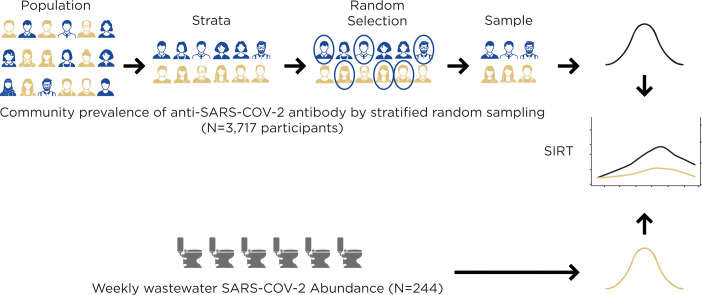
Abstract
Robust epidemiological models relating wastewater to community disease prevalence are lacking. Assessments of SARS-CoV-2 infection rates have relied primarily on convenience sampling, which does not provide reliable estimates of community disease prevalence due to inherent biases. This study conducted serial stratified randomized samplings to estimate the prevalence of SARS-CoV-2 antibodies in 3717 participants, and obtained weekly samples of community wastewater for SARS-CoV-2 concentrations in Jefferson County, KY (USA) from August 2020 to February 2021. Using an expanded Susceptible-Infected-Recovered model, the longitudinal estimates of the disease prevalence were obtained and compared with the wastewater concentrations using regression analysis. The model analysis revealed significant temporal differences in epidemic peaks. The results showed that in some areas, the average incidence rate, based on serological sampling, was 50 % higher than the health department rate, which was based on convenience sampling. The model-estimated average prevalence rates correlated well with the wastewater (correlation = 0.63, CI (0.31,0.83)). In the regression analysis, a one copy per ml-unit increase in weekly average wastewater concentration of SARS-CoV-2 corresponded to an average increase of 1–1.3 cases of SARS-CoV-2 infection per 100,000 residents. The analysis indicates that wastewater may provide robust estimates of community spread of infection, in line with the modeled prevalence estimates obtained from stratified randomized sampling, and is therefore superior to publicly available health data.
Abbreviations: COVID-19, coronavirus disease 2019; LMPHW, Louisville Metro Department of Public Health and Wellness; ODE, ordinary differential equation; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Spike IgG, immunoglobulin G response to SARS-CoV-2 specific spike; SIR, susceptible, infected, recovered; SIRT, susceptible, infected, recovered, seropositive; WBE, wastewater-based epidemiology
Keywords: COVID-19, Epidemiology, Sewer, Stratified randomized sampling, Wastewater-based epidemiology
Graphical abstract
1. Introduction
Since early in the coronavirus disease 2019 (COVID-19) pandemic, wastewater sampling has emerged as a rapid, convenient, and economical tool for assessing the presence and temporal changes in the concentration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in communities (Wu et al., 2020). Approximately 34–52 % of COVID-19 infected patients shed detectable SARS-CoV-2 in their feces up to 16–27 days from the onset of symptoms (Zhang et al., 2021), which can be detected by passive and anonymous community wastewater monitoring. Thus, although wastewater-based epidemiology (WBE) appears to be a promising new source of community disease prevalence data, to be fully informative it requires careful calibration to a reliable clinical reference. During the COVID-19 pandemic, clinical testing was conducted to estimate the community incidence and prevalence, which relied heavily on non-probability or convenience sampling. Although it is necessary to readily track infection rates in real-time, data from non-probability sampled populations are inherently biased and unlikely to provide reliable estimates of the prevalence and incidence of infection (Bilal et al., 2021; Yiannoutsos et al., 2021). Moreover, data from testing only individuals with symptoms are unlikely to gauge prevalence, as many infected individuals show no symptoms, and such data are likely to be always enriched in individuals suspecting infections or experiencing symptoms. Therefore, data reported by local health authorities fail to meaningfully address the need for reliable estimates of spatiotemporal infections, and do not account for individuals who are asymptomatic or have not volunteered for diagnostic testing.
Systematic serological surveys with spatiotemporal resolutions offer opportunities for better surveillance of infectious diseases (Metcalf et al., 2016). A systematic assessment of community-wide spread of infection and immunity could be obtained by randomized sampling, and stratified to include individuals of different ages, sexes, and socioeconomic statuses, as well as those living in different geographic areas (Pollán et al., 2020). In the COVID-19 pandemic, randomized surveys have been conducted over relatively short periods of time in the United States: across the states of California, Georgia, Indiana, Oregon, and Rhode Island (Menachemi et al., 2020; Chan et al., 2021; Yiannoutsos et al., 2021; Layton et al., 2022; Sullivan et al., 2022). Although such a snapshot measure of the cumulative infection of serology is an accurate way of estimating past prevalence of infection in communities, the lack of repeated measurements reduces the utility of these results in relation to wastewater measurements. Since wastewater results are not cumulative and change over time, the discrete wastewater measurements have the potential to identify changes in infection rates and geographic hot spots.
Previous wastewater and serological surveys at a community scale have focused on hepatitis A and E viruses (Martinez Wassaf et al., 2014; Yanez et al., 2014). Existing SARS-CoV-2 wastewater to community COVID-19 case models have been fitted to results obtained from convenience clinical sampling, models that may underrepresent community trends (Cao and Francis, 2021; Hoar et al., 2022; Li et al., 2022; Nourbakhsh et al., 2022; Xiao et al., 2022), limited random samples of communities via SARS-CoV-2 in nasal swabs (Layton et al., 2022) or blood bank serological surveys (Saththasivam et al., 2021; Nourbakhsh et al., 2022). Longitudinal stratified serological sampling could provide robust surveillance estimates that are required for the evaluation of wastewater fidelity.
The purpose of this study was to compare the amount of SARS-CoV-2 in wastewater with community COVID-19 prevalence and seroprevalence after adjusting for spatial and temporal heterogeneity. To accomplish this, this study compared the rates of SARS-CoV-2 infection obtained from serial, stratified random serological sampling in conjunction with serial sampling of virus levels in community wastewater using statistical prevalence modeling that was adjusted for both uncertainty in seropositivity measurements and heterogeneity in temporal and spatial epidemic trends. The simultaneous analysis of these contemporaneous datasets enabled a quantitative comparison of both sampling approaches. Thus, this study design, to the best of our knowledge, represents the most reliable and economical monitoring and surveillance effort for infectious agents attempted to date.
2. Methods
2.1. Temporal probability-based seroprevalence of COVID-19
The study was conducted in the Louisville/Jefferson County metropolitan area in Kentucky (KY), USA, which has a population of approximately 767,000 individuals and represents the largest urban population center in the state. Four probability-designed testing efforts were conducted, each lasting approximately one week. These waves were separated by a 1–3 month window. The first wave of testing commenced on June 10, 2020, and the last wave concluded on February 11, 2021. Households were sampled using an address-based sampling frame derived from the US Postal Service delivery files (Iannacchione, 2011). The addresses in the county were stratified by geography and race before sampling. For each wave, between 18,000 and 36,000 invitations to participate in the study were mailed to the addresses, and the sampled adults were asked to complete a screening interview and schedule an appointment for testing. The response rate was approximately 3 % and with this, the potential non-response bias was examined by comparing the infection rates to those reported in official case reports from the county. The survey sample estimates were two to three times higher for each wave, which suggested that these estimates were less biased than alternative sources of data. The study data were collected and managed using the Research Electronic Data Capture (REDCap) tools hosted at the University of Louisville (Harris et al., 2009; Harris et al., 2019). Refer to Supplement A for further details.
2.2. Clinical COVID-19 positive case rates
Administrative data pertaining to daily counts of publicly reported COVID-19 infected individuals by street address from July 6, 2020, to February 28, 2021, were provided by the Louisville Metro Department of Public Health and Wellness (LMPHW) under a Data Transfer Agreement. The official statistics of the LMPHW were considered as the convenience sample. Refer to Supplement B for further details.
2.3. SARS-CoV-2 (N1) concentration in wastewater
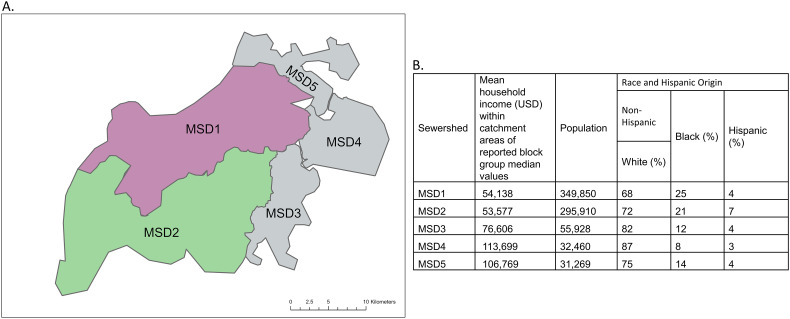
Influent 24-h composite wastewater samples (N = 244) were obtained from five water quality treatment plants, corresponding to the sampling sectors, one to four times per week from August 17, 2020, to February 22, 2021, to detect the presence and concentration of SARS-CoV-2 (N1) (Rouchka et al., 2021; Yeager et al., 2021). Some of the sewershed dynamics have been provided in previously published reports from the study area, including a review of fecal strength indicators (pepper mild mottle virus and cross-assembly phage), if the sewer system is a combined sewer that also receives rainwater, and flow rates (Rouchka et al., 2021; Yeager et al., 2021; Holm et al., 2022a; Holm et al., 2022b). This earlier research formed the basis of using wastewater concentrations to model infection trajectories in the community instead of normalized values. The five water quality treatment plant subpopulations jointly comprised approximately 97 % of the county's population. This allowed for the capture and separation of different wastewater regions within a large county (Fig. 1 ). Refer to Supplement C for more details.
Fig. 1.
A) Distribution of wastewater sewersheds in Jefferson County, Kentucky (USA), different colors correspond to different spatial strata in the analysis, and B) The salient demographic features of each sewershed.
2.4. Estimating prevalence based on community seroprevalence testing
Serostatus was determined as a qualitative assessment by measuring the levels of the SARS-CoV-2 spike protein specific immunoglobulin G (Spike IgG) antibodies in peripheral blood samples using previously reported methods (Hamorsky et al., 2021). Seroprevalence can detect the IgG antibodies of COVID-19 patients up to 300 days following infection (Alfego et al., 2021; Centers for Disease Control and Prevention, n.d.). There was a low number of participants with a positive for Spike IgG antibodies for the discrete MSD3, MSD4, and MSD5 sewersheds (MSD3–5, N = 31 positive participants; Table 1 ). To ensure an adequate balance of the demographic profiles of the subpopulation in the stratified analysis, the data from the three smallest areas were pooled together, resulting in three spatial strata.
Table 1.
Seroprevalence of SARS-CoV-2 by wave and location (95 % credible interval).
| Number of participants | Number of participants positive for SARS-CoV-2 spike protein specific IgG antibodies | Estimated posterior average seroprevalence per 105 people (95 % credible interval | Estimated posterior average prevalence per 105 people (95 % credible interval) | |
|---|---|---|---|---|
| Overall | ||||
| MSD1 | 1998 | 88 | 4702 (2330, 7074) | 78 (1, 155) |
| MSD2 | 1063 | 70 | 6253 (3400, 9105) | 136 (18, 254) |
| MSD3–5 | 621 | 31 | 6231 (3113, 9348) | 174 (14, 334) |
| Other | 35 | 3 | ||
| Total | 3717 | 192 | 17,186 (12,340, 22,032) | 388 (175, 601) |
| Wave 1 | ||||
| MSD1 | 295 | 10 | 2153 (1651, 2656) | 16 (1,31) |
| MSD2 | 121 | 3 | 2230 (1691, 2769) | 20 (1, 38) |
| MSD3–5 | 88 | 2 | 2210 (1708, 2712) | 12 (0, 24) |
| Other | 2 | 0 | ||
| Total | 506 | 15 | 6593 (5701, 7485) | 48 (21, 75) |
| Wave 2 | ||||
| MSD1 | 935 | 22 | 3240 (2109, 4371) | 51 (2,100) |
| MSD2 | 372 | 15 | 3953 (2151, 5756) | 91 (20, 163) |
| MSD3–5 | 271 | 5 | 3698 (2175, 5221) | 90 (13, 165) |
| Other | 15 | 0 | ||
| Total | 1593 | 42 | 10,891 (8274, 13,508) | 232 (117, 347) |
| Wave 3 | ||||
| MSD1 | 480 | 32 | 4461 (2437, 6485) | 106 (1,211) |
| MSD2 | 342 | 21 | 6003 (3098, 8909) | 192 (38, 346) |
| MSD3–5 | 134 | 3 | 6032 (3120, 8944) | 217 (23, 411) |
| Other | 9 | 1 | ||
| Total | 965 | 57 | 16,496 (11,911, 21,081) | 515 (246, 784) |
| Wave 4 | ||||
| MSD1 | 288 | 24 | 7342 (2720, 11,963) | 119 (0, 238) |
| MSD2 | 228 | 31 | 10,416 (5335, 15,498) | 222 (18, 425) |
| MSD3–5 | 128 | 21 | 10,506 (4591, 16,420) | 332 (19, 645) |
| Other | 9 | 2 | ||
| Total | 653 | 78 | 28,264 (19,200, 37,328) | 673 (281, 1065) |
IgG = immunoglobulin G.
The model that was used to estimate the prevalence from seropositivity is a modification of the classical susceptible-infected-recovered (SIR) ecological model used in epidemics (Britton, 2010), hereafter referred to as SIRT (the additional compartment ‘T’ denotes seropositivity). The tracking of the seropositivity status appears necessary as most individuals do not build detectable levels of antibodies until sometime after infection (Alfego et al., 2021). The SIRT model uses a system of ordinary differential equations (ODE) to describe the time evolution of the proportions of susceptible (S), infected (I), recovered (R), and seropositive (T) individuals in a large population. Further details are provided in Supplement D.
To apply the SIRT model to estimate disease prevalence, the study adapted the ODE-based survival analysis method proposed recently (KhudaBukhsh et al., 2020; Di Lauro et al., 2022). Following the work of KhudaBukhsh et al. (2020), the ODE trajectories S t, I t, R t, and T t were treated as respective probabilities, where a randomly selected individual from a large population is, at time t, susceptible, infected, recovered, or seropositive. In this model, the results of all individual antibody-based tests conducted at time t was considered as independent binary variables, with the probability of a positive test given by T t* = T t + (1 − spe)(1 − T t), where spe is the specificity level of the diagnostic test (100 % sensitivity level of the test is assumed). Given that at time t, n t individuals are tested with k t testing positive, the corresponding log-likelihood function is LL t(θ) ∝ k t log T t* + (n t − k t) log(1 − T t*), where θ denotes the vector of the SIRT model parameters that require estimation. The Bayesian method based on Markov chain Monte Carlo was used to estimate θ to properly capture prior information and account for various sources of uncertainty. With the estimated values of the parameters available, the ODE of the SIRT model was applied to calculate the average estimated prevalence over time.
2.5. Correspondence between estimates of SARS-CoV-2 prevalence from community seroprevalence test sampling and wastewater measurements
Using the contemporaneous wastewater concentrations and estimated prevalence, a regression-based correspondence model was derived between the average wastewater concentration and prevalence of COVID-19 in Jefferson County, both in aggregated and stratified sewershed locations. The prevalence rates were calculated based on the SIRT model estimates using census data and geo-coding techniques to estimate the actual infection counts in the sewersheds. The analysis was based on a linear regression model for the prevalence rate and negative binomial regression for infection counts.
2.6. Ethics
For the seroprevalence and data on COVID-19 infected individuals provided by the LMPHW under a Data Transfer Agreement, the University of Louisville Institutional Review Board approved this as Human Subjects Research (IRB number: 20.0393). For the wastewater data, the University of Louisville Institutional Review Board classified this as non-human subjects research (reference #: 717950).
2.7. Role of the funding source
The funders of this study had no role in study design, data collection, data analysis, data interpretation, or writing of this report.
3. Results
3.1. Seroprevalence and prevalence in Louisville/Jefferson County
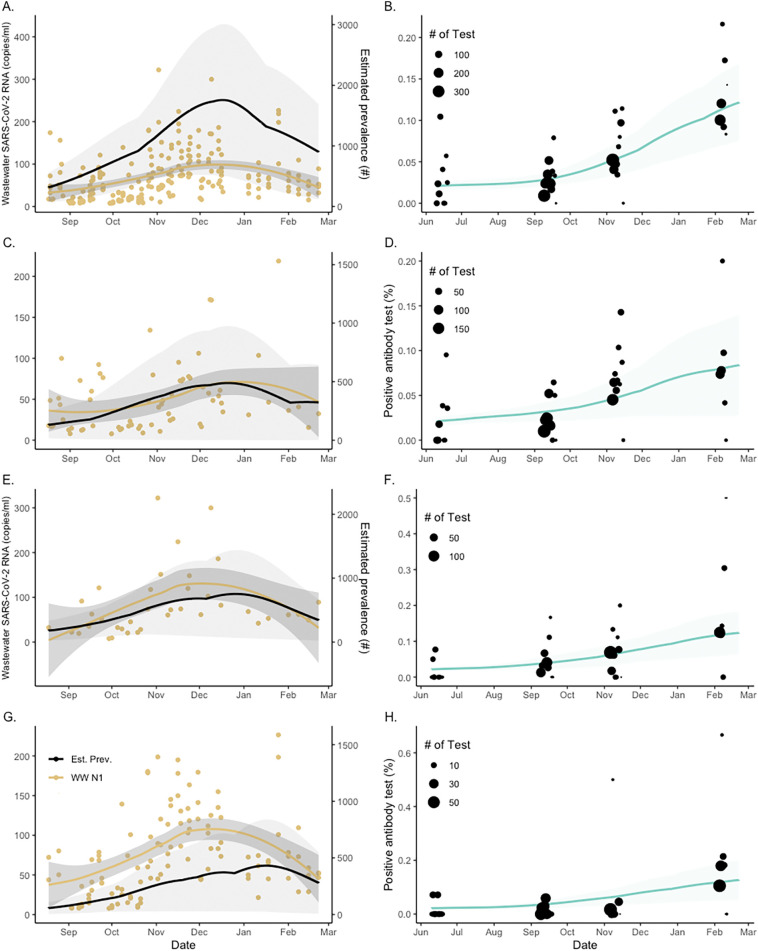
Table 1 shows the total number of adults that were tested; percentage of patients who tested positive for the SARS-CoV-2 Spike IgG antibodies in the four waves and three areas of data collection; and the SIRT model estimated values of average seroprevalence and prevalence in cases per 100,000 people. The average seroprevalence estimates appeared to largely follow the pattern of empirical values, increasing over different testing waves, as more individuals were first infected and acquired antibodies in response to the virus. However, this was not the pattern of the model-based prevalence estimates that are presented in the last column. The results show that the estimates of prevalence, indicated by the fourth wave of the epidemic, declined in sewershed MSD1 but expanded in MSD3–5, which experienced an overall higher average prevalence during the testing period than the other sewersheds. Fig. 2 presents the model-based prevalence predictions (left panels) and the aggregated and stratified model-based seroprevalence fit (right panels). The different prevalence trends in terms of epidemic peak sizes and timings in different sewersheds are clearly visible in the plots. The high variability in the observed positivity rates (marked by dots in the right panels) was reflected in the wide credible bounds for model-based prevalence predictions (marked by shaded areas in the left panels).
Fig. 2.
Sampled seroprevalence and wastewater concentrations of SARS-CoV-2 in Jefferson County (Panels A–B) and sewersheds (Panels C–H) at specific dates between 2020 and 2021. Sewershed areas are stratified as MSD1 (C–D), MSD2 (E–F), and MSD3–5 (G–H). Left panels show changes in the SARS-CoV-2 (N1) concentration in wastewater (indicated by discrete yellow dots; solid black lines represent the model-estimated prevalence; and the shaded area corresponds to 95 % credible bounds). Right panels show the percent seroprevalence in study participants (represented by black dots indicating sample-size weighted observations; the green line is the best fit of the model median prediction; and the shaded area corresponds to the 95 % credible bounds).
3.2. Correspondence between the estimated disease prevalence and wastewater concentrations
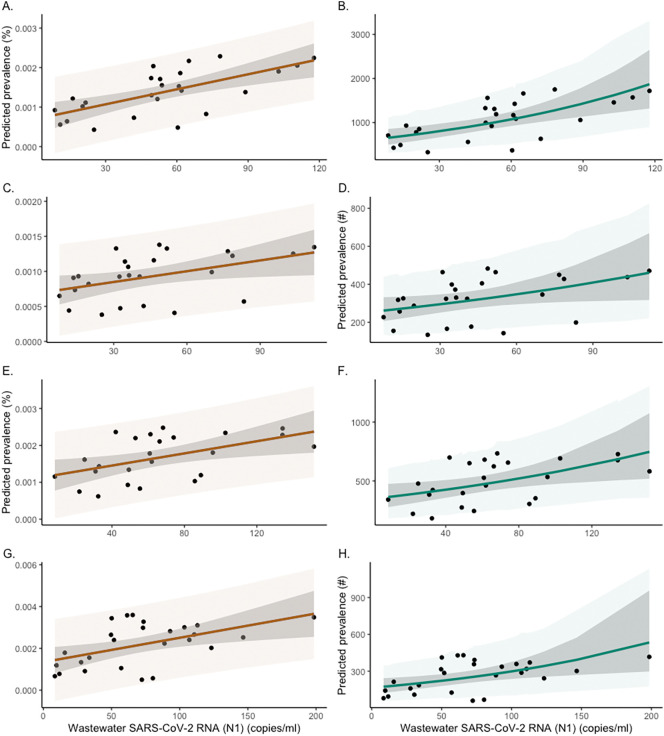
SARS-CoV-2 was detected in 90 % of the wastewater samples. Fig. 2 shows the stratified serial plots of mean weekly wastewater concentrations (refer to Supplement D for discussions on flow normalization) superimposed on the corresponding prevalence estimates in the left panels. The community wastewater concentrations and prevalence estimates showed good qualitative agreement over time. To quantify the extent of agreement, two types of Bayesian regression analyses were performed. In the first analysis, the aggregate and stratified SIRT model estimates of the percentage prevalence were regressed on observed wastewater concentrations using simple linear regression. In the second analysis, the Bayesian negative binomial (NB) regression model was used to regress the model-predicted prevalence counts on the same set of wastewater measurements. The respective data and model predictions are shown in Fig. 3 , where weekly aggregated data were used for better data stability.
Fig. 3.
Relationship between predicted weekly prevalence of SARS-CoV-2 infections and wastewater concentration of SARS-CoV-2 (N1) in Jefferson County (Panels A–B) and sewersheds MSD1 (C–D), MSD2 (E–F), and MSD3–5 (G–H). Left panels show the linear regression between the weekly prevalence percentage and SARS-CoV-2 (N1) concentrations in wastewater (indicated by dots). Right panels show the negative binomial regression (NB) between the adjusted prevalence (count per 100,000 people) and weekly SARS-CoV-2 (N1) concentrations (indicated by dots). The shaded area is the 95 % credible interval (darker) and 95 % prediction interval (lighter).
For the aggregated data from Jefferson County (Fig. 3, top left panel), the results of a simple linear regression model showed a strong correlation of 0.63 (posterior CI = (0.31, 0.83)) between the prevalence of SARS-CoV-2 and average wastewater SARS-CoV-2 concentrations. In the aggregate linear regression model, the estimated slope coefficient corresponded to a relative prevalence increase of 1.27 cases per 100,000 people (posterior CI = (0.67, 1.88)) for every unit increase in wastewater concentration. The rates for the different sewershed areas are presented in Table 2 . Similar results were obtained from the NB regression, which is more appropriate for directly modeling the infection counts. For the aggregated data, the NB regression model (Fig. 3, top right panel)) gives a log-scale regression coefficient of 0.0097 (posterior CI = (0.00452, 0.0151)), which corresponded to a prevalence increase of approximately 1.01 case per 100,000 people for a one-unit increase in wastewater concentration. The remaining sewershed zone rates from the NB regression are listed in Table 2. Bayesian regression models are based on flat (non-informative) prior distributions of model parameters.
Table 2.
Posterior mean values derived from the regression analysis of prevalence versus mean wastewater concentrations (Fig. 3). The posterior means for the simple and negative binomial (NB) regression models are based on the Markov chain Monte-Carlo analysis (detailed in Supplemental D).
| Linear regression model |
NB regression model |
Correlation coefficient (95 % credible interval) | |
|---|---|---|---|
| Posterior slope (95 % credible interval) |
Posterior slope (95 % credible interval) |
||
| Jefferson County Aggregated | 1.269 × 10−5 (6.693 × 10−6, 1.876 × 10−5) |
9.692 × 10−3 (4.527 × 10−3, 1.514 × 10−2) |
0.631 (0.312, 0.833) |
| MSD1 | 5.068 × 10−6 (5.050 × 10−7, 9.392 × 10−6) |
5.463 × 10−3 (5.150 × 10−4, 1.060 × 10−3) |
0.416 (0.032, 0.718) |
| MSD2 | 8.279 × 10−6 (2.020 × 10−6, 1.482 × 10−5) |
5.082 × 10−3 (1.172 × 10−3, 9.090 × 10−3) |
0.464 (0.102, 0.744) |
| MSD3–5 | 1.156 × 10−5 (3.360 × 10−6, 1.985 × 10−5) |
5.951 × 10−3 (1.272 × 10−3, 1.062 × 10−2) |
0.475 (0.126, 0.739) |
3.3. Comparison with administratively reported estimates
In addition to comparing the SIRT-based prevalence estimates to the observed wastewater concentrations of SARS-CoV-2, we also compared the corresponding incidence estimates with publicly reported new COVID-19 cases in Jefferson County. Weekly counts were chosen to smooth the administrative reporting variability and weekend reporting delays. The results of this analysis suggest that the model-based incidence estimates obtained from the observed seropositivity rates in the four waves of testing were significantly higher than the official incidence. The ratios of our model-estimated to officially reported cases were 1.47, 1.14, 1.48, and 1.80 for the aggregated county data for the stratified sewershed areas MSD1, MSD2, and MSD3–5 (see Supplement D, Fig. D3), respectively.
4. Discussion
In this study, we examined the relationship between wastewater SARS-CoV-2 concentrations and the prevalence of infection among community members. Rather than using publicly available infection rates, which are subject to bias due to convenience sampling, the prevalence of infection was estimated using repeated measurements of seropositivity in a randomized sampling of area populations. This study developed models for estimating both community prevalence and wastewater concentrations. Although not all infected populations shed SARS-CoV-2 in feces (Zhang et al., 2021), the wastewater data corresponded well to the community prevalence. Combined analyses of these models indicated that a one-unit prevalence increase in wastewater concentration corresponded to approximately 1 case per 100,000 people, which could be used to assess community-wide prevalence from wastewater data alone.
The comparison of administratively reported COVID-19 cases with the model used in this study suggests that convenience sampling significantly underestimated the rates of infection in the community. However, the extent of underestimation varied across different sewersheds with ratios ranging from 1.14 to 1.80. The model developed in this study is conceptually similar to the Susceptible-Exposed-Infected-Recovered (SEIR) model developed by Fernandez-Cassi et al. (2021), but includes several improvements to reduce model uncertainty using the quantification of a random seropositivity sample. Xiao et al. (2022) suggest that the ratio of wastewater viral copy-numbers to reported COVID-19 cases changes over time, whereas this model was adapted over time with a consistent transfer function. Although the analysis in this study only spans a single wave of COVID-19 infections, strong patterns of a linear relationship between wastewater and prevalence were seen in more recent data (which are not presented here) that have been collected over several waves of infection. These results do not support a log-log scale linear relationship between wastewater and prevalence as proposed by Layton et al. (2022) and Cao and Francis (2021). Previous convenience-based studies paired to wastewater may have underreported the correlation due to inadequate or no case data from portions of a community, underreported at-home COVID-19 antigen rapid self-test results, and the reliance on clinical syndromic surveillance. Hoar et al. (2022) have suggested a log10 change in SARS-CoV-2 wastewater load corresponding to a 0.6 log10 change in new COVID-19 laboratory-confirmed cases per day in a sewershed. Although their model focuses on predicting new cases, and hence is not directly comparable with the model in this study, a qualitative agreement was evident between both models, nevertheless.
The estimates from the use of SARS-CoV-2 Spike IgG antibodies to evaluate the community-wide prevalence of infection observed in this study was consistent with previous large studies (Pollán et al., 2020). The use of systematic serological surveys for calibrating wastewater measurements removes most of the selection biases observed in previous relationships (Cao and Francis, 2021; Hoar et al., 2022; Li et al., 2022; Nourbakhsh et al., 2022; Xiao et al., 2022). Although seropositivity provides estimates of past infection, an increase in seropositivity over a defined period is likely to be a reliable indicator of the spread of infection. Hence, over the course of the eight-month project, enriched contextual data were provided to city decision-makers and stakeholders to inform long-term trends in infection rates and concentrations of SARS-CoV-2 in local wastewater. A comparison of environmental data obtained from wastewater sampling with data obtained from in-person examinations allowed for the development of comprehensive and internally validated datasets; these datasets can be used for the assessment of the trajectory of COVID-19 in the community and identification of geographically defined sub-populations where the virus was lingering. Since approximately 97 % of the population of Jefferson County, KY, uses the sewage system, measurements of SARS-CoV-2 abundance in wastewater are largely representative of the community, which, when coupled with a randomized population-based seroprevalence sampling strategy, makes this an ideal dataset for estimating the correspondence between estimates of seroprevalence and WBE. While the model was developed using anti-spike protein antibody data in unvaccinated populations, it could be readily modified for vaccinated communities using anti-N-protein antibodies.
The major strength of this approach is the repeated, randomized sampling design used to estimate community-wide changes in seroprevalence paired with frequent wastewater sampling concentration changes, which enables the development of a mathematical function that models the relationships among the input variables. To our knowledge, this is the only effort to cross-validate paired, randomized, longitudinal seroprevalence with wastewater in specific geographic areas within a large metropolitan area. Finally, although this study reports on SARS-CoV-2 infection rates, it could also be readily extended for the measurement and validation of other viruses and bacteria, as well as other wastewater analytes, such as pharmaceutical or xenobiotic metabolites.
5. Limitations
Despite its many strengths, this study had some limitations. The shedding rate and duration of COVID-19 infected persons are individual-specific (Zhang et al., 2021). Additionally, fecal shedding of SARS-CoV-2 may only occur in about 50 % of individuals, or fewer (Zhang et al., 2021). Both seroprevalence and wastewater are a measure of a portion of the community that has been infected, and not necessarily active infections. The seroprevalence design in this study only considered the adult population, while wastewater included the entire community population that uses flush toilets in the county (the population that is approximately two years and older). The wastewater sampling excluded individuals who were not connected to the piped infrastructure, such as residents using septic tanks. Although this work has shown that wastewater is superior to administrative data, it still plausibly undercounts the true community infection rates. The broader generalization of the specific modeled relationship may be affected by sewer-system-dependent factors that are not fully understood (Hart and Halden, 2020). For example, the wastewater matrix includes complex chemistry and structural specifics that may affect the amount of virus that is recoverable. Replication in other locations would greatly assist in identifying the contributions of such differences.
6. Conclusion
This study proposes a novel analytical model to predict the SARS-CoV-2 disease prevalence using systematic, retrospective serological surveys with a spatiotemporal resolution to directly compare the concentrations of viral RNA in wastewater with disease prevalence at a sub-community scale. As a community-wide random sampling approach was used, it is likely that this survey-design model used less biased underlying data than previous studies, which were based entirely on convenience sampling; therefore, this model was less susceptible to the underreporting of community infections and more comparable to the population shedding feces in wastewater. The spatial estimates of this study were in agreement with the corresponding data of SARS-CoV-2 wastewater concentrations. These findings indicate that wastewater data could be used as a surrogate for the prevalence of COVID-19 and other pathogens.
CRediT authorship contribution statement
AB, TS, KEP, and RK conceived and developed the idea for the study. KEP supervised the serological assays, ABS and KK performed the serology assays and quality control assessments and uploaded the data to the REDCap database. TS and RH supervised the wastewater analyses. GAR and BC developed the modeling methodology and performed model-based data analyses, which were supported by CAR and KB. RHH wrote the first draft of the manuscript, and all authors contributed to the interpretation of the data and critical revisions of the article. MB accessed and verified the data reported herein. All authors had full access to all data in the study and had the final responsibility of the decision to submit for publication.
Data sharing
The seroprevalence and wastewater data used in the study can be accessed from the website https://doi.org/10.5281/zenodo.6247636. The computer code that implemented our model-based analysis will be made available immediately after publication. The code will be shared with researchers who provide a methodologically sound proposal approved by GAR and AB. Proposals should be directed to rempala.3@osu.edu and aruni.bhatnagar@louisville.edu; requesters will need to sign a data-access agreement.
Funding
This work was supported by contracts from the Centers for Disease Control and Prevention and the Louisville-Jefferson County Metro Government as a component of the Coronavirus Aid, Relief, and Economic Security Act, as well as grants from the James Graham Brown Foundation, Owsley Brown II Family Foundation, and the Welch Family. Serological work was also supported in part by the Jewish Heritage Fund and the Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases. Additionally, the methodology for the current work was partially developed under the National Sciences Foundation grant to Dr. Rempala (DMS-2027001). Krzysztof Banecki's research was co-funded by Warsaw University of Technology within the Excellence Initiative: Research University (IDUB) programme, and co-supported by Polish National Science Centre (2019/35/O/ST6/02484). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this study.
Acknowledgements
We thank the Louisville/Jefferson County Metropolitan Sewer District for their valuable collaboration in the wastewater sample collection process.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.158567.
Appendix A. Supplementary data
Supplementary material
Data availability
The seroprevalence and wastewater data used in the study can be accessed from the website https://doi.org/10.5281/zenodo.6247636.
References
- Alfego D., Sullivan A., Poirier B., et al. A population-based analysis of the longevity of SARS-CoV-2 antibody seropositivity in the United States. EClin. Med. 2021;36 doi: 10.1016/j.eclinm.2021.100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal U., Tabb L.P., Barber S., Diez Roux A.V. Spatial inequities in COVID-19 testing, positivity, confirmed cases, and mortality in 3 US cities: an ecological study. Ann. Intern. Med. 2021;174:936–944. doi: 10.7326/M20-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton T. Stochastic epidemic models: a survey. Math. Biosci. 2010;225:24–35. doi: 10.1016/j.mbs.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Cao Y., Francis R. On forecasting the community-level COVID-19 cases from the concentration of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Interim guidelines for COVID-19 antibody testing. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html
- Chan P.A., King E., Xu Y., et al. Seroprevalence of SARS-CoV-2 antibodies in Rhode Island from a statewide random sample. Am. J. Public Health. 2021;111:700–703. doi: 10.2105/AJPH.2020.306115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lauro F., KhudaBukhsh W.R., Kiss I.Z., Kenah E., Jensen M., Rempała G.A. Dynamic survival analysis for non-Markovian epidemic models. J. R. Soc. Interface. 2022;19:20220124. doi: 10.1098/rsif.2022.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., et al. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamorsky K.T., Bushau-Sprinkle A.M., Kitterman K., et al. Serological assessment of SARS-CoV-2 infection during the first wave of the pandemic in Louisville Kentucky. Sci. Rep. UK. 2021;11:18285. doi: 10.1038/s41598-021-97423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P.A., Taylor R., Minor B.L., et al. The REDCap consortium: building an international community of software platform partners. J. Biomed. Inform. 2019;95 doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020;172 doi: 10.1016/j.watres.2020.115473. [DOI] [PubMed] [Google Scholar]
- Hoar C., Chauvin F., Clare A., et al. Monitoring SARS-CoV-2 in wastewater during New York City's second wave of COVID-19: sewershed-level trends and relationships to publicly available clinical testing data. Environ. Sci. Water Res. Technol. 2022;8:1021–1035. doi: 10.1039/d1ew00747e. [DOI] [Google Scholar]
- Holm R.H., Mukherjee A., Rai J.P., et al. SARS-CoV-2 RNA abundance in wastewater as a function of distinct urban sewershed size. Environ. Sci. Water Res. Technol. 2022;8:807–819. doi: 10.1039/D1EW00672J. [DOI] [Google Scholar]
- Holm R.H., Nagarkar M., Yeager R.A., et al. Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods, Kentucky. FEMS Microbes. 2022 doi: 10.1093/femsmc/xtac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannacchione V.G. The changing role of address-based sampling in survey research. Public Opin. Q. 2011;75:556–575. doi: 10.1093/poq/nfr017. [DOI] [Google Scholar]
- KhudaBukhsh W.R., Choi B., Kenah E., Rempala G.A. Survival dynamical systems: individual-level survival analysis from population-level epidemic models. Interface Focus. 2020;10 doi: 10.1098/rsfs.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton B., Kaya D., Kelly C., et al. Evaluation of a wastewater-based epidemiological approach to estimate the prevalence of SARS-CoV-2 infections and the detection of viral variants in disparate Oregon communities at city and neighborhood scale. Environ. Health Perspect. 2022;130 doi: 10.1289/EHP10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mazurowski L., Dewan A., et al. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Sci. Total Environ. 2022;817 doi: 10.1016/j.scitotenv.2022.152958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Wassaf M.G., Pisano M.B., Barril P.A., et al. First detection of Hepatitis E virus in Central Argentina: environmental and serological survey. J. Clin. Virol. 2014;61:334–339. doi: 10.1016/j.jcv.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Menachemi N., Yiannoutsos C.T., Dixon B.E., et al. Population point prevalence of SARS-CoV-2 infection based on a statewide random sample—Indiana, April 25–29, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:960. doi: 10.15585/mmwr.mm6929e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf C.J., Farrar J., Cutts F.T., et al. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet. 2016;388:728–730. doi: 10.1016/S0140-6736(16)30164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourbakhsh S., Fazil A., Li M., et al. A wastewater-based epidemic model for SARS-CoV-2 with application to three Canadian cities. Epidemics. 2022;39 doi: 10.1016/j.epidem.2022.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollán M., Pérez-Gómez B., Pastor-Barriuso R., et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouchka E.C., Chariker J.H., Saurabh K., et al. The rapid assessment of aggregated wastewater samples for genomic surveillance of SARS-CoV-2 on a city-wide scale. Pathogen. 2021;10:1271. doi: 10.3390/pathogens10101271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saththasivam J., El-Malah S.S., Gomez T.A., et al. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P.S., Siegler A.J., Shioda K., et al. Severe acute respiratory syndrome coronavirus 2 cumulative incidence, United States, August 2020–December 2020. Clin. Infect. Dis. 2022;74 doi: 10.1093/cid/ciab626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., et al. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez L.A., Lucero N.S., Barril P.A., et al. Evidence of Hepatitis A virus circulation in central Argentina: seroprevalence and environmental surveillance. J. Clin. Virol. 2014;59:38–43. doi: 10.1016/j.jcv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Yeager R., Holm R.H., Saurabh K., et al. Wastewater sample site selection to estimate geographically-resolved community prevalence of COVID-19: a sampling protocol perspective. GeoHealth. 2021 doi: 10.1029/2021GH000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiannoutsos C.T., Halverson P.K., Menachemi N. Bayesian estimation of SARS-CoV-2 prevalence in Indiana by random testing. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2013906118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cen M., Hu M., et al. Prevalence and persistent shedding of fecal SARS-CoV-2 RNA in patients with COVID-19 infection: a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2021;12 doi: 10.14309/ctg.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The seroprevalence and wastewater data used in the study can be accessed from the website https://doi.org/10.5281/zenodo.6247636.