Abstract
The gene pth, encoding peptidyl-tRNA hydrolase (Pth), is essential for protein synthesis and viability of Escherichia coli. Two pth mutants have been studied in depth: a pth(Ts) mutant isolated as temperature sensitive and a pth(rap) mutant selected as nonpermissive for bacteriophage λ vegetative growth. Here we show that each mutant protein is defective in a different way. The Pth(Ts) protein was very unstable in vivo, both at 43°C and at permissive temperatures, but its specific activity was comparable to that of the wild-type enzyme, Pth(wt). Conversely, the mutant Pth(rap) protein had the same stability as Pth(wt), but its specific activity was low. The thermosensitivity of the pth(Ts) mutant, presumably, ensues after Pth(Ts) protein levels are reduced at 43°C. Conditions that increased the cellular Pth(Ts) concentration, a rise in gene copy number or diminished protein degradation, allowed cell growth at a nonpermissive temperature. Antibiotic-mediated inhibition of mRNA and protein synthesis, but not of peptidyl-tRNA drop-off, reduced pth(Ts) cell viability even at a permissive temperature. Based on these results, we suggest that Pth(Ts) protein, being unstable in vivo, supports cell viability only if its concentration is maintained above a threshold that allows general protein synthesis.
Peptidyl-tRNA (pep-tRNA) hydrolase (Pth) is an enzyme essential for the viability of Escherichia coli. It is believed that the role of Pth in cell metabolism is to cleave pep-tRNAs prematurely released from ribosomes (23). Recently, it has been reported that expression of short open reading frames in DNA sequences, named minigenes, is especially toxic to bacteria defective in Pth (7, 30). As translation of minigene transcripts results in premature release of pep-tRNA, this intermediate accumulates under Pth limitation, provoking cell death. A mutation in the gene encoding Pth, named pth(Ts), determines a thermosensitive phenotype in E. coli (3). Bacterial mutants harboring this mutation grow exponentially at 30°C, but upon a shift to 43°C, they accumulate pep-tRNA and undergo inhibition of protein synthesis (3, 17, 23). Another mutation, termed pth(rap), was identified by its conferring on cells the inability to maintain the vegetative growth of bacteriophage λ under conditions that allow exponential cell growth (11, 16). pth(Ts) and pth(rap) mutations correspond to amino acid alterations, Gly101 to Asp and Arg134 to His, respectively (11). These substitutions do not affect the proposed active site in the three-dimensional structure of the Pth protein, but they change highly conserved residues in the deduced Pth polypeptide sequence from different sources (6, 26). Both mutations cause considerable reduction in the Pth activity in cell extracts, and the Pth activity of the pth(Ts) mutant extracts is sensitive to incubation at high temperature (11, 25). Paradoxically, pth(Ts) mutant extracts do not show a defect in in vitro protein synthesis experiments conducted at 43°C (J. Hernández-Sánchez and G. Guarneros, unpublished data; 25).
We investigated mutant gene expression and the activities and stabilities of the mutant Pth proteins to understand the nature of the pth(Ts) and pth(rap) defects. Unlike pth(rap), the pth(Ts) mutation does not affect Pth specific activity but generates a highly unstable protein in vivo. Pth(Ts) protein concentration, already low at 32°C, is further reduced at 43°C. Overproduction of Pth(Ts) helps the mutant cells to survive at the nonpermissive temperature. We propose that the excess enzyme promotes general protein synthesis and, therefore, its own synthesis.
MATERIALS AND METHODS
Bacterial strains and viability experiments.
The E. coli K-12 strains used here are shown in Table 1.
TABLE 1.
Strains used in this work
| Strain | Genotype | Source and/or reference(s) |
|---|---|---|
| C600 | thr-1 leu-6 thi-1 supE44 tonA lacY1 | Our collection |
| C600 pth(Ts) | pth (Ts) zch::Tn10 | 11 |
| C600 rap | pth(rap) zch::Tn10 | 15 |
| CAG6068 | C600 dnaK756:Tn10 | 28 |
| CAG6069 | C600 dnaJ259:Tn10 | 28 |
| CAG764 | C600 grpE280:Tn10 | 28 |
| GG6070 | CAG6069 ΔTn10 | 22 and this work |
| GG6071 | GG6070 pth(Ts) zch::Tn10 | GG6070 with P1 [C600 pth(Ts) zch::Tn10]; Tcr; this work |
| CP78 | arg his leu thr thi | 25 |
| AA7852 | CP78 pth(Ts) | 25 |
| MC4100 | F−araD Δ(argF-lacZYA)U169 rpsL relA1 flbB deoC ptsF rbsR | 4 |
| SG12047 | C600 lon146::ΔTn10 | 13 |
| SG22094 | MC4100 Δlon clpP::cat resA::kan | S. Gottesman's collection |
| SG22098 | MC4100 clpP::cat | S. Gottesman's collection |
| SG22101 | MC4100 clpX::kan | S. Gottesman's collection |
| SG22099 | MC4100 clpA::kan | S. Gottesman's collection |
The viability experiments were performed by growing cells at 30°C to an optical density at 600 nm of in Luria-Bertani medium (LB) or LB supplemented with 100 μg of ampicillin per ml (LB-AP). Temperature was adjusted to 43°C by mixing 5-ml cultures at 30°C with 1.5 ml of medium at 90°C (see Fig. 3), and other antibiotics were added as indicated (see Fig. 4). At various times, samples were taken, diluted, plated on tryptone-broth agar, and incubated at 30°C to quantify CFU.
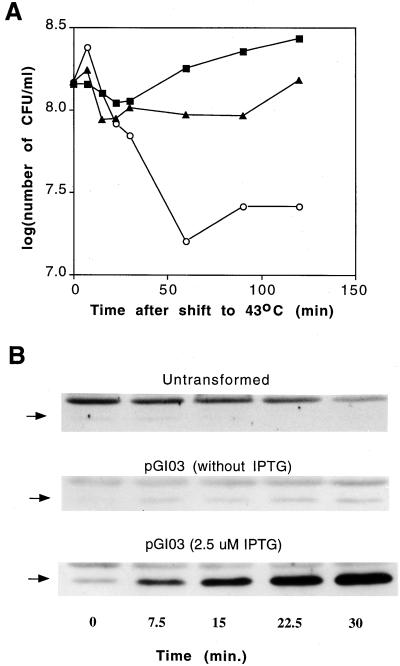
FIG. 3.
Effect of pth(Ts) gene overexpression on the survival of pth(Ts) mutant cells after shift to 43°C. (A) Colony-forming ability of strain AA7852 untransformed (open circles) or transformed with plasmid pGI03 with (closed squares) or without (closed triangles) IPTG added at 0 min. (B) Concentration of Pth(Ts) protein, as estimated by immunoblot analysis, in the cells at the indicated times after the temperature shift. The arrows indicate Pth(Ts) position.
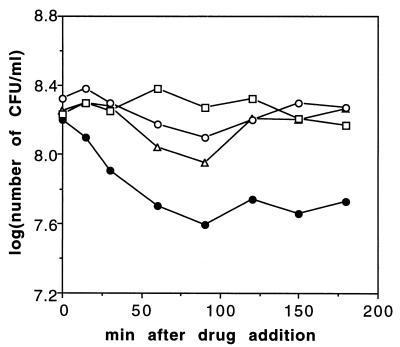
FIG. 4.
Effect of antibiotic addition on pth(Ts) mutant cell growth at 30°C. The figure shows colony-forming ability of AA7852 cells after addition of rifampin (100 μg/ml) (open circles), erythromycin (80 μg/ml) (open squares), rifampin plus chloramphenicol (50 μg/ml) (open triangles), or rifampin plus erythromycin (closed circles).
Plasmid construction.
The plasmids to overproduce Pth protein were constructed by cloning PCR fragments that contained the different pth alleles between the EcoRI and HindIII sites of pKQV4 (27). The primers used were 5′-CAGTGAATTCCGCGCCAG-3′ (upper) and 5′-GTAATGGAAATAAGCTTGCCTATTATAC-3′ (lower). Restriction sites are underlined, and boldface bases are changes relative to the previously reported sequence (11). pGI01 carries wild-type pth, pGI02 carries pth(rap), and pGI03 carries pth(Ts). In these constructs, pth transcription is controlled by the tac promoter and is inducible with isopropyl-β-d-thiogalactopyranoside (IPTG).
Concentration, stability, and half-life of Pth protein.
For the experiments the results of which are shown in Fig. 2 and Table 3, E. coli cells, untransformed or transformed with plasmids encoding the wild-type or mutant Pths, were grown to an optical density at 600 nm of 0.3 at 30°C in LB or LB-AP. IPTG (1 mM) was then added, and the cultures were further incubated for 6 h. Cells were harvested, lysed with Triton X-100 as described by Chen et al. (5), and dialyzed against buffer B (10 mM Tris-HCl [pH 7.6], 10 mM magnesium acetate, 20 mM NH4Cl) (21), and the extracts were electrophoresed as described below. In these experiments, 50% of the overproduced Pth(Ts) was lost as debris, but only 5% of Pth(wt) was lost. For the experiments the results of which are shown in Fig. 1 and Table 4, cells were grown to mid-log phase and then rifampin (500 μg/ml) and chloramphenicol (175 μg/ml) were added. Thereafter, samples taken at different times were lysed by being boiled in Laemmli sample buffer, resolved by electrophoresis on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels, and transferred onto nitrocellulose membranes (31). Immunoreactive proteins were detected with rabbit polyclonal anti-Pth serum (17) (diluted 1:8,000) and anti-rabbit peroxidase-labeled immunoglobulins (diluted 1:10,000) (ECL Western blotting kit; Amersham). Pth protein concentration (see Table 3) and stability (see Table 4) were estimated by densitometry using a standard curve with known concentrations of purified Pth in the same immunoblot assay. Each of these assays and those the results of which are given in Fig. 2 were done at least in duplicate; the calculated average deviation was lower than 20% of the mean.
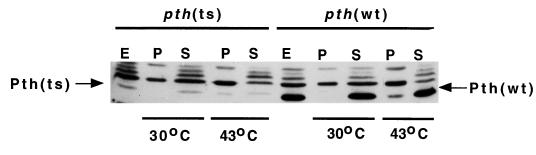
FIG. 2.
In vitro aggregation of Pth(Ts) protein. S30 extracts of the indicated strains were incubated for 20 min at 30 or 43°C and centrifuged at 10,000 × g for 10 min. Pth protein concentration was visualized by immunoblot assay in equivalent amounts of total extract (E), pellet (P), or supernatant (S) fractions. The intensity of the cross-reacted materials above Pth is stronger than that in Fig. 1 because of increased protein concentrations loaded on the gels.
TABLE 3.
| Strainc type | Untransformed (S30)
|
Transformed
|
|||||
|---|---|---|---|---|---|---|---|
| S30
|
DEAE
|
CMd sp act | |||||
| Concn | Sp act | Concn | Sp act | Concn | Sp act | ||
| pth(wt) | 0.60 (±0.15) | 6.83 | 18.00 (±3.2) | 8.33 | 120 (±14.4) | 8.75 | 6.40 |
| pth(rap) | 0.64 (±0.15) | 0.46 | 19.20 (±4.1) | 0.027 | 129 (±23.2) | 0.03 | 0.04 |
| pth(Ts) | 0.12 (±0.02) | 7.5 | 0.48 (±0.1) | 6.04 | 1.70 (±0.2) | 8.82 | |
The Pth concentration (nanograms of Pth per microgram of total protein) was estimated as described for Fig. 1. The numbers represent the means, and the values in parentheses are the estimated standard deviations in assays of four extracts.
In picomoles of substrate per minute per microgram of Pth protein.
For transformed strains, the pth alleles in the chromosome and in the constructs were identical: C600 pth(wt)/pGI01; C600 pth(rap)/pGI02 and C600 pth(Ts)/pGI03.
CM, carboxymethyl cellulose.
FIG. 1.
Variant Pth protein stabilities. Pth protein concentrations in C600 pth(wt), C600 pth(rap), or C600 pth(Ts) at various times were measured after rifampin and chloramphenicol addition (see Materials and Methods). The arrow indicates Pth(Ts) protein position.
TABLE 4.
Concentration and stability of Pth(Ts) protein in strains mutant for chaperones or proteases
| Straina | Relative concnb | Half-lifec (min) |
|---|---|---|
| C600 | 1.00 | 3.0 (±0.35) |
| MC4100 | 1.00 | 2.9 (±0.43) |
| C600 dnaK756 | 0.98 | 2.5 (±0.30) |
| C600 dnaJ259 | 4.50 | >16 |
| C600 grpE280 | 0.89 | 3.9 (±0.32) |
| MC4100 Δlon ΔclpP | 5.89 | >16 |
| MC4100 Δlon | 2.78 | >16 |
| MC4100 ΔclpP | 2.98 | >16 |
| MC4100 ΔclpX | 0.95 | 3.1 (±0.40) |
| MC4100 ΔclpA | 0.97 | 2.9 (±0.31) |
The wild-type and mutant strains, which contained the pth(wt) allele in the chromosome, were transformants for pGI03.
The relative concentration was obtained by dividing the Pth(Ts) protein concentration in each mutant strain by the concentration in the parental strain.
Half lives were estimated as described for Fig. 1. The numbers represent the means, and the values in parentheses are the estimated standard deviations.
Pth purification.
The cells were grown exponentially at 30°C in LB-AP containing 1 mM IPTG. Cells were sedimented and lysed as described above. The extracts were applied to a DEAE-cellulose column (DE52 Whatman; 1.6 by 5 cm). The flowthrough was collected and dialyzed against acetate buffer(10 mM, pH 6.4) followed by dialysis against morpholine ethanesulfonic acid (MES) buffer (10 mM, pH 6.4). The dialysate was applied to a carboxymethyl cellulose column (CM52 Whatman; 1.6 by 3 cm), and the retained protein was eluted with a linear 60 to 300 mM NaCl gradient. The fractions with the greatest concentration of protein were collected and dialyzed against Tris-HCl buffer (10 mM, pH 7.6)–30% glycerol. Pth activity was assayed essentially as described elsewhere (1). A 40-fold purification was achieved by the above two steps. The resulting product was virtually homogeneous as judged by SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown).
RESULTS
Expression and stability of Pth mutants.
The different phenotypes shown by the pth(Ts) and the pth(rap) mutant strains (11) suggested distinctive properties for the mutant Pth proteins. First, we tested this inference by determining the stability of the mutant proteins at 30°C by immunoblot analysis (see Materials and Methods). The results (Fig. 1) showed that the mutant Pth(Ts) differed in three ways from the Pth(wt) and Pth(rap) proteins; (i) it was fivefold less concentrated (compare 0-min lanes), (ii) its migration was retarded, and (iii) it was very unstable. The half-life of Pth(Ts) in vivo was estimated as 3 min at both 30 and 43°C, making it at least 120 times less stable than Pth(wt) or Pth(rap) proteins (data not shown). The difference in Pth concentrations could not be explained by the specific mRNA levels in the cells. mRNA concentration, as determined by reverse transcriptase-PCR, was even higher in the pth mutants than in the wild-type cell (data not shown).
Extracts prepared from pth(Ts) mutant cells, untransformed or transformed with pGI03 for overexpression of Pth(Ts), showed a reduction of 40% in Pth activity at 43°C (Table 2). This result does not agree with previous reports of 90% reduction under similar conditions (11, 25). This discrepancy may be due to so far unidentified differences in the extract preparations.
TABLE 2.
| Strain | S30 extract
|
DEAE flowthrough
|
||
|---|---|---|---|---|
| 30°C | 43°C | 30°C | 43°C | |
| C600 | 6.80 (±1.3) | 6.35 (±0.9) | 43.5 | 47.4 |
| C600 pth(Ts) | 1.57 (±0.4) | 0.98 (±0.3) | 5.5 | 3.3 |
| C600/pGI01 | 203 (±45) | 186 (±32) | 1,146 | 1,075 |
| C600 pth(Ts)/pGI03 | 10 (±2.5) | 6.34 (±0.8) | 43.2 | 32.8 |
Percent hydrolyzed substrate per minute per microgram of total protein. For S30, values are shown as means ± the estimated standard deviations in assays of four different S30 extracts; for DEAE flowthrough, values are means of three replicates for a single extract, and the average deviations were lower than 20% of the means.
The preparations were preincubated at the indicated temperatures for 20 min and assayed at 32°C as in reference 11.
We examined the behavior of the Pth(Ts) protein in crude extracts by immunoblot analysis. The results (Fig. 2) show that, upon incubation at 43°C, approximately half of the Pth(Ts) protein aggregated and sedimented upon low-speed centrifugation, whereas most of the Pth(wt) protein remained in solution (compare lanes 4 and 5 with 9 and 10). Neither Pth(wt) nor Pth(Ts) aggregated at 30°C (compare lanes 2 and 3 with 7 and 8). In addition, about half of Pth(Ts) protein was recovered in the precipitate by incubation of mutant cells at 43°C (data not shown).
Specific activities of the Pth variants.
It has been reported elsewhere that the Pth activity in wild-type extracts is 10 times that of either the Pth(rap) or Pth(Ts) mutant at 30°C (11, 25). To measure the relative activities of each of these Pth variants, we attempted to overproduce and purify the respective Pth preparations. The genes pth(wt), pth(Ts), and pth(rap) were cloned under the control of tac promoter, the lacIq repressor, and the ribosome binding site in the expression vector pKQV4 (see Materials and Methods). Upon IPTG induction, the yield of Pth(wt) and Pth(rap) proteins in crude extracts was 30-fold larger than that of the untransformed cells, but the yield of Pth(Ts) was only fourfold greater (Table 3; compare data columns 1 and 3). This low yield can be accounted for by both the instability and the propensity to aggregate of the Pth(Ts) protein as shown above. The specific activities for Pth(wt) and Pth(Ts) in transformed and untransformed S30 extracts as well as in the DEAE flowthrough were comparable. However, the specific activity of Pth(rap) protein was only a small fraction of that of the others: 0.3 to 0.45% for transformed extracts or 7% for S30 untransformed extracts. This last figure probably reflects an overestimation of the Pth(rap) activity caused by RNase degradation of the substrate. We found it difficult to purify the Pth(Ts) protein. The DEAE-cellulose step concentrated the protein only three to four times (Table 3, columns 3 and 5, row 3) compared to a sevenfold increase for the other two proteins. In addition, we were unable to recover the Pth(Ts) protein from carboxymethyl cellulose, as Pth(Ts) remained in the column. However, as shown above, the specific activity of this variant, calculated in the DEAE flowthrough, was similar to that of the wild-type protein (column 6, rows 1 and 3).
Effect of Pth(Ts) instability on cell viability.
It has been argued elsewhere that the sensitivity to temperature of the pth(Ts) mutant results from inactivation of Pth(Ts), accumulation of pep-tRNA, and arrest of protein synthesis (1, 23). As shown above, Pth(Ts) is highly unstable in vivo even at 30°C. It is likely that the mutant defect resulted from the chemical instability of the Pth(Ts) protein. If this hypothesis is correct, then an increase in the pth(Ts) expression rate in the cell would offset the rate of inactivation of Pth(Ts) protein and the cells would grow even at the nonpermissive temperature. To test this, cultures with different levels of Pth(Ts) protein were incubated at 43°C. We used the pth(Ts) mutant transformed with the pth(Ts) construct pGI03, uninduced or induced for pth(Ts) expression (Fig. 3A). The untransformed pth(Ts) mutant died rapidly at 43°C after an initial peak of growth (Fig. 3A) while a faint band of Pth(Ts) protein was visible up to 7.5 min. Then it decayed to undetectable levels (Fig. 3B, top panel). The untransformed pth(Ts) mutant cells did not lyse as judged by optical density (data not shown); therefore, the loss of Pth(Ts) protein at 43°C was not due to cell disruption. The decrease in protein did not result from a reduction in mRNA concentration; rather, the pth(Ts) mRNA levels increased twofold in 20 min at 43°C (data not shown). In the transformed uninduced culture, the viability pattern was not reduced as strikingly, and eventually the culture grew (Fig. 3A). In this case, the Pth(Ts) protein levels increased after 7.5 min and remained at an intermediate level up to 30 min (Fig. 3B, center panel) probably due to the leaky tac promoter expression even in the presence of lacIq repressor. The IPTG-induced culture showed a steady increase in Pth(Ts) protein (Fig. 3B, bottom panel) and a constant growth rate after 30 min (Fig. 3A). The Pth(Ts) protein concentration obtained within the first 30 min was probably responsible for promotion of cell growth at later times. We assume that there is a threshold in the Pth(Ts) concentration above which the enzyme allows protein synthesis, including its own synthesis, and mutant cell growth at 43°C.
To prove that the reduced stability of Pth(Ts) caused cell death even at the permissive temperature, we designed an experiment to arrest protein synthesis without a temperature shift and with no inhibition of protein decay and pep-tRNA drop-off. In a culture of pth(Ts) mutant cells, RNA and protein synthesis were prevented by addition of rifampin and erythromycin at 30°C. Samples were taken at various times, the antibiotics were diluted out, and cell viability was estimated at 30°C. The results show that the viability decreased to 25% in 90 min in the cultures treated with both rifampin and erythromycin, whereas it was weakly affected in cultures treated with either rifampin or erythromycin alone (Fig. 4). The same treatment did not affect viability in a strain harboring wild-type pth (data not shown). We hypothesize that the double antibiotic procedure prevents new Pth(Ts) from being synthesized. In addition, the effect on growth by decay of the already low Pth(Ts) concentration is aggravated by the stimulation by erythromycin of pep-tRNA release from ribosomes. Analogous double treatment with choramphenicol, an antibiotic which does not cause pep-tRNA drop-off (24), was much less effective in reducing viability.
The nature of the pth(Ts) defect.
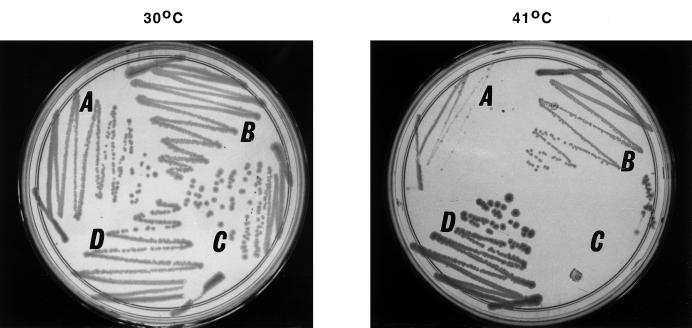
We have shown above that the protein Pth(Ts) migrates less than the wild-type protein in SDS-PAGE. This effect could be due either to a difference in the molecular weight or to a structural oddity of the Pth(Ts) protein. To investigate these alternatives, we analyzed the digestion products of the Pth proteins after Staphylococcus aureus V8 protease treatment or cyanogen bromide cleavage. In both reactions, the products were identical for the Pth(wt) and Pth(Ts) proteins (data not shown), suggesting that the abnormal migration is due to a structural alteration rather than to a variation in molecular weight. This notion was confirmed by further experiments. Proteases and chaperones have as their substrates misfolded or partially folded proteins that arise from different events (13). Thus, it seemed reasonable to study whether mutations in some of the genes encoding chaperones and proteases affected Pth(Ts) protein stability in the cell. The results indicated that defective dnaJ, lon, or clpP genes increased the Pth(Ts) protein concentration and stability (Table 4). The absence of the active dnaJ gene leads to the same modest Pth(Ts) degradation as the double absence of lon and clpP wild-type genes. The other assayed mutations, dnaK, grpE, clpA, and clpX, did not affect the protein concentration. The pth(Ts) dnaJ double mutant grew better at 41°C than did the pth(Ts) mutant (Fig. 5), as expected for a strain with increased levels of Pth(Ts) (see above).
FIG. 5.
Effect of dnaJ259 mutation on pth(Ts) mutant cell growth at 30 and 41°C. (A) C600 dnaJ259 pth(Ts). (B) C600 dnaJ259. (C) C600 pth(Ts). (D) C600. Cells from a single colony were streaked on LB-agar plates and incubated overnight at the indicated temperatures. In this case, 41°C was used as the nonpermissive temperature because the dnaJ259 strain is not viable at 43°C.
DISCUSSION
In the present work, we have investigated some properties of the Pth proteins from wild-type E. coli and the mutant pth(Ts) and pth(rap) strains. The specific activity of Pth(Ts) protein is normal but is extremely unstable in vivo, whereas the Pth(rap) protein is as stable as the wild-type protein but has very low specific activity. The fact that Pth(Ts) protein shows a propensity to aggregate may account for the strong temperature sensitivity of the activity observed in some cases (11, 25).
Based on the results obtained, we propose a sequence of events to explain the temperature-sensitive growth of the pth(Ts) mutant. The cell concentration of the Pth(Ts) protein is low, even at 30°C, but a temperature increase results in a further reduction (Fig. 3). No net Pth(Ts) synthesis occurs at 43°C, and the mutant cells die due to the defect in essential activity and accumulation of pep-tRNA. A controlled rise in pth(Ts) gene expression increased cell viability at 43°C (Fig. 3) probably because sufficient Pth(Ts) activity allows general protein synthesis and, therefore, its own synthesis above a threshold concentration.
Of course, protein synthesis occurs at 30°C because the pth(Ts) mutant is viable at this temperature. As the half-lives of Pth(Ts) at 30 and 43°C are identical (data not shown), an additional factor is responsible for the arrest of protein synthesis. The up-shift to 43°C may inhibit synthesis either by increasing the rate of pep-tRNA accumulation or by reducing the rate of pep-tRNA hydrolysis (23). Since the direct estimation of Pth(Ts) activity in vitro is not affected substantially at 43°C (Table 2), we assume that an increase in the rate of pep-tRNA generation is inhibitory. It has been shown elsewhere that the polypeptide elongation rate rises at 43°C (9); an equivalent overflow in the generation of pep-tRNA is expected. The pth(Ts) mutant transformed with a pth(Ts) construct (pGI03) or in combination with the mutation dnaJ259 is not temperature sensitive, presumably because the Pth(Ts) protein levels are above the threshold which allows protein synthesis.
We observed only 40% reduction of in vitro Pth(Ts) activity at 43°C (Table 2). This result is unexpected, as it has been reported elsewhere that the activity of Pth(Ts) extracts in hydrolyzing diacetyl-lysyl-tRNA is reduced about 10-fold upon incubation at 43°C (11, 25). However, in other reports Pth(Ts) activity was not temperature sensitive for the hydrolysis of different substrates (10). As 50% of Pth(Ts) protein aggregates upon incubation of crude extracts at 43°C (Fig. 2), we suspect that the temperature sensitivity observed in vitro depends on the degree of aggregation during the procedure to prepare extracts. About 50% aggregation of Pth(Ts) protein also occurs in vivo at 43°C in conditions where 90% of the Pth(wt) protein remains in solution (data not shown). However, once in cell extracts, the Pth(Ts) activity remains stable and active in promoting translation at 43°C (J. Hernández-Sánchez and G. Guarneros, unpublished data).
The lower viability of mutant cells can be provoked also at 30°C by combined rifampin-erythromycin treatment but not by either antibiotic alone. Since the pth transcripts are unstable (data not shown), inhibition of new rounds of transcription by rifampin, together with the instability of the Pth(Ts) protein, would rapidly reduce the enzyme concentration to levels incompatible with viability.
Both erythromycin and chloramphenicol interfere with protein biosynthesis, but erythromycin enhances the accumulation of pep-tRNA and chloramphenicol prevents it (23). It is likely that a combination of effects of rifampin and erythromycin intensify pth(Ts) mutant lethality. We conclude that Pth(Ts) protein instability is the primary cause of lethality.
In addition to protein instability, the pth(Ts) mutation determines abnormal protein migration, slightly slower than that of Pth(wt), in SDS-PAGE (Fig. 1). This abnormality may not be caused by an actual change in the molecular weight of the protein, as we did not observe electrophoretic differences in the products of partial proteolysis relative to those of Pth(wt) (data not shown). Rather, it may be the result of a structural property of the Pth(Ts) molecule. It is known that changes of a single amino acid residue in a protein cause abnormal polypeptide migration in SDS-PAGE (2). The pth(Ts) mutation changes Gly to Asp in a hydrophobic pocket on the surface of the Pth protein (26). As SDS binding is highly cooperative at nonpolar residues, a polar substitution at the Gly pocket could especially affect migration (2).
Pth(Ts) is at least 120-fold less stable than the wild-type enzyme in vivo at both 30 and 43°C (Fig. 1 and data not shown). Mutations in the gene encoding the ATP-dependent Lon protease and in the heat shock gene dnaJ resulted in increased Pth(Ts) protein stability (Table 4). These data suggest that Pth(Ts) is a target for the abnormal protein degradation system previously described (19). The DnaK-GrpE-dependent protein degradation system (29) does not appear to participate in Pth(Ts) protein degradation because neither dnaK756 nor grpE280 mutations affect Pth(Ts) stability (Table 4). In addition to Lon, it is likely that a ClpP-containing protease participates in Pth(Ts) protein degradation. A deletion in clpP, but not in clpX or in clpA genes, has a stabilizing effect on Pth(Ts) protein (Table 4). ClpP is a peptidase which, in combination with ClpA or ClpX ATPases, forms ATP-dependent proteases with unique substrate specificity (14). Overexpression of the groEL-S operon suppresses pth(Ts) mutant temperature sensitivity (18). This observation corresponds with chaperonin stabilization of abnormal proteins (12) and renders unlikely the possibility of Pth(Ts) degradation by GroEL-GroES (20).
Our calculations indicate that untransformed wild-type and pth(rap) mutant cells contains each 1,300 Pth molecules. We estimated this number from the result of Pth mass and the corresponding number of cells (0.6 ng [Table 3]; 1.3 × 107 cells). This number strongly disagrees with the value of 25 Pth molecules per cell estimated previously by Dutka et al. (8). The concentration of Pth(wt) and Pth(rap) proteins in cell extracts is similar (Table 3, first column), but the specific activity of Pth(wt) is at least 160-fold that of Pth(rap) (Table 3). Thus, the activity of one Pth(wt) molecule would be equivalent to that of 160 Pth(rap) molecules; as one molecule per cell is the lowest average estimate in a viable cell, and pth(rap) mutant cells are fully viable at 30°C, a value of 25 Pth molecules per pth(rap) mutant cell cannot be correct.
Overexpression of tRNALys suppresses the temperature sensitivity of the pth(Ts) mutant (18). Based on the data presented here, it is possible that tRNALys may affect the rate of Pth(Ts) protein synthesis or the stability of the mutant protein. More experiments are necessary to prove whether these hypotheses are correct.
ACKNOWLEDGMENTS
We thank Carol Gross and Fernando de Las Peñas for providing the chaperone mutants and Susan Gottesman for the protease-defective strains. We thank Luc Dendooven for critically reading the manuscript and an anonymous referee for careful editing of the submitted version.
This work was supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT) of Mexico and the Howard Hughes Medical Institute to G.G. L.R.C.-V. was the recipient of a loan fellowship from CONACyT.
REFERENCES
- 1.Anderson R P, Menninger J R. Test of the ribosome editor hypothesis. III. A mutant Escherichia coli with a defective ribosome editor. Mol Gen Genet. 1987;209:313–318. doi: 10.1007/BF00329659. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong D J, Roman A. Mutagenesis of human papillomavirus types 6 and 16 E7 open reading frames alters the electrophoretic mobility of the expressed proteins. J Gen Virol. 1992;73:1275–1279. doi: 10.1099/0022-1317-73-5-1275. [DOI] [PubMed] [Google Scholar]
- 3.Atherly A G, Menninger J R. Mutant E. coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nat New Biol. 1972;240:245–246. doi: 10.1038/newbio240245a0. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4534. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S M, Takiff H E, Barber A M, Dubois G C, Bardwell J C A, Court D L. Expression and characterization of RNase III and Era proteins. J Biol Chem. 1990;265:2888–2895. [PubMed] [Google Scholar]
- 6.De la Vega F M, Galindo J M, Gold I G, Guarneros G. Microbial genes homologous to the peptidyl-tRNA hydrolase-encoding gene of Escherichia coli. Gene. 1996;169:97–100. doi: 10.1016/0378-1119(95)00823-3. [DOI] [PubMed] [Google Scholar]
- 7.Dinçbas V, Heurgué-Hamard V, Buckingham R H, Karimi R, Ehrenberg M. Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J Mol Biol. 1999;291:745–759. doi: 10.1006/jmbi.1999.3028. [DOI] [PubMed] [Google Scholar]
- 8.Dutka S, Meinnel T, Lazennec C, Mechulam Y, Blanquet S. Role of the 1-72 base pair in tRNAs for the activity of Escherichia coli peptidyl-tRNA hydrolase. Nucleic Acids Res. 1993;21:4025–4030. doi: 10.1093/nar/21.17.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farewell A, Neidhardt F C. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganoza M C, Barraclough N, Wong J T. Purification and properties of an N-formylmethionyl-tRNA hydrolase. Eur J Biochem. 1976;65:613–622. doi: 10.1111/j.1432-1033.1976.tb10379.x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Villegas M R, De la Vega F M, Galindo J M, Segura M, Buckingham R H, Guarneros G. Peptidyl-tRNA hydrolase is involved in λ inhibition of host protein synthesis. EMBO J. 1991;10:3549–3555. doi: 10.1002/j.1460-2075.1991.tb04919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon C L, Sather S K, Casjens S, King J. Selective in vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J Biol Chem. 1994;269:27941–27951. [PubMed] [Google Scholar]
- 13.Gottesman S. Minimizing proteolysis in Escherichia coli: genetic solutions. Methods Enzymol. 1989;185:119–129. doi: 10.1016/0076-6879(90)85013-e. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 15.Guarneros G, Machado G, Guzman P, Garay E. Genetic and physical location of the Escherichia coli rap locus, which is essential for growth of bacteriophage lambda. J Bacteriol. 1987;169:5188–5192. doi: 10.1128/jb.169.11.5188-5192.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson D, Weil J. A mutant of Escherichia coli that prevents growth of phage λ and is bypassed by λ mutants in nonessential region of the genome. Virology. 1976;71:546–559. doi: 10.1016/0042-6822(76)90380-9. [DOI] [PubMed] [Google Scholar]
- 17.Hernández-Sánchez J, Valadez J G, Herrera J V, Ontiveros C, Guarneros G. Lambda bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J. 1998;17:3758–3765. doi: 10.1093/emboj/17.13.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heurgué-Hamard V, Mora L, Guarneros G, Buckingham R H. The growth defect in Escherichia coli deficient in peptidyl-tRNA hydrolase is due to starvation for Lys-tRNA(Lys) EMBO J. 1994;15:2826–2833. [PMC free article] [PubMed] [Google Scholar]
- 19.Jubete Y, Maurizi M R, Gottesman S. Role of the heat shock protein DnaJ in the lon-dependent degradation of naturally unstable proteins. J Biol Chem. 1996;271:30798–30803. doi: 10.1074/jbc.271.48.30798. [DOI] [PubMed] [Google Scholar]
- 20.Kandror O, Sherman M, Rhode M, Goldberg A L. Trigger factor is involved in GroEL-dependent protein degradation in Escherichia coli and promotes binding of GroEL to unfolded proteins. EMBO J. 1995;14:6021–6027. doi: 10.1002/j.1460-2075.1995.tb00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kossel H. Purification and properties of peptidyl-tRNA hydrolase from Escherichia coli. Biochim Biophys Acta. 1970;204:191–202. doi: 10.1016/0005-2787(70)90502-2. [DOI] [PubMed] [Google Scholar]
- 22.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menninger J R. Peptidyl-transfer RNA dissociates during protein synthesis from ribosomes of Escherichia coli. J Biol Chem. 1976;251:3392–3398. [PubMed] [Google Scholar]
- 24.Menninger J R. Accumulation of peptidyl-tRNA is lethal to Escherichia coli. J Bacteriol. 1979;137:694–696. doi: 10.1128/jb.137.1.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menninger J R, Walker C, Foon-Tan P, Atherly A G. Studies on metabolic role of peptidyl-transfer RNA hydrolase. I. Properties of a mutant E. coli with temperature-sensitive peptidyl-transfer RNA hydrolase. Mol Gen Genet. 1973;121:307–324. doi: 10.1007/BF00433230. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt E, Mechulam Y, Fromant M, Plateau P, Blanquet S. Crystal structure at 1.2 Å resolution and active site mapping of Escherichia coli peptidyl-transfer RNA hydrolase. EMBO J. 1997;16:4760–4769. doi: 10.1093/emboj/16.15.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauch M A, Spiegelman G B, Perego M, Johnson W C, Burbulys D, Hoch J A. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straus D, Walter W, Gross C. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 29.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl F U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenson T, Herrera J V, Kloss P, Guarneros G, Mankin A S. Inhibition of translation and cell growth by minigene expression. J Bacteriol. 1999;181:1617–1622. doi: 10.1128/jb.181.5.1617-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]