Dear Editor
Recently, more than 40,000 monkeypox cases in non-endemic countries around the world has been reported, most of which were in Europe and American. Monkeypox virus is mainly transmitted through direct contact, including close contact with respiratory secretions, skin damaged parts or contaminated items of infected people or animals [1,2]. Monkeypox outbreaks usually have a central point, and the original patient usually has a travelling history related to epidemic areas or a clear history of exposure to infectious sources (such as some rodents) [2]. However, the current monkeypox outbreak occurred in several non-endemic countries in a short time, and the vast majority of reported cases have neither been to the endemic areas in Africa nor direct contact with wild animals [1]. Furthermore, after the COVID-19, people's social distance increases, and the probability of contact transmission should be decreased. It is difficult to explain the current monkeypox epidemic with the usual transmission pathways. It's worth noting that, in a 2013 monkeypox outbreak in the Democratic Republic of the Congo, both intrahousehold transmission and interhousehold transmission were observed, although how interhousehold spread occurred was not certain [3]. There might be some unknown cryptic transmission pathway.
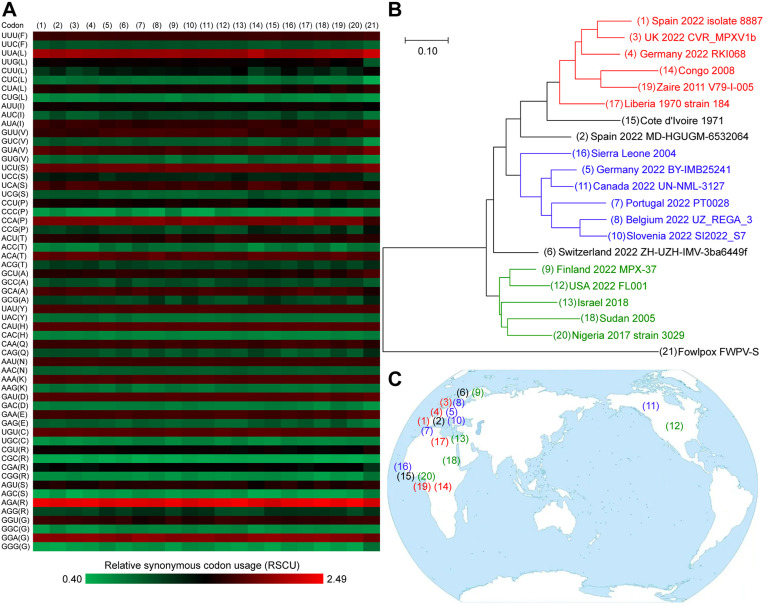
The source and transmission path of the current epidemic are still unclear. Traditional phylogenetic analyses use the whole genome sequence, hemagglutinin gene [4] or the surface glycoprotein gene (Supplementary Fig. 1). However the nucleotide differences of virus strains sampled from different places are too small to clarify an evolutionary clue. The relative synonymous codon usage (RSCU) bias can also reflect the origin of the virus [5]. The storage hosts of monkeypox viruses are generally primates, rodents or artiodactyls, while human beings are only the temporary host. The RSCU biases of different virus strains are usually close to those of their storage hosts [5]. Here we estimated the RSCU biases of 20 monkeypox virus strains (including 10 European strains, 2 North American strains, 7 African strains isolated from 1970 to 2017 and the 2018 Israel strain). The relationship among these sequences was calculated using the squared Euclidean distance (Fig. 1 A and Supplementary Table 1). Then a phylogenetic tree constructed by using the Euclidean distance method (Fig. 1B). It was found that the current outbreak in non-endemic countries may have at least three origins: Libya 1970 cluster, Sierra Leone 2004 cluster, and Sudan 2005 – Nigeria 2017 cluster (Fig. 1B). The RSCU bias of Canada 2022 strain was highly similar to that of Germany 2022 BY-IMB25241 strain; The USA 2022 FL001 strain was highly similar to Finland 2022 MPX-37 strain (Supplementary Table 2), indicating that the viruses in North America may come from Europe by travelling presumably. Interestingly, the two virus strains both collected from Germany belong to two clusters. The distribution of virus clusters was staggered, which proves that current epidemic is multi-originated with complex transmission paths (Fig. 1C).
Fig. 1.
Cluster analysis of monkeypox viruses based on relative synonymous codon usage (RSCU) bias. (A) Heat map of RSCU, excluding tryptophan codon UGG, start codon AUG, stop codons UAA, UGA and UAG. (1)–(20) are monkeypox viruses; (21) is a fowlpox virus, as an outgroup. (B) Phylogenetic tree constructed by using the Euclidean distance method with the Neighbor-Joining (NJ) model. Three distinct clusters are marked with red, blue and green respectively. Bar represents 0.10 Euclidean distance. (C) Geographical distribution of 20 monkeypox virus strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The sudden and unexpected appearance of monkeypox simultaneously in several non-endemic countries suggests that there might have been undetected transmission for some unknown duration of time followed by recent amplifier events [6]. So far, no animal reservoir for monkeypox viruses has been identified in non-endemic regions. Therefore it is unlikely a zoonotic spillover from animals to humans, and instead due to multiple importations of the viruses from different endemic regions, and subsequent cryptic inter-human transmissions that were undetected [6]. However, zoonotic transmission cannot be ruled out completely. Some animals are highly susceptible to monkeypox virus, such as American Sciuridae and European squirrels [7]. Interestingly, in 2015, a poxvirus strain was isolated in a renal transplant patient [8]. Unexpectedly, the genome sequencing results found that the closest relative of this poxvirus, identified in North America, was a poxvirus (strain Yoka) collected from a mosquito pool from central Africa in 1972 [8]. The possible transmission of monkeypox by Aedes mosquito requires further investigations.
In conclusion, the current monkeypox outbreak in non-endemic countries may have at least three origins: Libya 1970 cluster, Sierra Leone 2004 cluster, and Sudan 2005 – Nigeria 2017 cluster. The geographical distribution of virus clusters was in cross, indicating that it was multi-originated and the transmission paths may be very complex. Multiple importations of the viruses from different endemic regions may occur and some animals might participate in these multiple transcontinental transmissions. It is very important to determine the origin and transmission path of the virus, so as to achieve targeted prevention and treatments.
Authors' contributions
Prof. Shu Yuan: Conceptualization; Data collection; Analysis; Writing-original draft; Writing-review & editing. Si-Cong Jiang: Data collection; Analysis; Writing-review & editing. Zhong-Wei Zhang: Data collection; Analysis; Writing-review & editing. Xin-Yue Yang: Data collection; Analysis; Writing-review & editing. Yu-Fan Fu: Data collection; Analysis; Writing-review & editing. Zi-Lin Li: Analysis; Writing-review & editing. Jing Hu: Analysis; Writing-review & editing.
Funding
This work was supported by the Sichuan Province Youth Science and Technology Innovation Team (20CXTD0062 to S. Yuan) and the Applied Basic Research Program of Sichuan Province (2020YJ0410 to Z.-W. Zhang).
Declaration of competing interest
Nothing to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2022.102444.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Perez Duque M., Ribeiro S., Martins J.V., et al. Ongoing monkeypox virus outbreak, Portugal, 29 April to 23 May 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunge E.M., Hoet B., Chen L., et al. The changing epidemiology of human monkeypox-A potential threat? A systematic review. PLoS Neglected Trop Dis. 2022;16 doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nolen L.D., Osadebe L., Katomba J., et al. Extended human-to-human transmission during a monkeypox outbreak in the Democratic Republic of the Congo. Emerg Infect Dis. 2016;22:1014–1021. doi: 10.3201/eid2206.150579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antinori A., Mazzotta V., Vita S., et al. Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200421. May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 6.Alakunle E.F., Okeke M.I. Monkeypox virus: a neglected zoonotic pathogen spreads globally. Nat Rev Microbiol. 2022;20:507–508. doi: 10.1038/s41579-022-00776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haddad N. The presumed receptivity and susceptibility to monkeypox of European animal species. Infect Dis Now. 2022;52:294–298. doi: 10.1016/j.idnow.2022.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakis N.S., Li Y., Abraham J.L., et al. Novel poxvirus infection in an immune suppressed patient. Clin Infect Dis. 2015;61:1543–1548. doi: 10.1093/cid/civ643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.