performance. (A) The overall operation of the strategy for SARS-CoV-2 RNA detection. (B) Evaluation of end-point fluorescence intensity in the presence of different concentrations of SARS-CoV-2 RNA (1 nM, 100 pM, 10 pM, 1 pM, 100 fM, 10 fM, 1 fM, 100 aM, 10 aM, 1 aM and blank). (C) The linear relationship between Δ FL and the logarithm of the target concentration. Error bar means SD (n = 3).
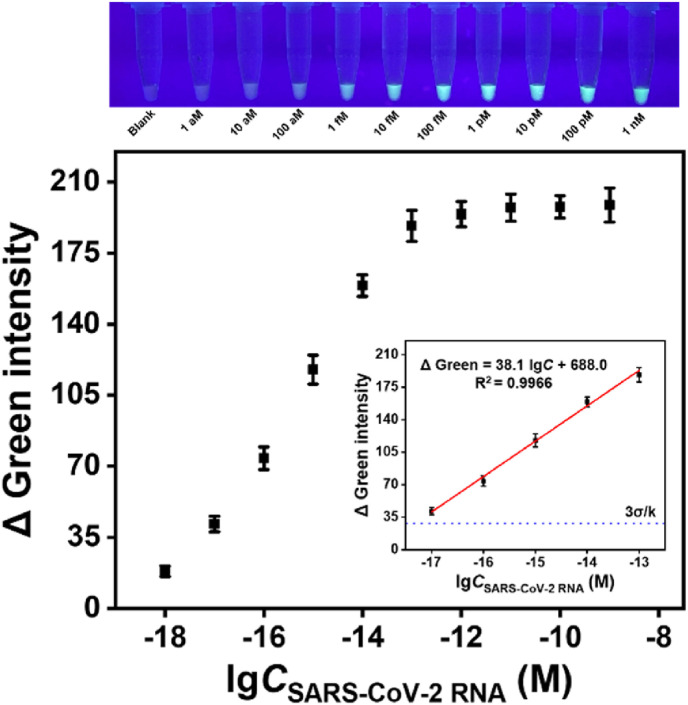
For POC detection, it is better to easily give results by a simple method. For this purpose, we used a smartphone-assisted analysis method for detecting the target RNA. Images of all samples under UV light are recorded by a smartphone camera and analyzed in RGB (Red, Green, Blue) values using the free “Palette Cam” app (Fig. 4 , Fig. S4). Compared with the Δ Red and Δ Blue, Δ Green exhibited a better linear relationship (Δ Green = 38.1 lgC + 688.0, R2 = 0.9966) and a wider linear range (10 aM–100 fM) between the logarithm of target concentration, so Δ Green was preferred as the reference signal. The detection limit can be calculated as low as 4.81 aM (∼3 copies/μL) using 3σ/k. Thus, it is very promising for the POC detection of viruses without complex instruments.
Fig. 4.
Analytical performance (using “Palette Cam” app). The linear relationship between Δ Green intensity and the logarithm of the target concentration. Inset: Visual fluorescence intensity of SARS-CoV-2 RNA (different concentrations) involved in Cas12a-assisted RTF-EXPAR reaction under UV light irradiation. Error bar means SD (n = 3).
3.5. Specificity and practicability evaluation
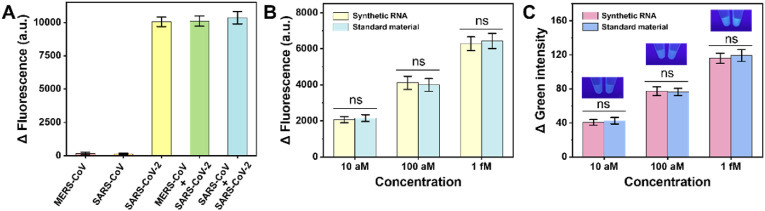
To validate the specificity of our method for SARS-CoV-2 RNA detection, we tested our protocol against two coronaviruses, which possess a certain similarity in sequence compared with SARS-CoV-2 (SARS-CoV, MERS-CoV). As expected, the addition of other coronavirus sequences did not cause an increase in fluorescence intensity, and the method could specifically detect SARS-CoV-2 RNA (Fig. 5 A).
Fig. 5.
Specificity and practicability evaluation. (A) Evaluation of the specificity of the detection strategy. The concentrations of MERS-CoV and SARS-CoV: 10 pM. The concentration of SARS-CoV-2: 100 fM. (B) Δ Fluorescence, (C) Δ Green analyses of SARS-CoV-2 RNA transcribed in vitro standard material. Error bar means SD (n = 3). ns, nonsignificant.
The above investigations used synthetic short RNA fragment as the target, however, in practical applications, the target is the RNA of the whole genome. Ultimately, to further demonstrate that the proposed detection method can be applied to COVID-19 detection, the SARS-CoV-2 RNA transcribed in vitro standard material was utilized as the detection target. Both the fluorescence (Fig. 5B) and smartphone (Fig. 5C) results showed that the signals of the standard material and the synthetic target RNA were comparable, demonstrating that the proposed strategy possesses the promise of being applied to the actual sample detection. Unfortunately, no clinical trials of the proposed method have been realized. In the days to come, we will always be looking for clinical samples to test this strategy.
4. Conclusion
In summary, we developed a new Cas12a-assisted RTF-EXPAR assay that enables the fast, sensitive, simple, and accurate detection of SARS-CoV-2. It was demonstrated that the assay can be performed within 40 min, requiring only isothermal control. The detection limit can be even 3.77 aM (∼2 copies/μL) when using endpoint fluorescence output and even 4.81 aM (∼3 copies/μL) when using the smartphone-assisted analysis method (RGB analysis). Cas12a-assisted also improves detection accuracy. These features support the future practicability of Cas12a-assisted RTF-EXPAR in POC molecular detection. While the method we constructed enables ultrasensitive detection, it does require multiple manual steps. However, we believe that in future work, we can solve the interference of multi-step manual operations on detection by building an integrated device for automatic distributed injection and detection. In addition, pfAgo capable of high-temperature resistance can be introduced, which is expected to realize one-step operation.
CRediT authorship contribution statement
Xiao-Min Hang: Conceptualization, Methodology, Project administration, Software, Visualization, Writing – original draft. Hui-Yi Wang: Conceptualization, Project administration, Software, Validation, Visualization, Writing – review & editing. Peng-Fei Liu: Methodology, Software, Validation, Visualization, Writing – review & editing. Kai-Ren Zhao: Software, Validation, Visualization, Writing – review & editing. Li Wang: Resources, Writing – review & editing, Supervision, Data curation, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (No. 21707053) and Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. SJCX22_1873).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114683.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., Hicks P., Manzoni T.B., Oniyide O., Ramage H., Mathew D., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., D'Andrea K., Kuthuru O., Dougherty J., Pattekar A., Kim J., Han N., Apostolidis S.A., Huang A.C., Vella L.A., Kuri-Cervantes L., Pampena M.B., Betts M.R., Wherry E.J., Meyer N.J., Cherry S., Bates P., Rader D.J., Hensley S.E. Cell. 2021;184(7):1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J.G., Iturbe L.O., Duprey J.L.H., Carter I.R., Southern C.D., Rana M., Whalley C.M., Bosworth A., Beggs A.D., Hicks M.R., Tucker J.H.R., Dafforn T.R. Proc. Natl. Acad. Sci. U.S.A. 2021;118(35) doi: 10.1073/pnas.2100347118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.E., Lee P.W., Trick A.Y., Park J.S., Chen L., Shah K., Mostafa H., Carroll K.C., Hsieh K., Wang T.H. Biosens. Bioelectron. 2021;190 doi: 10.1016/j.bios.2021.113390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhu D., Huang T., Yang Z., Liu B., Sun M., Chen J.X., Dai Z., Zou X. Anal. Chem. 2021;93(37):12707–12713. doi: 10.1021/acs.analchem.1c02588. [DOI] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Luo R., Li S., Li H., Qin Y., Zhou D., Liu H., Gong X., Chang J. Anal. Chem. 2020;92(19):13336–13342. doi: 10.1021/acs.analchem.0c02642. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., Veer B., Brink S., Wijsman L., Goderski G., Romette J., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Eurosurveillance. 2020;25(3) [Google Scholar]
- Cox D.B., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzburg S.C.A., Wu W.Y., Mohanraju P., Swartjes T., Alkan F., Gorodkin J., Staals R.H.J., van der Oost J. Nucleic Acids Res. 2020;48(6):3228–3243. doi: 10.1093/nar/gkz1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Somoza R.A., Wang L., Welter J.F., Li Y., Caplan A.I., Liu C.C. Angew. Chem. Int. Ed. 2019;58(48):17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.G., Knop M., Anders S. Sci Transl Med. 2020;12(556) doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice M., De Falco M., Zappi D., Antonacci A., Scognamiglio V. Biosens. Bioelectron. 2022;205 doi: 10.1016/j.bios.2022.114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Puig H., Lee R.A., Najjar D., Tan X., Soenksen L.R., Angenent-Mari N.M., Donghia N.M., Weckman N.E., Ory A., Ng C.F., Nguyen P.Q., Mao A.S., Ferrante T.C., Lansberry G., Sallum H., Niemi J., Collins J.J. Sci. Adv. 2021;7(32) doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshan M.A., Amani A., Faridi-Majidi R. ACS Appl. Mater. Interfaces. 2021;13(13):14816–14843. doi: 10.1021/acsami.0c22381. [DOI] [PubMed] [Google Scholar]
- Feng W., Peng H., Xu J., Liu Y., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Babiuk S., Zhang H., Le X.C. Anal. Chem. 2021;93(37):12808–12816. doi: 10.1021/acs.analchem.1c03456. [DOI] [PubMed] [Google Scholar]
- Glökler J., Lim T.S., Ida J., Frohme M. Crit. Rev. Biochem. Mol. Biol. 2021;56(6):543–586. doi: 10.1080/10409238.2021.1937927. [DOI] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang X.M., Liu P.F., Tian S., Wang H.Y., Zhao K.R., Wang L. Biosens. Bioelectron. 2022;211 doi: 10.1016/j.bios.2022.114393. [DOI] [PubMed] [Google Scholar]
- Hanna T.P., Evans G.A., Booth C.M. Nat. Rev. Clin. Oncol. 2020;17(5):268–270. doi: 10.1038/s41571-020-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., St Reid P.M., Punyadeera C., Chang L., Gendelman H.E. Nat. Mater. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Bae P.K., Kim H., Song Y.J., Yi S.Y., Kwon J., Seo J.S., Lee J., Jo H.S., Park S.M., Park H.S., Shin K.S., Chung S., Shin Y.B. Biosens. Bioelectron. 2021;191 doi: 10.1016/j.bios.2021.113406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Macdonald J. Biosens. Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. ACS. Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Liu J.K., Nie X.Q., Zhao G.P., Wang J. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. Trends Biotechnol. 2019;37(7):730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Liang M., Li Z., Wang W., Liu J., Liu L., Zhu G., Karthik L., Wang M., Wang K.F., Wang Z., Yu J., Shuai Y., Yu J., Zhang L., Yang Z., Li C., Zhang Q., Shi T., Zhou L., Xie F., Dai H., Liu X., Zhang J., Liu G., Zhuo Y., Zhang B., Liu C., Li S., Xia X., Tong Y., Liu Y., Alterovitz G., Tan G.Y., Zhang L.X. Nat. Commun. 2019;10:3672. doi: 10.1038/s41467-019-11648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.F., Zhao K.R., Liu Z.J., Wang L., Ye S.Y., Liang G.X. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112954. [DOI] [PubMed] [Google Scholar]
- Matheson N.J., Lehner P.J. Science. 2020;369(6503):510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Anal. Chem. 2020;92(24):16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. PLOS Biol. 2006;4(7) doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel P., Hu C., Mao H. ACS Sens. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M.S., Paliwoda R.E., Zhang H., Le X.C. Anal. Chem. 2018;90(18):11033–11039. doi: 10.1021/acs.analchem.8b02788. [DOI] [PubMed] [Google Scholar]
- Schneider L., Blakely H., Tripathi A. Electrophoresis. 2019;40:2706–2717. doi: 10.1002/elps.201900167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R.F. Science. 2020;369(6504):608–609. doi: 10.1126/science.369.6504.608. [DOI] [PubMed] [Google Scholar]
- Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Lab Chip. 2021;21(9):1634–1660. doi: 10.1039/d0lc01156h. [DOI] [PubMed] [Google Scholar]
- Tauh T., Lee S.M., Meyler P., Mozel M., McLennan M., Hoang L. Can. J. Anesth. 2021;68(10):1569–1571. doi: 10.1007/s12630-021-02035-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Tu Y.P., Iqbal J., O'Leary T. ELife. 2021;10 doi: 10.7554/eLife.65726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- van Dongen J.E., Berendsen J.T., Steenbergen R.D., Wolthuis R.M., Eijkel J.C., Segerink L.I. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang X., Wu Y., Guo M., Gu C., Dai C., Kong D., Wang Y., Zhang C., Qu D., Fan C., Xie Y., Zhu Z., Liu Y., Wei D. Nat. Biomed. Eng. 2022;6(3):276–285. doi: 10.1038/s41551-021-00833-7. [DOI] [PubMed] [Google Scholar]
- Wei Y., Yang L., Zhang X., Sui D., Wang C., Wang K., Shan M., Guo D., Wang H. Drug Metab. Dispos. 2018;46(5):525–531. doi: 10.1124/dmd.117.078444. [DOI] [PubMed] [Google Scholar]
- Wu X., Tay J.K., Goh C.K., Chan C., Lee Y.H., Springs S.L., Wang D.Y., Loh K.S., Lu T.K., Yu H. Biomaterials. 2021;274 doi: 10.1016/j.biomaterials.2021.120876. [DOI] [PubMed] [Google Scholar]
- Xia S., Chen X. Cell Discov. 2020;6(1):37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K.R., Wang L., Liu P.F., Hang X.M., Wang H.Y., Ye S.Y., Liu Z.J., Liang G.X. Sens. Actuators B Chem. 2021;346 [Google Scholar]
- Zhou B., Ye Q., Li F., Xiang X., Shang Y., Wang C., Shao Y., Xue L., Zhang J., Wang J., Ding Y., Chen M., Wu Q. Sens. Actuators B Chem. 2022;351 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.