Abstract
The accurate measurement of serological response to SARS-CoV-2 vaccination is needed to correlate responses with effective protective immunity. The World Health Organization (WHO) has created an international standard to allow harmonization of immune response assessment to an arbitrary unit across different commercial assays; however, the accuracy of reporting of SARS-CoV-2 spike antibody titers in international standard units (BAU or IU/mL) from commercial assays is not well studied. Here, we report the performance comparison of four quantitative commercial assays testing for SARS-CoV-2 spike immunoglobins using the WHO's international standard. Sera, EDTA-plasma and heparinized plasma collected from individuals who are vaccine naïve or received BNT162b2 (Pfizer/BioNTech), mRNA-1273 (Moderna) or ChAdOx1-S (Oxford-AstraZeneca) were tested using Abbott Architect AdviseDx SARS-CoV-2 IgG II, DiaSorin LIAISON SARS-CoV-2 TrimericS IgG, Roche Elecsys Anti-SARS-CoV-2 S and GenScript cPass SARS-CoV-2 surrogate virus neutralization assays. The sensitivities ranged from 90% to 100%, and specificities from 88% to 100%. These four assays had excellent agreement (0.79–0.93) and correlation (0.87–0.97); however, Passing-Bablok regression analysis indicated that data generated by these assays were not comparable. Our data suggests that natural SARS-CoV-2 infection elicited a greater antibody response compared to vaccines, evident by a significantly higher neutralizing antibody titer in unvaccinated individuals who seroconverted.
Keywords: COVID-19, SARS-CoV-2, Quantitative serological assays, WHO international standard, Antibodies, Immunoglobulin
Abbreviations
- BAU
binding arbitrary unit
- CLIA
chemiluminescent immunoassay
- ELISA
enzyme-linked immunosorbent assays
- Ig
immunoglobulin
- IS
International Standard
- IU
international unit
- SARS-CoV 2
severe acute respiratory syndrome coronavirus 2
- WHO
World Health Organization
1. Introduction
During the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, many serology assays were rapidly deployed into the market [1,2]. These serologic assays utilize different chemistry [lateral flow immunoassays, enzyme-linked immunosorbent assays (ELISA), chemiluminescent immunoassay (CLIA)], target different antigens (spike glycoprotein, nucleocapsid) or immunoglobulin classes, and provide qualitative, semiquantitative or quantitative results [3], [4], [5], [6].
Antibody tests can be used to confirm retrospective infection in cases of high clinical suspicion (when diagnostic testing may not have been performed, or was previously negative), for diagnosis of late-onset post-infection complications, such as the multisystem inflammatory syndrome in children [7,8]. Additionally, population-based seroprevalence can be used to evaluate the success of infection control measures at the community level [9,10], and estimate asymptomatic/non-reported symptomatic SARS-Cov-2 infection rates. Several SARS-CoV-2 vaccines based on mRNA or live adenovirus vector expressing the spike (S) glycoprotein have been approved and are widely available globally [11,12]. As such, an increasing proportion of the world population has received at least one dose of vaccine [13,14]. Subsequently, serologic assays have become essential tools to measure vaccine induced humoral responses to the spike protein to better understand antibody longevity and effectiveness.
However, our current understanding of the effectiveness of vaccines to decrease transmission of new variants of concern, the duration of vaccine elicited immunity, and its correlation to protection against breakthrough infections are inadequate [15], [16], [17], [18]. The challenge of comparing results between studies is the lack of harmonization of quantitative standards, as commercial assay manufacturers use their own arbitrary units.
The World Health Organization (WHO) Expert Committee on Biological Standards established an international standard (IS) for SARS-CoV-2 immunoglobulin based on pooled human plasma from convalescent patients with the aim to facilitate accurate calibration of serological assays for SARS-CoV-2 immunoglobulin detection [19,20]. The preparation is assigned the concentration of 1000 international unit per milliliter (IU/mL) for neutralizing antibody activity and is considered equivalent to 1000 binding antibody units per milliliter (BAU/mL), an arbitrary unit that can be used in comparison studies of binding antibody assays. Therefore, both units are considered numerically equivalent [19].
Recently, some commercial serological assays for SARS-CoV-2 have implemented conversion factors to allow antibody titers to be reported in international units; however, the accuracy of reporting in WHO IS units by these commercial assays is not well characterized.
Here, we evaluated serologic reporting of three commercial automated binding antibody immunoassays and one surrogate virus neutralization ELISA for anti-SARS-CoV-2 antibodies in WHO established IS units using clinically collected plasma or serum from vaccine recipients or vaccine naïve individuals. We describe the demographic characteristics of the donors and studied SARS-CoV-2 antibody levels associated with vaccination status.
2. Methods
2.1. Study participants and sample acquisition
Thirty SARS-CoV-2 antibody positive EDTA plasma, sera, and heparinized plasma samples, and 30 serologically negative samples of each type (based on previous Abbott Architect AdviseDx SARS-CoV-2 IgG II results) were used in this study. The WHO IS for Anti-SARS-CoV-2 Immunoglobulin was purchased from the National Institute for Biological Standards and Control (NIBSC, Hertfordshire, UK).
2.2. SARS-CoV-2 serologic testing
Abbott Architect AdviseDx SARS-CoV-2 IgG II Quant, DiaSorin TrimericS IgG, Roche Elecsys Anti-SARS-CoV-2 S, and GenScript cPass SARS-CoV-2 surrogate virus neutralization assays were used as per manufacturer recommendations to assess antibody titer [21], [22], [23], [24].
2.3. Diagnostic sensitivity and specificity calculations
Assay diagnostic sensitivities and specificities were calculated using clinically defined anti-SARS-CoV-2 positive samples as the reference standard in SPSS v26 (IBM SPSS Statistics for Windows, Armonk, NY, USA). Clinically positive samples were defined as any sample determined as anti-SARS-CoV-2 positive by three of the four assays used in this study.
2.4. Statistical analysis
Data were summarized by percentages and frequencies for categorical variables, and medians and interquartile ranges (IQRs) for continuous variables. Categorical variables were compared using the Chi-square test or Fisher's exact test where appropriate; the continuous variables (age and antibody titers) were compared using Mann-Whitney U test or Kruskal-Wallis test with Dunn's multiple comparison correction. Assay agreements were analyzed with Cohen's Kappa and Spearman rank correlations. We defined kappa scores of 0.6 to 0.69 as moderate, 0.7 to 0.8 as good and above 0.8 as excellent agreement [25]. Assay results were compared with Passing-Bablok linear regression and Bland-Altman plots using STATA v16 (StataCorp LLC, College Station, TX, USA) [26]. All other statistical analyses were performed using SPSSv25 (IBM SPSS Statistics for Windows, Armonk, NY, USA)[27]. Graphs were generated in GraphPad Prism v9 (GraphPad Software, San Diego, CA, USA).
2.5. Ethics
Ethics approval was granted by the University of Alberta research ethics board (study reference numbers Pro00101916 and Pro00113764).
3. Results
Anti-SARS-CoV-2 spike seropositive individuals were older [Median age 46 vs 34, (P < 0.05)] and more likely to have received one or two doses of mRNA-1273 or BNT162b2 vaccines (P < 0.001) (Table 1 ).
Table 1.
Donor demographic characteristics and SARS-CoV-2 vaccination status by serology status (n = 180).
| SARS-CoV-2 Serology Status | P-value* | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibody negativen = 90 | Antibody positiven = 90 | |||||||||||||
| Age, Median (IQR) | 34 | (9, | 54) | 46 | (9, | 71) | 0.5 | |||||||
| Age Group, n (%) | under 18 | 29 | 32.2% | 31 | 34.4% | 0.003 | ||||||||
| 18–25 | 7 | 7.8% | 0 | 0.0% | ||||||||||
| 26–45 | 22 | 24.4% | 14 | 15.6% | ||||||||||
| 46–55 | 11 | 12.2% | 9 | 10.0% | ||||||||||
| 56–65 | 12 | 13.3% | 8 | 8.9% | ||||||||||
| 66–75 | 5 | 5.6% | 14 | 15.6% | ||||||||||
| above 75 | 4 | 4.4% | 14 | 15.6% | ||||||||||
| Sex, n (%) | Female | 45 | 50.0% | 35 | 38.9% | 0.15 | ||||||||
| Male | 15 | 16.7% | 25 | 27.8% | ||||||||||
| Unknown | 30 | 33.3% | 30 | 33.3% | ||||||||||
| Vaccination Status | Unvaccinated | 73 | 81.1% | 1 | 1.1% | <0.001 | ||||||||
| 1 Dose | 9 | 10.0% | 38 | 42.2% | ||||||||||
| 2 Doses | 2 | 2.2% | 20 | 22.2% | ||||||||||
| Unknown | 6 | 6.7% | 31 | 34.4% | ||||||||||
| Vaccine dose 1 Type (brand) | AstraZeneca | 3 | 3.3% | 5 | 5.6% | <0.001 | ||||||||
| Moderna | 1 | 1.1% | 12 | 13.3% | ||||||||||
| Pfizer/BioNTech | 7 | 7.8% | 41 | 45.6% | ||||||||||
| Not Immunized | 79 | 87.8% | 32 | 35.6% | ||||||||||
| Vaccine dose 2 type (brand) | Moderna | 0 | 0.0% | 8 | 8.9% | <0.001 | ||||||||
| Pfizer/BioNTech | 2 | 2.2% | 23 | 25.6% | ||||||||||
| Not Immunized | 88 | 97.8% | 59 | 65.6% | ||||||||||
Serology status was defined based on the first Abbott Architect RBD results used for sample selection.
Mann-Whitney U test for Age and Chi-square test or Fisher's exact test for all categorical variables, unknown or not immunized groups not included in the analyses.
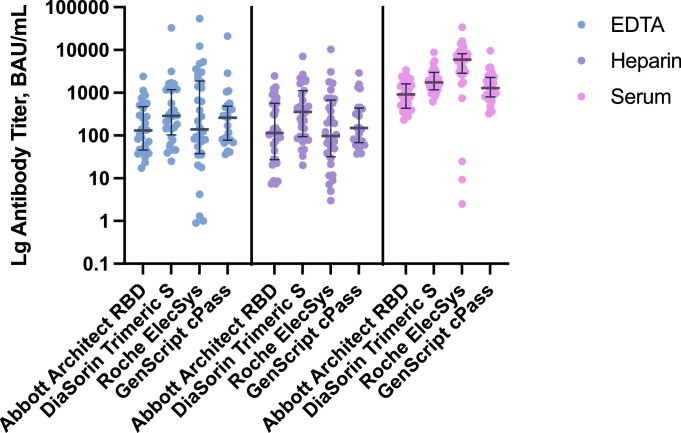
The Roche Elecsys assay reported the highest number of SARS-CoV-2 positives (10–13% more than Abbott RBD) across all sample types and the widest range of antibody titers. DiaSorin trimeric S assay reported the highest minimal titers, while Abbott RBD had the narrowest dynamic range (Table 2 , Fig. 1 ). Eighty-eight percent (25/30) of EDTA plasma and 75% (24/32) of heparin plasma were interpreted as positive on both Abbott RBD and GenScript cPass assays, with 100% concordance for serum positivity.
Table 2.
Comparison of test measurements produced by Abbott RBD, DiaSorin Trimeric S, Roche Elecsys and GenScript cPass assays in EDTA plasma, heparin plasma and serum samples.
| Abbott RBD | DiaSorin TrimericS | Roche Elecsys | GenScript cPass | P-value | |
|---|---|---|---|---|---|
| EDTA Plasma, n = 60 | |||||
| Positive, n | 30 | 31 | 34 | 25 | |
| Titer, BAU/mL | 0.30 | ||||
| Min | 17.2 | 39 | 0.9 | 36.1 | |
| Median, IQR | 130.4 (47.2, 431.9) | 291 (140, 1190) | 138.6 (38.3, 1764.4) | 259.2 (77, 453.6) | |
| Max | 2405.9 | 33,100 | 54,001.7 | 21,021.3 | |
| Heparin Plasma, n = 60 | |||||
| Positive, n | 32 | 29 | 32 | 24 | |
| Titer, BAU/mL | 0.10 | ||||
| Min | 7.4 | 46 | 3.0 | 37.0 | |
| Median, IQR | 114.8 (27.3, 561.7) | 419 (145, 1120) | 97.6 (34, 640.5) | 150.1 (69.6, 433.4) | |
| Max | 2467.7 | 7060 | 10,293.1 | 2913.8 | |
| Serum, n = 60 | |||||
| Positive, n | 30 | 30 | 33 | 30 | |
| Titer, BAU/mL | <0.0001 | ||||
| Min | 231.6 | 614 | 2.5 | 320.3 | |
| Median, IQR | 923 (465.8, 1670) | 1750 (1170, 3000) | 5917.7 (3010.3, 8154.3) | 1279 (806.3, 2267.5) | |
| Max | 6056.0 | 8770 | 33,954.5 | 9616.9 | |
P-values generated from comparing medians using the Kruskal-Wallis test, significant at < 0.05.
Min: minimum.
Max: maximum.
Fig. 1.
SARS-CoV-2 antibody titer reported in binding antibody unit per mL by Architect RBD, DiaSorin TrimericS, Roche Elecsys and GenScript cPass in EDTA plasma, heparin plasma and serum. The dots depict titers of individual samples, and the lines represent median titer values and interquartile ranges.
Inter-assay agreement between the Abbott RBD, DiaSorin TrimericS and Roche Elecsys assays was excellent, with kappa scores ranging from 0.88 to 0.93; the agreement between GenScript cPass and the other three assays was slightly lower (k = 0.78 −0.88) (Table 3 ). The four assays correlated very well with Spearmen's rho above 0.87; however, Passing-Bablok regression revealed the average measured values between any two of the four assays were significantly different (Table 3, Figure S1).
Table 3.
Agreements and correlation of SARS-CoV-2 antibody titer in positive samples between Abbott RBD, DiaSorin Trimeric S, Roche Elecsys and GenScript cPass assays.
| Assay pair | Cohen's k | Spearman's rho (95% CI) | Passing-Bablok average difference (95%CI) |
|---|---|---|---|
| RBD/Trimeric S | 0.93 (95% CI 0.88, 0.99)€ | 0.97 (95%CI 0.96, 0.98)* | −74.95 (−10,101.94, −9952.04)⁎⁎ |
| RBD/Elecsys | 0.92 (95% CI 0.87, 0.98)€ | 0.87 (95%CI 0.81, 0.92)* | −2280.84 (−13,052.84, −8491.16)⁎⁎ |
| RBD/cPass | 0.86 (95% CI 0.78, 0.93)€ | 0.94 (95%CI 0.90, 0.96)* | 573.27 (−12,507.97, −13,654.50)⁎⁎ |
| Trimeric S/Elecsys | 0.88 (95% CI 0.81, 0.95)€ | 0.90 (95%CI 0.86, 0.93)* | −2205.89 (−11,716.78, −7304.99)⁎⁎ |
| Trimeric S/cPass | 0.88 (95% CI 0.81, 0.95)€ | 0.94 (95%CI 0.91, 0.96)* | 650.93 (−2231.08, −3532.94)⁎⁎ |
| Elecsys/cPass | 0.78 (95% CI 0.69, 0.87 s)§ | 0.90 (95%CI 0.84, 0.93)* | 3113.32 (−8083.30, −14,309.94)⁎⁎ |
Correlation is significant at p < 0.01.
Average differences are significant at p < 0.05. Differences are measured in BAU/mL.
Cohen's kappa score of 0.7–0.8 was defined as good.
above 0.8 as excellent agreement.
Direct testing of the WHO IS showed the Roche Elecsys was able to detect antibodies in up to 1000-fold dilutions and quantify titers as low as 1 BAU/mL (Table S1). Abbott RBD was able to identify IgG in dilutions corresponding to titers of 100 and 10 BAU/mL, while DiaSorin TrimericS and GeneScript cPass were able to detect and quantify titers as low as 100 BAU or IU/mL (Table S1).
All assays except GenScript cPass (89.8%) had sensitivities of 100% when compared to clinically defined positive samples (reference standard; Table 4 ). The specificity of Abbott RDB, DiaSorin TrimericS and Roche Elecsys ranged from 88% to 98% while GenScript cPass was 100% (Table 4).
Table 4.
Sensitivity and specificity of RBD, TrimericS, Elecsys and cPass calculated by using user-defined clinically positive reference standard.
| N = 180 | Compared to clinically positive Samples (3/4 assays positive) | |||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | TP | TN | FP | FN | |
| Abbott Architect RBD | 100.0% | 95.7% | 88 | 88 | 4 | 0 |
| DiaSorin Trimeric S | 100.0% | 97.8% | 88 | 90 | 2 | 0 |
| Roche Elecsys | 100.0% | 88.0% | 88 | 81 | 11 | 0 |
| GenScript cPass | 89.8% | 100.0% | 79 | 92 | 0 | 9 |
Of the 90 samples originally chosen as positives, two (2.2%) tested positive on only two platforms and were re-classified as clinically negative. Nine (10%) and 79 (87.8%) tested positive on three and four platforms, respectively. Two previous Abbott RBD negative heparin plasma samples were reclassified as positive by Abbott RBD (Table 2). Overall, SARS-CoV-2 antibody titers were found to be five- to ten-fold higher in serum compared to plasma sample types and significant differences (P < 0.0001) were observed (Table 1, Table S2–3).
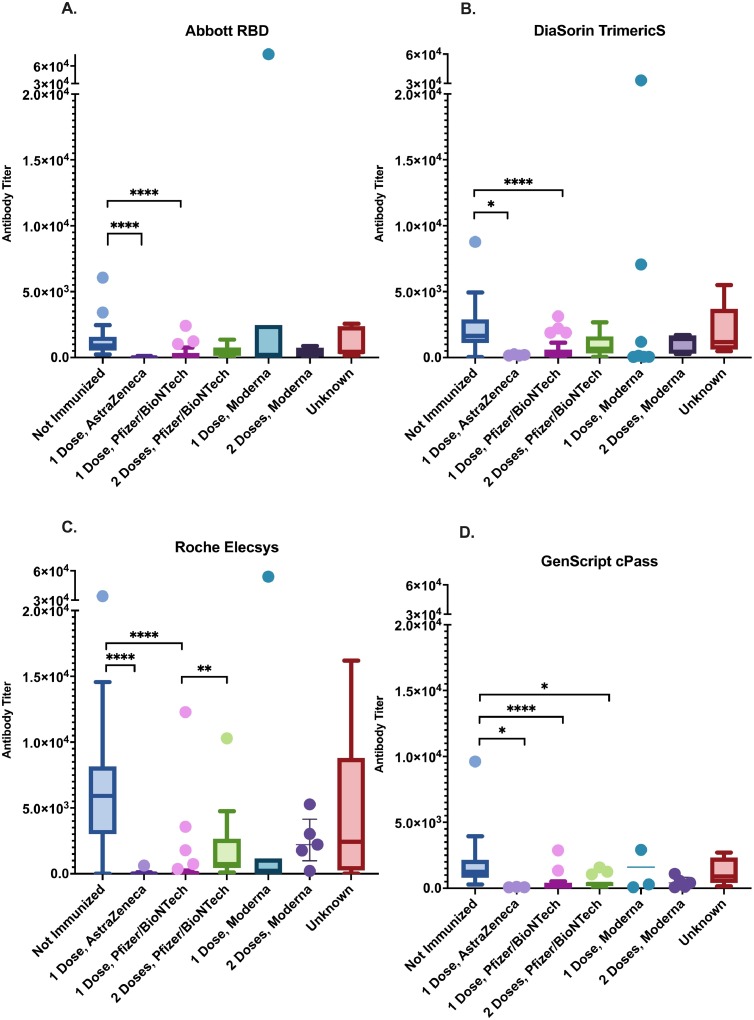
Antibody titers in vaccine naïve participants, who were presumably infected by SARS-CoV-2 naturally, were higher than vaccinated participants who received one dose of either ChAdOx1-S or BNT162b2, quantified by all four assays (Fig. 2 ). Roche Elecsys reported a dosage dependent increase of antibody titer induced by BNT162b2 vaccine which was not found by the other three assays (Fig. 2). Neutralizing antibody levels were found to be significantly higher in seropositive vaccine naïve individuals than those who received ChAdOx1-S or BNT162b2 vaccines by GenScript cPass regardless of doses received (Fig. 2).
Fig. 2.
Anti-SARS-CoV-2 antibody titers in vaccinated and vaccine naïve participants quantified in BAU/mL by the four serologic assays under study.
4. Discussion
Overall, we observed excellent correlation (r = 0.87–0.97) and agreement (k 0.78–0.93) between the three binding antibody assays and the neutralization antibody ELISA. These assays are suitable to be used clinically; however, data generated in standardized WHO IS units are not comparable, as Passing-Bablok regression revealed significant differences in proportional measurements between assays.
Abbott RBD and DiaSorin TrimericS had the best correlation, whereas Roche Elecsys and GenScript cPass results differed the most. The difference in assay chemistry, antigen specificity, and antibody class targeted (IgG vs. total Ig vs. pan-Ig neutralizing antibodies) may explain the small differences in diagnostic sensitivities and specificities, especially between GenScript cPass and Roche Elecsys S, because neutralizing antibodies are only a small proportion of the total antibodies measured by Elecsys S. It may also explain the observed differences in dynamic range (Roche Elecsys vs Abbott RBD) and minimal titer detected (DiaSorin TrimericS).
As the WHO IS is comprised of pooled plasma from eleven different SARS-CoV-2 convalescent patients [and therefore contains various isoforms of antibodies (IgA, IgM and IgG) targeting numerous epitopes on the spike protein, with some neutralizing ability [28], it is not surprising that assay platforms did not produce equivalent results given their different antibody class targets and epitope targets (RBD or trimeric spike protein). However, despite these differences, the numerical values of the positive dilutions produced by these assays are very similar to the expected WHO IS titer (Table S1).
To date, very few comparison studies exist that assess SARS-CoV-2 commercial assay performance against the WHO IS. Studies not using standardized unit BAU/mL found that numerical results of the Abbott RBD, DiaSorin Trimeric and Roche Elecsys S test kits are not interchangeable [29] and not surprisingly, the sensitivity (93.6–96%), correlation (r = 0.8–0.9) and agreement (k = 0.6–0.8) between the four assays was lower than we observed in this study (Table 2 and Table 3)[30, 31]. In line with our findings, Bradley et al., reported equivalent correlation between the Abbott RBD assays to Roche Elecsys S (r = 0.83 vs 0.87); however, we observed an improved correlation with GenScript cPass using standardized IU/mL (r = 0.86 vs 0.94) rather than% neutralization [32]. In fact, 100% concordance of the Abbott RBD, Roche Elecsys S and GenScript cPass results was achieved in both Bradley et al., and our study, highlighting the importance of using the WHO IS when comparing methods (Table S1). Other groups described similar but slightly lower correlations (range 0.76–0.8) with Roche Elecsys S, Abbott RBD, and DiaSorin TrimericS to a virus neutralization test and correlation ranges from 0.5 to 0.9 between these three tests using the WHO IS units [33], [34], [35].
Average proportional differences of measured values between the four serological assays were revealed using Passing-Bablok regression analysis. Consistent with our findings, Perkmann et al., and Lukaszuk et al., found that Abbott RBD, DiaSorin TrimericS, Roche Elecsys had high proportional errors using Passing-Bablok regression, suggesting that results of these assays are not interchangeable [31,36]. An increased effort by the manufacturers to standardize assay output across platforms would be helpful to achieve cross-utility. Until then, comparison of results between assays should be interpreted with care.
We found a higher titer of neutralizing antibodies measured by the GenScript cPass in vaccine naïve seropositive individuals compared to those who received ChAdOx1-S or BNT162b2 regardless of dose (Fig. 2). The number of mRNA-1273 recipients were too small to draw any solid conclusions. This observation suggests that natural infection induces higher levels of antibody compared to vaccination. The ChAdOx1-S and the two mRNA vaccines were formulated to encode only the full-length spike protein [21,37,38], therefore individuals with natural infections likely generate polyclonal antibodies against multiple epitopes of the whole virus (including neutralizing antibodies) at a higher level. Moreover, the vaccine naïve participants belong to a young age group (Median age=9): age is another factor that may contribute to the increase in neutralizing antibody level observed, as children were shown to produce a robust neutralizing antibody response after SARS-CoV-2 infection [39]. In general, children with any upper respiratory infection may have higher viral loads than adults [40]. Recent research has suggested that natural infection generates longer antibody duration against future COVID-19 illnesses than two doses of vaccine in the previously unexposed individuals. Vaccinated individuals with prior infection have the highest level and longest duration of antibody response, which remains high after 12 months post vaccination [41,42]. However, more evidence is needed to gain a better understanding of the protective immunity induced by natural infection, or combinations of vaccine, and what risk factors may influence the immunity against breakthrough infections. Further standardization of serological assays would allow analysis using pooled data across studies to facilitate that.
Because the three types of samples used in this study were not paired (i.e. not collected from the same individuals), higher antibody titers observed in serum specimens cannot be considered an intrinsic characteristic of the specimen type, but rather reflects the characteristics of serological responses in the donors [people of all ages (plasma) vs. those under 25 (serum)]. In samples with higher levels of antibody, serum reported a broader range of antibody level detected between the four assays compared to plasma specimens. There is likely a larger intrinsic error range at the upper limits of the quantifiable range, where the assays were saturated, which is where more of the serum specimen titers fall compared to plasma sample types (Fig. 1).
One of the limitations of this study is that we were unable to link prior SARS-CoV-2 infection directly to vaccine naïve participants who tested positive for SARS-CoV-2 spike or neutralizing antibody, as nucleic acid amplification test (NAAT) results and anti-nucleocapsid serologic results were unavailable; however, the majority (24/32, 75%) of specimens were from children under 12 who were (at the time of specimen collection) ineligible for vaccination as per provincial immunization guidelines. This suggests the observed seroconversion was not due to vaccination but rather natural infection. Furthermore, the duration from prior SARS-CoV-2 infections to time of sample collection was unclear in unimmunized seropositive individuals. Therefore, the higher anti-SARS-CoV-2 immunoglobulin level observed in vaccine naïve participants (Fig 2) could be due to the difference in sample collection time and may not necessarily indicate that better protection is elicited by natural infection compared to vaccination. Lastly, the study is limited by a relatively small sample size and samples collected prior to September 2021 thus more recent samples are needed to evaluate assay performance on serologic responses to new variants (e.g. omicron) that harbor multiple mutations in the RBD.
In summary, we compared the quantitative reporting of SARS-CoV-2 spike immunoglobulin titers in WHO established international standard units by Abbott RBD, DiaSorin TrimericS, Roche Elecsys S and GenScript cPass in serum and plasma types. We found these assays were in excellent agreement and are suitable to be used clinically, but the data generated in standardized WHO IS units are not interchangeable. SARS-CoV-2 natural infections appeared to induce a greater level of neutralizing antibodies than vaccinations. Additional work to harmonize serologic assays for SARS-CoV-2 using the WHO international standard would be beneficial.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Sean C. Taylor at GenScript USA inc. for providing the GenScript cPass SARS-CoV-2 neutralization antibody detection kits and the technologists at Public Health Laboratory Calgary, Alberta Precision Laboratories for specimen retrieval and generating the Abbott RBD and DiaSorin Trimeric S test results. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105292.
Appendix. Supplementary materials
References
- 1.West R.M., Kobokovich A., Connell N., Gronvall G.K. Antibody (Serology) tests for COVID-19: a case study. mSphere. 2021;6 doi: 10.1128/mSphere.00201-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto L.A., Shawar R.M., O'Leary B., Kemp T.J., Cherry J., Thornburg N., et al. A trans-governmental collaboration to independently evaluate SARS-CoV-2 serology assays. Microbiol Spectr. 2022;10 doi: 10.1128/spectrum.01564-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theel E.S. SARS-CoV-2 Serologic Testing. 4th edition. ed 2021.
- 4.Van Caeseele P., Canadian Public Health Laboratory N. Bailey D. Canadian society of clinical C, Forgie SE, association of medical M, et al. SARS-CoV-2 (COVID-19) serology: implications for clinical practice, laboratory medicine and public health. CMAJ. 2020;192 doi: 10.1503/cmaj.201588. E973-E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vengesai A., Midzi H., Kasambala M., Mutandadzi H., Mduluza-Jokonya T.L., Rusakaniko S., et al. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst. Rev. 2021;10:155. doi: 10.1186/s13643-021-01689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F., et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N. Engl. J. Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong D.S.Y., Fragkou P.C., Schweitzer V.A., Chemaly R.F., Moschopoulos C.D., Skevaki C., et al. How to interpret and use COVID-19 serology and immunology tests. Clin. Microbiol. Infec. 2021;27:981–986. doi: 10.1016/j.cmi.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West R., Kobokovich A., Connell N., Gronvall G.K. COVID-19 antibody tests: a valuable public health tool with limited relevance to individuals. Trends Microbiol. 2021;29:214–223. doi: 10.1016/j.tim.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerner A.M., Eisinger R.W., Lowy D.R., Petersen L.R., Humes R., Hepburn M., et al. The COVID-19 serology studies workshop: recommendations and challenges. Immunity. 2020;53:1–5. doi: 10.1016/j.immuni.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieu E., Ritchie H., Ortiz-Ospina E., Roser M., Hasell J., Appel C., et al. A global database of COVID-19 vaccinations. Nat. Hum. Behav. 2021;5:947–953. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- 14.Mathieu E., Ritchie H., Ortiz-Ospina E., Roser M., Hasell J., Appel C., et al. Author correction: a global database of COVID-19 vaccinations. Nat. Hum. Behav. 2021;5:956–959. doi: 10.1038/s41562-021-01160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peeling R.W., Heymann D.L., Teo Y.Y., Garcia P.J. Diagnostics for COVID-19: moving from pandemic response to control. Lancet. 2022;399:757–768. doi: 10.1016/S0140-6736(21)02346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciotti M., Ciccozzi M., Pieri M., Bernardini S. The COVID-19 pandemic: viral variants and vaccine efficacy. Crit. Rev. Clin. Lab. Sci. 2022;59:66–75. doi: 10.1080/10408363.2021.1979462. [DOI] [PubMed] [Google Scholar]
- 17.Galipeau Y., Greig M., Liu G., Driedger M., Langlois M.A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee E., Oh J.E. Humoral immunity against SARS-CoV-2 and the impact on COVID-19 pathogenesis. Mol. Cells. 2021;44:392–400. doi: 10.14348/molcells.2021.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristiansen P.A., Page M., Bernasconi V., Mattiuzzo G., Dull P., Makar K., et al. WHO international standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397:1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F., Althaus T., Tan C.W., Costantini A., Chia W.N., Van Vinh Chau N., et al. WHO international standard for SARS-CoV-2 antibodies to determine markers of protection. Lancet Microbe. 2022;3 doi: 10.1016/S2666-5247(21)00307-4. e81-e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonelli F., Blocki F.A., Bunnell T., Chu E., De La O.A., Grenache D.G., et al. Evaluation of the automated LIAISON((R)) SARS-CoV-2 TrimericS IgG assay for the detection of circulating antibodies. Clin. Chem. Lab Med. 2021;59:1463–1467. doi: 10.1515/cclm-2021-0023. [DOI] [PubMed] [Google Scholar]
- 23.Riester E., Findeisen P., Hegel J.K., Kabesch M., Ambrosch A., Rank C.M., et al. Performance evaluation of the Roche Elecsys Anti-SARS-CoV-2 S immunoassay. J. Virol. Methods. 2021;297 doi: 10.1016/j.jviromet.2021.114271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor S.C., Hurst B., Charlton C.L., Bailey A., Kanji J.N., McCarthy M.K., et al. A new SARS-CoV-2 dual-purpose serology test: highly accurate infection tracing and neutralizing antibody response detection. J. Clin. Microbiol. 2021:59. doi: 10.1128/JCM.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHugh M.L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 26.StataCorp. Stata statistical software release 15. 2017.
- 27.Corp I. IBM SPSS statistics for windows, version 25.0. 2017.
- 28.Mattiuzzo GB, E.M.; Hassall, M.; Routley, S.; Richardson, S.; Bernasconi, V.; Kristiansen, P.; Harvala, H.; Roberts, D.; Semple, M.G.; Turtle, L.C.W.; Openshaw PJM and Kenneth Baillie on behalf of the ISARIC4C Investigators, Lise Sofie Haug Nissen-Meyer, Arne Broch Brantsæter, Helen Baxendale, Eleanor Atkinson, Peter Rigsby, David Padley, Neil Almond, Nicola J. Rose, Mark page and the collaborative study participants. Establishment of the WHO international standard and reference panel for anti-SARS-CoV-2 antibody. In: WHO the Expanded Programme on Immunization of the Department of Immunization VaB, editor.z. p. 60.
- 29.Di Meo A., Miller J.J., Fabros A., Brinc D., Hall V., Pinzon N., et al. Evaluation of three Anti-SARS-CoV-2 serologic immunoassays for post-vaccine response. J. Appl. Lab Med. 2022;7:57–65. doi: 10.1093/jalm/jfab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung K., Shin S., Nam M., Hong Y.J., Roh E.Y., Park K.U., et al. Performance evaluation of three automated quantitative immunoassays and their correlation with a surrogate virus neutralization test in coronavirus disease 19 patients and pre-pandemic controls. J. Clin. Lab Anal. 2021;35:e23921. doi: 10.1002/jcla.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkmann T., Perkmann-Nagele N., Koller T., Mucher P., Radakovics A., Marculescu R., et al. Anti-Spike Protein Assays to Determine SARS-CoV-2 antibody levels: a Head-to-Head comparison of five quantitative assays. Microbiol. Spectr. 2021;9 doi: 10.1128/spectrum.00247-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley B.T., Bryan A., Fink S.L., Goecker E.A., Roychoudhury P., Huang M.L., et al. Anti-SARS-CoV-2 antibody levels measured by the AdviseDx SARS-CoV-2 assay are concordant with previously available serologic assays but are not fully predictive of sterilizing immunity. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00989-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egger A.E., Irsara C., Holzer B., Winkler C., Bellmann-Weiler R., Weiss G., et al. Borderline and weakly positive antibody levels against the S-protein of SARS-CoV-2 exhibit limited agreement with virus neutralization titres. J. Clin. Virol. Plus. 2022;2 doi: 10.1016/j.jcvp.2021.100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saker K., Escuret V., Pitiot V., Massardier-Pilonchery A., Paul S., Mokdad B., et al. Evaluation of commercial Anti-SARS-CoV-2 antibody assays and comparison of standardized titers in vaccinated health care workers. J. Clin. Microbiol. 2022;60 doi: 10.1128/JCM.01746-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Infantino M., Pieri M., Nuccetelli M., Grossi V., Lari B., Tomassetti F., et al. The WHO international standard for COVID-19 serological tests: towards harmonization of anti-spike assays. Int. Immunopharmacol. 2021;100 doi: 10.1016/j.intimp.2021.108095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukaszuk K., Kiewisz J., Rozanska K., Dabrowska M., Podolak A., Jakiel G., et al. Usefulness of IVD kits for the assessment of SARS-CoV-2 antibodies to evaluate the humoral response to vaccination. Vaccines (Basel) 2021:9. doi: 10.3390/vaccines9080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., et al. Interim results of a phase 1-2a trial of Ad26.COV2.S Covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P., Cai J., Jia R., Xia S., Wang X., Cao L., et al. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerg Microbes Infect. 2020;9:1254–1258. doi: 10.1080/22221751.2020.1772677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlton C.L., Babady E., Ginocchio C.C., Hatchette T.F., Jerris R.C., Li Y., et al. Practical Guidance for Clinical Microbiology Laboratories: viruses Causing Acute Respiratory Tract Infections. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall V., Foulkes S., Insalata F., Kirwan P., Saei A., Atti A., et al. Protection against SARS-CoV-2 after Covid-19 vaccination and previous infection. N. Engl. J. Med. 2022;386:1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne G., Hames T., Scotton C., Gent N., Johnsen A., Anderson R.M., et al. Does infection with or vaccination against SARS-CoV-2 lead to lasting immunity? Lancet Respir. Med. 2021;9:1450–1466. doi: 10.1016/S2213-2600(21)00407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.