Abstract
23S ribosomal RNA (rRNA) of Escherichia coli 50S large ribosome subunit contains 26 post-transcriptionally modified nucleosides. Here, we determine the extent of modifications in the 35S and 45S large subunit intermediates, accumulating in cells expressing the helicase inactive DbpA protein, R331A, and the native 50S large subunit. The modifications we characterized are 3-methylpseudouridine, 2-methyladenine, 5-hydroxycytidine, and nine pseudouridines. These modifications were detected using 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesul-fonate (CMCT) treatment followed by alkaline treatment. In addition, KMnO4 treatment of 23S rRNA was employed to detect 5-hydroxycytidine modification. CMCT and KMnO4 treatments produce chemical changes in modified nucleotides that cause reverse transcriptase misincorporations and deletions, which were detected employing next-generation sequencing. Our results show that the 2-methyladenine modification and seven uridines to pseudouridine isomerizations are present in both the 35S and 45S to similar extents as in the 50S. Hence, the enzymes that perform these modifications, namely, RluA, RluB, RluC, RluE, RluF, and RlmN, have already acted in the intermediates. Two uridines to pseudouridine isomerizations, the 3-methylpseudouridine and 5-hydroxycytidine modifications, are significantly less present in the 35S and 45S, as compared to the 50S. Therefore, the enzymes that incorporate these modifications, RluD, RlmH, and RlhA, are in the process of modifying the 35S and 45S or will incorporate these modifications during the later stages of ribosome assembly. Our study employs a novel high throughput and single nucleotide resolution technique for the detection of 2-methyladenine and two novel high throughput and single nucleotide resolution techniques for the detection of 5-hydroxycytidine.
Graphical Abstract
INTRODUCTION
The Escherichia coli (E. coli) ribosome, which migrates in a sucrose gradient as a particle with a sedimentation coefficient of 70S, consists of the 30S small and the 50S large subunit. The 30S subunit comprises one 16S ribosomal RNA (rRNA) molecule and 21 ribosomal proteins (r-proteins).1 The 50S large subunit is made up of two rRNA molecules, 23S and 5S, and 33 r-proteins.1 In E. coli, the 16S and 23S rRNAs are post-transcriptionally modified. The rRNA modifications have been shown to modulate the catalytic activity of the ribosome, stabilize the proper rRNA structure formation, and the correct rRNA-r-protein interactions.2–5 Moreover, enzymes performing RNA post-transcriptional modifications have been suggested to be RNA folding chaperones.6,7 Hence, both the enzymes that perform the modifications and the modifications themselves could modulate the ribosome assembly. Determining the precise points during ribosome assembly, the rRNA modifications are incorporated into intermediates populated under different cellular conditions, is important for a complete understanding of the ribosome assembly process.
E. coli 23S rRNA contains 26 post-transcriptional modifications.8,9 The most common modification in E. coli 23S rRNA is uridine (U) to pseudouridine (ψ) isomerization. There are nine ψ nucleotides in 23S rRNA, 748, 957, 1915, 1921, 2508, 2584, 2461, 2608, and 2609, and one 3-methyl ψ (m3ψ), 1919.8 Additionally, E. coli 23S rRNA contains 13 methylated nucleosides: 1-methylguanine (m1G) 747, 5-methyluridine (m5U) 749, 6-methyladenine (m6A) 1620, 2-methylguanine m2G 1837, m5U 1943, 5-methylcytidine (m5C) 1966, m6A 2034, 7-methylguanine (m7G) 2073, 2′O-methylguanine (Gm) 2255, m2G 2449, 2′O-methylcytidine (Cm) 2502, 2-methyladenine (m2A) 2507, and 2′O-methyluridine Um 2556.8 Lastly, E. coli 23S rRNA contains one 5-hydroxycytidine (OH5C) 2505 and one dihydrouridine (D) 2453.8,9
Both in vitro and in vivo experiments were employed to determine the time points during large subunit ribosome assembly when modifications are incorporated into 23S rRNA. The in vitro experiments used purified methyltransferases and investigated the ability of these enzymes to methylate naked 23S rRNA,10–14 a partially deproteinated 50S precursor,15 the 50S large subunit,16,17 or the 70S ribosome.16,18 Methyltransferases that modified the naked 23S rRNA were considered to act early during large subunit ribosome assembly, the methyltransferase that modified the partially deproteinated 50S precursor was considered to act during the intermediate stages of large subunit, and the methyltransferases that acted only in the 50S and 70S particles were considered to act during the late stages of large subunit ribosome assembly. The in vitro studies collectively determined the time points at which eight methyltransferases carry out their function during the large subunit assembly process.
To determine when the remaining modifications were incorporated into 23S rRNA and to investigate whether in vivo the modifying enzymes acted at different stages of large subunit ribosome assembly compared to in vitro, three studies investigated the extent of 23S rRNA modifications in the large subunit’s intermediates isolated from cells. Leppik et al. isolated a 40S large subunit intermediate from cells lacking the DEAD-box RNA helicase DeaD.19 Employing gel electrophoresis primer extension, they compared the extents of the Ψ 1915, 1919, and 1921 incorporation into the 40S intermediate, the native 50S, and the 70S ribosome. The comparison demonstrated that the Ψ isomerase RluD, which converts U 1915, U 1919, and U 1921 to Ψ, acts late during large subunit ribosome assembly.19
Siibak et al. isolated large subunit intermediates, accumulating in cells treated with chloramphenicol and erythromycin.20 Both antibiotics produced two large subunit intermediates that migrated as particles with sedimentation coefficients of 35S and 45S. The extent of 10 Ψ-isomerizations and 13 nucleoside methylations on the 35S, 45S intermediates, and 50S large subunit was investigated using high-performance liquid chromatography and gel electrophoresis primer extension. Next, they grouped the modifications into three classes. The modifications that occurred at the same extent in the 35S and 50S particles were considered to be placed during early stages of large subunit ribosome assembly. The modifications that were largely missing from the 35S intermediates but were placed at the same extent in the 45S intermediates and the 50S large subunit were considered to occur during the intermediate stages of large subunit ribosome assembly.20 Lastly, the modifications that were largely missing in the intermediates and present only in the 50S large subunit were considered to occur during the late stages of large subunit ribosome assembly.20
More recently, Popova et al. and Rabuck-Gibbsons et al. isolated large subunit intermediates from wild-type cells and cells lacking the DEAD-box RNA helicase SrmB.21,22 Using quantitative mass spectrometry (MS), they determined the time points during large subunit assembly that the incorporations of four Ψ nucleotides, m3Ψ, 14 methylated nucleosides, and OH5C occur.21,22
Interestingly, a subset of 23S rRNA modifications are incorporated at different time points during large subunit ribosome assembly in cells grown under different conditions.19,20,22 Also, differences on the time points of the modifications were observed between the in vivo accumulated intermediates and the in vitro particles.19,20,22 Combining the above observations further demonstrates that the intermediates’ rRNA structure, r-protein, and maturation factor compositions modulate the time points during large subunit ribosome assembly at which the enzymes incorporate the modifications into 23S rRNA. More importantly, these comparisons demonstrate the need for identifying and characterizing novel classes of intermediates to better understand the interconnection between different ribosome assembly events and rRNA modifications.
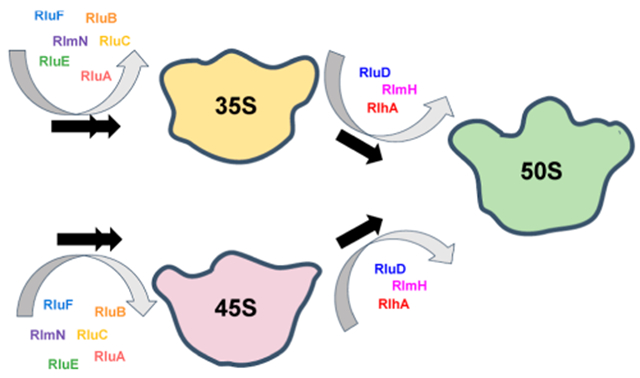
Here, we investigate the extent of twelve 23 rRNA modifications, nine Ψ nucleotides, m3Ψ, m2A, and OH5C, in the 35S and 45S large subunit intermediates accumulated in E. coli cells, expressing R331A DbpA, and compare the extent of these modifications to those of native 50S large subunits. DbpA is a DEAD-box RNA helicase implicated in peptidyl transferase center maturation.23–26 Based on the sucrose gradient migration and 5′ end 23S rRNA processing, the 35S is an early-stage large subunit assembly intermediate.27 Similarly, based on sucrose gradient migration, 5′ end 23S rRNA processing, and r-protein composition, the 45S is a late-stage large subunit assembly intermediate.27–29 Furthermore, the 35S and 45S intermediates belong to two independent pathways of large subunit ribosome assembly.27 Therefore, the experiments outlined here describe when the modification enzymes act on two different pathways and stages of large subunit assembly.
If a modification is present in the 35S or 45S intermediate to the same extent as in the native 50S large subunit, the enzyme performing the modification acts in cells before the 35S or 45S intermediate is populated. On the other hand, if a modification is missing in the 35S or 45S intermediate but is present in the native 50S large subunit, the enzyme performing the modification is in the process of acting in the 35S or 45S intermediate, or it will act during the later stages of large subunit ribosome assembly once the 35S or 45S intermediate have further matured.
In addition, the 35S and 45S intermediates act as markers of the time points, during 50S large subunit ribosome assembly, when modifications are incorporated. Since the 35S intermediate is an early-stage large subunit ribosome assembly intermediate,27 on the pathway, in which the 35S intermediate is populated, a modification present in the 35S intermediate is incorporated during the early stages of large subunit ribosome assembly. Furthermore, a modification missing in the 35S intermediate is incorporated during the intermediate or late stages of large subunit ribosome assembly. On the other hand, the 45S intermediate is a late-stage large subunit ribosome assembly intermediate.27–29 Thus, on the pathway in which the 45S intermediate is populated, a modification present in the 45S intermediate is incorporated during the early or intermediate stages of large subunit ribosome assembly. A modification missing in the 45S intermediate is incorporated during the late stages of large subunit ribosome assembly.
The technique we employ to simultaneously detect the Ψs, m3Ψ, m2A, and OH5C modifications involves 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) and alkaline treatment of rRNA combined with reverse transcriptase misincorporation and deletion counting. The reverse transcriptase misincorporations and deletions were determined by next-generation sequencing (NGS). Our technique is similar to the previously used techniques for Ψ detections, which involve CMCT30 or bisulfite treatment.31
CMCT treatment has been used extensively to detect Ψ formations. CMCT forms adducts with Ψ at positions N1 and N3, G at position N1, and U at position N3.32 Treatment of RNA with alkaline solution removes the CMCT adducts from G and U.33 On the other hand, the CMCT adduct at the N3 position of Ψ is resistant to alkaline treatment.33 The CMCT adduct at the N3 position of Ψ has been shown to produce reverse transcriptase stops and misincorporations.30,34 Recently, NGS counting of stops or a combination of stops and misincorporations, produced by CMCT treatment of RNA, has been used to determine the U to Ψ conversion.30,34 The bisulfite treatment of RNA combined with NGS was employed to simultaneously detect Ψ, m5C, and m1A.31 In the bisulfite experiments, the Ψ nucleotides were detected by deletion counting, while m5C and m1A modifications were detected by misincorporation counting.
Our results indicate that Ψ 748, Ψ 957, Ψ 2461, Ψ 2508, Ψ 2584, Ψ 2608, Ψ 2609, and m2A 2507 were present in the 35S and 45S intermediates to the same extent as in the native 50S large subunit. Hence, enzymes performing these modifications act before the 35S and 45S intermediates are assembled in the cell. On the other hand, Ψ 1915, Ψ 1921, m3Ψ1919, and OH5C 2505 modifications are considerably less present in the 35S and 45S intermediates when compared with the native 50S large subunit. Taken together, these results demonstrate that the enzymes performing these modifications are in the process of acting in the 35S and 45S intermediates or will act at the later stages of large subunit assembly.
In addition, by treating 23S rRNA with KMnO4 and counting the misincorporations and deletions of the reverse transcriptase, we are able to accurately detect OH5C. Therefore, employing our techniques, the OH5C could be identified in an RNA molecule using CMCT modification and validated using KMnO4 modification. Historically OH5C has been detected in RNA using MS,21,22,35,36 which is not a high throughput and single nucleotide resolution technique. Our techniques allow the detection of OH5C at a single nucleotide resolution and in a high throughput manner.
MATERIALS AND METHODS
Large Subunit Particle Isolation.
The isolation of large subunit particles from cells were performed, as previously described with a few minor changes.27 The ribosomal particles were isolated from E. coli BLR (DE3) plysS ΔdbpA/kanR cells. The cells were transformed with the pET3a vector bearing the coding sequence of the wild-type DbpA or R331A DbpA constructs. The cells were grown at 37 °C in 200 mL of media containing 10 g/L tryptone, 1 g/L yeast extract, 10g/L NaCl, 100 μg/mL carbenicillin, and 34 μg/mL chloramphenicol. When the cell reached an optical density at 600 nm of approximately 0.3, the cell growth was arrested by adding 200 mL of ice. Next, the cells were pelleted by centrifugation.
The cell pellet was resuspended in a buffer consisting of 20 mM HEPES-KOH (pH 7.5), 30 mM NH4Cl, 1 mM MgCl2, 4 mM BME, and 300 μg of lysozyme and incubated in ice for 30 min. Next, the mixture was flash frozen in liquid nitrogen and thawed at room temperature. This step was repeated three times to ensure complete cell lysis. The lysed cells were treated with 20 units of DNase I (RNase free) for 90 min on ice to digest the DNA. Subsequently, the cell lysate was cleared by centrifugation. The cleared lysate was either directly loaded on a 20–40% sucrose gradient or flash frozen in small aliquots and stored in −80 °C.
The 20–40% gradients were made in a buffer consisting of 20 mM HEPES KOH (pH 7.5), 150 mM NH4Cl, 1 mM MgCl2, and 4 mM BME. The gradients were prepared using the Biocomp Gradient Master. To separate the particles, the cleared lysate was layered on top of the gradient, and the gradient was spun at 32,000 revolution per minute using SW 32 rotor for 16 h at 4 °C. The gradient was fractionated using the Teledyne R1 fraction collector combined with the SYR-101 syringe pump and collected in ultraviolet transparent 96 well plates from Corning. Next, the 96-well plates’ absorbance at 260 nm was read using the SpectraMax plate reader from Molecular Devices. The fractions containing the particles were pooled together and were either directly used for the chemical modification experiments or flash frozen in small aliquots and stored in −80 °C.
In a sucrose gradient, the 45S particle is clearly separated both from the 35S intermediate and 50S large subunit.27–29 On the other hand, the 35S intermediate travels near and under the 30S small subunit.27 Therefore, the isolated 35S particle sample also contains the native 30S small subunit.27 The NGS data for the 35S particles were aligned both to 23S rRNA and 16S rRNA. Lastly, all the 50S particles in this study were isolated from E. coli cells, expressing the wild-type DbpA.
CMCT Treatment of rRNA.
The CMCT treatment of RNA was performed, as previously described with a few modifications.37,38 The proteins were removed from the ribosomal particles by phenol/chloroform extraction, and the rRNA was concentrated by ethanol precipitation. Approximately 5 μg of rRNA from each particle was treated with 170 mM CMCT in a buffer containing 50 mM bicine (pH 8.3), 7 M urea, and 4 mM EDTA for 30 min at 37 °C in a total reaction volume of 120 μL. The CMCT reaction was arrested by the addition of 120 μL of 0.3 M sodium acetate buffer at pH 5.5. Next, the rRNA was ethanol precipitated, resuspended in 100 μL of sodium carbonate buffer (pH 10.4), incubated at 37 °C for 2.5 h and then at 65 °C for 30 min. The reaction was stopped with the addition of 100 μL of 0.3 M sodium acetate buffer at pH 5.5. The rRNA was ethanol precipitated and later used for NGS library preparation. The control reactions went through the exact same process as the experimental sample, the only exception being that they were not exposed to CMCT.
KMnO4 Treatment of rRNA.
30S and 50S particles used for the KMnO4 treatment were isolated from cells, expressing the wild-type DbpA. The proteins were removed from the particles using phenol/chloroform extraction followed by ethanol precipitation. About 5 μg of rRNA was treated with 0.12 mM KMnO4 in a total volume of 25 μL and in the presence of 30 mM of sodium acetate (pH 4.4). The reaction was allowed to continue for 3 or 6 min and was stopped by the addition of 2 μL of 14.3 M 2-mercaptoethanol.39,40 The rRNA was subsequently concentrated and desalted using ethanol precipitation and used for NGS library preparation. The control reactions went through the exact same process as the experimental sample, the only exception being that they were not exposed to KMnO4.
NGS Library Preparation and Sequencing.
The NGS sequencing library was performed following the randomer workflow of the selective 2′-hydroxyl acylation analyzed by primer extension and the mutational profiling (SHAPE-MaP) protocol.41 In brief, we used nine nucleotide long random primers for the reverse transcriptase reaction. The Supper-Script II in the presence of 6 mM Mn2+ was used to perform the reverse transcriptase reaction. The NEB second strand synthesis kit was used to convert single-stranded DNA to double-stranded DNA, while the Illumina Nextera XT kit was used to tag the sequencing adaptors. The sequencing was performed using the MiSeQ 2 × 150 paired-end Illumina platform.
Mutation Rate Determination.
The mutation rates at specific nucleotides were calculated using ShapeMapper 1.2.41 ShapeMapper 1.2 determines the mutations by counting the number of misincorporations and deletions at a specific nucleotide position. Subsequently, the mutation rate is calculated by dividing the number of total misincorporations plus deletions at a specific nucleotide by the read depth at that nucleotide.41 Thus, the mutation rate as calculated by ShapeMapper 1.2 is the fraction of misincorporated and deleted nucleotides at a specific nucleotide position.41
The NGS reads were aligned to the sequence of 23S rRNA rrlB gene or 16S rRNA rrsB gene of DE3 (BL21) cells. Lastly, the ShapeMapper 1.2 suggested parameters for the randomer workflow and Illumina Nextera XT kit were used to calculate the mutation rate at each nucleotide position.41
Determination of rRNA Modifications Using CMCT.
Two different biological samples were treated with CMCT and NaHCO3, while one control sample was treated only with NaHCO3. The background corrected mutation rate was calculated using the equation below.
| (1) |
In eq 1, BCm(i)(p)(rn) is the background corrected mutation rate, for nucleotide i, of particle p for replicated rn. m(CMCT)(i)(p)(rn) is the ShapeMapper1.2 calculated mutation rate for the CMCT- and NaHCO3-treated sample, nucleotide i, particle p, and replicated rn. m(i)(p) is the ShapeMapper1.2 calculated mutation rate for the control samples treated only with NaHCO3 for nucleotide i and particle p.
In the manuscript’s figures, the nucleotides that differ in different ribosome genes and the nucleotides that have a mutation rate of higher than 5% in the control sample are considered unmodified and are not shown.41
The average background corrected mutation rate was calculated using the equation given below
| (2) |
In eq 2, avg(i)(p) is the average mutation rate background corrected for biological replicates 1 and 2 for nucleotide i of particle p. BCm(i)(p)(r1) and BCm(i)(p)(r2) are background corrected mutation rates for the two CMCT- and NaHCO3-treated biological replicates for nucleotide i and particle p.
The standard deviation error between two different biological replicates is calculated using the equation given below
| (3) |
In eq 3, std(i)(p) is the standard deviation errors for the nucleotide i of particle p treated with CMCT and NaHCO3. Avg(i)(p) is the average mutation rate background corrected from eq 2, and BCm(i)(p)(r1) and BCm(i)(p)(r2) are the background corrected mutation rates for the biological replicates 1 and 2 calculated from eq 1.
OH5C Determination Using KMnO4 Treatment.
In order to determine OH5C modifications using KMnO4 treatment, we calculated the background corrected mutation rate using eq 1. For these calculations, the experimental sample was the sample treated with KMnO4, while the control sample was the untreated sample. In the manuscript’s figures, the nucleotides which vary in different E. coli ribosomal genes and the nucleotides that have a mutation rate of higher than 5% in the control sample are not shown.41 Supporting Information (Table S1) shows the mutation rates of the 23S rRNA nucleotide OH5C 2505 exposed to KMnO4 for 3 or 6 min and the control sample that never saw KMnO4.
RESULTS AND DISCUSSION
Ψ Isomerases RluA, RluB, RluC, RluE, and RluF act before the 35S and 45S intermediates are populated in the cell, while Ψ isomerase RluD and methyltransferase RlmH are in the process of acting in the 35S and 45S intermediates or will act during the later stages of large subunit assembly.
The data in Figure 1A show the NGS mutation rates for U nucleotides after CMCT and NaHCO3 treatment of 23S rRNA from the native 50S large subunit. The mutation rates are background corrected by subtracting the mutation rates of 23S rRNA treated only with NaHCO3 from the 23S rRNA mutation rates treated with CMCT and NaHCO3 (Equation 1). ShapeMapper 1.2,41 which counts mutations as the sum of the deletions and misincorporations, was used to determine the mutation rates at each nucleotide position. There are nine Ψ nucleotides in 23S rRNA. The nine Ψ nucleotides have mutation rates considerably higher than the other Us in 23S rRNA. Thus, counting deletions and misincorporations, we accurately detect the Ψ nucleotides in the 23S rRNA of the native 50S large subunit.
Figure 1.
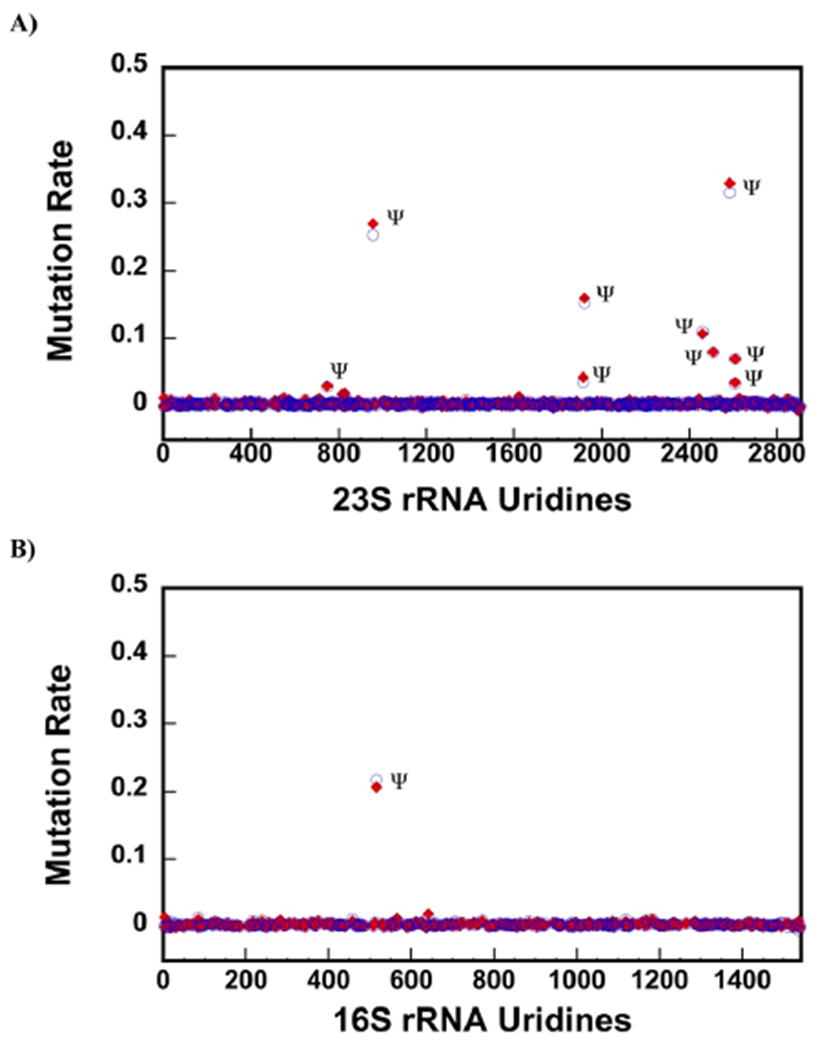
CMCT followed by NaHCO3 treatment detects nine Ψ nucleotides in 23S rRNA of the 50S and the only Ψ in 16S rRNA of the 30S. (A,B) Background corrected rates of mutations were calculated as explained in the Materials and Methods section of the paper. Only the U residues are shown in the x-axes. The blue circles and the red diamonds are the data from two different biological samples. There are nine Ψ nucleotides in 23S rRNA of the 50S large subunit. We are able to detect all of them (A). The mutation rates for m3Ψ, as explained in the Results and Discussion section of the paper, are not shown in this figure (A). There is only one Ψ in 16S rRNA, which our chemical modification and mutation calculation methods also detect (B).
Interestingly, methylation of Ψ 1919 at position N3, similar to the CMCT modification of Ψ at position N3, produces reverse transcriptase deletions and misincorporations (Supporting Information, Table S2). The CMCT combined with NaHCO3 treatment cannot modify the N3 methylated Ψ 1919 but does modify the N3 unmethylated Ψ 1919. As a consequence, the mutation rates we observe at position 1919 in the CMCT- and NaHCO3-treated samples are the sum of the mutation rates of methylation and CMCT adduct formation at the Ψ 1919 N3 position. If a fraction of Ψ 1919 nucleotides is unmethylated, we should observe a higher mutation rate on CMCT- and NaHCO3-treated 23S rRNA compared to 23S rRNA that was exposed only to NaHCO3. However, because Ψ 1921 is modified by CMCT, and ShapeMapper 1.2 clusters the mutations occurring in the same read at the 3′ end mutation position,42 a number of mutations occurring at Ψ 1919 in the CMCT-treated samples are counted by Shape-Mapper 1.2 as Ψ 1921 mutations. This decreases the mutation rate at the Ψ 1919 nucleotide for the CMCT-treated samples when compared with the control sample (Supporting Information, Table S2). Consequently, the background corrected mutation rates at nucleotide 1919 are negative numbers (Supporting Information, Table S2) and are not shown in Figure 1A.
The data in Figure 1B show the background corrected mutation rates for 16S rRNA U nucleotides from the 30S small subunit treated with CMCT followed NaHCO3 (Equation 1). There is one known Ψ in 16S rRNA, which is at position 516. We are able to correctly determine the 16S rRNA’s Ψ. Furthermore, similar to 23S rRNA, we do not observe false positive Ψ nucleotides in 16S rRNA.
Figure 2A shows the background corrected average mutation rate for the Ψ nucleotides 748, 957, 2461, 2508, 2584, 2608, and 2609 of 23S rRNA from different particles. The average mutation rates were calculated, as explained in the Materials and Methods section (eq 2). The average mutation rates for these nucleotides are very similar between the 35S and 45S intermediates and the native 50S large subunit (Table 1); hence, these Ψ isomerizations are performed before the 35S and 45S are populated in cells. The isomerizations are performed as follows: U 748 to Ψ by RluA;43 nucleotides U 957, 2508, and 2584 to Ψ by RluC;44 U 2461 to Ψ by RluE;45 U 2608 to Ψ by RluF;45 and U 2609 to Ψ by RluB.45 Thus, in the cells expressing R331A DbpA, the RluA, RluB, RluC, RluE, and RluF Ψ isomerases act before the 35S and 45S intermediates assemble.
Figure 2.
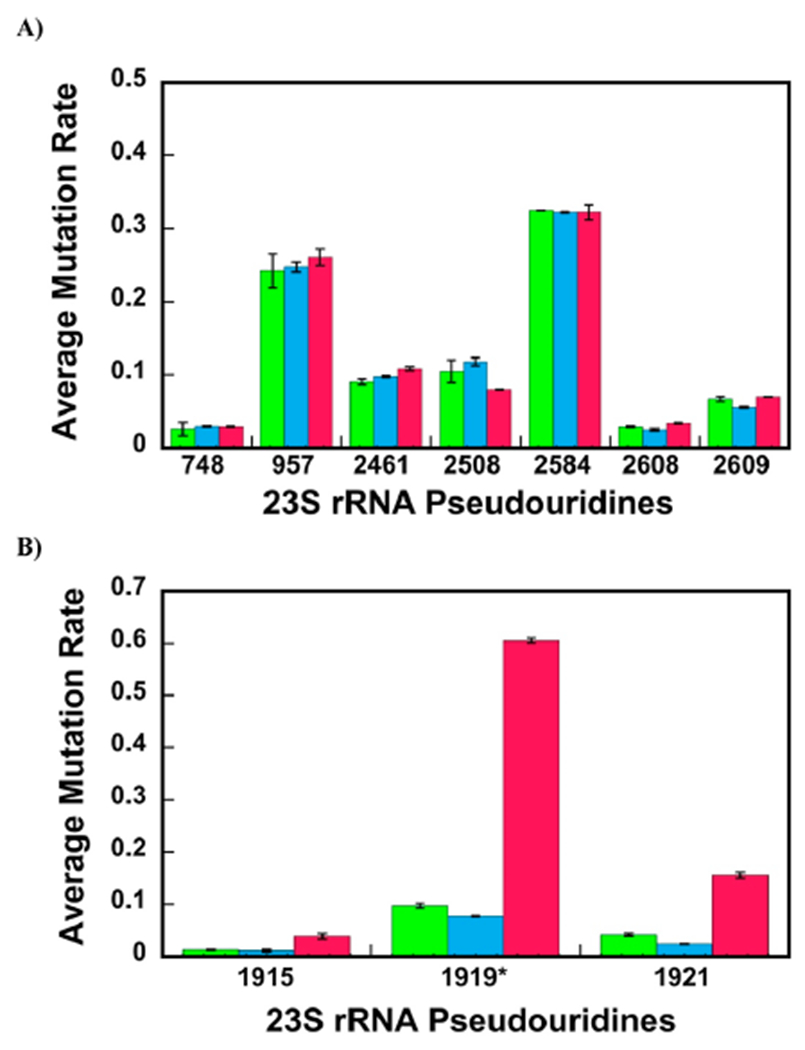
Majority of Ψ modifications are present in the 35S and 45S intermediates. The calculation of average mutation rates for the Ψs detections were performed, as explained in the Materials and Methods section of the paper. (A,B) Average mutation rates for specific nucleotides of the 35S and 45S intermediates and native 50S large subunit are shown in green, blue, and red, respectively. The data are the averages of two biological replicates, and the errors are the standard deviation from the averages. (A) Nucleotides U 748, 957, 2461, 2508, 2584, 2608, 2609 isomerizations to Ψ occur before the 35S and 45S intermediates are populated in cells. The extents of the nucleotides U 748, 957, 2461, 2508, 2584, 2608, and 2609 isomerizations to Ψ are similar for the 35S, 45S, and 50S particles. Therefore, these isomerizations occur before the 35S and 45S intermediates are formed. (B) Nucleotides U 1915, 1919, and 1921 are in the process of being isomerized to Ψ in the 35S and 45S intermediates or will be isomerized to Ψ during the later stages of large subunit ribosome assembly. The nucleotides U 1915, 1919, and 1921 are extensively more isomerized to Ψ in the native 50S large subunit than that in the 45S and 35S intermediates. As explained in the manuscript, the average mutation rate for m3Ψ 1919 (1919*) was calculated from the two CMCT- and NaHCO3-treated samples without subtracting the mutation rate of NaHCO3 only treated control sample. Thus, no background correction was performed on the m3Ψ 1919 nucleotide data.
Table 1.
Modified Nucleotide Compositions of 23S rRNA in the Intermediates and Native 50S Large Subunit
| Mutation Ratesc |
||||
|---|---|---|---|---|
| Modifieda Nucleotide | Enzymeb | 35S | 45S | 50S |
| Ψ 748 | RluA | 0.026 ± 0.009 | 0.030 ± 0.001 | 0.029 ± 0.001 |
| Ψ 957 | RluC | 0.242 ± 0.023 | 0.247 ± 0.007 | 0.261 ± 0.012 |
| Ψ 1915 | RluD | 0.013 ± 0.000 | 0.011 ± 0.002 | 0.039 ± 0.005 |
| dm3 Ψ 1919 | RluD/RlmH | 0.097 ± 0.005 | 0.077 ± 0.001 | 0.605 ± 0.004 |
| Ψ 1921 | RluD | 0.042 ± 0.003 | 0.024 ± 0.001 | 0.156 ± 0.006 |
| Ψ 2461 | RluE | 0.090 ± 0.004 | 0.098 ± 0.002 | 0.108 ± 0.003 |
| OH5C 2505 | RlhA | 0.070 ± 0.002 | 0.108 ± 0.001 | 0.265 ± 0.006 |
| m2A 2507 | RlmN | 0.083 ± 000 | 0.097 ± 0.001 | 0.090 ± 0.008 |
| Ψ 2508 | RluC | 0.105 ± 0.016 | 0.118 ± 0.006 | 0.080 ± 0.000 |
| Ψ 2584 | RluC | 0.324 ± 0.000 | 0.322 ± 0.001 | 0.322 ± 0.010 |
| Ψ 2608 | RluF | 0.0291 ± 0.001 | 0.025 ± 0.002 | 0.034 ± 0.001 |
| Ψ 2609 | RluB | 0.067 ± 0.003 | 0.055 ± 0.001 | 0.070 ± 0.000 |
The modified nucleotides investigated in this study.
Enzymes incorporating the modifications.
Mutation rates were calculated as explained in the Materials and Methods section of the paper (eq 1). The values shown are the averages of two different biological samples. The errors are the standard deviations from the averages (eqs 2 and 3).
The RluD enzyme performs the U 1919 to Ψ isomerization, while the RlmH enzyme methylates Ψ 1919 at position N3. As explained in the manuscript, for nucleotide 1919, the mutation rates were not background corrected, and values shown are averages and standard deviations for uncorrected mutation rates.
Figure 2B shows the background corrected average mutation rates for nucleotides 1915 and 1921 and uncorrected average mutation rate for the 1919 nucleotide from the 35S, 45S, and 50S. The average mutation rates for 1915 and 1921 nucleotides are considerably higher for the native 50S large subunit than the 35S and 45S intermediates (Table 1, Supporting Information, Table S3). Therefore, the nucleotide U 1915 and 1921 isomerizations to Ψ occur after the 35S and 45S intermediates are populated in cells. RluD isomerizes nucleotides U 1915 and 1921 to Ψ.46 Thus, RluD is in the process of acting in 1915 and 1921 nucleotides of the 35S or 45S intermediates or will act in these nucleotides during later stages of large subunit ribosome assembly.
The average mutation rate at nucleotide 1919 of 23S rRNA is also significantly smaller in the 35S and 45S intermediates than that in the native 50S large subunit (Figure 2B, Table 1, Supporting Information, Tables S2 and S3). As explained above, for the nucleotide 1919, we count the combined mutation rates of the methylated and CMCT-modified Ψ at position N3. Consequently, all the mutations we observe for nucleotide 1919 are at a minimum U nucleotides isomerized to Ψ nucleotides. RluD performs U 1919 to Ψ isomerization,46 and we conclude that RluD is in the process of performing the U 1919 to Ψ isomerization in the 35S and 45S intermediate or will perform this isomerization during the later stages of large subunit ribosome assembly. RlmH methylates U 1919 only after RluD has isomerized this U to Ψ.18 Thus, RlmH, similar to RluD, is in the process of acting in the 35S and 45S intermediates or will act during the later stages of large subunit ribosome assembly.
Next, we compared our results of Ψ isomerizations with those of previously investigated large subunit intermediates. The nucleotides U 1915, 1919, and 1921 are isomerized to Ψ during the late stages of large subunit ribosome assembly in wild-type cells,22 cells lacking the SrmB protein,22 and cells lacking the DEAD-box RNA helicase DeaD.19 On the other hand, experimental measurements performed with particles, accumulated in cells treated with chloramphenicol20 and erythromycin,20 demonstrate that the nucleotides U 1915, 1919, and 1921 are isomerized to Ψ during the middle-to-late stages of large subunit ribosome assembly. The determination that in the 35S intermediate, the nucleotide U 1915, 1919, and 1921 isomerizations to Ψ have occurred at a significantly smaller extent than that in the native 50S large subunit (Table 1, Supporting Information, Table S3, Figure 2B), indicating that, on the pathway, in which the 35S intermediate is populated, the nucleotides U 1915, 1919, and 1921 isomerizations to Ψ occur during the intermediate stages of large subunit ribosome assembly, similar to cells treated with chloramphenicol20 and erythromycin,20 or, alternatively, during the late state of large subunit ribosome assembly, similar to wild-type cells,22 cells lacking SrmB,22 and cells lacking DeaD.19
Moreover, in the 45S intermediate, the Ψ modifications at positions 1915, 1919, and 1921 have occurred at significantly smaller extents than that in the native 50S large subunit. Therefore, on the pathway, in which the 45S intermediate is populated, nucleotide U 1915, 1919, and 1921 isomerizations to Ψ occur, similar to the experimental measurements performed on the intermediates from wild-type cells, cells lacking the SrmB protein, and cells lacking the DeaD protein, during the late stages of large subunit ribosome assembly.
In vitro experiments,18 experimental measurements performed on intermediates from wild-type cells,22 and cells lacking the SrmB protein22 have shown that the methylation of Ψ 1919 by RlmH occurs during the very late stages of large subunit ribosome assembly. Since our data show that the extent of combined m3Ψ and Ψ modifications at position 1919 is small in the 35S and 45S intermediates when compared with the native 50S large subunit (Figure 2B), we conclude that on the pathway, in which the 35S intermediate is populated, the m3Ψ modification occurs during the intermediate or late stages of large subunit ribosome assembly, while on the pathway, in which the 45S intermediate is populated, similar to the in vitro experiments,18 experimental measurements performed on wild-type cells22 and cells lacking the SrmB protein,22 the m3Ψ modification occurs during the late-stages of large subunit ribosome assembly.
RlhA has not completed its function in the 35S and 45S intermediates, while RlmN acts before the 35S and 45S intermediates are assembled in cells.
In addition to Ψ, the retention of CMCT adduct after CMCT and alkaline treatment has been observed for 2-methylthio-N6-isopentenyladenosine (ms2i6A)38 and OH5C.36 The positions of ms2i6A and OH5C modifications by CMCT remains unknown.36,38 Here, we show that under our experimental conditions, OH5C 2505 and m2A 2507 retain the CMCT adducts after CMCT and NaHCO3 treatment, and these adducts produce reverse transcriptase misincorporation and deletion (Figure 3A). The detection of OH5C 2505 and m2A 2507 using CMCT treatment was performed similar to ψ detection experiments and as explained in the Materials and Methods section.
Figure 3.
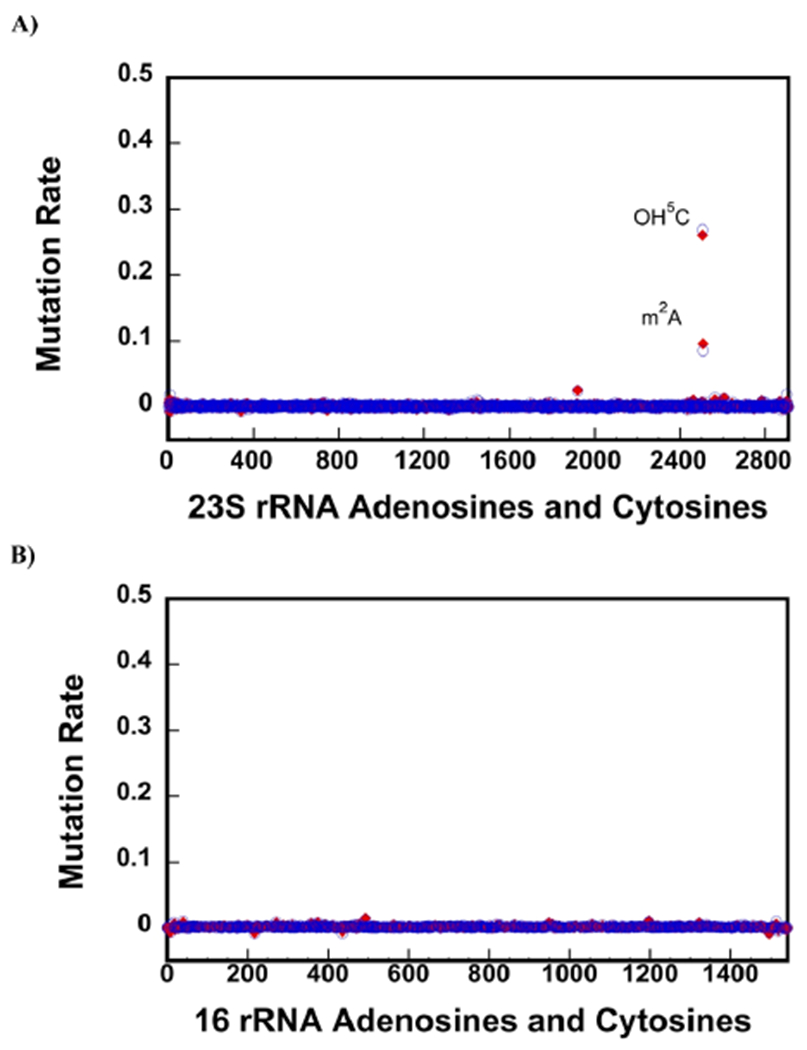
CMCT treatment followed by NaHCO3 exposure accurately detects OH5C and m2A modifications. (A,B) Background corrected mutation rates were calculated, as explained in the Materials and Methods section of the paper. Only the A and C residues are shown in the x-axes. The blue circles and the red diamonds are the data collected on two different biological samples. 23S rRNA of the native 50S large subunit contains one OH5C (2505) and m2A (2507). The background corrected mutation rates of OH5C 2505 and one m2A 2507 residues are significantly higher than the other C and A residues in 23S rRNA (A). Thus, employing CMCT and NaHCO3 treatment, we are able to detect both OH5C and m2A modifications and do not observe false positive OH5C or m2A modifications. There are no OH5C and m2A modifications in 16S rRNA of the 30S, and we do not observe false positive OH5C and m2A modifications in this RNA molecule (B).
The mutation rates for OH5C 2505 and m2A 2507 for CMCT-treated 23S rRNA from the native 50S large subunit are significantly larger than the mutation rates for the other A and C bases in the native 50S’s 23S rRNA (Figure 3A). Furthermore, we do not observe false positive OH5C or m2A modifications in the CMCT-treated native 50S’s 23S rRNA. In the 16S rRNA, there are no known OH5C or m2A modifications, and we do not observe an increase on the mutation rates for the CMCT-treated 16S rRNA from the native 30S small subunit (Figure 3B). In summary, the CMCT treatment of RNA combined with NGS accurately detects the OH5C and m2A modifications. Future experiments will determine the precise chemical structures of CMCT-modified OH5C and m2A nucleotides.
Next, we compared the extent of OH5C and m2A modifications in 35S, 45S, and 50S particles (Figure 4, Table 1, Supporting Information, Table S3). Significantly, less OH5C modifications are present in the 35S and the 45S intermediates when compared with the native 50S large subunit. Thus, the RlhA enzyme,35 which places the hydroxyl group at the position C5 of C 2505, is in the process of acting in the 35S and 45S intermediates or acts during the later stages of large subunit ribosome assembly. On the other hand, the mutation rates at position m2A 2507 are very similar for 35S, 45S, and 50S particles (Figure 4, Table 1 and Supporting Information, Table S3). This demonstrates the enzyme that incorporates this modification, RlmN,47 has performed its action before the 35S and 45S intermediates are populated in cells.
Figure 4.
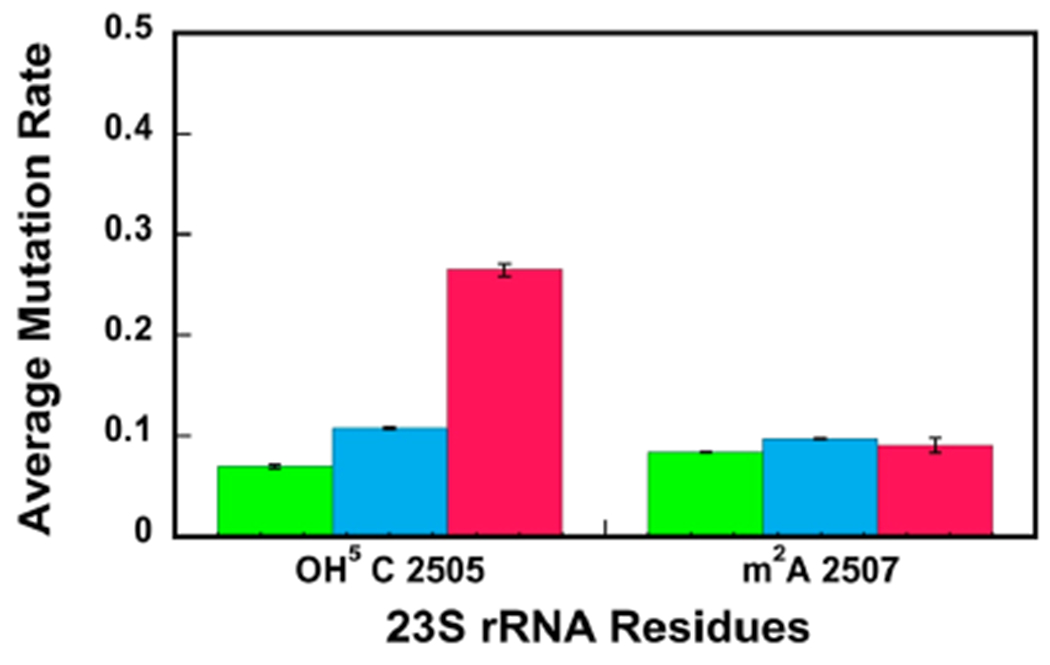
OH5C is not present at a significant level in the 35S and 45S intermediates’ 23S rRNA, whereas m2A is incorporated into 23S rRNA before the 35S and 45S intermediates are populated in cells. The average mutation rates for OH5C and m2A were calculated, as explained in Materials and Methods section of the paper. The average mutation rates for the 35S and 45S intermediates and the native 50S large subunit are shown in green, blue, and red, respectively. The data are the averages of two biological replicates, and the errors are the standard deviations from the averages. The extent of the OH5C modification in the 35S and 45S intermediates is significantly smaller than the extent of the OH5C modification in the native 50S. On the other hand, the extent of m2A modification in the 35S and 45S intermediates is similar to the extent of m2A modification in the native 50S.
Experimental measurements performed on intermediates isolated from wild-type cells demonstrated that C 2505 hydroxylation occurs during intermediate stages of large subunit ribosome assembly.22 In cells lacking the SrmB protein, the hydroxylation of C 2505 occurs during the late stages of large subunit ribosome assembly.22 OH5C is largely missing from the 35S intermediate (Table 1, Supporting Information, Table S3 and Figure 4). This observation indicates that on the pathway, in which the 35S intermediate is populated, the hydroxylation of C 2505 occurs during the intermediate stages of large subunit assembly, similar to the wild-type cells, or late stages of large subunit ribosome assembly, similar to cells lacking the SrmB protein.22 The OH5C modification is present in the 45S intermediate at a significantly less extent than that in the native 50S large subunit (Table 1, Supporting information, Table S3 and Figure 4). Consequently, on the pathway, in which the 45S intermediate is populated, the OH5C modification occurs during the late stages of large subunit ribosome assembly, similar to the cells lacking the SrmB protein.22
Experimental measurements performed on intermediates from wild-type cells,22 cells lacking the SrmB protein,22 cells treated with chloramphenicol,20 and cells treated with erythromycin20 show that the m2A 2507 modification is performed during the early stages of large subunit ribosome assembly. Our determination that the m2A 2507 modification is present at the same extent as in the native 50S large subunit and in the 35S and 45S intermediates (Table 1, Supporting Information, Table S3, and Figure 4) indicates that the m2A 2507 modification occurs during the early stages of large subunit ribosome assembly on the pathway, in which the 35S particle is populated, and during the early or intermediate stages of large subunit ribosome assembly on the pathway, in which the 45S intermediate is populated.
Detection of OH5 C Using KMnO4 Oxidation.
With the goal of developing another high throughput technique for the detection of OH5 C modifications, in addition to CMCT treatment, we treated the rRNA with KMnO4. KMnO4 is an oxidizing agent that has been used extensively to investigate the DNA structure and modifications.48 Our hypothesis was that oxidation of OH5C by KMnO4 would form molecules, which produce reverse transcriptase misincorporations and deletions.
The background corrected mutation rates of C nucleotides from native 50S 23S rRNA treated with 0.12 mM KMnO4 for 3 or 6 min are shown in Figure 5A (eq 1 and Materials and Methods). The mutation rates of OH5C 2505 are considerably higher than the mutation rates of the other C nucleotides in 23S rRNA. Hence, using KMnO4 we can accurately determine the OH5C nucleotides in 23S rRNA, and we do not observe false positive C mutations.
Figure 5.
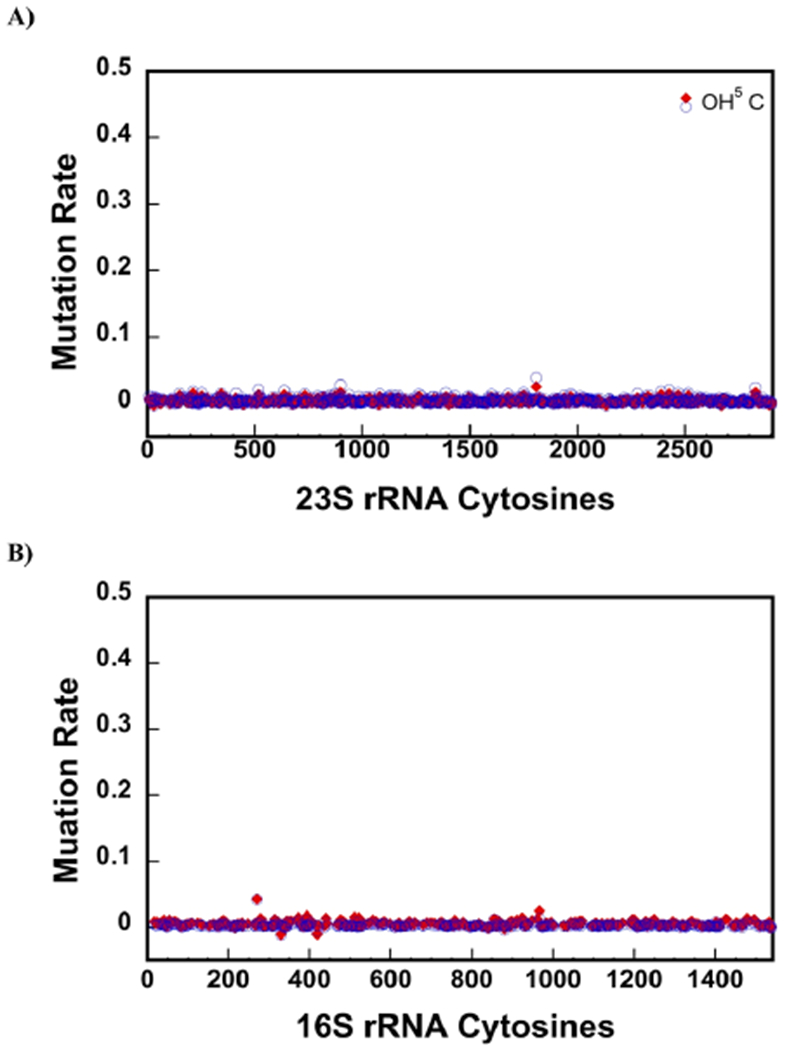
KMnO4 treatment accurately detects the OH5C. Mutations rates after treating the sample with KMnO4 were calculated, as explained in the Materials and Methods section of the paper. Only C nucleotides are shown in the x-axes. The red diamonds and blue circles are the data from treating the rRNA with KMnO4 for 3 or 6 min, respectively. (A) Mutation rates of 23S rRNA from the native 50S large subunit treated with KMnO4. KMnO4 treatment detects the single OH5C modification in 23S rRNA of the native 50S large subunit. (B) Mutation rate of 16S rRNA from 30S treated with KMnO4. No OH5C is present in 16S rRNA of the 30S, and we do not observe false positive OH5C in this molecule.
The background corrected mutation rates of C nucleotides from native 50S 23S rRNA treated with 0.12 mM KMnO4 for 3 or 6 min are shown in Figure 5A (eq 1 and Materials and Methods). The mutation rates of OH5C 2505 are considerably higher than the mutation rates of the other C nucleotides in 23S rRNA. Hence, using KMnO4 we can accurately determine the OH5C nucleotides in 23S rRNA, and we do not observe false positive C mutations.
The 16S rRNA from the native 30S small subunit was also treated with 0.12 mM KMnO4 for 3 or 6 min. There are no OH5C modifications in 16S rRNA, and we do not observe false positive OH5C in 16S rRNA using KMnO4 treatment (Figure 5B). Together, the data obtained in 23S rRNA and 16S rRNA molecules demonstrate that KMnO4 treatment of RNA combined with mutation and deletion counting accurately determines OH5C modifications.
CONCLUSIONS
We have determined in this study the extent of twelve 23S rRNA modifications on two large subunit ribosome assembly intermediates, the 35S and 45S, from two different stages and pathways of the 50S large subunit ribosome assembly. A number of modifications occur at the same stages of large subunit ribosome assembly on the pathways investigated here, the pathways investigated under different cellular conditions,19–22 and/or in vitro18 assembly pathways. This observation suggests that the rRNA structural motifs, the r-proteins’, and maturation factors’ compositions, which a number of modification enzyme recognize, are present at similar time points during large subunit ribosome assembly occurring under various cellular conditions and in vitro. Furthermore, while there are more than 170 known RNA modifications,49 and many of these modifications have been implicated in antibiotic resistance,50 cancer,50 and neurodegenerative diseases,51 only a subset of RNA modifications can be detected in a high throughput and single nucleotide manner.30,31,52,53 The simultaneous detection of Ψ, m3Ψ, m2A, and OH5C modifications in a high throughput and in a single nucleotide resolution manner described in this study is readily generalizable to other RNA molecules. Lastly, MS, which is not a high throughput technique, has been used to detect OH5C on RNA.9,22,35,36,54 Here, we have developed two techniques, involving CMCT and KMnO4 treatments combined with misincorporations and deletion counting, to detect OH5C modification in RNA in a single nucleotide and high throughput manner.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Riley C. Gentry for collecting the NGS data.
Funding
This work was supported in part by the National Institute of General Medical Sciences (grant R01-GM131062 to E.K).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00096.
Mutation rates of nucleotide OH5C 2505 untreated and treated with KMnO4; mutation rates for nucleotide m3 Ψ 1919 treated with CMCT plus NaHCO3 or only NaHCO3; and mutation rates’ confidence intervals of 23S rRNA-modified nucleotides (PDF)
Accession Codes
E. coli DbpA Uniprot entry: P21693. E. coli RluA Uniprot entry: P0AA37. E. coli RluC Uniprot entry: P0AA39. E. coli RluD Uniprot entry: P33643. E. coli RlmH Uniprot entry: P0A818. E. coli RlhA Uniprot entry: P76104. E. coli RlmN Uniprot entry: P36979. E. coli RluC Uniprot entry: P0AA39. E. coli RluF Uniprot entry: P32684. E. coli RluB Uniprot entry: P37765
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.2c00096
The authors declare no competing financial interest.
The NGS raw files and ShapeMapper 1.2 output files were deposited on Gene Expression Omnibus. The GEO accession code is GSE196821.
Contributor Information
Eda Koculi, Department of Biochemistry and Molecular Biology, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland 21205, United States.
Samuel S. Cho, Department of Physics and Department of Computer Science, Wake Forest University, Winston–Salem, North Carolina 27109, United States.
REFERENCES
- (1).Noeske J; Wasserman MR; Terry DS; Altman RB; Blanchard SC; Cate JHD High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol 2015, 22, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shajani Z; Sykes MT; Williamson JR Assembly of bacterial ribosomes. Annu. Rev. Biochem 2011, 80, 501–526. [DOI] [PubMed] [Google Scholar]
- (3).Kaczanowska M; Rydén-Aulin M Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev 2007, 71, 477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chow CS; Lamichhane TN; Mahto SK Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol 2007, 2, 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Decatur WA; Fournier MJ rRNA modifications and ribosome function. Trends Biochem. Sci 2002, 27, 344–351. [DOI] [PubMed] [Google Scholar]
- (6).Gc K; Gyawali P; Balci H; Abeysirigunawardena S Ribosomal RNA Methyltransferase RsmC Moonlights as an RNA Chaperone. Chembiochem 2020, 21, 1885–1892. [DOI] [PubMed] [Google Scholar]
- (7).Keffer-Wilkes LC; Veerareddygari GR; Kothe U RNA modification enzyme TruB is a tRNA chaperone. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ofengand J; Campo MD (2004) Modified Nucleosides of Escherichia coli Ribosomal RNA, EcoSal Plus 1, DOI: 10.1128/ecosalplus.4.6.1 [DOI] [PubMed] [Google Scholar]
- (9).Havelund JF; Giessing AMB; Hansen T; Rasmussen A; Scott LG; Kirpekar F Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J. Mol. Biol 2011, 411, 529–536. [DOI] [PubMed] [Google Scholar]
- (10).Hansen LH; Kirpekar F; Douthwaite S Recognition of nucleotide G745 in 23 S ribosomal RNA by the rrmA methyltransferase. J. Mol. Biol 2001, 310, 1001–1010. [DOI] [PubMed] [Google Scholar]
- (11).Sergiev PV; Lesnyak DV; Bogdanov AA; Dontsova OA Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J. Mol. Biol 2006, 364, 26–31. [DOI] [PubMed] [Google Scholar]
- (12).Purta E; O’Connor M; Bujnicki JM; Douthwaite S YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol 2008, 383, 641–651. [DOI] [PubMed] [Google Scholar]
- (13).Lesnyak DV; Sergiev PV; Bogdanov AA; Dontsova OA Identification of Escherichia coli m2G methyltransferases: I. the ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol 2006, 364, 20–25. [DOI] [PubMed] [Google Scholar]
- (14).Purta E; O’Connor M; Bujnicki JM; Douthwaite S YgdE is the 2’-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol 2009, 72, 1147–1158. [DOI] [PubMed] [Google Scholar]
- (15).Sergiev PV; Serebryakova MV; Bogdanov AA; Dontsova OA The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol 2008, 375, 291–300. [DOI] [PubMed] [Google Scholar]
- (16).Caldas T; Binet E; Bouloc P; Costa A; Desgres J; Richarme G The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem 2000, 275, 16414–16419. [DOI] [PubMed] [Google Scholar]
- (17).Bügl H; Fauman EB; Staker BL; Zheng F; Kushner SR; Saper MA; Bardwell JCA; Jakob U RNA methylation under heat shock control. Mol. Cell 2000, 6, 349–360. [DOI] [PubMed] [Google Scholar]
- (18).Ero R; Peil L; Liiv A; Remme J Identification of pseudouridine methyltransferase in Escherichia coli. RNA 2008, 14, 2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Leppik M; Peil L; Kipper K; Liiv A; Remme J Substrate specificity of the pseudouridine synthase RluD in Escherichia coli. FEBS J 2007, 274, 5759–5766. [DOI] [PubMed] [Google Scholar]
- (20).Siibak T; Remme J Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA 2010, 16, 2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Popova AM; Williamson JR Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc 2014, 136, 2058–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Rabuck-Gibbons JN; Popova AM; Greene EM; Cervantes CF; Lyumkis D; Williamson JR SrmB Rescues Trapped Ribosome Assembly Intermediates. J. Mol. Biol 2020, 432, 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nicol SM; Fuller-Pace FV The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 11681–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fuller-Pace FV; Nicol SM; Reid AD; Lane DP DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993, 12, 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tsu CA; Kossen K; Uhlenbeck OC The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA 2001, 7, 702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Diges CM; Uhlenbeck OC Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001, 20, 5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gentry RC; Childs JJ; Gevorkyan J; Gerasimova YV; Koculi E Time course of large ribosomal subunit assembly in E. coli cells overexpressing a helicase inactive DbpA protein. RNA 2016, 22, 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Arai T; Ishiguro K; Kimura S; Sakaguchi Y; Suzuki T; Suzuki T Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U. S. A 2015, 112, No. E4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Elles LMS; Sykes MT; Williamson JR; Uhlenbeck OC A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009, 37, 6503–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhou KI; Clark WC; Pan DW; Eckwahl MJ; Dai Q; Pan T Pseudouridines have context-dependent mutation and stop rates in high-throughput sequencing. RNA Biol. 2018, 15, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Khoddami V; Yerra A; Mosbruger TL; Fleming AM; Burrows CJ; Cairns BR Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ho NW; Gilham PT Reaction of pseudouridine and inosine with N-cyclohexyl-N’-beta-(4-methylmorpholinium)-ethylcarbodiimide. Biochemistry 1971, 10, 3651. [PubMed] [Google Scholar]
- (33).Naylor R; Ho NWY; Gilham PT Selective chemical modifications of uridine and pseudouridine in polynucleotides and their effect on the specificities of ribonuclease and phosphodiesterases. J. Am. Chem. Soc 1965, 87, 4209–4210. [DOI] [PubMed] [Google Scholar]
- (34).Heiss M; Kellner S Detection of nucleic acid modifications by chemical reagents. RNA Biol. 2017, 14, 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kimura S; Sakai Y; Ishiguro K; Suzuki T Biogenesis and iron-dependency of ribosomal RNA hydroxylation. Nucleic Acids Res. 2017, 45, 12974–12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Fasnacht M; Gallo S; Sharma P; Himmelstoss M; Limbach PA; Willi J; Polacek N Dynamic 23S rRNA modification ho5C2501 benefits Escherichia coli under oxidative stress. Nucleic Acids Res. 2022, 5O, 473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bakin AV; Ofengand J Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol 1998, 77, 297–309. [DOI] [PubMed] [Google Scholar]
- (38).Durairaj A; Limbach PA Improving CMC-derivatization of pseudouridine in RNA for mass spectrometric detection. Anal. Chim. Acta 2008, 612, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Fritzsche EJ; Hayatsu H; Igloi GL; Iida S; Kössel H. The use of permanganate as a sequencing reagent for identification of 5-methylcytosine residues in DNA. Nucleic Acids Res. 1987, 15, 5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kouzine F; Wojtowicz D; Baranello L; Yamane A; Nelson S; Resch W; Kieffer-Kwon K-R; Benham CJ; Casellas R; Przytycka TM; Levens D Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst. 2017, 4, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Smola MJ; Rice GM; Busan S; Siegfried NA; Weeks KM Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 2015, 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Siegfried NA; Busan S; Rice GM; Nelson JAE; Weeks KM RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wrzesinski J; Nurse K; Bakin A; Lane BG; Ofengand J A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA(phe). RNA 1995, 1, 437–48. [PMC free article] [PubMed] [Google Scholar]
- (44).Conrad J; Sun D; Englund N; Ofengand J The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA. Biol. Chem 1998, 273, 18562–18566. [DOI] [PubMed] [Google Scholar]
- (45).Campo MD; Kaya Y; Ofengand J Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 2001, 7, 1603. [PMC free article] [PubMed] [Google Scholar]
- (46).Raychaudhuri S; Conrad J; Hall BG; Ofengand J A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 1998, 4, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Toh S-M; Xiong L; Bae T; Mankin AS The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 2008, 14, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bui CT; Rees K; Cotton RGH Permanganate oxidation reactions of DNA: perspective in biological studies. Nucleos Nucleot. Nucleic Acids 2003, 22, 1835–1855. [DOI] [PubMed] [Google Scholar]
- (49).Boccaletto P; Machnicka MA; Purta E; Piątkowski P; Bagiński B; Wirecki TK; de Crécy-Lagard V; Ross R; Limbach PA; Kotter A; Helm M; Bujnicki JM MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Sanchez MIGL; Cipullo M; Gopalakrishna S; Khawaja A; Rorbach J Methylation of Ribosomal RNA: A Mitochondrial Perspective. Front. Genet 2020, 11, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chatterjee B; Shen CJ; Majumder P RNA Modifications and RNA Metabolism in Neurological Disease Pathogenesis. Int. J. Mol. Sci 2021, 22, 11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Helm M; Motorin Y Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet 2017, 18, 275–291. [DOI] [PubMed] [Google Scholar]
- (53).Clark WC; Evans ME; Dominissini D; Zheng G; Pan T tRNA base methylation identification and quantification via high-throughput sequencing. RNA 2016, 22, 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Andersen TE; Porse BT; Kirpekar F A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA 2004, 10, 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


