Abstract
Adaptation to inhibitory concentrations of the antifungal agent fluconazole was monitored in replicated experimental populations founded from a single, drug-sensitive cell of the yeast Candida albicans and reared over 330 generations. The concentration of fluconazole was maintained at twice the MIC in six populations; no fluconazole was added to another six populations. All six replicate populations grown with fluconazole adapted to the presence of drug as indicated by an increase in MIC; none of the six populations grown without fluconazole showed any change in MIC. In all populations evolved with drug, increased fluconazole resistance was accompanied by increased resistance to ketoconazole and itraconazole; these populations contained ergosterol in their cell membranes and were amphotericin sensitive. The increase in fluconazole MIC in the six populations evolved with drug followed different trajectories, and these populations achieved different levels of resistance, with distinct overexpression patterns of four genes involved in azole resistance: the ATP-binding cassette transporter genes, CDR1 and CDR2; the gene encoding the target enzyme of the azoles in the ergosterol biosynthetic pathway, ERG11; and the major facilitator gene, MDR1. Selective sweeps in these populations were accompanied by additional genomic changes with no known relationship to drug resistance: loss of heterozygosity in two of the five marker genes assayed and alterations in DNA fingerprints and electrophoretic karyotypes. These results show that chance, in the form of mutations that confer an adaptive advantage, is a determinant in the evolution of azole drug resistance in experimental populations of C. albicans.
The evolution of resistance to toxicants by pathogens in agriculture and medicine (24) is the result of natural selection acting on genetic variability, the ultimate source of which is mutation. The rate at which new genetic variability arises in a population, and the sequence in which mutations that confer an adaptive advantage occur, may determine the rate and extent to which a population adapts to the presence of a toxicant. The sequence in which beneficial mutations occur is a matter of chance. The purpose of this study was to determine the role of chance in the evolution of azole resistance in experimental populations of the pathogenic yeast Candida albicans in which mutation was the only source of new genetic variability.
C. albicans is both a ubiquitous commensal and an important opportunistic human pathogen causing common ailments such as thrush and vaginitis, as well as chronic conditions in immunocompromised patients (10). Not surprisingly, repeated azole therapy for chronic oral infections in AIDS patients has been associated with an increase in azole resistance. Despite the identification of a mating-type-like locus in C. albicans (6), there is no known mechanism of sexual recombination (5, 13). C. albicans is nevertheless well known for its ability to adapt. For example, the emergence of azole resistance has been reported in several matched series of clonal isolates from patients undergoing azole treatment (reviewed by White et al. [26]).
We measured adaptation to inhibitory concentrations of the antifungal agent fluconazole in replicated experimental populations founded from a single, drug-sensitive cell of C. albicans and reared over 330 generations. The experimental evolution protocol that we used to investigate the emergence of drug resistance in C. albicans is similar in principle to that used in studies of general adaptation in large populations of viruses (27), bacteria (1, 8, 25), and other fungi (4, 31). The experimental evolution approach offered three main advantages in identifying the source and dynamics of adaptive change in C. albicans. First, population size and origin and numbers of cell generations were controlled and known with certainty. This level of control allowed the effect of genetic drift to be minimized by keeping population size large throughout the experiment (>1 million individuals in each population). Second, since a single cell was the progenitor of all populations, no outside genotypes entered these populations, and there was no genetic exchange between individual yeast cells within these populations, mutation (used here in the broadest sense to include all heritable genetic changes) was the only possible source of genetic variability. Under these conditions, mutations that confer an adaptive advantage in the presence of the drug have the opportunity to increase in frequency in response to natural selection during the course of the experiment. Third, environmental conditions were controlled. Throughout the experiment, drug concentration was adjusted to inhibit substantially the growth of the fungus but not enough to result in failure to reach a high concentration of cells during stationary phase at the end of each daily growth cycle. Our intent was to create an environment in which the evolution of drug resistance was likely to occur.
In this study, our objective was to determine whether the course of evolution was the same or different among the replicate populations starting from identical initial genotype and conditions. We monitored azole cross-resistance, expression of four genes known to confer fluconazole resistance, and several markers with no known relationship to drug resistance in the experimental populations.
MATERIALS AND METHODS
Strains and culture conditions.
Twelve populations of C. albicans were founded from a single colony of a strain isolated from an oral swab from a human immunodeficiency virus-positive patient at The Toronto Hospital. The populations were serially propagated for 330 generations (∼100 days) in RPMI 1640 medium (11). Every day, 1 ml from each overnight culture was serially transferred into 9 ml of fresh medium and cells were grown at 35°C with constant agitation. Six populations (N1 to N6) were grown without drug. Six populations (D7 to D12) were grown in twice their most recently measured MIC of fluconazole (Roerig-Pfizer Inc., New York, N.Y.). The drug concentrations in the experiment were never reduced, although three of the populations (D7, D10, and D12) showed a drop in MIC during the experiment. In MIC tests for populations D8 and D10 from generation 260, significant growth continued above the MIC to 64 μg/ml; these two populations were then grown with fluconazole at 128 μg/ml. Isolates were stored in 1 ml of glycerol citrate (3% trisodium salt, 40% glycerol) at −70°C.
Determination of MICs.
During the experiment, the MIC of fluconazole was determined for each population at each sampling time in at least four replicates by the broth microdilution method using the National Committee for Clinical Laboratory Standards (M27-A) protocol (11). In addition, MICs of fluconazole (Pfizer) and ketoconazole and itraconazole (Janssen Pharmaceutica, Beerse, Belgium) were determined, by the same protocol, for five single-colony samples and one mass-culture sample from the progenitor (T118-0), N4 at generation 330 (N4-330), D7 to D12 at 330, D7 at 260, D10 at 200, and D12 at 260. The MIC of fluconazole was also determined in replicate at pH 4 (9), otherwise according to the standard National Committee for Clinical Laboratory Standards protocol, for one mass-culture sample from the same populations for which MICs of the three drugs were determined.
Isolation of total yeast RNA and RNA electrophoresis.
Yeast cells were grown to the logarithmic growth phase in 5 ml of yeast-peptone-glucose (1% yeast extract, 2% Bacto Peptone, and 2% d-glucose) at 30°C with constant shaking. RNA was prepared using glass beads (2) for the same samples for which MICs of the three drugs were determined. Northern blots were prepared according to a published protocol (19).
PCR amplification of C. albicans DNA probes.
Amplifications from genomic DNA of probes for the following genes were done as described by Sanglard et al.: CDR1 and MDR1 (also referred to as BENr) (19), CDR2 (18), and ERG11 (also referred to as CYP51A1) (17).
DNA probe hybridizations and Northern blot quantification.
Northern blots were probed sequentially. Blots were also probed with TEF3, which served as an internal control for loaded quantities of RNA (19). Membranes were analyzed with an InstantImager (Packard Instrument Co., Meriden, Conn.) for quantitative analysis of the signals.
Sequencing.
The entire open reading frame of the ERG11 gene was sequenced from the progenitor, from N4-330, and from D7 to D12 at generation 330, D7-260, D10-200, and D12-260. The open reading frame was PCR amplified using primers and conditions described by Sanglard et al. (17), and two additional internal sequencing primers were designed, 5′-GAGCATAACCTGGAGAAACTAA-3′ and 5′-TTGAGCAGCATCACGTCTCC-3′. Sequencing reactions were prepared using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Perkin-Elmer), according to the manufacturer's instructions, with an initial 95°C denaturation for 2 min for cycle sequencing on the GeneAmp PCR System 9600 (Perkin-Elmer). The reactions were analyzed on the ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems, Perkin-Elmer).
Detection of loss of heterozygosity.
The 12 experimental populations at generation 330 and the generation of the MIC peak for the three drug populations that subsequently dropped in MIC were assayed for loss of heterozygosity in five marker genes heterozygous in the progenitor (Table 1). For each population, one mass-culture sample and one single-colony sample were assayed. The marker genes were amplified as follows. PCR mixtures (20 μl) contained 0.5 μM (each) primer, 200 μM deoxynucleoside triphosphates, 1× PCR Buffer II with 2 mM MgCl2 (Perkin-Elmer, Norwalk, Conn.), 0.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), and 10 μl of a 100-fold dilution of genomic DNA prepared according to the method of Scherer and Stevens (22). Amplifications were carried out in a Perkin-Elmer GeneAmp System 9600 Thermocycler programmed for an initial denaturation at 95°C for 8 min followed by 35 cycles of denaturation at 95°C; primer annealing at 50 to 58°C; and extension at 72°C for 20, 30, and 60 s, with a 5-min extension at 72°C on the final cycle.
TABLE 1.
Primers and probes for heterozygosity analysis
| Probe (GenBank accession no.) | Sequence | Chromosome (SfiI fragment) | Temp (°C) |
|---|---|---|---|
| C2F17a (Y07665) | ACTAATCTATCGAGAGAACG (amp)c | 3 | 50 |
| GTCAGATGGTACGGACAAG (amp) | 50 | ||
| ACTTGGCCGACGAAG (hyb)c | 48 | ||
| ACTTGGTCGACGAAG (hyb) | 44 | ||
| CPH1b (U15152) | AGCATTATCATTCCATTACGACGAGT (amp) | 1 | 50 |
| TTTATATAGGTTGGGGTGGCGAC (amp) | 50 | ||
| TCTTCTGTGTTTCCATG (hyb) | 44 | ||
| TCTTCTGAGTTTCCATG (hyb) | 45 | ||
| ARG4 (L25051) | TCACGGCAATTCTTGAACGAG (amp) | 7 (G) | 50 |
| GCTAAAGCACCAGATCCTAATGGAG (amp) | 50 | ||
| GATGGGCTCATTGGT (hyb) | 44 | ||
| GATGGGCACATTGGT (hyb) | 45 | ||
| GATGGGCCCATTGGT (hyb) | 50 | ||
| C15F2a (Y07668) | TAGTTAGTTTGCCTTGTTCC (amp) | 2 | 50 |
| GAGAGCTACGTGAGCTCGTG (amp) | 50 | ||
| GCCCGCTGGTCATTGT (hyb) | 54 | ||
| GCCCGCTGATCATTGT (hyb) | 51 | ||
| PDE1b (L12045) | TAATATGCTAGGTGGGGGTTCCTT (amp) | 5 (M) | 58 |
| GCCCAATATGCCTAGTTTCAAAATC (amp) | 58 | ||
| CTTTATAATCAAACCAC (hyb) | 36 | ||
| CTTTATAATTAAACCAC (hyb) | 33 |
Sequences are from the work of Gräser et al. (5).
Oligonucleotides hybridize to the reverse complement of the accessioned sequence. Other oligonucleotides hybridize to the accessioned sequence.
Primers for amplification are designated “(amp)” with the corresponding annealing temperature. Oligonucleotide probes are designated “(hyb)” with the corresponding hybridization-wash temperature.
Southern blots of PCR-amplified marker genes (loci) were prepared by standard methods (16). Allele-specific oligonucleotide probes were designed for each locus based on DNA sequence data and were end labeled with [γ-32P]ATP (3,000 Ci/mmol; DuPont-NEN, Markham, Ontario, Canada). The allele-specific probes (Table 1) were hybridized for 2 h to the Southern blots within 5°C of the midpoint melting temperature (Tm) of the probe according to the protocol of Cowen et al. (3a) and Saville et al. (20). Three washes of 20 s each were done at the same temperature as the hybridization. The blots were then exposed to X-ray film (BIOMAX MR Kodak film) at room temperature for 1 h.
Fingerprinting.
DNA fingerprinting with probe 27A was performed by the protocol of Scherer and Stevens (23), for the same samples that were assayed for loss of heterozygosity.
Electrophoretic karyotyping.
Chromosome-sized DNAs were separated using CHEF-DR II pulsed-field electrophoresis systems (Bio-Rad Laboratories). The preparation of CHEF (contour-clamped homogeneous electric field) samples followed the procedure described on the C. albicans Information Page (http://alces.med.umn.edu/candida/methods.html). Chromosome separation on pulsed-field gels followed condition 2 of the protocol. Probes specific to the terminal SfiI fragments (3) of each chromosome were hybridized to Southern blots of electrophoretic karyotypes (Table 2). HindIII (GIBCO BRL) and NotI (New England BioLabs) digestion of agarose-embedded CHEF DNA samples was performed by standard methods (New England BioLabs 1996–1997 catalog). CHEF conditions for the HindIII-digested samples were as described by Rustchenko et al. (14). CHEF conditions for the NotI-digested samples were as described by Iwaguchi et al. (7). The intergenic spacer of the nuclear ribosomal DNA (rDNA) repeat was PCR amplified with standard primers (28) and hybridized to Southern blots of the restriction enzyme-digested samples.
TABLE 2.
Amplification primers for probes specific for SfiI fragments of each chromosome
| Probe (accession no.) | Sequence | Chromosome (SfiI fragment) | Annealing temp (°C) |
|---|---|---|---|
| ACT1 (X16377) | GGA CTT GTG TTG TTA TCT GG | 1 (L) | 50 |
| AGA GAA AAT ACC CGT GAC G | |||
| HGT1 (Y16834) | CAG TCT TCA TAC ATA CCC CGG | 1 (S) | 50 |
| CAG GAT TTT CAC GAT CAC C | |||
| SAS2 (1946R)a | TTC TGA AGG ATC AAT TGC C | 2 (U) | 50 |
| AAA CCG ATT TAT CGT CAA G | |||
| GCN1 (S14D5)a | CGC TTG TGG AAG TAC TTG GG | 2 (A) | 58 |
| GCT TGT CGT CAG ACT CTT GTA AGG | |||
| ILV2 (265169F08.y1.seq)a | ATA GCA GTA TTG GCA GCA GC | 3 (O) | 55 |
| ACC ATC ACA TGC ATT GAG TC | |||
| YER132 (p79F)a | GCC ATC ATC GAC AGT ATC C | 3 (P) | 50 |
| CCC AAT AAG TAT GTT TGC C | |||
| YJR61 (p356R)a | TTC CGC TAC GGT ATT CTC | 4 (H) | 50 |
| AAC CAG ACG TTA GTT GTC G | |||
| HIS4 (AJ003115) | CCA GAC AGG AGA ATC CAA CG | 4 (BB) | 55 |
| CCT AGA GCT TGG TGA ACC AGC | |||
| HEX1 (L26488) | GTG AAG CTG CTT TAT GGT CGG A | 5 (I) | 58 |
| AGT ATC ATC GAG ACT CGC GTG ACT | |||
| GCD11 (1997R)a | TAC CAC CTT ATG GAA GTG GAC | 5 (M) | 60 |
| GGT TCT ACT CCA ACT GGT GC | |||
| TSM1 (G4C4R)a | CTT CCT GAC ATT GCC TGA AG | 6 (C) | 55 |
| CCA CCA GAT ATG AGT GAC CC | |||
| ALS1 (L25902) | AAC ATG TAC TGT GAA CGA CGC | 6 (O) | 60 |
| AAT GGA CGA TAA CCA GCA GG | |||
| p321 (gel39821)a | TGC AAC ATT GAT GGC AGC | 7 (C) | 60 |
| TGC AGT TGG AGT CTC AGT CG | |||
| p143 (gel40106)a | GCG CGC TTC CAT TAC TAG G | 7 (G) | 60 |
| ACG TTC AGC AGA GAA CCA GG | |||
| INO1 (L22737) | ATC TAA TGT GGC TAC GGT GC | R (D) | 55 |
| CCT TAG TGA CAG AAT TGG ACG | |||
| ADE1 (L12463) | CCA CCG ACA GAA TTT CTG C | R (M, S) | 55 |
| GGC TAC ACC ATC TTT ACC AGC |
Accession number for the Stanford Candida albicans Sequencing Project (http://alces.med.umn.edu/candida/geneinfo.html). Other accession numbers are for GenBank.
Determination of doubling times.
Doubling times during the exponential growth phase were measured for the progenitor, one population grown without drug at generation 330, the six populations evolved in the presence of drug at generation 330, and the generation of the MIC peak for the three populations propagated with drug that subsequently dropped in MIC. Doubling times were determined in replicate in RPMI 1640 and yeast-peptone-glucose media at 35°C with constant agitation. The concentration of cells was monitored with a spectrophotometer (Beckman Du-64 spectrophotometer; Beckman Instruments, Inc.).
RESULTS
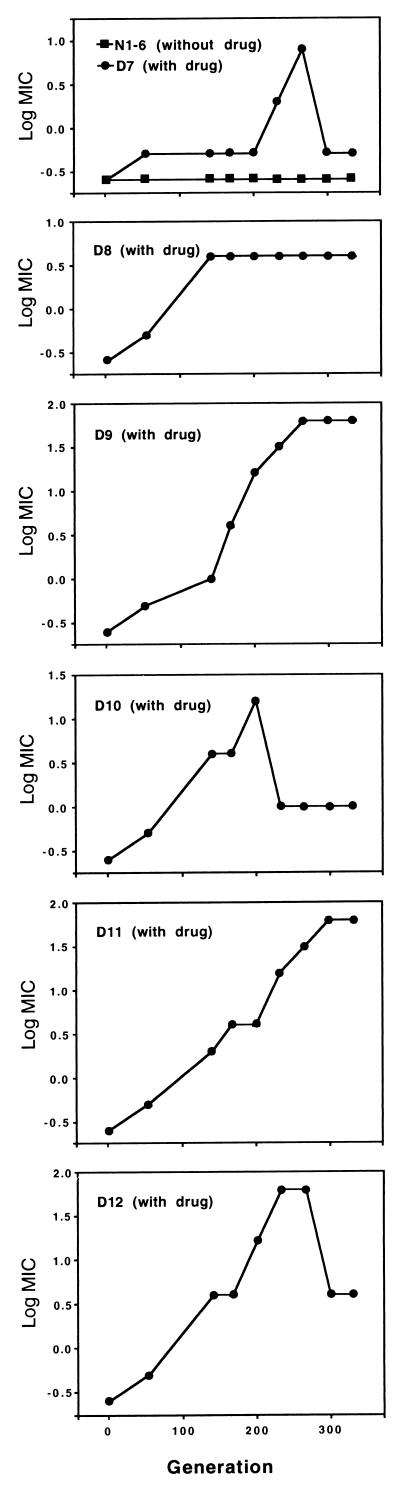
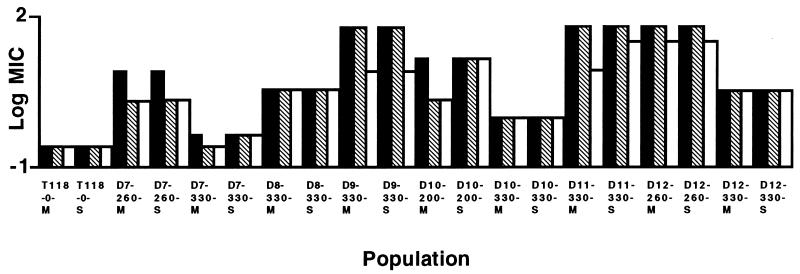
We monitored adaptation to inhibitory concentrations of fluconazole over 330 generations of experimental evolution in 12 replicate populations founded from a single azole-susceptible cell of C. albicans (fluconazole MIC, 0.25 μg/ml). Six populations were grown with fluconazole at twice their most recently measured MIC, and six were propagated without drug. There was no change in MIC for any of the six populations grown without drug (Fig. 1). Among the six populations grown with fluconazole (Fig. 1), two populations (D9 and D11) achieved the highest level of fluconazole resistance (MIC, 64 μg/ml) measured in this standard test and retained this level to generation 330. One population (D8) achieved an intermediate level of resistance (MIC, 4.0 μg/ml) and remained at this level until generation 330. Two populations (D10 and D12) achieved increased resistance (MIC, 16.0 and 64.0 μg/ml, respectively) and then showed a decrease (MIC, 1.0 and 4.0 μg/ml, respectively). One population (D7) achieved a small increase in resistance (MIC, 0.5 μg/ml), sharply increased resistance at generation 260 (MIC, 8.0 μg/ml), and then a decrease at generation 330 to the previous level. In all cases, the level of resistance achieved at generation 330 was stable over three transfers on azole-free solid medium. Resistance, relative to the progenitor, was retained over 50 generations of further experimental evolution in the absence of drug (Fig. 2), with one exception (D7-330-M). The increase in fluconazole MIC was accompanied by a corresponding increase in resistance to itraconazole and ketoconazole (data not shown). When MICs were determined at pH 4, the basic relationship of MICs among the populations was not altered, but MICs of fluconazole were slightly higher at pH 4 than at pH 7, as was observed in some cases by Marr et al. (9). All populations contained ergosterol in their cell membranes and were amphotericin sensitive (data not shown).
FIG. 1.
Adaptation to fluconazole in experimental populations. Twelve populations were established from a single cell of an azole-susceptible strain of C. albicans. The populations were propagated in RPMI 1640 medium for 330 generations. Six populations (D7 to D12) were grown in twice their most recently measured MIC of fluconazole, and six (N1 to N6) were grown without drug.
FIG. 2.
The stability of acquired resistance in 50 generations (∼15 days) in RPMI 1640 medium without fluconazole. Susceptibility to fluconazole was determined at generations 0 (solid), 25 (hatched), and 50 (unfilled). M refers to mass cultures; S refers to a single-colony isolate.
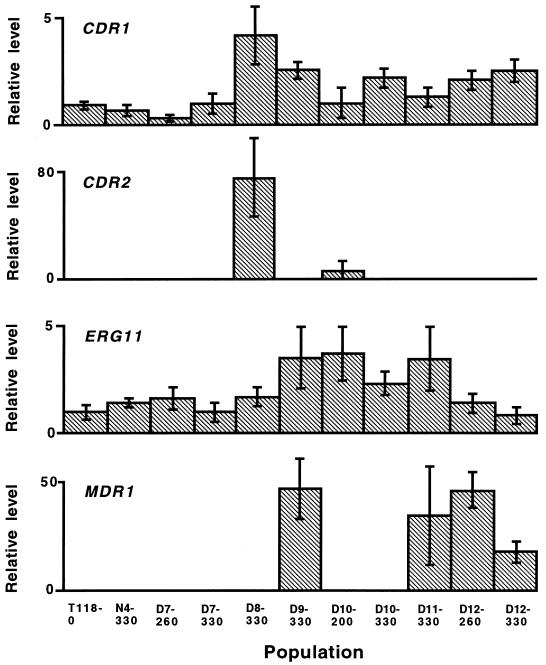
Each population grown in the presence of drug acquired resistance in a different way, overexpressing a unique combination of four genes known to be important in fluconazole resistance (26): the gene encoding the target enzyme of azoles in the ergosterol biosynthesis pathway, ERG11; two ATP-binding cassette transporters, CDR1 and CDR2; and a major facilitator, MDR1 (Fig. 3). While the CDR gene products pump many azoles from the cell, the MDR1 product specifically pumps fluconazole. There was highly significant variation in mRNA levels among the experimental populations for CDR1 and ERG11 (Kruskal-Wallis test, P < 0.001). CDR2 was strongly expressed in one population (D8-330), was expressed at a low level in another population (D10-200), and was not detected in the remaining nine populations. Four populations (D9-330, D11-330, D12-260, and D12-330) strongly expressed the MDR1 gene, while MDR1 mRNA was not detected in seven populations. Other factors contributing to azole resistance must be operating in the populations overexpressing MDR1 to account for the azole-cross-resistant phenotypes, as the efflux pump encoded by this gene appears to specifically pump fluconazole (26). No mutations were detected in the nucleotide sequence of ERG11 in the replicate populations. This was confirmed for all populations under selection by functional expression of the ERG11 alleles in Saccharomyces cerevisiae (17).
FIG. 3.
Relative mRNA levels of four C. albicans genes involved in azole resistance. Bars represent standard deviations for each sample (n = 6 replicate measurements). Variation among the population samples was highly significant (Kruskal-Wallis test, P < 0.001).
In addition to four genes known to be important in azole resistance, we also monitored molecular markers with no known relation to drug resistance: polymorphic nucleotide sites in five DNA regions known to be heterozygous in the progenitor genotype and the 27A DNA fingerprint widely used to type clinical isolates of C. albicans (23). No changes in neutral markers or DNA fingerprint were detected in any of the populations not exposed to drug, while several changes including loss of heterozygosity in two of the five genes known to be heterozygous in the progenitor were detected in several populations evolved in the presence of fluconazole. The changes were as follows: C15F2, position 174, AG to AA in D7-260; and PDE1, position 1046, CT to TT in D7-330 and CT to CC in D10-200 (single-colony isolate only). Changes in the DNA fingerprint were detected in two of nine samples from populations evolved in the presence of drug. The changes were the gain of a band in D7-260 and the loss of a band in D12-260 (single-colony isolate only).
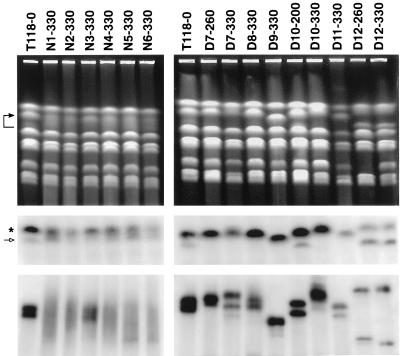
Southern hybridizations of electrophoretic karyotypes revealed numerous chromosomal changes in the evolved populations relative to the ancestral isolate (Fig. 4). The most variable chromosome was chromosome R, which contains the genes coding for rDNA. Both probes for chromosome R showed the same hybridization results: the progenitor (T118-0) had one distinct strongly hybridizing band and one weakly hybridizing band of unknown origin, the six populations evolved without drug at generation 330 each had a smear, and the drug populations each had one or two distinct bands of variable size (Fig. 4).
FIG. 4.
Electrophoretic karyotypes. (Top panels) Ethidium-stained gels. The arrow indicates chromosome R in isolate T118-0; the bracket indicates range of chromosome R sizes in other isolates. (Middle panels) Southern hybridization with INO1 (GenBank accession no. L22737), a probe specific for chromosome R. The asterisk indicates the strongly hybridizing band in T118-0; the open arrow indicates the weakly hybridizing band of unknown origin in T118-0. (Bottom panels) rDNA as visualized by Southern hybridization of HindIII digests with the intergenic spacer region of the rDNA.
Further experiments examined the nature of the variation in chromosome R. HindIII liberates the tandem rDNA array from each chromosome R homologue as a single fragment (14). Southern hybridization of HindIII digests with the intergenic spacer region of the rDNA demonstrated variation in the size of the rDNA arrays among the experimental populations (Fig. 4). The variation in size of the rDNA array corresponded to the size variation of chromosome R. NotI has a single cutting site within the rDNA unit (7). Southern hybridization of NotI digests with the intergenic spacer region of the rDNA showed that there were no changes in the rDNA unit size among the experimental populations (data not shown).
In the chromosomes other than chromosome R, there was no variation detected for chromosomes 1, 2, 3, 6, and 7 with either of the probes specific to the terminal SfiI fragments for each chromosome. With the exception of chromosome R, the C. albicans chromosomes are numbered 1 through 7, largest to smallest (3). For chromosome 4, there was no variation detected with the probe for SfiI fragment BB, while hybridization with the probe specific for SfiI fragment H showed a change for populations D10-330 and D7-330. The probe hybridized to one distinct band in all other populations, but there were two distinct bands for D10-330, one the same size as that in the other populations and the other, of equal intensity, at the same position as chromosome 5. D7-330 had one strongly hybridizing band of the expected size and also a weakly hybridizing band at the same position as chromosome 2. For chromosome 5, both probes hybridized to one distinct band with no size variation for all population samples except D12-260, in which the band was the same size as chromosome 2. In the sample from D12-260, there was no band corresponding to that of chromosome 5 in the other samples (Fig. 4).
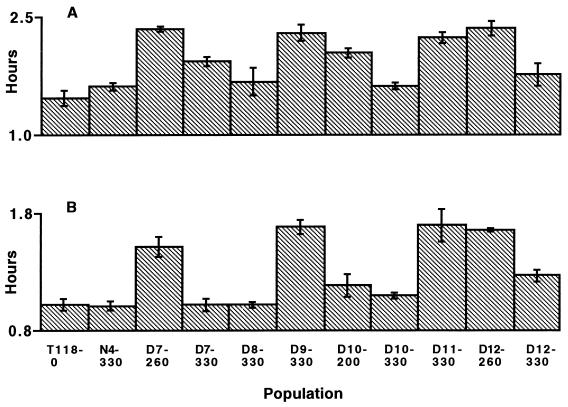
Increased doubling time in the absence of drug was detected in samples grown in both RPMI 1640 and yeast-peptone-glucose media from population D7 at generation 260, D9 at 330, D11 at 330, D12 at 260, and D12 at 330 (Fig. 5). Variation among population samples was highly significant (Kruskal-Wallis test, P < 0.001).
FIG. 5.
Doubling times of populations during the exponential growth phase. (A) Doubling times in RPMI 1640 medium. (B) Doubling times in yeast-peptone-glucose medium. Bars represent standard deviation for each sample (n = 2 replicate measurements). Variation among the population samples was highly significant (Kruskal-Wallis test, P < 0.001).
DISCUSSION
This experimental evolution study was designed to examine the role of chance in the evolution of azole resistance in C. albicans. Since the replicate populations were founded from a single genotype, any variation among populations is due to mutation occurring during the experiment. The populations were propagated in controlled, uniform environments in which the drug concentration was adjusted to provide the fungus with the best chance of developing resistance. This experimental system allows natural selection to operate; the populations are exposed to an environment, and any heritable properties enhancing fitness in that environment can respond to natural selection. All populations exposed to fluconazole adapted to the presence of drug, showing increased azole cross-resistance. The emergence of drug resistance in the initially identical populations followed strikingly different trajectories (Fig. 1) associated with different patterns of expression of four genes implicated in azole resistance (Fig. 3) and doubling times in the absence of drug (Fig. 5).
These results suggest that random processes determine the trajectory of change in drug resistance in response to selection. These chance events may include mutation and mitotic recombination in any of the several genes affecting drug resistance. We interpret the changes as the result of selective sweeps in which the frequency of the mutant type with an adaptive advantage increases to a high level in a population.
In these clonal populations, additional collateral changes in the genome have the opportunity to hitchhike along with the mutation under selection. Genetic markers with no known relation to drug resistance were assayed in the experimental populations, including loss of heterozygosity in five marker genes heterozygous in the progenitor and the 27A DNA fingerprint. No changes were detected in any of the populations grown without drug, while several changes, including loss of heterozygosity in two of the five genes known to be heterozygous in the progenitor and changes in the DNA fingerprint, were detected in four of nine samples from populations evolved in the presence of fluconazole. With the exception of chromosome R, there were no chromosomal changes detected in any of the populations that were not exposed to drug. In contrast, in populations evolved in the presence of the drug, there was one chromosomal change in three of nine population samples, a different change in each sample. This shows that the response to the drug in C. albicans is accompanied by an increased frequency of genomic changes. Genomic changes including gene conversion, mitotic crossing over, and chromosome rearrangements have been implicated as a source of genetic variation in this asexual yeast (15, 21).
The genomic changes in the populations evolved in the presence of drug showed no consistent association with drug resistance, in contrast to the findings of Perepnikhatka et al. (12). In chromosome R, the high degree of variability of sizes among the populations exposed to the drug was not directional; some populations evolved rDNA arrays of increased size while others decreased in size. Furthermore, the populations grown without drug also exhibited a high degree of variability in the size of the rDNA arrays within each population, with no change in drug resistance throughout the experiment.
In population D7, the appearance of neutral changes indicates two selective sweeps (the increase in frequency of a genotype containing a mutation conferring a selective advantage), one, approaching generation 260, that was accompanied by the loss of heterozygosity in region C15F2 and the gain of a fragment in the DNA fingerprint, and the other, approaching generation 330, that was accompanied by the loss of heterozygosity in PDE1. The first sweep could not have been to complete fixation, because the neutral changes at generation 260 were reversed to the ancestral states at generation 330. This cannot be due to reversion because, once heterozygosity at a specific nucleotide site is lost, it is very unlikely to be regained by point mutation. Interclonal competition must have occurred in population D7 (1).
Loss of size variants of chromosome R provides further evidence for selective sweeps in populations evolved in the presence of drug (D7 to D12) but not in populations evolved in the absence of drug (N1 to N6). D7 to D12 each had one or two sharp bands of chromosome-sized DNA containing the rDNA repeat (chromosome R), which varied in size among them (Fig. 4). In contrast, N1 to N6 each had a smear, rather than the distinct chromosome R band. The only possible explanation is that there is a mixture of chromosome R sizes in each of the lines N1 to N6 (single-colony isolates from these populations each had one or two sharp chromosome R bands of variable size [data not shown]). This variation was eliminated in populations D7 to D12 due to selective sweeps, resulting in one or two sharp bands. The progenitor, which was isolated from a single cell, had one sharp band plus a second weakly hybridizing band of unknown origin for chromosome R. The reduction in size variation in chromosome R in D7 to D12 cannot be explained by genetic drift since population size was large (minimum number of individuals was 106).
In the presence of an antimicrobial agent, a resistant genotype is at an advantage compared to less resistant genotypes. But in the absence of drug, the resistant genotype may be at a disadvantage. A significant cost of resistance, evident as an increase in doubling time in the absence of drug, was detected in samples from population D7 at generation 260, D9 at 330, D11 at 330, D12 at 260, and D12 at 330 (Fig. 5). In two (D7 and D12) of the three populations (D7, D10, and D12) showing a decrease in MIC during the course of the 330 generations, this cost was mitigated as doubling time decreased. The decreases in MIC should therefore not be interpreted as a reduction in overall fitness in the presence of the drug. The three populations demonstrating a decrease in MIC during the experiment continued to grow at the higher drug concentration established at their MIC peak. Fitness in experimental populations under the selection imposed by the drug may involve a complex interplay between various resistance mechanisms and cell growth parameters.
Interpreting our results in the context of Sewall Wright's adaptive landscape (29, 30), fitness can be represented as altitude on a landscape with topography. Alleles at different loci interact to determine the fitness of a population, and multiple adaptive peaks are separated by maladaptive valleys. In this experiment, the only source of genetic variation was mutation defined in the broadest sense to include changes in nucleotide sequence, gene conversion, mitotic crossing over, and chromosome rearrangements. There was no immigration into populations and no genetic exchange between individuals. Genetic drift was minimal as the populations were all large. Under these conditions, all populations responded to selection by gaining altitude on the adaptive landscape, but they climbed different slopes associated with different molecular mechanisms of drug resistance. We do not yet know if the different resistance mechanisms would give enhanced drug resistance and fitness when combined together in the same genotype—whether or not populations will converge on one global adaptive peak or diverge to series of isolated peaks with further experimental evolution. How do these results pertain to natural populations? In the host, evolution of C. albicans is complicated by the status of the immune system, the physical niche in the body, drug treatment history, immigration, genetic drift, and competition with other microbes. In addition to these factors, our results show that chance is an important factor in the evolution of drug resistance.
ACKNOWLEDGMENTS
This work was supported by a grant-in-aid from Pfizer Canada Inc. and Research Grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to L.M.K. and J.B.A. and by grant no. 3100-045716 from the Swiss Research National Foundation to D.S. L.E.C. was supported by an NSERC Postgraduate Scholarship.
We thank Claire Wickens for technical assistance.
REFERENCES
- 1.Arjan J A, Visser M, Zeyl C W, Gerrish P J, Blanchard J L, Lenski R E. Diminishing returns from mutation supply rate in asexual populations. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M. Preparation of yeast RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 13.12.2–13.12.3. [Google Scholar]
- 3.Chu W S, Magee B B, Magee P T. Construction of an SfiI macrorestriction map of the Candida albicans genome. J Bacteriol. 1993;175:6637–6651. doi: 10.1128/jb.175.20.6637-6651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Cowen L E, Sirjusingh C, Summerbell R C, Walmsley S, Richardson S, Kohn L M, Anderson J B. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob Agents Chemother. 1999;12:2930–2938. doi: 10.1128/aac.43.12.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferea T L, Botstein D, Brown P O, Rosenzweig R F. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hull C M, Johnson A D. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 7.Iwaguchi S, Homma M, Tanaka K. Clonal variation of chromosome size derived from the rDNA cluster region in Candida albicans. J Gen Microbiol. 1992;138:1177–1184. doi: 10.1099/00221287-138-6-1177. [DOI] [PubMed] [Google Scholar]
- 8.Lenski R E, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marr K A, Rustad T R, Rex J H, White T C. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob Agents Chemother. 1999;43:1383–1386. doi: 10.1128/aac.43.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merz W G. Candida albicans strain delineation. Clin Microbiol Rev. 1990;3:321–334. doi: 10.1128/cmr.3.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 12.Perepnikhatka V, Fischer F J, Niimi M, Baker R A, Cannon R D, Wang Y K, Sherman F, Rustchenko E. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol. 1999;181:4041–4049. doi: 10.1128/jb.181.13.4041-4049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallie M, Bastide J M. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rustchenko E P, Curran T M, Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol. 1993;175:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rustchenko-Bulgac E P, Sherman F, Hicks J B. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J Bacteriol. 1990;172:1276–1283. doi: 10.1128/jb.172.3.1276-1283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 19.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saville B J, Kohli Y, Anderson J B. mtDNA recombination in a natural population. Proc Natl Acad Sci USA. 1998;95:1331–1335. doi: 10.1073/pnas.95.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherer S, Magee P T. Genetics of Candida albicans. Microbiol Rev. 1990;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherer S, Stevens D A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor M, Feyereisen R. Molecular biology and evolution of resistance to toxicants. Mol Biol Evol. 1996;13:719–734. doi: 10.1093/oxfordjournals.molbev.a025633. [DOI] [PubMed] [Google Scholar]
- 25.Travisano M, Mongold J A, Bennett A F, Lenski R E. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267:87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- 26.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wichman H A, Badgett M R, Scott L A, Boulianne C M, Bull J J. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 28.Williams D W, Wilson M J, Lewis M A, Potts A J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright S. Character change, speciation, and the higher taxa. Evolution. 1982;36:427–443. doi: 10.1111/j.1558-5646.1982.tb05065.x. [DOI] [PubMed] [Google Scholar]
- 30.Wright S. Surfaces of selective value revisited. Am Nat. 1988;131:115–123. [Google Scholar]
- 31.Zeyl C, Bell G. The advantage of sex in evolving yeast populations. Nature. 1997;388:465–468. doi: 10.1038/41312. [DOI] [PubMed] [Google Scholar]