Abstract
Artificial intelligence (AI) methods for the automatic detection and quantification of COVID-19 lesions in chest computed tomography (CT) might play an important role in the monitoring and management of the disease. We organized an international challenge and competition for the development and comparison of AI algorithms for this task, which we supported with public data and state-of-the-art benchmark methods. Board Certified Radiologists annotated 295 public images from two sources (A and B) for algorithms training (, source A), validation (, source A) and testing (, source A; , source B). There were 1,096 registered teams of which 225 and 98 completed the validation and testing phases, respectively. The challenge showed that AI models could be rapidly designed by diverse teams with the potential to measure disease or facilitate timely and patient-specific interventions. This paper provides an overview and the major outcomes of the COVID-19 Lung CT Lesion Segmentation Challenge — 2020.
Keywords: Medical image segmentation, COVID-19, Challenge
Graphical abstract
1. Introduction
The SARS-CoV-2 pandemic has had a devastating impact on the global healthcare systems. As of May 28, 2021, more than 169 million people have been infected in the world with over 3.5 million deaths (Hopkins, 2021). COVID-19 is known to affect nearly every organ system, including the lungs, brain, kidneys, liver, gastrointestinal tract, and cardiovascular system. The manifestations of the disease in the lung may be early indicators of future problems. These manifestations have been intensively reported in the adult populations and occasionally in pediatric subjects (Nino et al., 2021, Zang et al., 2021, Larici et al., 2020, Ojha et al., 2020, Wan et al., 2020). Since the early days of the pandemic, lung imaging has been critical for both the early identification and management of individuals affected by COVID-19 (Rubin et al., 2020). Imaging also provides invaluable support for the evaluation of patients with long COVID and after the acute sequelae of the diseases. Repeated waves of infection and changes in the disease course require data, including imaging, classification, quantification, and response tools, as well as standardized reliable interpretation as the global society struggles to provide widely available vaccines and faces evolving challenges such as new mutations of the virus.
The most common lung imaging modalities utilized for the evaluation of SARS-CoV-2 infections are chest radiographs (CXR) and chest computerized tomography (CT) with ultrasound (US) being used more sparingly. Chest CT is the reference modality that most accurately demonstrates the acute lung manifestations of COVID-19 (Bao et al., 2020, Zhu et al., 2020). As observed in CT, the most common findings in the chest of the affected individuals were ground-glass opacities (GGO) and pneumonic consolidations. Other manifestations include interstitial abnormalities, crazy paving pattern, halo signs, pleural abnormalities, bronchiectasis, bronchovascular bundle thickening, air bronchograms, lymphadenopathy, and pleural/pericardial effusions. The sensitivity of chest CT to detect these abnormalities in subjects with confirmed COVID-19 was widely variable and somewhat subjective, reported in the range of 44%–97% (median 69%) (Merkus and Klein, 2020).
Beside its role in the identification of patterns of SARS-CoV-2 infections, lung CT is also important in the determination of the severity of COVID-19 (Wan et al., 2020, Bao et al., 2020, Zhu et al., 2020, Cao et al., 2020, Bernheim et al., 2020). The presence, location and extension of the lung abnormalities are critical factors for the clinical management of patients to potentially facilitate decisions towards more timely and personalized medical interventions. Quantification of lesions may further provide the tracking of disease progression and response to therapeutic countermeasures. Thus, improving COVID-19 treatment starts with a clearer understanding of the patient’s disease state, which must include accurate identification, delineation and quantification of lung lesions and disease phenotypes and patterns.
A prior lack of global data collaboration limited clinicians and scientists in their ability to quickly and effectively understand COVID-19 disease, its severity and outcomes. As access to data has improved, quality annotations have remained a limiting factor in the development of useful artificial intelligence (AI) models derived from machine learning and deep learning (LeCun et al., 2015). Thus, a multitude of AI approaches have been developed, published and indicated great potential for clinical support, but they were often overfit, being trained using proprietary data or from a single site (Kang et al., 2020, Wang et al., 2020b, Fan et al., 2020, Oulefki et al., 2021, Shan et al., 2021, Ippolito et al., 2021). Alternatively, federated approaches allow algorithms to access data from multiple sites without the need of sharing raw data, but through this paradigm access is granted to a single algorithm and consortium, with sharing of model weights instead of raw data (Yang et al., 2021, Dayan et al., 2021). In particular, deep neural networks were used for the identification and segmentation of abnormal lung regions affected by SARS-CoV-2 infection. These can be grouped into two main classes: classification models that extract the affected region inside the lung area by comparison with data from healthy subjects (Bai et al., 2020, Li et al., 2020, Mei et al., 2020, Wang et al., 2020a), and segmentation models that directly extract the abnormal lung areas according to patterns in the image and (typically using fully convolutional networks) (Fan et al., 2020, Shan et al., 2021, Huang et al., 2020, Zhang et al., 2020, Zhou et al., 2021).
Without access to public data and an adequate platform to evaluate and compare their performance, AI approaches risk being overtrained, irreproducible, and ultimately clinically not useful. Thus, public efforts are needed to accelerate the understanding of the role of AI towards informing manifestations and qualifying impact of health crises such as the COVID-19 pandemic.
The COVID-19 Lung CT Lesion Segmentation Challenge 2020 (COVID-19-20) created the public platform to evaluate emerging AI methods for the segmentation and quantification of lung lesions caused by SARS-CoV-2 infection from CT images. This effort required a multi-disciplinary team science partnership among global communities in a broad variety of often disparate fields, including radiology, computer science, data science and image processing. The goal was to rapidly team up to combine multi-disciplinary expertise towards the development of tools to simultaneously both define and address unmet clinical needs created by the pandemic. The COVID-19-20 platform provided access to multi-institutional, multinational images originating from patients of different ages and gender, and with variable disease severity. The challenge team provided the ability to quickly label a public dataset, allowing radiologists to rapidly add precise annotations. Open access was offered to the annotated CTs of subjects with PCR-confirmed COVID-19, and to a baseline deep learning pipeline based on MONAI (Project MONAI, 2021) that could serve as a starting point for further algorithmic improvements. The challenge was hosted on a widely used competition website (covid-segmentation.grand-challenge.org) for easy and secure data access control. This paper presents an overview of this challenge and competition, including the data resources and the top ten AI algorithms identified from a highly competitive field of participants who tested the data in December 2020.
2. Submissions
The challenge was launched on November 2, 2020. The training and validation data were released and 1,096 teams registered before the training phase was closed on December 8, 2020. The 225 teams that completed the validation phase were given access to the test data. Ninety-eight teams from 29 countries on six continents completed the test phase. Fig. 1 shows the countries of origin of the 98 teams. Test results were released on December 18, 2020, and the statistical ranking of the top ten teams (see Section 3) was unveiled during a virtual mini symposium on January 11, 2021.1 Fig. 2 shows the demographic information for the team leaders, i.e., age group, sex, highest educational degree, student status and job category, and algorithmic characteristics for the 98 submissions that completed the training, validation and test phases. We requested participants to disclose whether they used external data for training their algorithms or if they used a general-purpose pre-trained network for initialization (e.g., a network pre-trained for another lung disease). The use of public networks pre-trained for the segmentation of COVID-19 lesions was not allowed (e.g., Clara_train_covid19_ct_lesion_seg2 ).
Fig. 1.
The countries of origin of the 98 teams that completed the training, validation and test phases of the challenge.
Fig. 2.
Demographic information of the leaders of the 98 teams that completed the training, validation and test phases of the challenge. The top row shows the age group (left), student status (middle) and sex (right) of the participant. The middle row shows the highest degree (left) and job category (right). Bottom row shows the algorithm characteristics for the 98 submissions that completed the training, validation and test phases of the challenge. We report if algorithms were fully-automated (left), used external data for training (middle) or used a general pre-trained network for initialization (right).
Participants uploaded the results on the validation and test data to the hosting website for evaluation. Only (semi-)automated methods were allowed. Submission of manual annotations was prohibited. For validation, the number of submissions from each user was limited to once-a-day for the purpose of refining their algorithms based on the live performance indicators on the challenge validation leaderboard.3 Submission of results on the test data was collected without showing the leaderboard and the last submission was used for final ranking. The test phase was open only to participants who had already submitted their results on the validation set. The leaderboard and final ranking are public and hosted on the challenge website.4
3. Materials & results
3.1. Data sources
This challenge utilized data from two public resources on chest CT images, namely the “CT Images in COVID-19”5 (Harmon et al., 2020) (Dataset 1) and “COVID-19-AR”6 (Desai et al., 2020) (Dataset 2) available on The Cancer Imaging Archive (TCIA) (Clark et al., 2013). CT images were acquired without intravenous contrast enhancement from patients with positive Reverse Transcription Polymerase Chain Reaction (RT-PCR) for SARS-CoV-2. Dataset 1 originated from China, while dataset 2 was acquired from the US population. In total, we used 295 images, including 272 images from Dataset 1 and 23 images from Dataset 2. Of these images, 199 and 50 from Dataset 1 were used for training and validation, respectively. We therefore refer to Dataset 1 as the “seen” data source that participants used to train and validate their algorithms during the first phase of the challenge. The test set contained 23 images each from Datasets 1 and 2 (46 images in total). Hence, Dataset 2 was only used in the testing phase, and we refer to it as the “unseen” data source.
Descriptive statistics, such as x-, y-, and z-spacings and voxel volume in both data sources are shown in Fig. 3. We also show the differences in COVID-19 lesion volumes annotated between the two data sources.
Fig. 3.
Data variability between “seen” and “unseen” sources; (a) Illustration of the differences in the voxel spacing and voxel volume grouped by training, validation, and test sets. (b) Differences in COVID-19 lesion volumes across the image data sources. (c) Normalized histograms showing the CT intensity distributions of the “seen” and “unseen” data sources in Hounsfield units (HU). Note, −1000 HU corresponds to air, and 750 to cancellous bone (Patrick et al., 2017).
3.2. Annotation protocol
All images were automatically segmented by a previously trained model for COVID lesion segmentation (Yang et al., 2021) that is publicly available.7 These segmentations were subsequently used as a starting point for board certified radiologists (RS, JZ, JM) who equally divided the dataset between them and manually adjudicated and corrected the segmentations. Therefore, the annotation for each case was confirmed by one radiologist. The annotation tool used was ITKSnap8 (Yushkevich et al., 2006) showing multiple reformatted views of the CT scans, and allowing manipulations and corrections of the initial automated segmentation results in three dimensions.
All abnormal lung lesions related to COVID-19 were included. Lesions comprised of consolidation, GGO (focal or diffuse), septal thickening, crazy paving, and bronchiectasis, consistent with Bao et al. (2020). Note, that diffuse GGOs were still readily identifiable by the radiologists and defined as abnormal.
3.3. Evaluation metrics
We used the three evaluation metrics described below. These metrics were both used to evaluate the performance of different algorithms, and to establish the interobserver variability.
-
1.
Dice Coefficient (Dice). A common evaluation metric of segmentation accuracy defined as the overlap between the volume of the ground truth segmentation and the predicted segmentation volume : .
-
2.
Normalized Surface Dice (NSD). Similarly to Dice, it provides the normalized measure of agreement between the surface of the prediction and the surface of the ground (Andrearczyk et al., 2021). We chose a threshold of 1 mm to define an “acceptable” derivation between the ground truth surface and the predicted surface.
-
3.
Normalized Absolute Volume Error (NAVE). The volume of COVID-19 lesion burden inside the patient’s lung can play an important role for clinical assessment (Sun et al., 2020). Therefore, a measure was chosen that assesses the agreement between the predicted and ground truth lesion volumes, defined as . Note, we used the negative of this value for ranking purposes as higher values indicate better performance in our ranking approach.
3.4. Interobserver performance
As a benchmark for comparing the AI algorithms with human performance on the lesion segmentation task, we measured the human interobserver agreement. We compared the annotations utilized in Yang et al. (2021) from 245 of the 272 cases from Dataset 1 used in the challenge with the ones obtained by our radiologists. The interobserver agreement showed mean ±standard deviation (median) of Dice, NSD, and NAVE of 0.702 ±0.172 (0.756), 0.538 ±0.147 (0.563), and 0.601 ±1.969 (0.180), respectively.
3.5. Statistical ranking method
Recent work on ranking analysis for biomedical imaging challenges has shown that ranking results can vary significantly depending on the chosen type of metric and ranking scheme (Maier-Hein et al., 2018). Most biomedical challenges use approaches such as “aggregate-then-rank” or “rank-then-aggregate”, which do not account for statistical differences between algorithms (Maier-Hein et al., 2018, Wiesenfarth et al., 2021). These findings motivated the development of a challenge ranking toolkit9 (Wiesenfarth et al., 2021) that we employed for our evaluation. This toolkit utilizes statistical hypothesis testing applied to each possible pair of algorithms. This allows us to better assess the differences between the evaluated metrics.
Following the notation of Wiesenfarth et al. (2021), our challenge contained tasks (Dice, NSD, NAVE on each “seen” and “unseen” test data). The test cases for each task are denoted as . In our case, for each task. A bootstrap approach is used to evaluate the ranking stability of the different algorithms. This means that ranking is performed repeatable on bootstrap samples, see Fig. 4. The statistical test employed to determine the consensus ranking is the one-sided Wilcoxon signed rank test with a significance level of , adjusted for multiple testing according to Holm (Wiesenfarth et al., 2021).
Fig. 4.
Blob plot visualization of the ranking variability via bootstrapping. An algorithm’s ranking stability is shown across the different tasks, illustrating the ranking uncertainty of the algorithm in each task. For more details see Wiesenfarth et al. (2021).
Each of the m tasks contributed equality to the final consensus using the Euclidean distance between averaged ranks across tasks. We ranked the 98 submitted algorithms using the proposed statistical consensus ranking algorithm to determine the top-10 methods, including the challenge winning algorithm.
3.6. Summary of top-10 algorithms
We show the final ranking of the top-10 performing algorithms in
Table 1. All top-10 algorithms were fully-automated methods, and all were based on some variation of the U-Net (Ronneberger et al., 2015, Çiçek et al., 2016), a fully convolutional network (Long et al., 2015) for image segmentation based on the popular encoder–decoder design with skip connections (Long et al., 2015, Drozdzal et al., 2016). U-Net has dominated the field of biomedical image segmentation in recent years (Haque and Neubert, 2020) and most challenge participants opted to use one of its implementations. In particular the nnU-Net open-source framework10 (Isensee et al., 2021), which has shown success in multiple biomedical image segmentation challenges, was a popular choice for challenge participants. The U-Net architectures included 2D, 3D, high- and low-resolution configurations. One team used the open-source platform MONAI11 (#68). The majority of algorithms used challenge data only with one method including additional unlabeled data from the public TCIA source (#53), which was done with pseudo labels in a semi-supervised approach. The majority directly targeted the segmentation of COVID-19 lesions, while one participant (#31) targeted multiple outputs, including body and lung masks.
Table 1.
Top-10 finalists after statistical ranking. “Value” represents the average rank the algorithm achieved across all tasks. We also show if methods were automated, used external data for training, the input data dimensions used in the algorithms, and the network architecture.
| Rank | Value | ID # | Fully automated |
Extra data |
Pre-trained | Ensemble | Data dimension |
Network architecture |
Authors | Country |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.6 | 53 | ✓ | ✓ | ✗ | ✗ | 3D | nnU-Net | S. Hu et al. | China |
| 2 | 6.0 | 38 | ✓ | ✗ | ✗ | ✓ | 3D | nnU-Net | F. Isensee et al. | Germany |
| 3 | 7.7 | 65 | ✓ | ✗ | ✗ | ✓ | 2D/3D | nnU-Net | C. Tang | USA |
| 4 | 8.4 | 58 | ✓ | ✗ | ✗ | ✓ | 3D | nnU-Net | Q. Yu et al. | China |
| 5 | 8.5 | 31 | ✓ | ✗ | ✗ | ✓ | 3D | nnU-Net | J. Sölter et al. | Luxembourg |
| 6 | 9.2 | 50 | ✓ | ✗ | ✗ | ✓ | 2D/3D | nnU-Net | T. Zheng & L. Zhang | Japan |
| 6 | 9.2 | 68 | ✓ | ✗ | ✓ | ✗ | 2D/3D | VGG16 Hybrid, MONAI |
V. Liauchuk et al. | Belarus |
| 8 | 9.4 | 95 | ✓ | ✗ | ✗ | ✓ | 3D | nnU-Net | Z. Zhou et al. | China |
| 9 | 10.6 | 29 | ✓ | ✗ | ✗ | ✗ | 3D | nnU-Net | J. Moltz et al. | Germany |
| 10 | 11.3 | 15 | ✓ | ✗ | ✗ | ✗ | 3D | U-Net | B. Oliveira et al. | Portugal |
A popular loss function for biomedical image segmentation is the Dice loss (Milletari et al., 2016). In this challenge, most finalists utilized it together with additional cross entropy, top-k (Lyu et al., 2020), and focal loss (Lin et al., 2017). An important strategy for winning image segmentation is model ensembling, the fusion of predictions from several independently trained models. Here, most methods used 5-fold cross validation and model ensemble to arrive at a consensus prediction.
A full description of the top-10 finalists’ algorithms by their authors is given in the Supplementary Material.
3.7. Ranking results
Table 2 shows the mean and standard deviation of the Dice coefficients for the top-10 performing algorithms on test cases from the “seen” and “unseen” data sources. Top algorithms performed relatively similar to each other, but all showed a marked decrease when being evaluated on the “unseen” data (Table 2).
Table 2.
Dice coefficients of the top-10 algorithms on (left) all test data, (middle) “seen” data (Dataset 1), and (right) “unseen” test data (Dataset 2).
| All test cases: |
“Seen” test cases: |
“Unseen” test cases: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID # | mean | std | median | ID # | mean | std | median | ID # | mean | std | median | ||
| 53 | 0.666 | 0.236 | 0.754 | 38 | 0.740 | 0.195 | 0.797 | 53 | 0.598 | 0.264 | 0.700 | ||
| 58 | 0.658 | 0.242 | 0.741 | 53 | 0.734 | 0.182 | 0.782 | 95 | 0.593 | 0.258 | 0.677 | ||
| 95 | 0.658 | 0.237 | 0.729 | 31 | 0.729 | 0.190 | 0.769 | 58 | 0.588 | 0.263 | 0.724 | ||
| 38 | 0.654 | 0.268 | 0.763 | 65 | 0.729 | 0.186 | 0.778 | 15 | 0.581 | 0.264 | 0.670 | ||
| 15 | 0.649 | 0.242 | 0.716 | 58 | 0.728 | 0.195 | 0.789 | 68 | 0.570 | 0.276 | 0.703 | ||
| 68 | 0.646 | 0.251 | 0.753 | 95 | 0.723 | 0.193 | 0.783 | 38 | 0.569 | 0.302 | 0.729 | ||
| 31 | 0.645 | 0.265 | 0.753 | 68 | 0.723 | 0.196 | 0.779 | 50 | 0.562 | 0.279 | 0.692 | ||
| 65 | 0.644 | 0.258 | 0.754 | 29 | 0.722 | 0.187 | 0.711 | 31 | 0.561 | 0.300 | 0.685 | ||
| 50 | 0.639 | 0.252 | 0.733 | 15 | 0.717 | 0.197 | 0.751 | 65 | 0.559 | 0.291 | 0.686 | ||
| 29 | 0.634 | 0.259 | 0.705 | 50 | 0.716 | 0.194 | 0.773 | 29 | 0.545 | 0.289 | 0.647 | ||
Fig. 5 shows boxplots of the top-10 performing algorithms for each of the tasks. In general, methods present more outliers on the “unseen” test dataset. Fig. 6 shows a typical example from the “seen” test data source. The 1st (#53) and 2nd (#38) ranked algorithms achieved a mean Dice coefficient >0.734 Dice on the “seen” dataset. Fig. 6 shows that most of the COVID-19 related lesions were well segmented by the automated algorithms. In contrast, Fig. 7 shows a challenging case from the “unseen” test data source. Both top-performing algorithms (#53 and #38) generated a false-positive segmentation region at a normal lung vessel while missing the real lesion. Their performance dropped to a Dice coefficient <0.598 on the “unseen” dataset. To illustrate the general performance of the top-10 algorithms on the individual test cases, Fig. 8 shows podium plots (Eugster et al., 2008) with the performance of different algorithms on the same test case connected by a line. Due to the limited test set size for task (), there was mostly no statistically significant difference found between the top-10 algorithms across the six tasks apart for #50 being significantly better than #29 on task “NSD (unseen)” at the 5% significance level.
Fig. 5.
Top-10 algorithms performance measured for the tasks used in the challenge, namely the Dice coefficient (top row), Normalized Surface Dice (middle row), and Normalized Absolute Volume Error (bottom row) on the “seen” (a, c, e) and “unseen” test datasets (b, d, f), respectively. Algorithms are ranked based on their performance from left to right individually for each task.
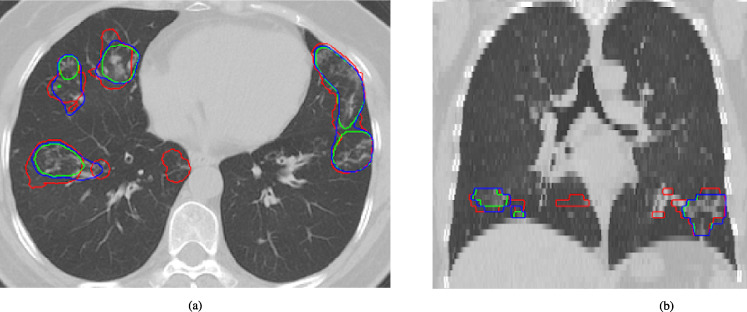
Fig. 6.
Example test case from the “seen” data source (Dataset 1). The performance of the top algorithms #53 and #38 is shown in green and blue, respectively. Ground truth annotations are shown in red (a: axial view, b: coronal view).
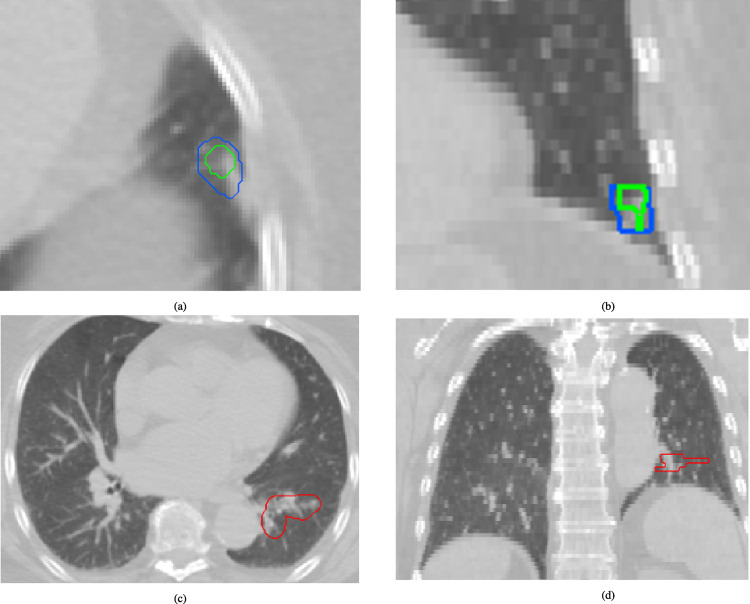
Fig. 7.
Example test case from the “unseen” data source. (a: axial view, b: coronal view) Top algorithms #53 and #38, shown in green and blue, respectively, both predict a false-positive lesion at the locations of a normal lung vessel. At the same time they missed the real lesion in red (c: axial view, d: coronal view).
Fig. 8.
Podium plots for “seen” (a) and “unseen” (b) test data. The participating algorithms are color-coded. Each colored dot shows the Dice coefficient achieved by the respective algorithm. The same test cases are connected by a line. The lower part of the charts displays the relative frequency for a given algorithm to achieve a podium place, i.e. rank achieved by a given algorithm.
4. Discussion
4.1. Performance of algorithms
Automatic AI algorithms showed great potential to accurately segment the lung COVID-19 lesions from CT images. In the validation phase, 87 out of 225 methods achieved superior Dice coefficients than the interobserver criteria (0.702), with the top team achieving a Dice coefficient of 0.771 (9.8% improvement). However, their level of robustness is inferior to the radiologist’s performance: the top team gets a Dice coefficient of 0.666 on the test data12 (5.1% decrease). This discrepancy could be due to various reasons. One reason could be the domain shift as half of the test data is from an “unseen” source that has not been used in the training or validation phases. Another reason could be the limited number of allowed submissions for the testing phase, which mitigates the possibility for overfitting to the test data. Moreover, the limited number of training data could also affect algorithm performance.
The evaluation of the analysis of top-10 algorithms revealed that the ensemble of segmentation from various individual automated methods plays an important role compared to other factors such as the complexity of the network architecture, the learning rate, losses, etc. Most 10 top teams used model ensembles to reduce outliers and improved their performance by collecting the consensus segmentation from separately trained models. This observation also shows that the training pipeline can potentially be further improved based on novel concepts like AutoML (Yang et al., 2019, Zoph et al., 2018) or neural architectures search (Yu et al., 2020, Zhu et al., 2019, Liu et al., 2018) algorithms.
4.2. Use of external training data
Only one of the top 10 teams, which was the winning team of the challenge, used external data in their final solution. Using this semi-supervised training approach, they obtained an improvement of 4.27% and 0.86% Dice coefficient on the training and validation data, respectively. Another team did similar work in a student–teacher manner and saw improvement in the validation score. However, they submitted their final results without using the external data after noticing partial overlap between the chosen unlabeled external dataset and the provided training data. Both teams demonstrate that using external data, even unlabeled, could improve the segmentation performance. While this finding clearly calls for larger training datasets, it also shows the great potential of semi-supervised methods to achieve more robust solutions, especially for the healthcare domain where the annotation cost is much higher than in other fields (Tajbakhsh et al., 2020).
4.3. U-net dominance
All top-10 teams used a 2D/3D U-Net variant with at most minor modifications. While this seems to conflict with hundreds of yearly publications creating new network architectures, it also shows that most existing deep learning algorithms lack the robustness offered by model ensembles to handle large data variations (e.g., voxel spacing, contrast, etc.) when training data are limited. nnU-Net (Isensee et al., 2021) was adopted by 5 out of the 10 teams to build an end-to-end solution while another team used MONAI.13 Unsurprisingly, these findings show that the majority of participants employed well-validated, open-source resources.
4.4. Data variability and generalizability gap
The challenge was designed to use “seen” and “unseen” data sources and thus evaluate the generalizability of AI algorithms in front of variable clinical protocols. Our data sources varied in provenience (China and US), scanner manufacturers (various, as typical in routine clinical practice) and imaging protocols (voxel spacing). Fig. 3 illustrates that the volumes of the annotated COVID lesions have similar distributions on the two data sources. However, there are substantial differences in the voxel spacing used for CT reconstruction in the data (“seen” cases have a 5 mm z-spacing, while the “unseen” cases were in the 3 mm range). To overcome these differences, most participants used common data normalization strategies, such as resampling all data to a constant voxel spacing (Isensee et al., 2021). Still, these differences in voxel spacing, together with variability in scanner manufacturers and imaging protocols, were likely the main contributors to the generalization gap seen in the performance of algorithms on the “unseen” test cases. Additional factors were related to the variability of manifestations of the disease in the lungs. For example, in the challenging case from the “unseen” test data source shown in Fig. 7, the top-performing algorithms generated false-positive predictions at a normal lung vessel while missing to segment the real lesion. Domain shifts like the ones observed in the data used in this challenge are still proving to be challenging for current AI models — an observation in line with other recent works discussing the shortcomings of AI models for the diagnosis of COVID-19 (Roberts et al., 2021, Wynants et al., 2020). Disease phase variability may also have broadened the features of what defines a standard or expected set of features. Early disease may not look like later disease cycles on CT, which may have also increased model noise.
4.5. Potential for clinical use
Segmentation and classification models have been postulated to impact diagnosis in outbreak settings with delayed or unavailable PCR, however the point of care classification of COVID-19 versus other pneumonia such as influenzae, could prove of some value during flu season in specific outbreak settings as an epidemiologic tool or as a red flag for patient isolation at the scanner, by early identification, thus expediting or prioritizing interpretation using more conventional radiologist review and verification. AI models have also been proposed to assist in triage or selection of resource-limited therapeutics or critical care, prognostication or prediction of outcomes, or as one data element of a multi-modal model combining clinical, laboratory and imaging data. Standardized response criteria for clinical trials can provide a level “playing field”, thus uniformly defining effects of medical and other countermeasures, or specific scenarios for patient-specific therapies. Specific phenotypes may respond to certain therapies, for example. Imaging AI could thus play a role in determining the optimal disease phase for steroid administration or monoclonal antibodies, or even characterize the presence of different disease manifestations according to variant or underlying comorbidity, although many of these clinical or research utilities are quite speculative. AI models in COVID-19 have been justifiably criticized for a lack of generalize-ability, lack of clinical testing and validation, impracticality of model design, “me-too” models and studies, and easy replaceability of functionality with standard clinical tools. Potential clinical impact has yet to match the excitement from the data science and computational community nor realize the promise at the outset of the pandemic. Federated learning and open-source tools and modeling may help address this, especially for specific research questions for clinical trials or radiologist-sparse settings.
4.6. Limitations
The challenge organizers aimed to create a fair and robust evaluation platform for (semi-)automatic AI algorithms. This was a timely effort completed with limited resources, thus several factors could potentially be improved in retrospect. For example, 295 annotated CT images from two different data sources were used in the challenge, which may be suboptimal data quantity for training deep learning algorithms, as performance metrics improve with size of datasets. However, the challenge set a benchmark for the development and evaluation of AI methods to segment lung lesions in COVID-19, the first of its kind to our knowledge, which was reflected by the large number of participants. It is advisable to add more data in future challenges, even if the data are non-annotated as the results of this challenge indicated.
Another limitation may be the data annotation. Each case was annotated by one radiologist who rectified the prediction from a publicly available COVID lesion segmentation AI model.14 Although these initial predictions may be considered as a suggestion from an expert, which is a typical workflow for many AI data annotation solutions, a second verification from another human expert would likely further improve the annotation quality.
Finally, the statistical consensus ranking algorithm over multiple tasks, although it overcomes the limitations of ranking based on single evaluation metrics, is computed only at the image level. The ranking does not provide a measurement of the algorithm on the lesion level, thus without consideration of each lesion’s clinical relevance. Such information, which was nor available in our data, could be important for clinical diagnosis and tracking of disease progress. It could also provide a more granular interpretation of the strengths and weaknesses of each algorithm, and a guidance on how to improve them.
5. Conclusions
The COVID-19 Lung CT Lesion Segmentation Challenge — 2020 provided the platform to develop and evaluate AI algorithms for the detection and quantification of lung lesions from CT images. AI models help in the visualization and measurement of COVID specific lesions in the lungs of infected patients, potentially facilitating more timely and patient-specific medical interventions. Over one thousand teams registered to participate in the challenge participating in this challenge reflecting the engagement of the global scientific community to combat COVID-19. The AI models could be rapidly trained and showed good performance that was comparable to expert clinicians. However, robustness to “unseen” data decreased in the testing phase, indicating that larger and more diverse data may be beneficial for training. A more granular interpretation of the strengths and weaknesses of each algorithm might highlight pathways on the road towards a future where AI and deep learning might help standardize, quantify, assess disease response, select patients or therapies, or predict outcomes. But first steps first, as the scientific community builds multi-disciplinary teams to develop new tools and methodology to walk before we run. As more AI applications are being introduced in the biomedical space, it is essential to adequately validate and compare the functionality of these applications through challenges as proposed in this paper.
Code availability
The baseline deep learning pipeline based on MONAI is available at https://github.com/Project-MONAI/tutorials/tree/master/3d_segmentation/challenge_baseline.
The model and software to automatically segment COVID lesions in chest CT is available at https://ngc.nvidia.com/catalog/models/nvidia:clara_train_covid19_ct_lesion_seg.
The software used by radiologist to correct the automatically generated lesion annotations was ITK-Snap and is available at http://www.itksnap.org/pmwiki/pmwiki.php.
Evaluation scripts used for ranking the algorithms will be made available on at https://covid-segmentation.grand-challenge.org.
The ranking software used is available at https://github.com/wiesenfa/challengeR.
CRediT authorship contribution statement
Holger R. Roth: Conceptualized, Co-organized the challenge, Drafted, Performed the statistical analysis of the algorithms testing results. Ziyue Xu: Conceptualized, Co-organized the challenge, Drafted. Carlos Tor-Díez: Conceptualized, Co-organized the challenge, Drafted. Ramon Sanchez Jacob: Provided expert annotations of the images to be used during training, Validation, Testing phases of the challenge. Jonathan Zember: Provided expert annotations of the images to be used during training, Validation, Testing phases of the challenge. Wenqi Li: Conceptualized, Co-organized the challenge, Drafted, Developed the baseline deep learning pipeline. Sheng Xu: Provided the initial data and expert annotations for analyzing the interobserver performance. Baris Turkbey: Provided the initial data and expert annotations for analyzing the interobserver performance. Evrim Turkbey: Provided the initial data and expert annotations for analyzing the interobserver performance. Dong Yang: Provided the automatically generated segmentations. Ahmed Harouni: Provided the automatically generated segmentations. Nicola Rieke: Conceptualized, Co-organized the challenge, Drafted. Shishuai Hu: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Fabian Isensee: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Claire Tang: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Qinji Yu: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Jan Sölter: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Tong Zheng: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Vitali Liauchuk: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Ziqi Zhou: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Jan Hendrik Moltz: Provided expert annotations of the images to be used during training, Validation, Testing phases of the challenge. Bruno Oliveira: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Yong Xia: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Klaus H. Maier-Hein: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Qikai Li: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Andreas Husch: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Luyang Zhang: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Vassili Kovalev: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Li Kang: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Alessa Hering: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. João L. Vilaça: Participated in the challenge, achieved a top-10 rank, Provided the results analyzed in this article. Mona Flores: Conceptualized, Co-organized the challenge, Drafted. Daguang Xu: Conceptualized, Co-organized the challenge, Drafted. Bradford Wood: Provided the initial data and expert annotations for analyzing the interobserver performance. Marius George Linguraru: Conceptualized, Co-organized the challenge, Drafted.
Acknowledgments
We would like to thank all participants for contributing to the challenge and this paper. We would also like to thank Stephanie Harmon, Maxime Blain, Michael Kassin, Nicole Varble, and Amel Amalou who were part of the organizing committee. AH & JS would like to acknowledge the Luxembourg National Research Fund, FNR. Finally, we would like to thank NVIDIA for providing GPU prizes to the challenge winners. All authors contributed expertise and edited the contents of this article. The final manuscript was approved by all authors.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.media.2022.102605.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
.
Data Availability
The leaderboards showing the results of all participating algorithms and data used in phase I of the challenge are available at https://covid-segmentation.grand-challenge.org.
References
- Andrearczyk Vincent, Oreiller Valentin, Depeursinge Adrien. Springer Nature; 2021. Head and neck tumor segmentation: First challenge, HECKTOR 2020, held in conjunction with MICCAI 2020, lima, peru, october 4, 2020, proceedings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Harrison X, Wang Robin, Xiong Zeng, Hsieh Ben, Chang Ken, Halsey Kasey, Tran Thi My Linh, Choi Ji Whae, Wang Dong-Cui, Shi Lin-Bo, et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Cuiping, Liu Xuehuan, Zhang Han, Li Yiming, Liu Jun. Coronavirus disease 2019 (COVID-19) CT findings: a systematic review and meta-analysis. J. Am. College Radiol. 2020;17(6):701–709. doi: 10.1016/j.jacr.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim Adam, Mei Xueyan, Huang Mingqian, Yang Yang, Fayad Zahi A, Zhang Ning, Diao Kaiyue, Lin Bin, Zhu Xiqi, Li Kunwei, et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Yinghao, Liu Xiaoling, Xiong Lijuan, Cai Kailin. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Med. Virol. 2020;92(9):1449–1459. doi: 10.1002/jmv.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiçek Özgün, Abdulkadir Ahmed, Lienkamp Soeren S, Brox Thomas, Ronneberger Olaf. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. 3D U-net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- Clark Kenneth, Vendt Bruce, Smith Kirk, Freymann John, Kirby Justin, Koppel Paul, Moore Stephen, Phillips Stanley, Maffitt David, Pringle Michael, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digital Imag. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan Ittai, Roth Holger R, Zhong Aoxiao, Harouni Ahmed, Gentili Amilcare, Abidin Anas Z, Liu Andrew, Costa Anthony Beardsworth, Wood Bradford J, Tsai Chien-Sung, et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021:1–9. doi: 10.1038/s41591-021-01506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai Shivang, Baghal Ahmad, Wongsurawat Thidathip, Jenjaroenpun Piroon, Powell Thomas, Al-Shukri Shaymaa, Gates Kim, Farmer Phillip, Rutherford Michael, Blake Geri, et al. Chest imaging representing a COVID-19 positive rural US population. Sci. Data. 2020;7(1):1–6. doi: 10.1038/s41597-020-00741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdzal Michal, Vorontsov Eugene, Chartrand Gabriel, Kadoury Samuel, Pal Chris. Deep Learning and Data Labeling for Medical Applications. Springer; 2016. The importance of skip connections in biomedical image segmentation; pp. 179–187. [Google Scholar]
- Eugster Manuel J.A., Hothorn Torsten, Leisch Friedrich. Exploratory and inferential analysis of benchmark experiments. Tech. Rep., No.30. 2008 [Google Scholar]
- Fan Deng-Ping, Zhou Tao, Ji Ge-Peng, Zhou Yi, Chen Geng, Fu Huazhu, Shen Jianbing, Shao Ling. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- Haque Intisar Rizwan I., Neubert Jeremiah. Deep learning approaches to biomedical image segmentation. Inf. Med. Unlocked. 2020;18 [Google Scholar]
- Harmon Stephanie A, Sanford Thomas H, Xu Sheng, Turkbey Evrim B, Roth Holger, Xu Ziyue, Yang Dong, Myronenko Andriy, Anderson Victoria, Amalou Amel, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nature Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins Johns. 2021. Home - johns hopkins coronavirus resource center. https://coronavirus.jhu.edu/. (Accessed 17 May 2021) [Google Scholar]
- Huang Lu, Han Rui, Ai Tao, Yu Pengxin, Kang Han, Tao Qian, Xia Liming. Serial quantitative chest CT assessment of COVID-19: a deep learning approach. Radiol. Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito Davide, Ragusi Maria, Gandola Davide, Maino Cesare, Pecorelli Anna, Terrani Simone, Peroni Marta, Giandola Teresa, Porta Marco, Franzesi Cammillo Talei, et al. Computed tomography semi-automated lung volume quantification in SARS-CoV-2-related pneumonia. Euro. Radiol. 2021;31(5):2726–2736. doi: 10.1007/s00330-020-07271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee Fabian, Jaeger Paul F, Kohl Simon AA, Petersen Jens, Maier-Hein Klaus H. NnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods. 2021;18(2):203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- Kang Hengyuan, Xia Liming, Yan Fuhua, Wan Zhibin, Shi Feng, Yuan Huan, Jiang Huiting, Wu Dijia, Sui He, Zhang Changqing, et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020;39(8):2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- Larici Anna Rita, Cicchetti Giuseppe, Marano Riccardo, Merlino Biagio, Elia Lorenzo, Calandriello Lucio, Del Ciello Annemilia, Farchione Alessandra, Savino Giancarlo, Infante Amato, et al. Multimodality imaging of COVID-19 pneumonia: from diagnosis to follow-up. A comprehensive review. Eur. J. Radiol. 2020 doi: 10.1016/j.ejrad.2020.109217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Yann, Bengio Yoshua, Hinton Geoffrey. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li Lin, Qin Lixin, Xu Zeguo, Yin Youbing, Wang Xin, Kong Bin, Bai Junjie, Lu Yi, Fang Zhenghan, Song Qi, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Tsung-Yi, Goyal Priya, Girshick Ross, He Kaiming, Dollár Piotr. Proceedings of the IEEE International Conference on Computer Vision. 2017. Focal loss for dense object detection; pp. 2980–2988. [Google Scholar]
- Liu Hanxiao, Simonyan Karen, Yang Yiming. 2018. Darts: Differentiable architecture search. arXiv preprint arXiv:1806.09055. [Google Scholar]
- Long, Jonathan, Shelhamer, Evan, Darrell, Trevor, 2015. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 3431–3440. [DOI] [PubMed]
- Lyu Siwei, Fan Yanbo, Ying Yiming, Hu Bao-Gang. Average top-k aggregate loss for supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2020 doi: 10.1109/TPAMI.2020.3005393. [DOI] [PubMed] [Google Scholar]
- Maier-Hein Lena, Eisenmann Matthias, Reinke Annika, Onogur Sinan, Stankovic Marko, Scholz Patrick, Arbel Tal, Bogunovic Hrvoje, Bradley Andrew P, Carass Aaron, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nature Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-07619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Xueyan, Lee Hao-Chih, Diao Kai-yue, Huang Mingqian, Lin Bin, Liu Chenyu, Xie Zongyu, Ma Yixuan, Robson Philip M, Chung Michael, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkus Peter J.F.M., Klein Willemijn M. The value of chest CT as a COVID-19 screening tool in children. Eur. Respir. J. 2020;55(6) doi: 10.1183/13993003.01241-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletari Fausto, Navab Nassir, Ahmadi Seyed-Ahmad. 2016 Fourth International Conference on 3D Vision (3DV) IEEE; 2016. V-net: Fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- Nino Gustavo, Zember Jonathan, Sanchez-Jacob Ramon, Gutierrez Maria J, Sharma Karun, Linguraru Marius George. Pediatric lung imaging features of COVID-19: a systematic review and meta-analysis. Pediatr. Pulmonol. 2021;56(1):252–263. doi: 10.1002/ppul.25070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha Vineeta, Mani Avinash, Pandey Niraj Nirmal, Sharma Sanjiv, Kumar Sanjeev. CT in coronavirus disease 2019 (COVID-19): a systematic review of chest CT findings in 4410 adult patients. Euro. Radiol. 2020;30:6129–6138. doi: 10.1007/s00330-020-06975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulefki Adel, Agaian Sos, Trongtirakul Thaweesak, Laouar Azzeddine Kassah. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit. 2021;114 doi: 10.1016/j.patcog.2020.107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick Sanjana, Birur N Praveen, Gurushanth Keerthi, Raghavan A Shubhasini, Gurudath Shubha, et al. Comparison of gray values of cone-beam computed tomography with hounsfield units of multislice computed tomography: an in vitro study. Indian J. Dental Res. 2017;28(1):66. doi: 10.4103/ijdr.IJDR_415_16. [DOI] [PubMed] [Google Scholar]
- Project MONAI . 2021. Project MONAI. https://monai.io/. (Accessed September 22, 2022) [Google Scholar]
- Roberts Michael, Driggs Derek, Thorpe Matthew, Gilbey Julian, Yeung Michael, Ursprung Stephan, Aviles-Rivero Angelica I, Etmann Christian, McCague Cathal, Beer Lucian, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3(3):199–217. [Google Scholar]
- Ronneberger Olaf, Fischer Philipp, Brox Thomas. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Rubin Geoffrey D, Ryerson Christopher J, Haramati Linda B, Sverzellati Nicola, Kanne Jeffrey P, Raoof Suhail, Schluger Neil W, Volpi Annalisa, Yim Jae-Joon, Martin Ian BK, et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology. 2020;296(1):172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Fei, Gao Yaozong, Wang Jun, Shi Weiya, Shi Nannan, Han Miaofei, Xue Zhong, Shen Dinggang, Shi Yuxin. Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction. Med. Phys. 2021;48(4):1633–1645. doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Dong, Li Xiang, Guo Dajing, Wu Lan, Chen Ting, Fang Zheng, Chen Linli, Zeng Wenbing, Yang Ran. CT quantitative analysis and its relationship with clinical features for assessing the severity of patients with COVID-19. Korean J. Radiol. 2020;21(7):859. doi: 10.3348/kjr.2020.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh Nima, Jeyaseelan Laura, Li Qian, Chiang Jeffrey N, Wu Zhihao, Ding Xiaowei. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020;63 doi: 10.1016/j.media.2020.101693. [DOI] [PubMed] [Google Scholar]
- Wan Shang, Li Mingqi, Ye Zheng, Yang Caiwei, Cai Qian, Duan Shaofeng, Song Bin. CT manifestations and clinical characteristics of 1115 patients with coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Acad. Radiol. 2020;27(7):910–921. doi: 10.1016/j.acra.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Jun, Bao Yiming, Wen Yaofeng, Lu Hongbing, Luo Hu, Xiang Yunfei, Li Xiaoming, Liu Chen, Qian Dahong. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020;39(8):2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- Wang Guotai, Liu Xinglong, Li Chaoping, Xu Zhiyong, Ruan Jiugen, Zhu Haifeng, Meng Tao, Li Kang, Huang Ning, Zhang Shaoting. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfarth Manuel, Reinke Annika, Landman Bennett A, Eisenmann Matthias, Saiz Laura Aguilera, Cardoso M Jorge, Maier-Hein Lena, Kopp-Schneider Annette. Methods and open-source toolkit for analyzing and visualizing challenge results. Sci. Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-82017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynants Laure, Van Calster Ben, Collins Gary S, Riley Richard D, Heinze Georg, Schuit Ewoud, Bonten Marc MJ, Dahly Darren L, Damen Johanna A, Debray Thomas PA, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. Bmj. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Dong, Roth Holger, Xu Ziyue, Milletari Fausto, Zhang Ling, Xu Daguang. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. Searching learning strategy with reinforcement learning for 3d medical image segmentation; pp. 3–11. [Google Scholar]
- Yang Dong, Xu Ziyue, Li Wenqi, Myronenko Andriy, Roth Holger R, Harmon Stephanie, Xu Sheng, Turkbey Baris, Turkbey Evrim, Wang Xiaosong, et al. Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from China, Italy, Japan. Med. Image Anal. 2021;70 doi: 10.1016/j.media.2021.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Qihang, Yang, Dong, Roth, Holger, Bai, Yutong, Zhang, Yixiao, Yuille, Alan L, Xu, Daguang, 2020. C2fnas: Coarse-to-fine neural architecture search for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 4126–4135.
- Yushkevich Paul A, Piven Joseph, Hazlett Heather Cody, Smith Rachel Gimpel, Ho Sean, Gee James C, Gerig Guido. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zang Si-Tian, Han Xu, Cui Qi, Chang Qing, Wu Qi-Jun, Zhao Yu-Hong. Imaging characteristics of coronavirus disease 2019 (COVID-19) in pediatric cases: a systematic review and meta-analysis. Transl. Pediatrics. 2021;10(1):1. doi: 10.21037/tp-20-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Kang, Liu Xiaohong, Shen Jun, Li Zhihuan, Sang Ye, Wu Xingwang, Zha Yunfei, Liang Wenhua, Wang Chengdi, Wang Ke, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Tongxue, Canu Stéphane, Ruan Su. Automatic COVID-19 CT segmentation using U-net integrated spatial and channel attention mechanism. Int. J. Imaging Syst. Technol. 2021;31(1):16–27. doi: 10.1002/ima.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Zhuotun, Liu Chenxi, Yang Dong, Yuille Alan, Xu Daguang. 2019 International Conference on 3D Vision (3DV) IEEE; 2019. V-nas: Neural architecture search for volumetric medical image segmentation; pp. 240–248. [Google Scholar]
- Zhu Jieyun, Zhong Zhimei, Li Hongyuan, Ji Pan, Pang Jielong, Li Bocheng, Zhang Jianfeng. CT imaging features of 4121 patients with COVID-19: A meta-analysis. J. Med. Virol. 2020;92(7):891–902. doi: 10.1002/jmv.25910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoph, Barret, Vasudevan, Vijay, Shlens, Jonathon, Le, Quoc V, 2018. Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 8697–8710.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
Data Availability Statement
The leaderboards showing the results of all participating algorithms and data used in phase I of the challenge are available at https://covid-segmentation.grand-challenge.org.