Abstract
The reversibility of the procancer effects of obesity was interrogated in formerly obese C57BL/6 mice that lost weight via a nonrestricted low-fat diet (LFD) or 3 distinct calorie-restricted (CR) regimens (low-fat CR, Mediterranean-style CR, or intermittent CR). These mice, along with continuously obese mice and lean control mice, were orthotopically injected with E0771 cells, a mouse model of triple-negative breast cancer. Tumor weight, systemic cytokines, and incidence of lung metastases were elevated in the continuously obese and nonrestricted LFD mice relative to the 3 CR groups. Gene expression differed between the obese and all CR groups, but not the nonrestricted LFD group, for numerous tumoral genes associated with epithelial-to-mesenchymal transition as well as several genes in the normal mammary tissue associated with hypoxia, reactive oxygen species production and p53 signaling. A high degree of concordance existed between differentially expressed mammary tissue genes from obese versus all CR mice and a microarray dataset from overweight/obese women randomized to either no intervention or a CR diet. Assessment of differentially methylated regions in mouse mammary tissues revealed that obesity, relative to the 4 weight loss groups, was associated with significant DNA hypermethylation. However, the anticancer effects of the CR interventions were independent of their ability to reverse obesity-associated mammary epigenetic reprogramming. Taken together, these preclinical data showing that the procancer effects of obesity are reversible by various forms of CR diets strongly support translational exploration of restricted dietary patterns for reducing the burden of obesity-associated cancers.
Keywords: Obesity, breast cancer, weight loss, methylation, metastasis
INTRODUCTION
More than 40% of adult US women are obese (1), and obesity at diagnosis is associated with worse breast cancer-specific survival in both pre- and postmenopausal women, regardless of hormone receptor status (2). Currently, no targeted therapies or specific treatment regimens exist for women with triple-negative breast cancer (TNBC), many of whom are also obese. Several epidemiological studies have examined the impact of postdiagnosis weight loss on breast cancer outcomes, and most have found that weight loss is associated with an increased risk of breast cancer recurrence and/or mortality (3–10). However, these findings are likely capturing a link between cancer progression and unintentional weight loss. Based on the data linking obesity with breast cancer progression, as well as results from lifestyle intervention trials in breast cancer survivors (11–13), many clinicians counsel postdiagnosis weight loss for obese breast cancer patients. However, most women find it challenging to achieve and maintain substantial weight loss, and optimal strategies for losing weight to improve breast cancer outcomes remain unclear.
Several weight loss trials with breast cancer recurrence or survival as the primary outcome are ongoing and, when completed in the coming years, are anticipated to further elucidate the impact of weight loss on obesity-associated breast cancer (14). Bariatric surgery is the only weight-loss intervention in the literature that consistently reduces the obesity-associated increase in breast cancer risk (15). However, bariatric surgery can be costly and poses independent risks, while its impact on breast cancer progression and metastases remains unclear. Therefore, low-cost, sustainable alternatives to bariatric surgery are needed. To date though, neither nonsurgical weight loss regimens, including low-fat diets and various forms of calorie restriction (CR) such as intermittent CR, nor healthful dietary patterns such as Mediterranean-style diets, have been well studied as interventions to reverse the procancer effects of obesity on TNBC.
We are using preclinical studies combined with transcriptomic and epigenomic analyses to identify mechanistic targets and nonsurgical intervention strategies for breaking the obesity-breast cancer link. We previously reported that (a) obesity increases spontaneous and transplanted Wnt-driven mammary tumor development and progression in association with upregulation of epithelial-to-mesenchymal transition (EMT) markers (16,17), while CR suppresses Wnt-driven tumor progression and inhibits EMT (16); (b) obesity reversal in chronically obese mice via a nonrestricted low-fat diet (LFD) fails to attenuate the tumor-promoting effects of obesity in a mouse model of Wnt-driven TNBC (18,19); and (c) obesity-elevated tumor growth in both obese and formerly obese (via nonrestricted LFD) mice is associated with aberrant increases in the expression and methylation of a network of proinflammatory genes in the normal mammary tissue (19). In the present study, we assess the impact of weight loss via 4 diet regimens, specifically a nonrestricted LFD and 3 forms of calorie restriction, on tumor growth, lung metastases, tumoral and mammary gland gene expression, and mammary methylation patterns, in the E0771 murine model of TNBC.
METHODS
Animal study
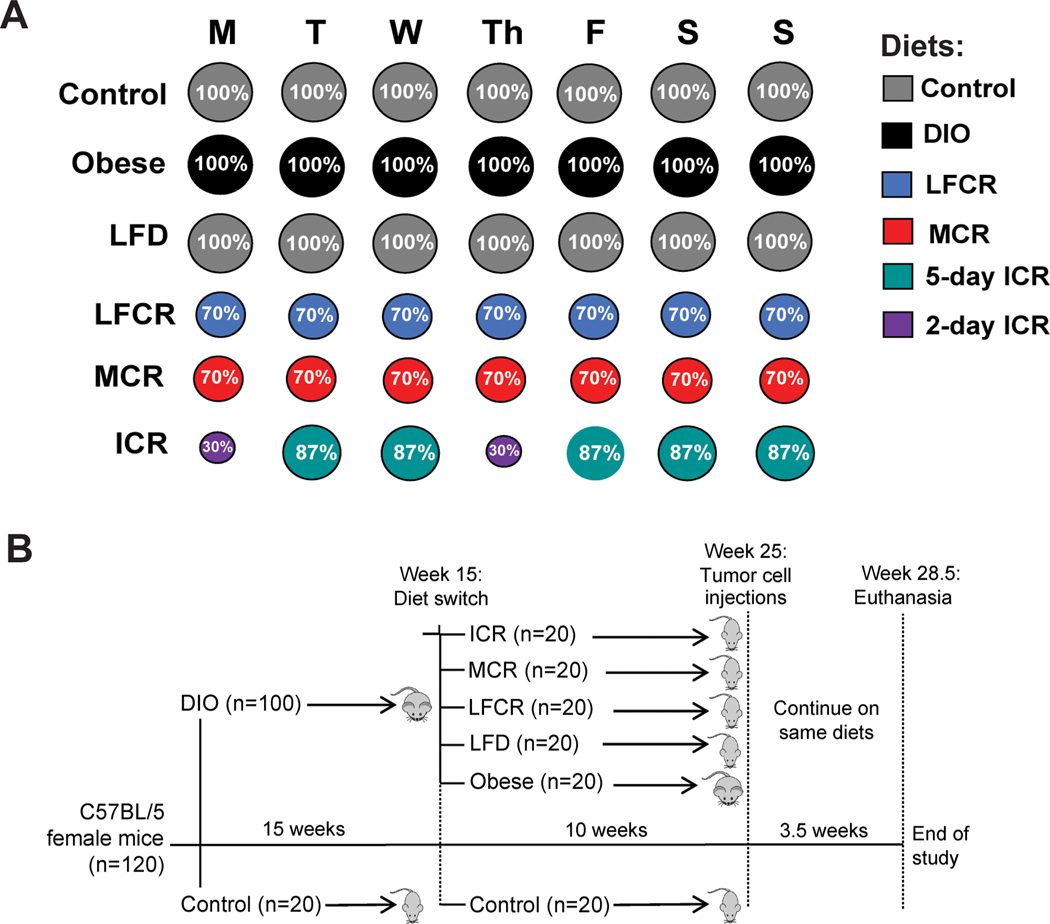
Animal studies and procedures were approved and monitored by the University of North Carolina Institutional Animal Care and Use Committee. Female, 6–8 week-old C57BL/6 mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA, USA). Mice were housed 2/cage and randomized (1:5) to receive either a low-fat (control; 10 kcal% fat) or diet-induced obesity (DIO; 60 kcal% fat) regimen, both fed ad libitum. All diets were from Research Diets, Inc., New Brunswick, NJ, USA. Body weight and food intake were measured weekly. After 15 weeks, mice receiving the control diet continued on that regimen ad libitum (control, n=20), while mice receiving the DIO regimen were randomized (n=20/group) to either remain on DIO diet ad libitum (obese) or switch to 1 of 4 weight loss regimens: 1) nonrestricted LFD; 2) low-fat, 30% calorie-restricted (LFCR); 3) Mediterranean-style, 30% calorie-restricted (MCR); or 4) intermittent calorie-restricted (ICR).
The amount of food for each CR diet provided 70% of the average kcal consumed by the control mice the previous week and was administered as daily aliquots to the CR mice. The ICR regimen consisted of a 13% CR diet for 5 days/week and a 70% CR, high-protein diet on 2 non-consecutive days/week. The MCR and ICR diet composition and schedule were modeled after the “daily energy restriction” and “intermittent energy and carbohydrate restriction” diets, respectively, utilized by Harvie and colleagues in randomized controlled trials (20,21). The intermittent restriction regimen was more readily adopted by the women in their trials than the more severe chronic CR regimen, which is challenging for many people to maintain (20,21). Figure 1A illustrates the schedule for each diet regimen following the week 15 diet switch, and Table 1 lists the nutrient profiles of all diets. Mice in the CR groups continued to be housed 2 per cage, and a cage divider was used to separate cage mates during food consumption. The mice remained on these diets through the end of study.
Figure 1.
Diet regimens and study design. (A) Following the week 15 diet switch, the control mice remained on the same low-fat control diet, and the diet-induced obesity (DIO) mice were randomized to either remain on the DIO diet (obese) or change to 1 of 4 weight loss regimens: non-restricted low-fat diet (LFD), low-fat calorie restricted (LFCR), Mediterranean-style calorie restricted (MCR), or intermittent calorie-restricted (ICR). The control, obese, and LFD mice were fed ad libitum, with the LFD group receiving the same low-fat diet as the control group. The LFCR and MCR groups were chronically 30% calorie restricted, receiving a daily food pellet of their respective diets equal to 70% of the previous week’s average daily control mouse kcal intake. On Mondays and Thursdays, the ICR mice received the high protein 2-day ICR diet in an amount equal to 30% of the previous week’s average daily control mouse kcal intake. On the other days of the week, the ICR mice received the 5-day ICR diet in an amount equal to 87% of the average daily control mouse kcal intake. See Table 1 for the macronutrient and fatty acid content of each diet. (B) At study initiation, the mice were randomized to 2 diets: DIO or control. After 15 weeks, the DIO mice were further randomized to remain on DIO (Obese) or switch to 1 of 4 weight loss diets: non-restricted LFD, LFCR, MCR, and ICR. The mice remained on these diets for 10 weeks and were then orthotopically injected with 3.5×104 E0771 mammary tumor cells. During the 3.5-week period between tumor cell injections and euthanization, the mice continued on the same diets.
Table 1.
Nutrient information on all diets
| ICR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Control | DIO | LFD | LFCR | MCR | 5-day | 2-day | Average | ||
| Kcal density of diet (kcal/g) | 3.85 | 5.21 | 3.85 | 3.78 | 4.13 | 4.31 | 3.36 | NA | |
|
| |||||||||
|
| |||||||||
| Macronutrient distribution (% of total kcal) | Carbohydrate | 70 | 20 | 70 | 57 | 41 | 44 | 27 | 39 |
| Protein | 20 | 20 | 20 | 29 | 29 | 25 | 50 | 32 | |
| Fat | 10 | 60 | 10 | 14 | 30 | 31 | 23 | 29 | |
|
| |||||||||
|
| |||||||||
| Average total daily kcal † | 8.78 | 10.5 | 8.26 | 6.10 | 6.10 | 7.50 | 2.61 | 6.10 | |
|
| |||||||||
|
| |||||||||
| Macronutrient kcal ‡ | Carbohydrate | 6.15 | 2.10 | 5.78 | 3.48 | 2.50 | 3.30 | 0.70 | 2.38 |
| Protein | 1.76 | 2.10 | 1.65 | 1.77 | 1.77 | 1.88 | 1.30 | 1.95 | |
| Fat | 0.88 | 6.30 | 0.83 | 0.85 | 1.83 | 2.33 | 0.60 | 1.77 | |
|
| |||||||||
|
| |||||||||
| Fatty acid distribution (% of total kcal) | SFA | 2.0 | 19.2 | 2.0 | 3.29 | 6.93 | 7.10 | 5.31 | 6.59 |
| MUFA | 3.0 | 21.0 | 3.0 | 4.16 | 15.0 | 15.6 | 11.5 | 14.4 | |
| PUFA | 5.0 | 19.8 | 5.0 | 6.55 | 8.04 | 8.34 | 6.21 | 7.73 | |
| n-3 FA | 0.46 | 1.0 | 0.46 | 0.69 | 0.60 | 0.62 | 0.46 | 0.57 | |
|
| |||||||||
|
| |||||||||
| Fatty acid kcal ¥ | SFA | 0.18 | 2.02 | 0.17 | 0.20 | 0.42 | 0.53 | 0.14 | 0.42 |
| MUFA | 0.26 | 2.21 | 0.25 | 0.25 | 0.92 | 1.17 | 0.24 | 0.90 | |
| PUFA | 0.44 | 2.08 | 0.41 | 0.40 | 0.49 | 0.63 | 0.16 | 0.50 | |
| n-3 FA | 0.04 | 0.11 | 0.04 | 0.04 | 0.04 | 0.05 | 0.01 | 0.04 | |
Average total kcal/mouse/day consumed
average kcal/mouse/day consumed from each macronutrient
average kcal/mouse/day consumed from different categories of fatty acid, all calculated from week 15 through the end of study. The Control and DIO mice were maintained on the above diets throughout the study. LFD, LFCR, MCR, and ICR mice were switched to the above diets after 15 weeks on the DIO diet, then remained on those diets through the end of study. LFCR and MCR mice received daily pellets equivalent to 70% of the average kcal/day consumed by the control mice in the previous week. ICR mice on average received the same amount of kcal/day as the LFCR and MCR mice. Abbreviations: DIO, diet-induced obesity; ICR, intermittent calorie restricted; kcal, kilocalorie; LFCR, low-fat calorie restricted; MCR, Mediterranean calorie restricted; MUFA, monounsaturated fatty acids; n-3 FA, omega-3 fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids.
At 25 weeks after their initial randomization, mice were orthotopically injected in the 4th mammary fat pad with 3.5×104 E0771 mammary tumor cells. Figure 1B illustrates the study design. In vivo tumor growth was monitored twice weekly with digital calipers. Body composition was assessed by quantitative magnetic resonance imaging (Echo Medical Systems, Houston, TX) in a random sample of 6 mice/group immediately before euthanization. All mice were euthanized when a tumor in 1 mouse from any group reached 1.5 cm in diameter. Tumors and nontumor-bearing 9th mammary glands were excised, weighed and divided into equal parts, with 1 half flash frozen in liquid nitrogen and stored at −80°C and the other half formalin fixed and paraffin embedded (FFPE). Lungs were excised, examined for evidence of macrometastases (none observed), and underwent FFPE prior to histological examination.
Serum analytes
One week before tumor injection, blood was collected from all mice by submandibular bleed after fasting 4–6 hours, and serum was processed and stored. In 5–8 mice/group, serum concentrations of a panel of metabolic hormones and growth factors was measured using a BioPlex Pro Mouse Diabetes Panel (BioRad, Hercules CA). Adiponectin, insulin-like growth factor (IGF)-1 and transforming growth factor beta 1 (TGF-β1) were measured with BioRad singleplex assays, while a panel of cytokines was measured using a BioPLex Pro Mouse Cytokine Panel (Bio-Rad). A BioRad MAGPIX multiplex analyzer was used to read all assays, as previously described (17).
Lung metastases
A veterinary pathologist, blinded to sample identities, examined a random sample of FFPE lung tissue from 6 mice/group for micrometastases, using 10 hematoxylin- and eosin-stained sections/FFPE lung tissue sample (100 μm apart). Lung metastasis burden was defined as the percent of positive sections per mouse.
Quantitative RT-PCR
Total RNA was isolated from flash-frozen tumor tissue and reverse transcribed as previously described (22). Tumor expression of EMT-related genes was assessed in 3–5 mice/group using a Mouse EMT RT2 Profiler PCR Array (Qiagen, Germantown, MD, USA), and read on a ViiA™7 RT-PCR System (Applied Biosystems, Waltham, MA, USA).
RNA sequencing analysis
Total RNA and genomic DNA were isolated from randomly selected flash-frozen distal mammary fat pad samples (n=3–5 mice/group) using TRIzol® Reagent (Sigma-Aldrich, St. Louis, MO, USA) according to manufacturer’s instructions. RNA quality was assessed by an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). RNA libraries were prepared for sequencing using the Illumina TruSeq Stranded Total RNA Library Preparation Kit and then sequenced using a 2×76 bases paired-end protocol on an Illumina HiSeq 2000 (Illumina Inc., San Diego, CA, USA). The reads were mapped to mouse genome (mm10) by TopHat (version 2.0.7). The number of fragments in each known gene from RefSeq database (UCSC Genome Browser 2013) was enumerated using HTSeq-count from HTSeq package (version 0.5.3p9). Differential expression was performed using DESeq2. Genes with an FDR-adjusted two-tailed P-value of <0.05 were considered differentially expressed. Differentially expressed genes (DEG) were assessed by principle components analysis (PCA) and entered into Ingenuity Pathway Analysis (IPA, Qiagen, Germantown, MD, USA) to identify significantly modulated canonical pathways, cellular functions, and predicted upstream regulators (P<0.05). The top functions were ranked by absolute z-score value (among those with P<0.05), while canonical pathways and upstream regulators were ranked by P-value.
DNA methylation analysis
Genome-wide methylation profiles in mammary tissue were determined by reduced representation bisulfite sequencing (RRBS). Genomic DNA (1 mg) was digested with MspI, and enrichment for CpG sites in CG-dense and CpG islands was achieved by selective collection and PCR amplification of MspI fragments with sizes of 40–170 bp. Sequence reads were mapped to the mouse genome (mm10) using Bismark Bisulfite Read Mapper. RNA and DNA library preparation and sequencing were performed at the UNC High-Throughput Sequencing Facility. An integrated analysis of the RNA-Seq and RRBS data was performed via Joint and Individual Variation Explained (JIVE) to identify genes displaying alterations in both DNA methylation and gene expression. Differentially methylated regions (DMR) were defined for each pairwise comparison as genomic sites with an average absolute value change in methylation of ≥15% and an FDR-adjusted two-tailed P-value of <0.05.
Gene Set Enrichment Analysis (GSEA)
To compare the gene expression profiles of normal mammary tissue from all weight loss groups, relative to the obese mice, with the transcriptomic changes that occur with human weight loss, we performed GSEA with a microarray dataset (provided by our collaborator Dr. Michelle Harvie) of normal breast tissue samples from premenopausal overweight or obese women who either continued their normal eating habits or underwent dietary energy restriction (864 kcal/day) for 1 menstrual cycle (GEO: GSE66159) (23,24). For this comparison, we generated several gene sets from our RNA sequencing data. First, for each pairwise comparison with the obese group, we identified genes that were significantly differentially expressed (adjusted p-value < 0.05) and subsequently selected the top 100 upregulated genes in each group compared with the obese group. Second, we identified all DEG (adjusted p-value < 0.05) from a comparison of all CR mice combined versus the obese group, excluding any genes that were also differentially expressed in the nonrestricted LFD versus obese mice comparison. Third, we identified DEG (adjusted p-value < 0.05) from a comparison of nonrestricted LFD mice versus obese mice, excluding any genes that were also differentially expressed in the all-CR versus obese mice comparison. Finally, mouse gene symbols were converted to the orthologous human gene symbol using Ensembl BioMart in order to align with the human microarray data (25). For GSEA analysis, the microarray expression data for the participants in the energy restriction and control groups was tested for enrichment of the gene sets described above that were generated from our mouse RNA sequencing data.
Statistical analysis
Animal study data are presented as mean ± SD. Differences between the experimental groups (excluding analyses of the RNA sequencing and RRBS data, as described in those sections) were analyzed by one-way ANOVA, followed by Tukey’s post hoc test, using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). P<0.05 was considered significantly different.
Data Availability
The RNA sequencing and RRBS data generated in this study are publicly available in the GEO repository (GSE202332).
RESULTS
CR promotes greater body-weight and fat losses than nonrestricted LFD in obese mice
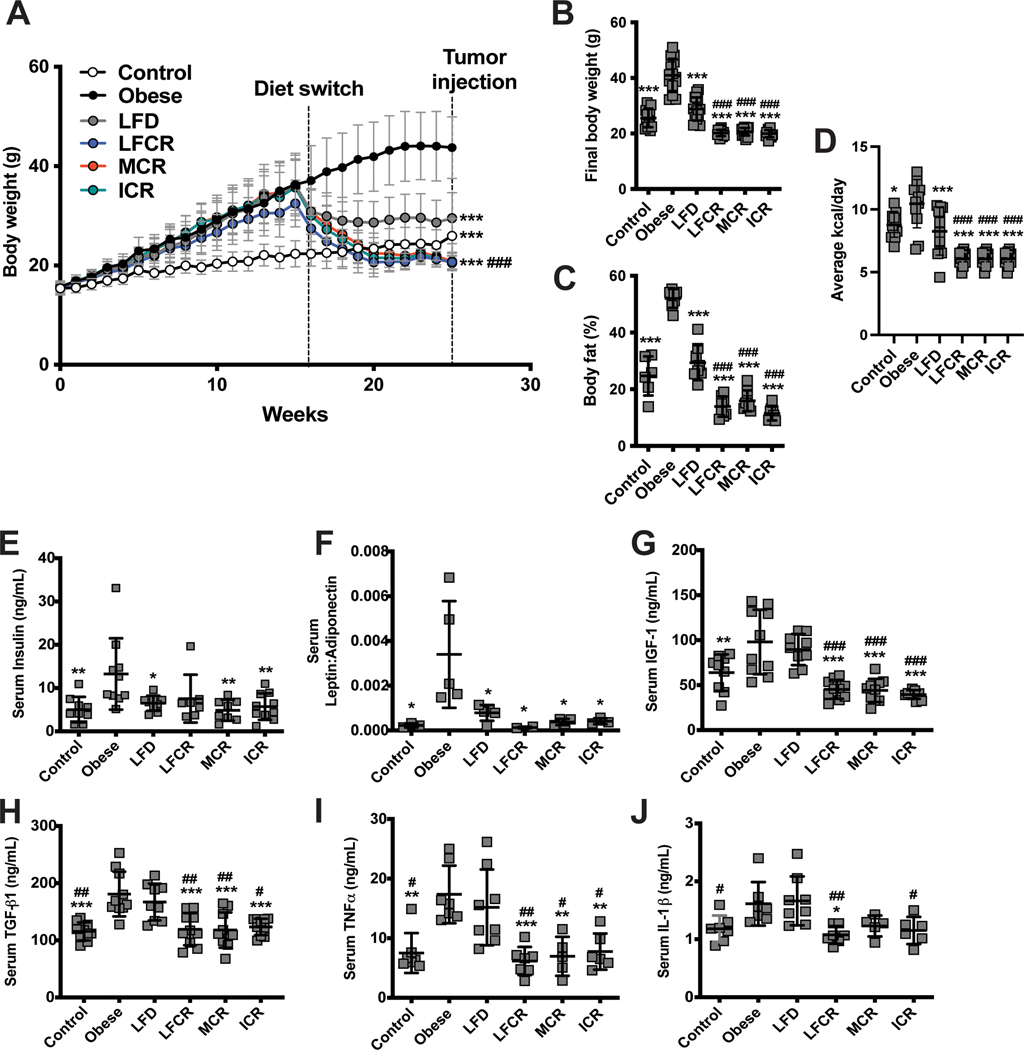
At the time of tumor cell injection, body weights were greater in the obese group than all other groups, were lesser in the 3 CR groups than the nonrestricted LFD group, and did not differ between the control and nonrestricted LFD groups (Figure 2A). At study endpoint, body weight and body fat percent were lower in the CR groups than all other groups, and lower in the nonrestricted LFD group than obese group (Figure 2B-C). The nutrient profiles of all diets are listed in Table 1. From week 15 through the study endpoint, the obese mice consumed a greater average kcal/day than all other groups, while the 3 CR groups consumed a lower average kcal/day than the control and LFD groups (Figure 2D).
Figure 2.
Calorie restriction promotes weight loss and reductions in adipokines in obese mice. (A) Weekly body weights were recorded for the control, obese, nonrestricted low-fat diet (LFD), low-fat calorie-restricted (LFCR), Mediterranean-style calorie-restricted (MCR), and intermittent calorie-restricted (ICR) mice. Final body weights (B) and body fat percentage (C) were measured just prior to euthanasia for all mice and 6 mice/group, respectively. (D) The average kcal/mouse/day consumed by each diet group was calculated for the time period between the end of week 15, when the diet switch occurred, and the end of study. (E) Insulin levels, (F) the leptin-adiponectin ratio, (G) insulin-like growth factor (IGF)-1 levels, (H) transforming growth factor beta 1 (TGF-β1) levels, (I) tumor necrosis factor alpha (TNFα) levels, and (J) interleukin 1 beta (IL-1β) levels were all measured in sera collected 1 week prior to tumor cell injections from 5–8 mice/group. *P<0.05, **P<0.01, ***P<0.001, all relative to the obese mice. #P<0.05, ##P<0.01, ###P<0.001, all relative to the nonrestricted LFD mice.
CR, but not nonrestricted LFD, resolves obesity-induced elevations in IGF-1 and proinflammatory cytokines
Table S1 lists the concentrations of all serum analytes measured. Serum insulin levels were higher in the obese group versus all groups except LFCR, and the serum leptin:adiponectin ratio was elevated in the obese group versus all other groups (Figure 2E-F). Serum IGF-1, TGF-β1, and TNFα concentrations were elevated in obese and nonrestricted LFD groups versus the CR groups, and they did not differ between obese and nonrestricted LFD mice (Figure 2G-I). Serum IL-1β levels were greater in nonrestricted LFD mice versus control, LFCR, and ICR mice and in obese versus LFCR mice (Figure 2J).
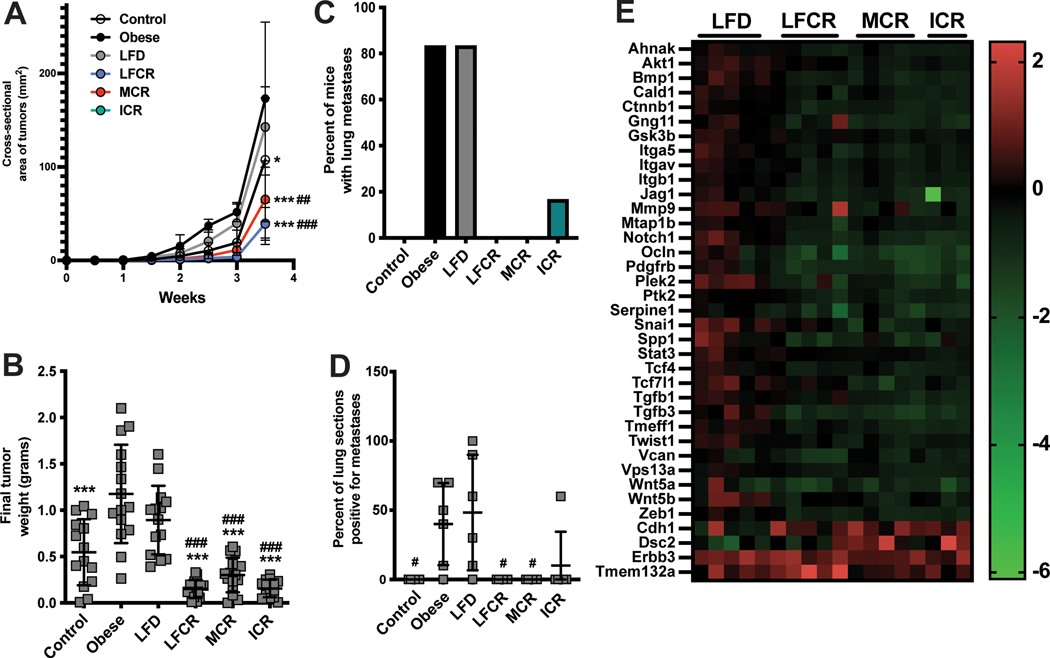
CR, but not nonrestricted LFD, reverses DIO effects on mammary tumor progression and metastasis
Figure 3A illustrates mammary tumor growth over time, demonstrating that the tumors in the obese and nonrestricted LFD mice had the steepest growth curves, particularly relative to the three CR groups. At the final palpation, all CR groups had significantly smaller tumors than the obese and LFD mice. Ex vivo tumor weight was greater in the obese group relative to all groups except nonrestricted LFD. Tumors were also smaller in the CR groups than nonrestricted LFD group (Figure 3B). Lung micrometastases occurred in 83% of mice in both the obese and nonrestricted LFD groups, 17% in the ICR group, and 0% in the other groups (Figure 3C). The nonrestricted LFD group also had greater lung metastasis burden, relative to the control, LFCR, and MCR groups. Metastasis burden was quantified as the percentage of the lung sections assessed that were positive for ≥1 micrometastatic lesion (Figure 3D).
Figure 3.
The protumor effects of obesity are reversed by weight loss via calorie restriction, but not a nonrestricted low-fat diet. (A) Tumor cross-sectional area (mm2) was calculated following biweekly palpations and is shown here over time for all control, obese, nonrestricted low-fat diet (LFD), low-fat calorie-restricted (LFCR), Mediterranean-style calorie-restricted (MCR), and intermittent calorie-restricted (ICR) mice. The ICR group is not visible on the graph because its growth curve is almost identical to the LFCR growth curve. (B) Tumor weight was measured ex vivo at the study endpoint in all mice. (C) Incidence of lung metastasis in each diet group was determined by examining the lungs of 6 mice/group for micrometastases. (D) Lung metastasis burden for each mouse was calculated as the percentage of lung sections positive for at least 1 micrometastasis, 6 mice/group assessed. (E) Tumor expression of genes related to the epithelial-to-mesenchymal transition were measured by quantitative RT-PCR array in all groups except control (n=3–5 mice/group). Expression levels in all 4 weight loss groups, relative to the obese group, of the 37 genes that differed between all CR mice combined and the obese group, the nonrestricted LFD group, or both (P<0.05) are shown. *P<0.05, ***P<0.001 relative to the obese mice. #P<0.05, ##P<0.01, ###P<0.001, relative to the nonrestricted LFD mice.
CR modifies tumoral expression of EMT-related genes
Tumoral expression of 37 genes in a panel of 84 EMT-related genes differed for all CR mice combined relative to the obese group, the nonrestricted LFD group, or both (P<0.05). While only 2 genes, Plek2 and Snai1, differed between the obese and nonrestricted LFD groups, these 2 groups both differed from the LFCR, MCR, and ICR groups in expression of 11, 15, and 18 genes, respectively (Figure 3E).
CR modifies normal mammary tissue expression of genes related to reactive oxygen species (ROS) production
To explore whether differences in normal mammary tissue signaling contribute to the differences in tumor progression, we performed RNA sequencing on nontumor-bearing mammary fat pad tissue from a subset of mice in each group. PCA plots of the sequencing data demonstrated within-group similarity in global gene expression among the obese and MCR mice. There was greater dissimilarity among the remaining groups (Figure S1).
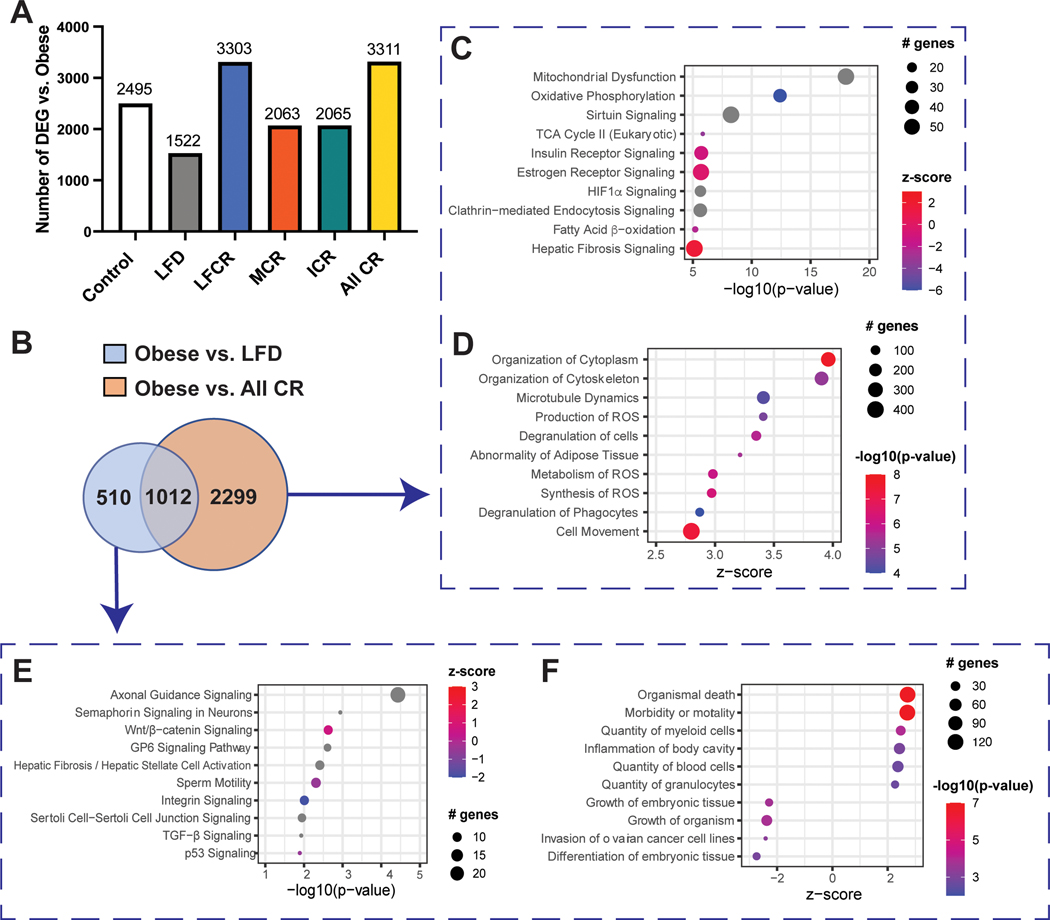
We then performed pairwise comparisons between the obese group and each other group, as well as all CR mice combined, to identify DEG. Compared with the obese group, the all-CR group had the greatest number of DEG (3311) and the nonrestricted LFD group had the lowest number (1522) (Figure 4A).
Figure 4.
Calorie restriction modulates gene expression in normal mammary tissue. (A) Differentially expressed genes (DEG, FDR adjusted P<0.05) were identified by RNAseq for obese mice versus every other group (control, nonrestricted low-fat diet (LFD), low-fat calorie-restricted (LFCR), Mediterranean-style calorie-restricted (MCR), and intermittent calorie-restricted (ICR) mice, n=3–5 mice/group) and all calorie-restricted (CR) mice combined. (B) Overlapping and non-overlapping DEG for the obese versus nonrestricted LFD and obese versus all-CR comparisons were determined. (C) The top ten Ingenuity Pathway Analysis (IPA) canonical pathways (ranked by -log10(p-value)) and (D) the top ten significant IPA functions (ranked by z-score) associated with obese versus all-CR DEG, excluding obese versus nonrestricted LFD DEG, are shown. (E) The top ten IPA canonical pathways (ranked by -log10(p-value)) and (F) the top ten significant IPA functions (ranked by z-score) associated with the obese versus nonrestricted LFD DEG, excluding obese versus all-CR DEG, are shown.
For the obese versus all-CR comparison, 2299 of 3311 (69%) DEG were not also differentially expressed between the obese and nonrestricted LFD mice (Figure 4B), while IPA analysis of these DEG indicated an enrichment in several canonical pathways related to metabolism, including “Mitochondrial Dysfunction” and “Sirtuin Signaling”. Most of these pathways had no z-score or higher activity in the CR mice (Figure 4C). We also found strong evidence of increased ROS production and metabolism in the mammary tissue of the obese mice, relative to all-CR mice (Figure 4D). Furthermore, the most significant IPA-predicted upstream regulator for this DEG set was TP53, which also had greater activity in the obese mice (Figure S2A) and is linked with ROS production (26). There was substantial overlap in the DEG linked to TP53, the “Mitochondrial Dysfunction” and “Sirtuin Signaling” pathways, and the “Synthesis of ROS” function (Figure S2B).
For the obese versus nonrestricted LFD comparison, 510 of 1522 DEG (34%) did not overlap with the obese versus all-CR comparison. IPA analysis of these DEG indicated an enrichment in the “p53 Signaling” pathway, with greater activity in the nonrestricted LFD than obese mice; no other metabolism-related pathways were among the top 10 pathways (Figure 4E). Functional analysis suggested increased immune activity in the obese mouse mammary tissue, with 3 of the top 10 enriched functions related to immune response (Figure 4F). There was no enrichment in any ROS-related functions, at any level of significance. TNF was among the 10 most significant IPA-predicted upstream regulators, but its negative z-score indicated greater activity in the nonrestricted LFD versus obese mice (Figure S2C).
Changes in mammary tissue DNA methylation are diet-dependent
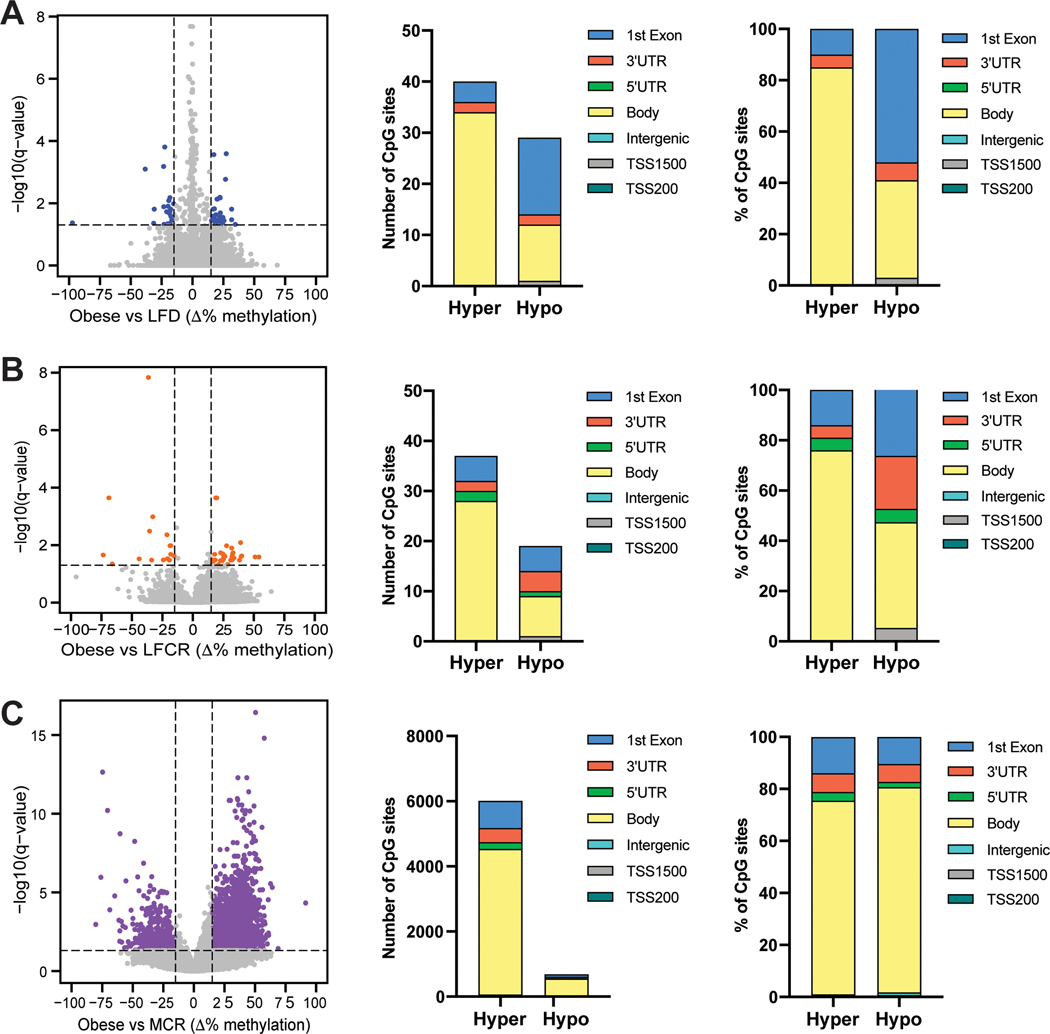
To assess whether reversal of obesity-induced epigenetic reprogramming mediates CR-induced changes in gene expression, we characterized global DNA methylation levels in the same normal mammary tissue samples that underwent RNA sequencing. Pairwise comparisons between the obese mice and each other group revealed 6693 DMR versus the MCR group and <100 DMR versus the remaining groups (Figure S3A), including no DMR versus the ICR group (Figure S3B). A graph of all methylation differences detected for the obese versus nonrestricted LFD comparison illustrates that only 69 of these differences met our DMR definition, and gene body hypermethylation predominated in the obese mice (Figure 5A). Among the 56 DMR for the obese versus LFCR comparison and the 6693 obese versus MCR DMR, the majority were again hypermethylated and within the gene body (Figure 5B-C). Overall, a pattern of DNA hypermethylation was observed in the mammary tissue of the obese mice, relative to all the weight loss groups, with the possible exception of ICR.
Figure 5.
Obesity reversal decreases global DNA methylation levels in normal mammary tissue. Pairwise comparisons of DNA methylation levels between (A) the obese and nonrestricted low-fat diet (LFD) groups, (B) the obese and low-fat calorie restricted (LFCR) groups, and (C) the obese and Mediterranean-style calorie restricted (MCR) groups are shown, expressed as the mean change in percent methylation and with a differentially methylated region (DMR) defined as a genomic site with an average absolute methylation value change of ≥15% and an FDR adjusted P<0.05 (n=3–5 mice/group). Locations of the DMR for each comparison, expressed as the number of DMR and percentage of DMR in each location, are also shown.
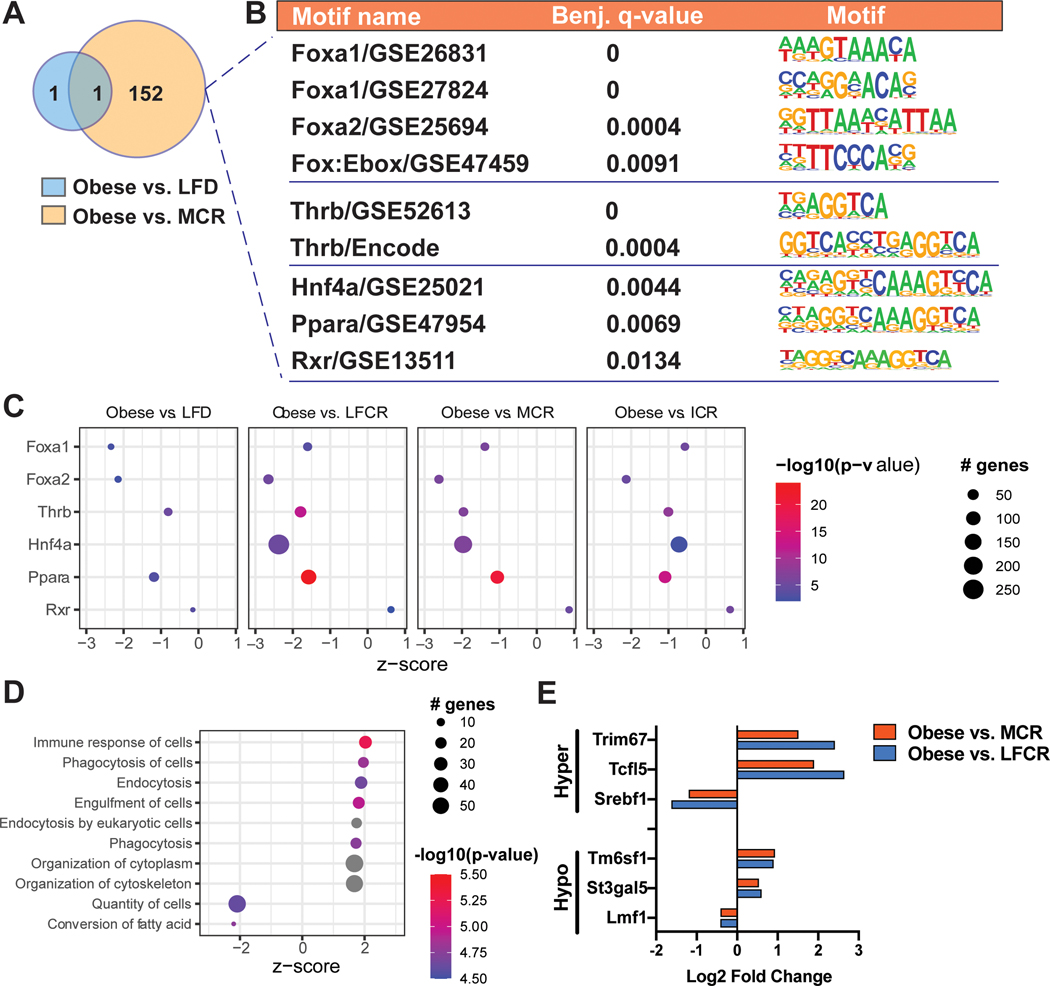
MCR decreases hypermethylation in binding motifs for obesity-linked transcription factors
We then focused on defining the functional relevance of these DMR, first utilizing Hypergeometric Optimization of Motif EnRichment (HOMER) analysis, which enabled the identification of enrichment in specific transcription factor binding motifs at the DMR. A single motif was exclusively enriched within the obese versus nonrestricted LFD DMR. In contrast, 152 motifs were exclusively enriched for the obese versus MCR comparison (Figure 6A; Table S2). Six of these motifs corresponded to transcription factors (TF) linked to energy balance, specifically forkhead (Fox)a1 and Foxa2, thyroid hormone receptor beta (Thrb), hepatocyte nuclear factor 4 alpha (Hnf4a), peroxisome proliferator-activated receptor alpha (Ppara), and retinoid X receptor (Rxr) (Figure 6B). At the DMR linked to these 6 motifs, there was consistent hypermethylation in the obese versus MCR mice (Figure S4). All 6 differentially methylated TFs were predicted upstream regulators for this comparison, with z-scores indicating greater activity in the MCR mice. Almost all of the TFs were also predicted upstream regulators for the obese versus nonrestricted LFD, LFCR, and ICR comparisons. The obese versus individual CR group comparisons had more overlapping genes and higher significance for each TF than the obese versus nonrestricted LFD comparison (Figure 6C).
Figure 6.
Methylation of obesity-linked transcription factor binding sites is reduced by Mediterranean-style calorie restricted (MCR) diet. (A-B) Hypergeometric Optimization of Motif EnRichment (HOMER) analysis was used to identify enrichment in transcription factor binding motifs at the differentially methylated regions (DMR) for the obese versus MCR and obese versus nonrestricted low-fat diet (LFD) comparisons. (C) Ingenuity Pathway Analysis (IPA) prediction of upstream regulators was conducted for obese versus nonrestricted LFD, obese versus nonrestricted low-fat calorie restricted (LFCR), obese versus MCR, and obese versus intermittent calorie restricted (ICR) comparisons. (D) The top ten significant IPA functions (ranked by z-score) associated with differentially expressed genes (DEG) that contained ≥1 DMR (DEG+DMR) for the obese versus MCR comparison are shown. (E) Relative expression levels of the overlapping DEG+DMR for the obese versus LFCR and obese versus MCR comparisons are shown.
We next examined the 270 DEG that also contained ≥1 DMR (DEG+DMR) for the obese versus MCR comparison, thereby assessing only DMR that occurred in parallel with differential gene expression. Functional analysis of these DEG+DMR primarily indicated an enrichment in immune response-related genes in the obese versus MCR mice (Figure 6D; Figure S5A). We determined that 6 DEG+DMR overlapped between the obese versus MCR and obese versus LFCR comparisons. The differential expressions for these DEG+DMR were similar in direction and magnitude and identical in location for both CR groups compared to obese mice (Figure 6E, Table S3). These genes were primarily involved in cell differentiation and lipid metabolism, including a downregulation in sterol regulatory element-binding protein 1 (Srebf1). Srebf1 was also a significant predicted upstream regulator for the obese versus MCR DEG comparison (Figure S5B). Four of these genes, Srebf1, Tripartite motif-containing protein 67 (Trim67), Transcription factor-like 5 protein (Tcfl5), and Transmembrane 6 superfamily member 1 (Tm6sf1), were also differentially expressed, but not differentially methylated, for the obese vs ICR comparison.
CR response in mouse mammary tissue parallels the CR response in breast tissue of women who are overweight
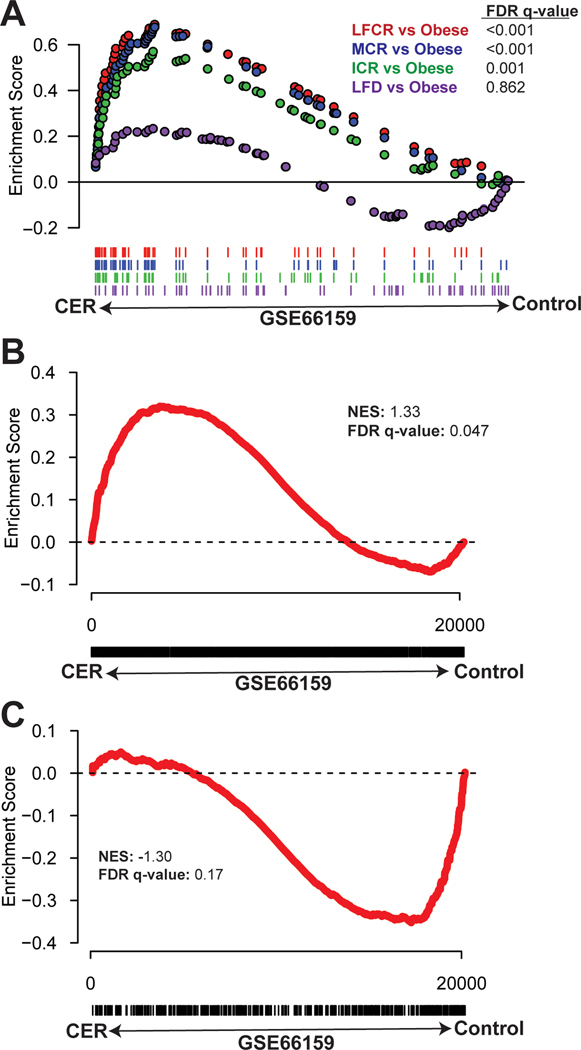
To determine the relevance of the CR-induced transcriptomic changes to human health, we next used GSEA to compare CR-responsive DEG from our data to normal human breast tissue samples following dietary energy restriction or no intervention (23,24). The top 100 upregulated genes from each CR group (but not the nonrestricted LFD group) versus the obese group were enriched in the human dietary energy restriction samples relative to the no intervention samples (Figure 7A). DEG exclusive to the all-CR versus obese comparison were also significantly enriched in the human dietary energy restriction samples relative to control (Figure 7B). In contrast, DEG exclusive to the nonrestricted LFD versus obese comparison were not enriched in either human phenotype (Figure 7C).
Figure 7.
Obesity reversal by calorie restriction promotes changes in the expression of mouse mammary tissue genes that are enriched in the breast tissue of calorie-restricted women. (A) The top 100 significant differentially expressed genes (DEG) from 4 pairwise comparisons (low-fat calorie-restricted (LFCR), Mediterranean-style calorie-restricted (MCR), intermittent calorie restricted (ICR), and nonrestricted low-fat diet (LFD) mice versus obese, n=3–5 mice/group) were assessed by Gene Set Enrichment Analysis (GSEA) for enrichment in a gene set from the breast tissue of women who were overweight/obese and randomized to a chronic energy restriction (CER) regimen versus usual diet (control) (GSE66159). GSEA was also used to assess (B) all DEG from the comparison of all-CR mice versus obese mice, excluding any DEG from the comparison of nonrestricted LFD versus obese mice, and (C) all DEG from the comparison of nonrestricted LFD versus obese mice, excluding any DEG from the all-CR versus obese comparison, for enrichment in the same human dataset.
DISCUSSION
Given the globally high rates of obesity and breast cancer, uncertainty regarding reversibility of the obesity-breast cancer link represents an important knowledge gap. Our study demonstrates, for the first time, that weight loss via 3 distinct CR regimens, but not a nonrestricted LFD, mitigates obesity’s procancer effects in a mouse model of TNBC.
One important limitation of the current study is the use of a single TNBC cell model due to budget constraints. However, the observed elevation in final tumor sizes in obese mice and formerly obese nonrestricted LFD mice, such that there is no statistical difference between the 2 groups, is consistent with our previous findings in different tumor models of breast cancer (18,19). By comparison, Makowski and colleagues demonstrated in the C3(1)-TAg transgenic mouse model of TNBC that switching to a nonrestricted LFD reverses the tumor-promoting effects of a high-fat diet (HFD) (27,28). However, their obesity induction period was substantially shorter than in our study, resulting in minimal weight differences between their control and diet-induced obese mice (27,28). Thus, the observed effects of their diet switch to a nonrestricted LFD was likely due to the change in dietary fat intake rather than obesity reversal. In another preclinical study, obesity-associated lung metastasis from a transplanted MMTV-PyMT mouse-derived mammary tumor cell line was reversed by weight loss via nonrestricted LFD (29). This study used a tail vein injection model of metastasis, which enabled them to focus on the metastatic microenvironment of the lung. In contrast, our study assessed how obesity and weight loss affect lung micrometastases from orthotopically injected mammary tumor cells, and we demonstrated that metastatic capacity in this model is not reduced by weight loss via nonrestricted LFD following chronic obesity, but is decreased by CR-induced weight loss. Consequently, the findings are complementary, together suggesting that the mammary gland microenvironment is an essential mediator of obesity’s lingering pro-metastatic effects after weight loss via LFD.
A number of factors may have contributed to the observed differences in tumor outcomes and metabolic markers between the LFD and CR groups. First, the LFD mice always had nonrestricted access to their food, while the CR mice received a daily food aliquot that they typically consumed within an hour, thereby subjecting them to an extended fasting period. A recent study by Das, et al. (30) suggests that the feeding schedule, and any fasting periods within that schedule, may be at least as important for tumor outcomes as the amount of kcal consumed and/or weight lost. They reported that obese mice fed a HFD on a time-restricted feeding schedule maintained a stable body weight, improved their glucose tolerance, and developed smaller E0771 mammary tumors and fewer lung metastases than obese mice fed a nonrestricted HFD. A second possible contributing factor was that our LFD mice consumed significantly more kcal/day than any of the CR groups during the weight loss period. Third, the LFD mice weighed significantly more than the CR mice, and had greater body fat levels, both at the time of tumor cell injection and at study endpoint.
In our murine model of TNBC, the global mammary gene expression of DIO mice, relative to all-CR mice, indicated extensive metabolic dysfunction, as most of the top differentially enriched pathways were metabolism-related. Functional evaluation also demonstrated enrichment in genes linked to ROS production and oxidative metabolism in obese versus all-CR mice. Typical effects of chronic positive energy balance on adipose tissue physiology include adipocyte hyperplasia and hypertrophy (31), with the latter leading to adipose tissue hypoxia, activating hypoxia-inducible factor (HIF)-1α signaling (32), as observed in our obese versus all-CR comparison. HIF-1α signaling, and hypoxic cell death, stimulates inflammation within adipose tissue, which involves infiltration of multiple proinflammatory immune cell types, including M1 macrophages. These immune cells produce inflammatory cytokines like TNFα, which was elevated in the serum of our obese and nonrestricted LFD mice, relative to the CR mice, and those cytokines activate macrophages, attracting additional immune cells (32,33). Hypoxia and proinflammatory cytokines both induce the production of ROS in the dysfunctional adipose tissue (33).
p53 was the most significant predicted upstream regulator for the obese versus all-CR comparison, with greater activity predicted in obese mice. In contrast, while p53 signaling was a top canonical pathway for the obese versus nonrestricted LFD comparison, greater activity was predicted in LFD mice. p53 is a key transcription factor responsible for sensing and coordinating appropriate cellular responses to metabolic stress. Under oxygen deprivation conditions, p53 directs cells towards nonoxidative energy generation and typically represses ROS synthesis (26). Evidence for the former in the obese mouse mammary tissue, relative to all-CR mice, was clear in the top 10 canonical pathways for this comparison, which included “Oxidative Phosphorylation”, “TCA Cycle”, and “Fatty Acid β-oxidation”. Each pathway was more active in CR mice, indicating inhibition of oxidative metabolism in obese mice. The continued elevation in ROS in obese mice, versus CR, despite high levels of p53 activity, may be due to p53’s failure to fully suppress ROS production. ROS can, in turn, activate Wnt and TGFβ signaling pathways in neighboring cancer cells, increasing their stemness and metastatic potential (33). Many stem- and EMT-related genes were upregulated in the obese and nonrestricted LFD mouse tumors, relative to all-CR mice, including Tgfb1, Tgfb3, and Wnt5a.
Previous studies have reported that energy balance affects DNA methylation in white adipose tissue (WAT). Wahl et al. (34) established robust associations in men and women between body mass index and changes in subcutaneous WAT DNA methylation, with methylation loci concentrated in genes involved in lipid metabolism, substrate transport, and inflammatory pathways. Similar patterns of differential methylation have been found by others in genes related to similar functions, as well as glycemic control (35–37). Significant weight loss following bariatric surgery reverses this obesity-induced epigenetic reprogramming (37,38). We therefore examined whether the differential effects observed following CR-induced versus nonrestricted LFD-induced weight loss on obesity-associated mammary tumor growth and lung metastasis may be mediated by epigenetic reprogramming in the normal mammary gland. Overall, our findings indicated that obesity is associated with mammary DNA hypermethylation, which has been linked to proinflammatory pathways (39). Surprisingly, among the pairwise comparisons, there were exponentially more DMR for the obese versus MCR group comparison, relative to all other comparisons not involving the MCR group. Preliminary findings from our recently completed study comparing DNA methylation, inflammatory markers, and tumor progression in obese versus formerly obese mice achieved by bariatric surgery (sleeve gastrectomy), using the same TNBC model, suggest surgical weight loss may exert similar anti-inflammatory and methylation effects, and comparable reductions in tumor growth, as observed here with MCR.
Hypermethylation at several transcription factor (TF) binding motifs was noteworthy in the mammary tissue from obese versus MCR mice. These TFs, which regulate lipid metabolism, adipocyte differentiation, and/or inflammation (40–44), were all predicted upstream transcriptional regulators for the comparison, with greater activation of each predicted in MCR mice. We also identified any genes that were both differentially expressed and contained a DMR at identical sites for the obese versus LFCR and MCR comparisons. Six genes, each involved in cell differentiation and lipid metabolism, met these criteria, including Srebf1, which is implicated in oxidative stress-induced inhibition of healthy WAT expansion (45). However, Srebf1 and 3 of the other genes were also differentially expressed, but not differentially methylated, in obese versus ICR mice. Consequently, our findings suggest that the anticancer effects of the CR interventions are likely independent of their ability to reverse epigenetic reprogramming.
Comparisons between the 3 CR diets found no significant differences in their ability to reverse obesity’s effects on tumor growth, likely due to their similar impact, relative to DIO or LFD, on p53-related metabolic, ROS-regulating, and oncogenic signaling pathways. These findings on tumor growth are consistent with previous animal studies that indicated that an ICR diet might be superior to a nonrestricted diet, and comparable to a chronic CR diet, in reducing mammary tumor growth (46–50). However, the design of these studies differs from ours in several ways, including an ICR regimen that rotated between equal time periods of nonrestricted and 50% CR feeding. Moreover, the CR diets in these studies were initiated at a young age without a prior obesity induction period, and metastatic progression was not assessed.
Several clinical trials have assessed the effects of weight loss via chronic CR versus ICR (the latter with a feeding schedule similar to our study design) on cancer-related biomarkers. Two studies from Harvie et al. (20,21) indicated that weight loss via an ICR diet reduces insulin resistance to a greater degree than a chronic CR regimen, but found no difference in their effects on inflammatory markers. Harvie et al. (24) subsequently compared breast tissue gene expression in premenopausal women before and after they followed an ICR diet for 1 menstrual cycle. This data was also compared to breast biopsy data from a previous chronic CR study (23). They found that 11 of 20 ICR subjects had significant changes in genes associated with metabolic pathways, and these changes were similar to the differences observed in the chronic CR study. The authors reported that, in general, the ICR diet induced more subtle and variable changes in breast gene expression than the chronic CR regimen (24). This description fits our findings regarding the effects of chronic versus intermittent CR on mammary gland gene expression, as more variability in gene expression occurred among the ICR mice than within the LFCR and MCR groups. We demonstrated that the gene sets from our 3 CR versus obese group comparisons, but not the nonrestricted LFD versus obese comparison, were all enriched in the human chronic CR gene set from the Harvie study (24), providing validation for the mouse to human translatability of our findings.
In sum, we established that weight loss via various forms of CR, including low-fat, Mediterranean-style, and intermittent CR regimens, but not a nonrestricted LFD, reverses obesity’s effects on mammary tumor growth and metastasis to the lung in a mouse model of TNBC. The persistence in tumor growth and metastasis observed in the formerly obese mice that were switched to nonrestricted LFD was consistent with our previous work using different models of TNBC (18,19) and was associated with unresolved obesity-associated abnormalities in mammary tissue metabolism and ROS production. In contrast, each of the CR regimens resolved these metabolic and oxidative perturbations. There was no clear evidence that sustained epigenetic reprogramming was involved. Given that substantial weight loss and maintenance is difficult for most people, further translational exploration of efficacious and sustainable restricted diet regimens are warranted, with the ultimate goal of developing mechanism-based, nonsurgical interventions to reduce the burden of obesity-related breast cancer.
Supplementary Material
PREVENTION RELEVANCE STATEMENT.
Obesity is an established risk and progression factor for triple negative breast cancer (TNBC). Given rising global rates of obesity and TNBC, strategies to reduce the burden of obesity-driven TNBC are urgently needed. We report the genomic, epigenetic and procancer effects of obesity are reversible by various calorie restriction regimens.
ACKNOWLEDGEMENTS
We would like to acknowledge the contributions of Andrew H. Sims, who passed away in 2021, to this project. His assistance with data analysis was much appreciated.
Financial support:
This study was supported by a grant from the Breast Cancer Research Foundation (S.D. Hursting) and R35 CA197627 (S.D. Hursting). L.W. Bowers and E.L. Rossi were supported by a grant from the National Cancer Institute (R25CA057726).
Footnotes
Conflicts of interest: The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Hales CM, Carroll MD, Fryar CD, Ogden CL. Prevalence of Obesity Among Adults and Youth: United States, 2015–2016. NCHS Data Brief. 2017;1–8. [PubMed] [Google Scholar]
- 2.Protani M, Coory M, Martin JH. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res Treat. 2010;123:627–35. [DOI] [PubMed] [Google Scholar]
- 3.Caan BJ, Kwan ML, Hartzell G, Castillo A, Slattery ML, Sternfeld B, et al. Pre-diagnosis body mass index, post-diagnosis weight change, and prognosis among women with early stage breast cancer. Cancer Causes Control. 2008;19:1319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols HB, Trentham-Dietz A, Egan KM, Titus-Ernstoff L, Holmes MD, Bersch AJ, et al. Body mass index before and after breast cancer diagnosis: associations with all-cause, breast cancer, and cardiovascular disease mortality. Cancer Epidemiol Biomarkers Prev. 2009;18:1403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Lu W, Zheng W, Gu K, Chen Z, Zheng Y, et al. Obesity and weight change in relation to breast cancer survival. Breast Cancer Res Treat. 2010;122:823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw PT, Ibrahim JG, Stevens J, Cleveland R, Abrahamson PE, Satia JA, et al. Postdiagnosis change in bodyweight and survival after breast cancer diagnosis. Epidemiology. 2012;23:320–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caan BJ, Kwan ML, Shu XO, Pierce JP, Patterson RE, Nechuta SJ, et al. Weight change and survival after breast cancer in the after breast cancer pooling project. Cancer Epidemiol Biomarkers Prev. 2012;21:1260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao P-P, Cai H, Peng P, Gu K, Su Y, Shu X-O, et al. Body mass index and weight change in relation to triple-negative breast cancer survival. Cancer Causes Control. 2016;27:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cespedes Feliciano EM, Kroenke CH, Bradshaw PT, Chen WY, Prado CM, Weltzien EK, et al. Postdiagnosis Weight Change and Survival Following a Diagnosis of Early-Stage Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang L, Hattori M, Fleming G, Jaskowiak N, Hedeker D, Olopade OI, et al. Impact of post-diagnosis weight change on survival outcomes in Black and White breast cancer patients. Breast Cancer Res. 2021;23:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demark-Wahnefried W, Rogers LQ, Gibson JT, Harada S, Frugé AD, Oster RA, et al. Randomized trial of weight loss in primary breast cancer: Impact on body composition, circulating biomarkers and tumor characteristics. Int J Cancer. 2020;146:2784–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn GL, Wang KA. Dietary fat reduction and breast cancer outcome: results from the Women’s Intervention Nutrition Study (WINS). Am J Clin Nutr. 2007;86:s878–881. [DOI] [PubMed] [Google Scholar]
- 13.Chlebowski RT, Blackburn GL, Thomson CA, Nixon DW, Shapiro A, Hoy MK, et al. Dietary fat reduction and breast cancer outcome: interim efficacy results from the Women’s Intervention Nutrition Study. J Natl Cancer Inst. 2006;98:1767–76. [DOI] [PubMed] [Google Scholar]
- 14.Chlebowski RT, Reeves MM. Weight Loss Randomized Intervention Trials in Female Cancer Survivors. J Clin Oncol. 2016;34:4238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feigelson HS, Caan B, Weinmann S, Leonard AC, Powers JD, Yenumula PR, et al. Bariatric Surgery is Associated With Reduced Risk of Breast Cancer in Both Premenopausal and Postmenopausal Women. Ann Surg. 2020;272:1053–9. [DOI] [PubMed] [Google Scholar]
- 16.Dunlap SM, Chiao LJ, Nogueira L, Usary J, Perou CM, Varticovski L, et al. Dietary energy balance modulates epithelial-to-mesenchymal transition and tumor progression in murine claudin-low and basal-like mammary tumor models. Cancer Prev Res (Phila). 2012;5:930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers LW, Rossi EL, McDonell SB, Doerstling SS, Khatib SA, Lineberger CG, et al. Leptin Signaling Mediates Obesity-Associated CSC Enrichment and EMT in Preclinical TNBC Models. Mol Cancer Res. 2018;16:869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Angel RE, Conti CJ, Wheatley KE, Brenner AJ, Otto G, Degraffenried LA, et al. The enhancing effects of obesity on mammary tumor growth and Akt/mTOR pathway activation persist after weight loss and are reversed by RAD001. Mol Carcinog. 2013;52:446–58. [DOI] [PubMed] [Google Scholar]
- 19.Rossi EL, de Angel RE, Bowers LW, Khatib SA, Smith LA, Buren EV, et al. Obesity-Associated Alterations in Inflammation, Epigenetics, and Mammary Tumor Growth Persist in Formerly Obese Mice. Cancer Prev Res. American Association for Cancer Research; 2016;9:339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvie MN, Pegington M, Mattson MP, Frystyk J, Dillon B, Evans G, et al. The effects of intermittent or continuous energy restriction on weight loss and metabolic disease risk markers: a randomized trial in young overweight women. International Journal of Obesity. Nature Publishing Group; 2011;35:714–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvie M, Wright C, Pegington M, McMullan D, Mitchell E, Martin B, et al. The effect of intermittent energy and carbohydrate restriction v. daily energy restriction on weight loss and metabolic disease risk markers in overweight women. Br J Nutr. 2013;110:1534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi EL, Dunlap SM, Bowers LW, Khatib SA, Doerstling SS, Smith LA, et al. Energy Balance Modulation Impacts Epigenetic Reprogramming, ERα and ERβ Expression, and Mammary Tumor Development in MMTV-neu Transgenic Mice. Cancer Res. 2017;77:2500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong KR, Sims AH, Harvie M, Chapman M, Dunn WB, Broadhurst D, et al. Biomarkers of dietary energy restriction in women at increased risk of breast cancer. Cancer Prev Res (Phila). 2009;2:720–31. [DOI] [PubMed] [Google Scholar]
- 24.Harvie MN, Sims AH, Pegington M, Spence K, Mitchell A, Vaughan AA, et al. Intermittent energy restriction induces changes in breast gene expression and systemic metabolism. Breast Cancer Res. 2016;18:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolard PM. Asking complex questions of the genome without programming. Methods Mol Biol. 2010;628:39–52. [DOI] [PubMed] [Google Scholar]
- 26.Humpton TJ, Vousden KH. Regulation of Cellular Metabolism and Hypoxia by p53. Cold Spring Harb Perspect Med. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin Y, Sundaram S, Essaid L, Chen X, Miller SM, Yan F, et al. Weight loss reduces basal-like breast cancer through kinome reprogramming. Cancer Cell Int. 2016;16:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaram S, Le TL, Essaid L, Freemerman AJ, Huang MJ, Galanko JA, et al. Weight Loss Reversed Obesity-Induced HGF/c-Met Pathway and Basal-Like Breast Cancer Progression. Front Oncol. 2014;4:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quail DF, Olson OC, Bhardwaj P, Walsh LA, Akkari L, Quick ML, et al. Obesity alters the lung myeloid cell landscape to enhance breast cancer metastasis through IL5 and GM-CSF. Nat Cell Biol. 2017;19:974–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das M, Ellies LG, Kumar D, Sauceda C, Oberg A, Gross E, et al. Time-restricted feeding normalizes hyperinsulinemia to inhibit breast cancer in obese postmenopausal mouse models. Nat Commun. 2021;12:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyengar NM, Hudis CA, Dannenberg AJ. Obesity and cancer: local and systemic mechanisms. Annu Rev Med. 2015;66:297–309. [DOI] [PubMed] [Google Scholar]
- 32.Wood IS, de Heredia FP, Wang B, Trayhurn P. Cellular hypoxia and adipose tissue dysfunction in obesity. Proc Nutr Soc. 2009;68:370–7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Liu S. Mechanistic insights of adipocyte metabolism in regulating breast cancer progression. Pharmacol Res. 2020;155:104741. [DOI] [PubMed] [Google Scholar]
- 34.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rönn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–813. [DOI] [PubMed] [Google Scholar]
- 36.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2015;44:1277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab. 2015;21:138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biology. 2015;16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samblas M, Milagro FI, Martínez A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics. 2019;14:421–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim WG, Cheng S. Thyroid hormone receptors and cancer. Biochimica et Biophysica Acta (BBA) - General Subjects. 2013;1830:3928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu JE, Han S-Y, Wolfson B, Zhou Q. The role of endothelial lipase in lipid metabolism, inflammation, and cancer. Histol Histopathol. 2018;33:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocrine Reviews. 2018;39:760–802. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R, Wang Y, Li R, Chen G. Transcriptional Factors Mediating Retinoic Acid Signals in the Control of Energy Metabolism. Int J Mol Sci. 2015;16:14210–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuno Y, Fukuhara A, Hashimoto E, Kobayashi H, Kobayashi S, Otsuki M, et al. Oxidative Stress Inhibits Healthy Adipose Expansion Through Suppression of SREBF1- Mediated Lipogenic Pathway. Diabetes. 2018;67:1113–27. [DOI] [PubMed] [Google Scholar]
- 46.Pape-Ansorge KA, Grande JP, Christensen TA, Maihle NJ, Cleary MP. Effect of moderate caloric restriction and/or weight cycling on mammary tumor incidence and latency in MMTV-Neu female mice. Nutr Cancer. 2002;44:162–8. [DOI] [PubMed] [Google Scholar]
- 47.Cleary MP, Hu X, Grossmann ME, Juneja SC, Dogan S, Grande JP, et al. Prevention of mammary tumorigenesis by intermittent caloric restriction: does caloric intake during refeeding modulate the response? Exp Biol Med (Maywood). 2007;232:70–80. [PubMed] [Google Scholar]
- 48.Rogozina OP, Bonorden MJL, Grande JP, Cleary MP. Serum insulin-like growth factor-I and mammary tumor development in ad libitum-fed, chronic calorie-restricted, and intermittent calorie-restricted MMTV-TGF-alpha mice. Cancer Prev Res (Phila). 2009;2:712–9. [DOI] [PubMed] [Google Scholar]
- 49.Dogan S, Rogozina OP, Lokshin AE, Grande JP, Cleary MP. Effects of chronic vs. intermittent calorie restriction on mammary tumor incidence and serum adiponectin and leptin levels in MMTV-TGF-α mice at different ages. Oncol Lett. 2010;1:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuno NK, Rogozina OP, Seppanen CM, Liao DJ, Cleary MP, Grossmann ME. Combination of intermittent calorie restriction and eicosapentaenoic acid for inhibition of mammary tumors. Cancer Prev Res (Phila). 2013;6:540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing and RRBS data generated in this study are publicly available in the GEO repository (GSE202332).