Abstract
Background:
Testicular germ cell tumors (TGCTs), histologically classified as seminomas and non-seminomas, are believed to arise from primordial gonocytes, with the maturation process blocked when are subjected to DNA methylation reprogramming. Single-nucleotide polymorphisms (SNPs) in DNA methylation machinery and folate-dependent one-carbon metabolism genes have been postulated to influence the proper establishment of DNA methylation.
Material and Methods:
In this pathway-focused investigation we evaluated the association between 273 selected tag SNPs from 28 DNA methylation-related genes and TGCT risk. We carried out association analysis at individual SNP and gene-based level using summary statistics from the Genome Wide Association Study meta-analysis recently conducted by the international Testicular Cancer Consortium on 10,156 TGCT cases and 179,683 controls.
Results:
In individual SNP analyses, seven SNPs, four mapping within MTHFR, were associated with TGCT risk after correction for multiple testing (q-value≤0.05). Queries of public databases showed that three of these SNPs were associated with MTHFR changes in enzymatic activity (rs1801133) or expression level in testis tissue (rs12121543, rs1476413). Gene-based analyses revealed MTHFR (q-value=8.4×10−4), MECP2 (q-value=2×10−3) and ZBTB4 (q-value=0.03) as the top TGCT-associated genes. Stratifying by tumor histology, four MTHFR SNPs were associated with seminoma. In gene-based analysis MTHFR was associated with risk of seminoma (q-value=2.8×10−4), but not with non-seminomatous tumors (q-value=0.22).
Conclusions:
Genetic variants within MTHFR, potentially having an impact on the DNA methylation pattern, are associated with TGCT risk.
Impact:
This finding suggests that TGCT pathogenesis could be associated to the folate cycle status, and this relation could be partly due to hereditary factors.
Introduction
Testicular cancer is the most common malignancy among men aged 15–40 years of European ancestry. Since the mid-20th century, testicular cancer incidence rates have been increasing in many countries and are predicted to further increase over the next decades (1,2).
Testicular germ cell tumors (TGCTs) account for the 98% of all testicular cancers and are histologically classified as seminoma and non-seminomatous tumors. The latter include embryonal carcinomas, teratomas, choriocarcinoma, and yolk sac tumors. Mixed germ cell tumors, composed of two or more germ cell tumor types, are typically classified as non-seminomas since they have similar molecular features and prognosis (3,4). Established TGCT risk factors include age, ethnicity, contralateral testicular cancer, larger adult height, cryptorchidism and positive family history (5).
A strong genetic component has been described in TGCT, with an estimated 37% heritability in twin studies (6). Genome-wide association studies (GWAS) have identified multiple independent common variants associated with TGCT risk, strongly suggesting that the genetic susceptibility for TGCT is not due to a few major high-penetrance genes, but rather to combined multiple genetic variants with modest-to-small effect sizes (7,8).
Both seminoma and non-seminomatous germ cell tumors are believed to arise from primordial gonocytes that have failed to differentiate normally into pre-spermatogonia in early fetal life (9). Accordingly, these immature fetal germ cells accumulate within the seminiferous tubule forming pre-invasive neoplastic lesions called germ-cell neoplasia in situ (GCNIS). The current pathogenetic model for TGCT is based on the hypothesis that the GCNIS cell could begin to proliferate at puberty and eventually acquire malignant potential (10,11).
During early embryonic development, gonocytes arrested in mitosis undergo extensive epigenetic remodelling including the genome-wide erasure of DNA methylation markers and de novo re-establishment of a parental imprinting pattern that is completed prior to birth (12). Studies have shown that the genome of GCNIS in the human adult testis exhibits global DNA methylation erasure (13,14), a common feature of primordial gonocytes (15,16).
Striking differences in methylation profiles between TGCT subtypes have been described: non-seminomatous tumors show aberrantly increased promoter methylation, whereas in seminomas the genome is mostly maintained in an unmethylated state (13,14,17,18). This finding suggests that DNA methylation could be important for the subtype-specific pathogenesis of TGCTs.
The proper establishment of DNA methylation patterns requires the activity of several proteins which together comprise the DNA methylation machinery. These proteins are responsible for: i) active removal of methyl groups (DNA demethylases or “erasers”); ii) establishment of the de-novo methylation and maintenance of the methylation pattern during DNA replication (DNA methyltransferases or “writers”); and iii) reading the methylation pattern by binding the 5-methylcytosine bases (methyl-CpG binding proteins or “readers”) (19). Methyl groups, essential for methylation reactions, are uniquely provided by the universal methyl donor S-adenosylmethionine, synthesized through the folate-dependent one-carbon metabolism (20).
To our knowledge, the expression pattern of genes codifying for the one-carbon metabolism enzymes has not been investigated in human TGCT tissue. However, a number of studies aimed at characterizing the expression of the genes of the DNA methylation machinery in the TGCT tissue, particularly of the DNA methyltransferases (DNMTs) and of the DNA demethylases of the TET family. The expression of DNMT1 has been described as upregulated in embryonal carcinomas in comparison with seminomas and teratomas (21), and the DNMT3L protein was specifically expressed in embryonal carcinomas but completely absent in seminomas (22). Moreover, lower DNMT3A and DNMT3B mRNA levels and a higher expression of the TET2 protein were observed in seminoma compared to non-seminomatous tumors (23). Evaluation of the expression profile of the TET enzymes showed increased levels especially of TET1, but also of TET2 and TET3 mRNAs in both seminomas and mixed TGCTs, compared to non-seminomatous tumors and the surrounding tumor-matched healthy testicular tissue (24).
It has been reported in literature that single-nucleotide polymorphisms (SNPs) around genes coding for proteins and enzymes involved in DNA methylation machinery and folate-dependent one-carbon metabolism can alter promoter activity and expression of the gene itself, thus influencing the establishment of individual methylation patterns (25–27).
We hypothesized that variants around and in genes involved in the DNA methylation machinery and in one-carbon metabolism can influence the risk of developing TGCT. We evaluated the associations between individual SNPs in DNA methylation-related genes and TGCT risk, and assessed their collective effect by performing gene-based analyses.
Material and Methods
Study population
The Testicular Cancer Consortium (TECAC; www.tecac.org) assembled multiple TGCT case-control studies conducted by more than 20 institutions from Europe and North America (8). All studies involved in the Consortium have collected blood or saliva samples, from which DNA has been extracted, and a selection of phenotype and questionnaire data on potential TGCT risk factors.
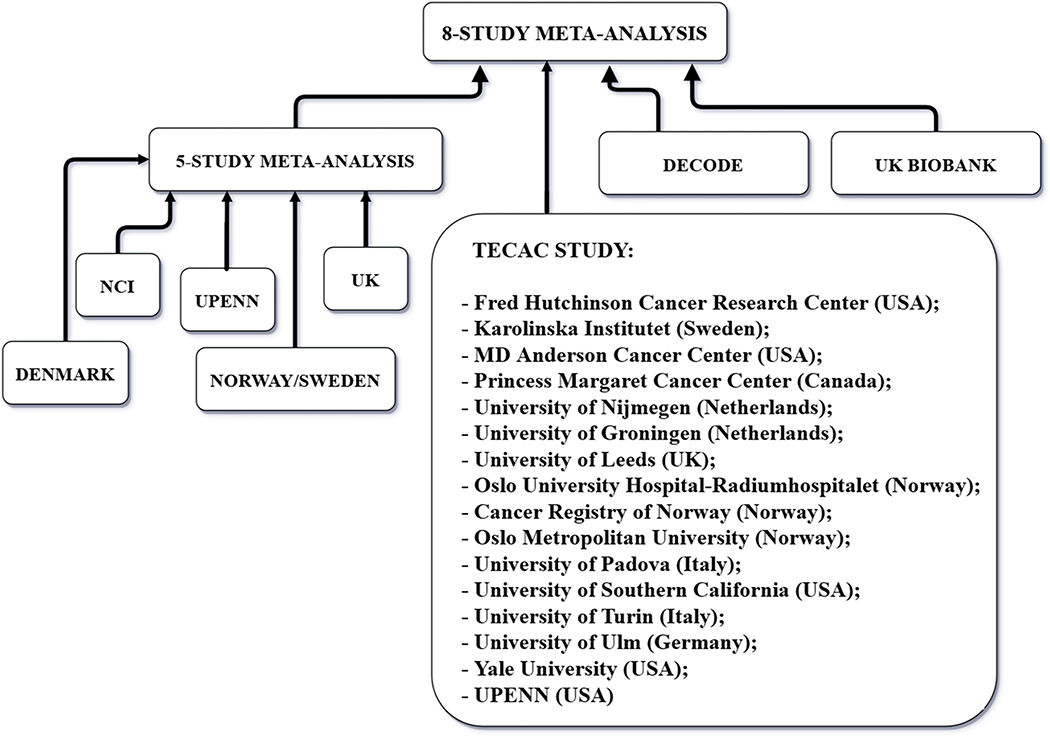
Data from eight sources were obtained by TECAC: (i-v) summary statistics from 5 independently conducted GWASes on TGCT (28–32) and previously published as a meta-analysis (33); (vi) individual level genotype data from the TECAC study involving 14 case-control studies conducted by the TECAC institution members in Europe and the United States, with genotyping centrally conducted at the Center for Applied Genomics at the Children’s Hospital of Philadelphia (13 studies) or MD Anderson Cancer Center (one study) using the Illumina Human Core array technology (8); (vii) the deCODE genetics company (deCODE genetics, RRID:SCR_003334) (34) study in Iceland; and (viii) the UK biobank study (UK Biobank, RRID:SCR_012815) (35). These studies were described in detail elsewhere (8). In total, the Consortium assembled 10,156 cases and 179,683 controls (Figure 1 and Table 1).
Figure 1.
Flow chart of the eight studies assembled by the Testicular Cancer Consortium. Cases and controls from these studies have been involved in the main analysis of the current work.
Table 1.
Number of TGCT cases and controls included for testing associations with SNPs in methylation-related genes by originating study, and for cases by histologic type, family history of TGCT, and history of cryptorchidism
| Studies | TGCT Cases | Controls (N) | ||||
|---|---|---|---|---|---|---|
| ALL (N) | Seminoma histology (N) | Non-seminomatous histology (N) | Family history (N) | Cryptorchidism (N) | ||
| GWAS-DENMARK | 183 | 88 | 55 | na | na | 363 |
| GWAS-NCI | 581 | 243 | 334 | 76 | 131 | 1,056 |
| GWAS-UPENN | 481 | 171 | 299 | 49 | 39 | 919 |
| GWAS- NORWAY/SWEDEN | 1,326 | 766 | 549 | na | na | 6,687 |
| GWAS-UK | 986 | 410 | 410 | 136 | 56 | 4,945 |
| TECAC STUDY | 5,602 | 2,456 | 2,760 | 95 | 295 | 5,006 |
| deCODE ICELAND | 300 | na | na | na | na | 151,991 |
| UK BIOBANK | 697 | 395 | 223 | na | na | 8,716 |
| Total | 10,156 | 4,529 | 4,630 | 356 | 521 | 179,683 |
N: number of subjects
GWAS: genome-wide association study
na: information not available by TECAC study: cases not included in stratified / restricted analysis
For most of these studies, information was available on the histological subtype classified as pure seminoma and non-seminomatous tumors (the latter including TGCTs with mixed histology), family history of TGCT, history of cryptorchidism and other selected key characteristics.
The current study was carried out on summary statistics data from the meta-analysis of the eight sources performed by the TECAC Consortium (8). Data from participants in each contributing study were collected and analyzed in accordance with the local ethical permissions and informed written consent.
Selection of genes and SNPs
To obtain a list of DNA methylation machinery and one-carbon metabolism genes, we conducted a search in public pathway catalogues in 2014, including BioCarta (BioCarta Pathways, RRID:SCR_006917), Reactome (Reactome, RRID:SCR_003485), KEGG (KEGG, RRID:SCR_012773) and NCI-PID (http://pid.nci.nih.gov/index.shtml) using the following queries: “DNA methylation”, “DNA methylation pathway”, “mechanisms of transcriptional repression by DNA methylation”, “epigenetic regulation of gene expression”, “folate cycle”, “one-carbon metabolism”, and “one carbon pool by folate”. We identified a preliminary list of protein-coding genes and checked the function of each gene manually using the public databases GENEcards (GeneCards, RRID:SCR_002773) and UniProtKB (UniProtKB, RRID:SCR_004426), keeping in the final list only genes strictly involved in the DNA methylation process.
We identified 28 DNA methylation pathway genes (Supplementary Table S1), classified into two groups based on the molecular mechanism in which they are involved: one-carbon metabolism (N=11 genes) and DNA methylation machinery (N=17 genes), the latter further classified in three subgroups: i) “writers” (N=4); ii) “erasers” (N=4); and iii) “readers” (N=9). No significant changes to this selection were identified when we repeated the gene search in 2021 in the Biocarta and the Reactome pathway databases.
For each gene, we selected a list of tag SNPs using Haploview 4.2 software (Haploview, RRID:SCR_003076), implemented with the Tagger pairwise method (Broad Institute, Cambridge, MA) applied to genotype data of the public database of the International HapMap Project (36). We used the phased genotype data (Human Genome Build 37p13) from the CEU (Utah Residents with Northern and Western European Ancestry) population, the sample that most closely resembles the subjects used in this study. We selected tag SNPs with the following characteristics: minor allele frequency of ≥5% to select only common variants inpersons of European ancestry, and an r2=0.8 as the linkage disequilibrium (LD) threshold. To include the 5′- and 3′-untranslated regulatory regions, tag SNP search was expanded by 10 kilobases up- and downstream of each gene sequence, as predicted clusters of transcription factor binding sites are most enriched in these sequences (37). Moreover, potential functional SNPs were included by searching in public databases, including Ensembl (Ensembl, RRID:SCR_002344), SNPedia (SNPedia, RRID:SCR_006125), and PubMed (PubMed, RRID:SCR_004846).
In total, 273 polymorphisms were selected for the current study. The SNPs were included as part of the custom content on the Illumina Human Bead Core array. The complete list of the candidate genes and the number of tag SNPs selected for each gene are provided in Supplementary Table S1.
Individual SNP analysis
Summary statistics of the association analysis of the selected tag SNPs and the risk of TGCT were provided by the TECAC Consortium. The estimates of the fixed-effect meta-analysis (overall summary p-values, odds ratios (ORs) and corresponding 95% confidence intervals (95% CI)) were obtained as previously described (8).
Four out of the 273 selected tag SNPs were neither genotyped nor imputed in any of the individual studies. We included only polymorphisms with available summary statistics from at least two of the eight studies, leading to the exclusion from the analysis of two other polymorphisms, one in MBD4 and one in DNMT3L, leaving a total of 267 tag SNPs in 28 genes for the final analytic data set (Supplementary Table S1).
We conducted stratified analyses for seminoma and non-seminomatous tumors in all studies except deCODE (which includes 3% of the total number of cases and 84.6% of the total number of controls); analyses restricted to the subgroup of cases with positive TGCT family history, or positive history of cryptorchidism were carried out on cases and controls of the NCI, UPENN and UK studies, and on a sub-set of the TECAC study for which this information were available.
Association p-values were adjusted for multiple comparisons using the Benjamini-Hochberg false discovery rate (FDR) method (38).
Gene-based analysis
Gene-based analysis was carried out using MAGMA (Multi-marker Analysis of GenoMic Annotation) v1.07b, which combines the individual SNP p-values to test the collective effect of multiple markers from a gene by properly incorporating LD between markers (39). In MAGMA, two types of gene test statistics are implemented. The SNP-wise Mean model is more attuned to the mean SNP association, though it is biased towards association in areas of higher gene LD. The SNP-wise Top model is more sensitive when only a small proportion of the analyzed SNPs in a gene show an association (39). We preferred this second approach and calculated a permutation-based p-value for each gene.
Analyses on MAGMA were conducted using the summary p-values for the associations between the tag SNPs and TGCT, and 100,000 permutations were computed for each gene. The European ancestry population from the 1000 Genomes Project Phase 3 (Build 37/European data only) was taken as the reference for LD patterns. Analyses were stratified by histological subtypes as in the individual SNP analyses, and further restricted to cases with a TGCT family history or a history of cryptorchidism. The Benjamini-Hochberg FDR method was used to adjust for multiple comparisons (38).
Functional assessment of SNPs and gene expression analysis in TGCT subtypes
The dbSNP database (dbSNP, RRID:SCR_002338) was interrogated to explore the potential functional consequences of the selected tag SNPs on gene expression and regulation, and on amino acid change (40). HaploReg v4.1 (HaploReg, RRID:SCR_006796) was used to evaluate their possible effects on protein binding sites and regulatory motifs (41). SNPnexus web server (SNPnexus, RRID:SCR_005192) was interrogated to predict the possible functional impact of each SNP at transcriptome and proteome levels and on regulatory elements (42). We also explored the MicroSNiPer (MicroSNiPer, RRID:SCR_009880) and miRNASNP-v3 (http://bioinfo.life.hust.edu.cn/miRNASNP) tools to assess a possible effect of SNPs on miRNA sequence and/or miRNA binding sites (43,44).
From SNiPA (https://snipa.helmholtz-muenchen.de/snipa3/) (45) we retrieved information on possible clinical significance and previously reported associations with other traits and human diseases. SNiPA also was applied, drawing on 1000 Genomes Project Phase 1 v.3 and Phase 3 v.5 data (1000 Genomes: A Deep Catalog of Human Genetic Variation, RRID:SCR_006828), to define the size of LD block spanning each SNP and to identify any proxy variants in high LD (r2≥0.8).
GTEx v7 (GTEx eQTL Browser, RRID:SCR_001618) was explored to predict the possible association with expression quantitative trait loci of each tag SNP and of each SNP in high LD with the tags in a sample of 322 normal adult testis tissues with donor genotypes available (46).
We analyzed publicly available gene expression datasets for genes showing different association patterns between seminoma and non-seminomatous tumors. Expression data from 43 seminoma and 68 non-seminomatous tumors were downloaded from the cBioPortal for Cancer Genomics (cBioPortal, RRID:SCR_014555) (47,48). We used the mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM) from the TGCA PanCancer Atlas dataset. Gene expression between the two histologic groups was compared using Wilcoxon-Mann-Whitney tests. Samples with z-scores above 2 and below −2 were excluded from the analysis.
Data Availability
All the data analysed in this study have been generated in a previous work (8), and are fully available in the supporting information of the current article.
Results
Table 1 reports the number of TGCT cases and controls for the eight studies involved in the meta-analysis, as well as the number of cases stratified by histologic subtype (not available for 3% of the cases), family history of TGCT (not available for 24.7% of cases and 93.4% of controls) and history of cryptorchidism (not available for 24.7% of cases and 93.4% of controls).
Individual SNP and gene-based analysis on all TGCT cases
The main analyses involved the evaluation of 273 tag SNPs from 28 DNA methylation-related genes in 10,156 cases and 179,683 controls. After correction for multiple testing, seven SNPs were associated with TGCT risk with q-values≤0.05, as reported in Table 2. The OR estimates ranged from 0.90 to 1.11 (Table 2). Four (rs1801133, rs12121543, rs1476413, rs13306556) were located in MTHFR (Gene ID: 4524), two (rs1734791, rs1624766) in MECP2 (Gene ID: 4204), and one (rs4796420) in ZBTB4 (Gene ID: 57659), none of which were associated with TGCT risk at genome-wide levels (8). With the exception of rs4796420, the heterogeneity for MTHFR and MECP2 polymorphisms among the eight studies was low. Considering the specific studies, no obvious study characteristic explaining the heterogeneity observed for the rs4796420 has been found. Complete results of all the analysed SNPs are reported in the Supplementary Table S2.
Table 2.
Individual SNP association results for the whole dataset
| SNP ID | GENE; location | Allele1/Allele2$ | Allele2 frequency | q-value | I2 | p-het# | Direction* | OR (95% CI)§ |
|---|---|---|---|---|---|---|---|---|
| rs1801133 | MTHFR; Exon #4 | A/G | 0.66 | 3.6×10−4 | 8.6 | 0.36 | −−−−−−+− | 0.90 (0.87–0.94) |
| rs1734791 | MECP2; Intronic | A/T | 0.15 | 7.8×10−3 | 0 | 0.99 | +++++?++ | 1.09 (1.05–1.14) |
| rs12121543 | MTHFR; Intronic | A/C | 0.75 | 0.02 | 0 | 0.94 | +?++++++ | 1.09 (1.04–1.14) |
| rs1476413 | MTHFR; Intronic | T/C | 0.73 | 0.02 | 0 | 0.99 | ++++++++ | 1.08 (1.03–1.13) |
| rs4796420 | ZBTB4; Intronic | A/T | 0.79 | 0.02 | 71.7 | 8×10−4 | +−+0++−+ | 1.09 (1.04–1.14) |
| rs1624766 | MECP2; Intronic | T/C | 0.20 | 0.02 | 0 | 0.84 | +++++?++ | 1.07 (1.03–1.12) |
| rs13306556 | MTHFR; Intronic | T/C | 0.66 | 0.05 | 0 | 0.98 | ++++++++ | 1.11 (1.04–1.19) |
Allele1: Reference allele; Allele2: Effect allele
P for heterogeneity test
Summary of effect directions of the single studies of the meta-analysis. “+” indicates a positive (increased) effect of the alternative allele on risk of TGCT, while “−” indicates a negative (decreased) effect of the alternative allele on risk of TGCT. “0” indicate null effect and “?” indicates missing effect. Study order: TECAC study, deCODE, UK, NCI, Denmark, Norway/Sweden, UPENN, UK biobank
OR: Odds Ratio; CI: Confidence Interval
In the gene-based analysis, three of the 28 analysed genes showed an association with TGCT risk, with a q-value below 0.05: MTHFR (q-value=8.4×10−4), MECP2 (q-value=2×10−3) and ZBTB4 (q-value=0.03) (Table 3).
Table 3.
Genes associated with risk of TGCT based on analysis of all SNPs in each gene
| GENE | N SNPs* | perm-p# | q-value§ |
|---|---|---|---|
| MTHFR | 13 | 3×10−5 | 8.4×10−4 |
| MECP2 | 4 | 1.4×10−4 | 2×10−3 |
| ZBTB4 | 7 | 3.2×10−3 | 0.03 |
| AHCY | 4 | 0.05 | 0.35 |
| MBD3L1 | 5 | 0.07 | 0.35 |
| SHMT1 | 8 | 0.1 | 0.35 |
| MAT1A | 14 | 0.1 | 0.35 |
| DNMT3L | 16 | 0.1 | 0.35 |
| DNMT1 | 8 | 0.17 | 0.42 |
| MAT2B | 9 | 0.18 | 0.42 |
| DNMT3B | 8 | 0.19 | 0.42 |
| ZBTB38 | 4 | 0.19 | 0.42 |
| UHRF1 | 13 | 0.20 | 0.42 |
| MTRR | 25 | 0.25 | 0.46 |
| TET2 | 9 | 0.26 | 0.46 |
| MBD2 | 10 | 0.27 | 0.46 |
| CBS | 17 | 0.30 | 0.50 |
| TET3 | 11 | 0.44 | 0.69 |
| MBD3 | 4 | 0.53 | 0.75 |
| BHMT | 13 | 0.53 | 0.75 |
| DNMT3A | 18 | 0.56 | 0.75 |
| MAT2A | 4 | 0.59 | 0.75 |
| TET1 | 7 | 0.65 | 0.76 |
| MBD2 | 7 | 0.65 | 0.76 |
| CTCF | 3 | 0.76 | 0.85 |
| MTR | 8 | 0.81 | 0.87 |
| MBD4 | 3 | 0.94 | 0.96 |
| GADD45b | 8 | 0.96 | 0.96 |
Number of SNPs tested within a gene
Gene level p-value computed by MAGMA after 100,000 permutations
Gene level q-value calculated on permutation p-value
Stratified analyses
The analyses stratified by histologic subtype included 4,529 seminomas and 4,630 non-seminomatous germ cell tumors.
After adjustment for multiple testing, MTHFR SNPs rs1801133, rs12121543, rs6541003 and rs1476413 were associated with seminoma with a q-value≤0.05 (q-values=1.6×10−4; 0.02; 0.03; and 0.05; respectively). Three of these SNPs were also among those top-ranked in non-stratified individual SNP analysis (see above). P value for heterogeneity and I2 index calculation revealed no substantial heterogeneity among studies (Supplementary Table S3). Complete results of this analysis are available in Supplementary Table S4.
None of the SNPs were associated with the risk of non-seminomatous tumors with a q-value≤0.05 (Supplementary Table S5); furthermore, none of the top-ranked SNPs were included in the top positions of the main analysis (Supplementary Table S2).
As shown in Table 4, gene-based analyses stratified by histological subtype revealed an association between MTHFR and seminoma risk (q-value=2.8×10−4), and no clear evidence of an association with non-seminomatous tumors for any of the 28 selected genes (Table 4).
Table 4.
Gene-based analysis stratified by histologic subtype
| Seminoma cases (N=4,529) vs. controls (N=27,693) | Non-seminomatous cases (N=4,630) vs. controls (N=27,693) | ||||
|---|---|---|---|---|---|
| GENE | perm-p# | q-value§ | GENE | perm-p# | q-value§ |
| MTHFR | 1×10−5 | 2.8×10−4 | DNMT1 | 7.9×10−3 | 0.22 |
| AHCY | 0.02 | 0.2 | MTRR | 0.02 | 0.35 |
| DNMT3L | 0.02 | 0.2 | MBD3 | 0.05 | 0.48 |
| SHMT1 | 0.05 | 0.34 | MBD3L1 | 0.09 | 0.64 |
| ZBTB38 | 0.06 | 0.36 | MTHFR | 0.2 | 0.70 |
| ZBTB4 | 0.07 | 0.40 | ZBTB4 | 0.26 | 0.70 |
| MAT1A | 0.10 | 0.40 | DNMT3B | 0.26 | 0.70 |
| MBD3L1 | 0.12 | 0.41 | MAT1A | 0.27 | 0.70 |
| MECP2 | 0.13 | 0.41 | TET1 | 0.28 | 0.70 |
| CBS | 0.20 | 0.55 | CBS | 0.30 | 0.70 |
| MAT2A | 0.27 | 0.61 | BHMT | 0.32 | 0.70 |
| MTRR | 0.28 | 0.61 | MECP2 | 0.37 | 0.70 |
| TET1 | 0.28 | 0.61 | UHRF1 | 0.38 | 0.70 |
| MBD1 | 0.38 | 0.76 | MBD1 | 0.39 | 0.70 |
| DNMT3B | 0.41 | 0.77 | MAT2B | 0.39 | 0.70 |
| UHRF1 | 0.46 | 0.77 | AHCY | 0.40 | 0.70 |
| MBD3 | 0.47 | 0.77 | SHMT1 | 0.48 | 0.75 |
| MAT2B | 0.51 | 0.79 | DNMT3L | 0.48 | 0.75 |
| TET3 | 0.56 | 0.83 | TET3 | 0.57 | 0.84 |
| MBD2 | 0.61 | 0.84 | MTR | 0.61 | 0.84 |
| DNMT1 | 0.67 | 0.84 | TET2 | 0.63 | 0.84 |
| BHMT | 0.68 | 0.84 | DNMT3A | 0.68 | 0.87 |
| TET2 | 0.69 | 0.84 | ZBTB38 | 0.72 | 0.88 |
| GADD45b | 0.76 | 0.86 | CTCF | 0.78 | 0.91 |
| DNMT3A | 0.77 | 0.86 | MAT2A | 0.85 | 0.95 |
| MBD4 | 0.88 | 0.94 | MBD2 | 0.95 | 0.99 |
| MTR | 0.90 | 0.94 | MBD4 | 0.95 | 0.99 |
| CTCF | 0.96 | 0.96 | GADD45b | 0.99 | 0.99 |
Gene level p-value computed by MAGMA after 100,000 permutations
Gene level q-value on permutation p-value
Analyses restricted to men with a positive family history of TGCT and those with a history of cryptorchidism were carried out on 356 and 521 cases, respectively, from the TECAC, NCI, UPENN and UK studies, which were compared with the 11,927 controls included in the same studies. In individual SNP analysis restricted to history of cryptorchidism four polymorphisms, all mapping in MECP2, were excluded since their summary statistics results were available for one study only (TECAC). Then, two hundred and sixty-three SNPs were used in this analysis.
In individual SNP analysis restricted to cases with family history for TGCT and to those with history of cryptorchidism, no SNP was associated with risk of TGCT after correction for multiple testing (Supplementary Tables S6 and S7).
In gene-based analyses, no gene was associated with TGCT risk, though AHCY and SHMT1 were two of the top-three most strongly associated genes both in analysis restricted to cases with family history for TGCT, and in analysis restricted to those with history of cryptorchidism (Supplementary Table S8).
Functional assessment of top SNPs and expression analysis in TGCT subtypes
Functional annotations of the tag SNPs most strongly associated with the TGCT risk in the main analysis are listed in Table 5. Six of the seven top variants were intronic, whereas rs1801133 was located in the MTHFR coding region. Rs1801133 was found to be a missense variant causing an amino acid substitution (p.Ala222Val) and defined as damaging by the in silico prediction tools Sift and PolyPhen, since it mapped in a highly-conserved sequence. The evaluation of the putative function of the seven top SNPs on regulatory motifs revealed that four of them were predicted to map to protein-binding sites, whereas all but rs1476413 could alter binding motifs for transcription factors. No variants were predicted to alter microRNA target sequences or CpG islands. Additional two tools (MicroSNiPer and miRNASNP-v3) to investigate possible microRNA binding sites in or near each SNP revealed the same results.
Table 5.
Functional annotation of tag SNPs in MTHFR, MECP2 and ZBTB4 associated with risk of TGCT identified in the individual SNP analysis
| SNP ID (GENE) | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic | rs1801133 (MTHFR) | rs12121543 (MTHFR) | rs1476413 (MTHFR) | rs13306556 (MTHFR) | rs1734791 (MECP2) | rs1624766 (MECP2) | rs4796420 (ZBTB4) |
| Consequence | Coding, missense | Intron variant | Intron variant | Intron variant | Intron variant | Intron variant | Intron variant |
| Amino acid change | Ala222Val | None | None | None | None | None | None |
| Proteins bound | CEBPB, HDAC8, POL2 | na | CEBPB | na | na | ZNF263 | GATA2, POL24H8, TAL1, POL2 |
| Motifs changed | Cphx | STAT | na | PLAG1 | DMRT1, GATA, HDAC2 | Arid5a, Foxj2 | HNF4, Pax-4 |
| Sift Prediction | damaging, high confidence | na | na | na | na | na | na |
| PolyPhen Prediction | probably damaging | na | na | na | na | na | na |
| Variant annotation | Methotrexate response - dosage, efficacy, toxicity / adverse drug reactions (adr); Carboplatin response – efficacy; Cyclophosphamide response – toxicity / adr; Gastrointestinal stroma tumor; MTHFR deficiency, thermolabile type |
na | na | na | na | na | na |
| Variant association (trait/p-value) | Homocysteine levels# p-value<4×10−104; <8×10−35; <1×10−19 Red cell distribution width p-value<1×10−23 Serum folate level§ p-value<4×10−19; <3×10−11 High altitude adaptation p-value<6×10−9 |
na* | Coronary artery disease p-value=2.28×10−5 |
Diastolic blood pressure via alcohol consumption interaction p-value<3×10−9 |
na | na | Educational attainment p-value<2×10−8 Lung function p-value<4×10−16 |
| LD block size | 1 bp | 3,669 bp* | 57,089 bp | 67,385 bp | 160,011 bp | 168,512 bp | 85,798 bp |
| Proxy SNPs in high LD (r2>0.8) | 1 variant | 3 variants* | 3 variants | 105 variants | 24 variants | 52 variants | 60 variants |
| Association with eQTLs in testis tissue | na | p-value=2.3×10−8 | p-value=5.9×10−11 | na | na | na | na |
| Association of high LD-SNPs (r2>0.8) with eQTLs in testis tissue | na | rs3818762 (r2=0.81) p-value=4.4×10−11 |
rs1023252 (r2=0.84) p-value=2.8×10−11 |
na | na | na | na |
na: not available; LD: linkage disequilibrium; eQTLs: expression quantitative trait loci
For functional analysis of rs12121543 on SNiPA tool, 1000 Genome Project Phase 1 v.3 data were used
Three independent studies;
Two independent studies
In theSNiPA database, the rs1801133 locus was associated with a range of diseases and human traits such as plasma homocysteine and folate levels, but also with response and efficacy to anticancer drugs such as carboplatin.
No other common variants were reported in the same LD block of rs1801133, whereas from 3 to 105 polymorphisms were in LD with the other six top SNPs.
In the sample of 322 normal adult testis tissues with available genotypes in the GTEx v7 database, the tagging SNPs rs12121543 and rs1476413 were associated with MTHFR expression quantitative trait loci (eQTLs) in human adult testis tissue (Table 5 and Supplementary Fig. S1, upper panel). Each SNP was in strong LD with another polymorphism which was associated with MTHFR eQTL in testis tissue: rs3818762 was tagged by rs12121543 (pairwise r2=0.81), whereas rs1023252 was a proxy for rs1476413 (pairwise r2=0.84) (Table 5 and Supplementary Fig. S1, lower panel). The C allele (major) of rs12121543 and the C allele (major) of rs1476413, both associated with decreased expression of MTHFR (Supplementary Fig. S1, upper panel), also were associated with an increased TGCT risk in the individual SNP analysis (Table 2).
Expression analyses by histologic subtypes were limited to MTHFR, which was associated with the risk of seminoma, but not with the risk of non-seminomatous tumors.
Since expression data on the adjacent non-neoplastic tissue were not available in the publicly available TGCT dataset, we carried out this analysis on expression data obtained in the tumor tissue. From this dataset we retrieved MTHFR expression data evaluated on 43 seminoma and 68 non-seminomatous tumor tissues. The p-value for comparison of MTHFR expression level between the two histologic subtypes was 0.098. Means of z-scores for seminoma and non-seminomatous tumors were −0.29 and −0.12, respectively.
Discussion
It has been suggested that epigenetic mechanisms may be important driving factors in the pathogenesis of testicular germ cell tumors. A recent large meta-analysis of GWAS on TGCT carried out by TECAC has identified genes critically involved in epigenetic reprogramming through chromatin remodelling and histone modifications, such as PRDM14 and the zinc finger protein genes ZFPM1, ZNF64, and ZNF217 (8). We used the genome-wide association dataset from the Testicular Cancer Consortium to conduct a pathway-focused study on polymorphisms within selected genes involved in DNA methylation, and found robust associations between variants in MTHFR and TGCT risk, with some having a possible functional role. We found associations, although weaker, for variants in MECP2 and ZBTB4.
MTHFR encodes the 5,10-methylenetetrahydrofolate reductase, an essential enzyme for the synthesis of the methyl donor S-adenosylmethionine. MTHFR is a well-studied gene, expressed in several human tissues: according to the Human Protein Atlas database, the highest levels have been reported in glandular cells of the epididymis (49). Mouse studies have revealed that MTHFR is expressed in fetal germ cells, from which the precursor GCNIS is thought to arise, and most highly during the phase of late de novo DNA methylation (50,51). However, no eQTL studies on human fetal germ cells are yet available; hence, it remains to be elucidated whether expression of MTHFR is particularly high also in the embryonic gonad of human males during the DNA re-methylation phase.
Common genetic variants of MTHFR have been studied in relation to several multifactorial disorders, e.g. cardiovascular diseases, pregnancy complications, congenital anomalies including neural tube defects, neuropsychiatric diseases, and cancer. Results of these studies have been conflicting, making the biological and clinical significance of these polymorphisms still uncertain (52). No MTHFR polymorphism has been associated with either congenital anomalies of the genitourinary system, that include both well-established (cryptorchidism) and suggested (hypospadias, inguinal hernia) risk factors for TGCT (53), or with the risk of TGCT itself.
Rs1801133, one of the most well-studied MTHFR polymorphisms, is a coding non-synonymous variant which substitutes a valine for an alanine at amino acid 222 in the catalytic domain, leading to the synthesis of a thermolabile isoform with reduced activity. As compared with the wild-type GG, the AA and GA genotypes are associated with only ~10–20% and ~65% enzyme efficiency, respectively, in converting folic acid into 5-methyltetrahydrofolate, the biologically active and usable form of folate. This mild MTHFR deficiency affects 5–20% of North Americans and Europeans (25). Our individual SNP analysis showed that the minor allele A, encoding the isoform of the enzyme with reduced level of activity, is associated with an increased risk of TGCT.
The association between rs1801133 with folate deficiency and high levels of homocysteine, a folate derivative, has been reported in many studies (54). Both conditions might induce epigenetic changes, leading to global DNA hypomethylation, DNA repair defects, and chromosomal instability, and have been also related to an increased risk of cancer (all types combined) (55). We hypothesize that the thermolabile isoform of MTHFR, coded by the rs1801133 minor allele A, might contribute to a hypomethylated environment by perturbating the folate cycle. Moreover, rs1801133 has been related to DNA hypomethylation in lymphocytes of healthy adults (56), which would be consistent with this hypothesis.
Two other MTHFR polymorphisms, the intronic variants rs12121543 and rs1476413, were associated with TGCT in the individual SNP analysis. The major alleles, associated with an increased risk of TGCT, were also associated with a decreased expression of MTHFR in testis tissue. Although these SNPs do not have the same deleterious effect on protein structure as rs1801133, they may exert modulating effects on MTHFR expression in testis tissue, with possible implications for the establishment of the DNA methylation patterns.
TGCT subtypes originate from the same preneoplastic cell; however, seminoma and non-seminomas exhibit different global DNA methylation patterns, with seminomas mostly hypomethylated and non-seminomatous tumors retaining high levels of DNA methylation (13). In the stratified analysis, we found that rs1801133 was specifically associated with seminomas, and not with non-seminomatous tumors. Similarly, in gene-based analysis stratified by histologic subtype MTHFR was found associated only with seminomas. We could hypothesize that common MTHFR variants, by causing decreased MTHFR expression or activity leading to lower amount of methyl groups produced, might be involved in the subtype-specific pathogenesis of hypomethylated seminomas. Our in silico analysis of whether MTHFR is downregulated in seminoma compared with non-seminomatous tumors, was necessarily limited by the amount of publicly available expression data, hence these analyses need replication in a larger series and a dedicated study design. In order to demonstrate whether MTHFR is differentially regulated in the tissue from which seminoma and non-seminomatous tumors originate, expression data obtained on adjacent normal tissue for the two histologic subtypes would be helpful.
The other genes emerging from the gene-based variant analysis that showed an association with TGCT are less well studied, and little is known about their involvement in cancer predisposition. Mutations in MECP2 (methyl-CpG-binding-protein 2) sequence have been related to congenital diseases and cancer (57). According to functional assessment, the top MECP2 SNPs associated with TGCT were predicted to alter regulatory motifs, suggesting they could influence MECP2 expression.
The main strength of this study is its very large sample size (for TGCT, a relatively rare malignancy), combined with a pre-selected panel of genes and a gene-based analysis with a specific focus on the DNA methylation machinery. TECAC, by pooling the efforts and resources of all its members, made it possible to analyze genome-wide data on more than 10,000 cases, which represents a crucial advantage since TGCT has a significant heritable basis due to multiple minor genetic factors. Another strength is the simultaneous modelling of the collective effect of multiple genetic variants within the same gene, as individual SNP effects might be too weak to be detected.
A limitation of our approach could be that we selected the tag SNPs only among polymorphisms that are in proximity to the genes. We recognize that regulation of gene expression can also be determined by intergenic non-coding SNPs kilobases away; however, as reported in literature (37), the majority of the regulatory regions are located 10 kilobases around each gene sequence. Thus, we are confident that our selection has captured most of the genetic variants potentially able to influence the expression of the genes. Another limitation is that the analyses were restricted only to genes known to be implicated in DNA methylation processes. It is known that epigenetic reprogramming is a very complex process involving other genes, such as those implicated in DNA repair, histone modifications and chromatin remodeling, or in microRNA biosynthesis and regulation. Additional studies are required, since the comprehensive examination of the association between genetic variants of the whole epigenetic machinery and TGCT risk is of interest, but outside the scope of this study. Moreover, some additional analyses aiming at studying more extensively the possible role of the folate cycle status in TGCT pathogenesis, with a special focus on the MTHFR gene, could not be performed in the context of the current study, but may be of interest for future research. First, mostly because of the lack of public databases with relevant data, we could not assess if decreased MTHFR expression is associated with altered DNA methylation in the normal, namely non-tumor, testicular tissue. Second, we could not explore if the identified MTHFR variants are predictive of chemotherapy response, as centrally gathered standardized information on therapy and response to treatment from the eight TGCT studies included in the TECAC GWAS meta-analysis was not available. Finally, while we could analyse seminomas and non-seminomas separately, further stratification by pure non-seminoma subtypes (i.e. cases with only one histological type out of choriocarcinoma, embryonal carcinoma, teratoma, and yolk cell carcinoma) was not possible because of the limited sample size due to the lack of information on the histological subtypes in some of the participating studies and the relative rarity of pure histology among non-seminomatous TGCTs.
In conclusion, in a large pathway-focused meta-analysis, we found that common polymorphisms in MTHFR, some of them potentially having an impact on the DNA methylation pattern, are associated with TGCT risk. This finding may contribute to support a potential involvement of epigenetic mechanisms in the pathogenesis of TGCT.
Supplementary Material
Acknowledgements
We thank the participants in the testicular cancer germ cell studies around the world who contributed to this study.
A. Ferlin would like to thank Dr. Maria Santa Rocca for technical assistance. J.A. Gietema would like to thank Nynke Zwart and Gerrie Steursma for their contributions to the study. J. Nsengimana would like to thank Louise Parkinson for coordinating the recruitment. R.A. Lothe and R.I. Skotheim are grateful to Anne C. Bakken, Sigrid M. Kraggerud and Sophie D. Fosså for their contributions. D.J. Vaughn would like to thank Dr. Linda Jacobs and Ms. Donna Pucci for their contributions to the local Penn TGCT study. K. Almstrup would like to thank Drs Ewa Rajpert-De Meyts and Ramneek Gupta for their contribution to the local study. T. Grotmol, T.B. Haugen, R. Karlsson, and F. Wiklund would like to thank Wenche Kristiansen for her contribution to the Norwegian/Swedish study.
This work was supported by: the National Institute of Health (www.nih.gov) (U01CA164947 to K.L. Nathanson, P.A. Kanetsky and S.M. Schwartz, CA114478 to K.L. Nathanson and P.A. Kanetsky, R01 CA104786 to T. Zheng); the Abramson Cancer Center at the University of Pennsylvania (www.pennmedicine.org/cancer) (P30 CA016520 to K.L. Nathanson and P.A. Kanetsky); the Villum Kann Rasmussen Foundation (www.vkrf.org), the Danish Strategic Research Council (www.ufm.dk/en/research-and-innovation/councils-and-commissions/former-councils-and-commissions/the-danish-council-for-strategic-research) (NABIIT grant), the Novo Nordisk Foundation (www.novonordiskfonden.dk/en/), the Danish Cancer Society (www.cancer.dk/research/), the Danish Childhood Cancer Foundation (www.boernecancerfonden.dk) (to K. Almstrup); the Cancer Research UK Programme (www.cancerresearchuk.org) (Award C588/A19167); the Institute of Cancer Research (www.icr.ac.uk), the Wellcome Trust Case Control Consortium (www.wtccc.org.uk) (to D.T. Bishop, J. Nsengimana, C. Loveday, and C. Turnbull); the National Cancer Institute (www.cancer.gov) (R01CA085914, contracts CN-67009 and PC-35142, to S.M. Schwartz, R01 CA102042 to V.K. Cortessis, K08CA263313 to K.T. Nead) and the Intramural Research Program of the National Cancer Institute (ZO1-CP-10144: “Clinical/Genetic Studies of Familial and Hereditary Cancer Syndromes” to M.H. Greene and M.N. Frone); the California Cancer Research Program (03–00174-30021, 99–0050-V-10260); the Robert E. and May R. Wright Foundation and the Whittier Foundation (www.whittiercf.org) (to V.K. Cortessis); the IMS, Inc support services contract (HHSN26120130003C to M.H. Greene and M.N. Frone); the Norwegian Cancer Society (www.kreftforeningen.no) (418975 – 71081 – PR-2006–0387, PK01–2007-0375); the Nordic Cancer Union (www.ncu.nu/) (S-12/07 to T. Grotmol, T.B. Haugen and F. Wiklund); the Swedish Cancer Society (www.cancerfonden.se) (2008/708, 2010/808, CAN2011/484, CAN2012/823 to T. Grotmol, T.B. Haugen and F. Wiklund); the Piedmont Region (www.regione.piemonte.it) (to L. Richiardi); the Italian Ministry for Education, University and Research (www.miur.gov.it) under the programme “Dipartimenti di Eccellenza 2018–2022” (D15D18000410001 to L. Richiardi); Dell’Elce Family Fund and Princess Margaret Cancer Foundation (www.thepmcf.ca) (to R.J. Hamilton); the Deutsche Krebshilfe grant (www.krebshilfe.de) (70113348 to D. Lessel); the South-Eastern Norway Regional Health Authority (www.helse-sorost.no/south-eastern-norway-regional-health-authority) (to R.A. Lothe and R.I. Skotheim); the Fred Hutchinson Cancer Research Center institutional funds (www.fredhutch.org) (to S.M. Schwartz); the Movember foundation (www.us.movember.com) (to C. Turnbull); the Cancer Prevention Research Institute of Texas (CPRIT scholar in Cancer Research RR190077 to K.T. Nead); the Fondazione Umberto Veronesi (www.fondazioneveronesi.it) (for a postdoctoral fellowship to C. Grasso).
The Testicular Cancer Consortium is comprised of investigator teams from around the world with the common aim to pool resources of all GWAS to identify new genetic markers of risk, further characterize the existing genomic regions with risk markers, examine potential maternal or parent-of-origin effects though case-parent triads and dyads and finally create a Consortium that formalizes the existing research alliances.
Abbreviations
- TGCT
testicular germ cell tumor
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association study
- GCNIS
germ-cell neoplasia in situ
- TECAC
testicular cancer consortium
- LD
linkage disequilibrium
- MAGMA
multi-marker analysis of genomic annotation
- FDR
false discovery rate
- eQTL
expression quantitative trait locus
Footnotes
Conflict of interest
The authors report no conflict of interest.
Testicular Cancer Consortium members
Kristian Almstrup3,4, Victoria K. Cortessis9, Alberto Ferlin12; Jourik A. Gietema14, Anna Gonzalez-Neira34, Mark H. Greene7, Tom Grotmol15, Robert J. Hamilton5, Peter A. Kanetsky33, Lambertus A. Kiemeney19, Davor Lessel20, Katherine A.McGlynn7, Katherine L. Nathanson2,30, Kevin T. Nead25, Jérémie Nsengimana26, Thorunn Rafnar35, Lorenzo Richiardi1, Stephen M. Schwartz8, Rolf I. Skotheim22,27, Clare Turnbull24,28, Fredrik Wiklund18, Tongzhang Zheng31.
34Human Cancer Genetics Programme, Spanish National Cancer Research Centre (CNIO), Madrid, Spain.
35deCODE Genetics/Amgen Inc., Reykjavik, Iceland.
References
- 1.Gurney JK, Florio AA, Znaor A, Ferlay J, Laversanne M, Sarfati D, et al. International trends in the incidence of testicular cancer: lessons from 35 years and 41 countries. EurUrol 2019;76:615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Znaor A, Skakkebaek NE, Rajpert-De Meyts E, Laversanne M, Kuliš T, Gurney J, et al. Testicular cancer incidence predictions in Europe 2010–2035: A rising burden despite population ageing. Int J Cancer 2020;147:820–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen H, Shih J, Hollern DP, Wang L, Bowlby R, Tickoo SK, et al. Integrated molecular characterization of testicular germ cell tumors. Cell Rep 2018;23:3392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akan S, Ediz C, Tavukcu HH, Ozgun A, Yilmaz O. The clinical significance of seminoma component in testicular mixed germ cell tumour. Urol Int 2020;104:489–96. [DOI] [PubMed] [Google Scholar]
- 5.Baird DC, Meyers GJ, Hu JS. Testicular cancer: diagnosis and treatment. Am Fam Physician 2018;97:261–8. [PubMed] [Google Scholar]
- 6.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA 2016;315:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Litchfield K, Loveday C, Levy M, Dudakia D, Rapley E, Nsengimana J, et al. Large-scale sequencing of testicular germ cell tumour (TGCT) cases excludes major TGCT predisposition gene. Eur Urol 2018;73:828–31. [DOI] [PubMed] [Google Scholar]
- 8.Pluta J, Pyle LC, Nead KT, Wilf R, Li M, Mitra N, et al. Identification of 22 novel susceptibility loci associated with testicular germ cell tumors. Nat Commun 2021;12:4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almstrup K, Hoei-Hansen CE, Wirkner U, Blake J, Schwager C, Ansorge W, et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res 2004;64:4736–43. [DOI] [PubMed] [Google Scholar]
- 10.Rajpert-De Meyts E, McGlynn KA, Okamoto K, Jewett MA, Bokemeyer C. Testicular germ cell tumours. Lancet 2016;387:1762–74. [DOI] [PubMed] [Google Scholar]
- 11.Batool A, Karimi N, Wu XN, Chen SR, Liu YX. Testicular germ cell tumor: a comprehensive review. Cell Mol Life Sci 2019;76:1713–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart KR, Veselovska L, Kelsey G. Establishment and functions of DNA methylation in the germline. Epigenomics 2016;8:1399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, et al. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 2008;21:1337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristensen DG, Nielsen JE, Jørgensen A, Skakkebæk NE, Rajpert-De Meyts E, Almstrup K. Evidence that active demethylation mechanisms maintain the genome of carcinoma in situ cells hypomethylated in the adult testis. Br J Cancer 2014;110:668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almstrup K, Nielsen JE, Mlynarska O, Jansen MT, Jørgensen A, Skakkebæk NE, et al. Carcinoma in situ testis displays permissive chromatin modifications similar to immature foetal germ cells. Br J Cancer 2010;103:1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen DG, Skakkebæk NE, Rajpert-De Meyts E, Almstrup K. Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int J Dev Biol 2013;57:309–17. [DOI] [PubMed] [Google Scholar]
- 17.Mirabello L, Kratz CP, Savage SA, Greene MH. Promoter methylation of candidate genes associated with familial testicular cancer. Int J Mol Epidemiol Genet 2012;3:213–27. [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen LH, Nielsen JE, Daugaard G, Hansen TV, Rajpert-De Meyts E, Almstrup K. Differences in global DNA methylation of testicular seminoma are not associated with changes in histone modifications, clinical prognosis, BRAF mutations or gene expression. Cancer Genet 2016;209:506–14. [DOI] [PubMed] [Google Scholar]
- 19.Weissman J, Naidu S, Bjornsson HT. Abnormalities of the DNA methylation mark and its machinery: an emerging cause of neurologic dysfunction. Semin Neurol 2014;34:249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friso S, Udali S, De Santis D, Choi SW. One-carbon metabolism and epigenetics. Mol Aspects Med 2017;54:28–36. [DOI] [PubMed] [Google Scholar]
- 21.Omisanjo OA, Biermann K, Hartmann S, Heukamp LC, Sonnack V, Hild A, et al. DNMT1 and HDAC1 gene expression in impaired spermatogenesis and testicular cancer. Histochem Cell Biol 2007;127:175–81. [DOI] [PubMed] [Google Scholar]
- 22.Minami K, Chano T, Kawakami T, Ushida H, Kushima R, Okabe H, et al. DNMT3L is a novel marker and is essential for the growth of human embryonal carcinoma. Clin Cancer Res 2010;16:2751–9. [DOI] [PubMed] [Google Scholar]
- 23.Lobo J, Guimarães R, Miranda-Gonçalves V, Monteiro-Reis S, Cantante M, Antunes L, et al. Differential expression of DNA methyltransferases and demethylases among the various testicular germ cell tumor subtypes. Epigenomics 2020;12:1579–92. [DOI] [PubMed] [Google Scholar]
- 24.Benešová M, Trejbalová K, Kučerová D, Vernerová Z, Hron T, Szabó A, et al. Overexpression of TET dioxygenases in seminomas associates with low levels of DNA methylation and hydroxymethylation. Mol Carcinog 2017;56:1837–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover PJ. Polymorphisms in 1-carbon metabolism, epigenetics and folate-related pathologies. J Nutrigenet Nutrigenomics 2011;4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murgatroyd C, Spengler D. Genetic variation in the epigenetic machinery and mental health. Curr Psychiatry Rep 2012;14:138–49. [DOI] [PubMed] [Google Scholar]
- 27.Inoue-Choi M, Nelson HH, Robien K, Arning E, Bottiglieri T, Koh WP, et al. Plasma S-adenosylmethionine, DNMT polymorphisms, and peripheral blood LINE-1 methylation among healthy Chinese adults in Singapore. BMC Cancer 2013;13:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanetsky PA, Mitra N, Vardhanabhuti S, Vaughn DJ, Li M, Ciosek SL, et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet 2011;20:3109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalgaard MD, Weinhold N, Edsgärd D, Silver JD, Pers TH, Nielsen JE, et al. A genome-wide association study of men with symptoms of testicular dysgenesis syndrome and its network biology interpretation. J Med Genet 2012;49:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher FR, Wang Z, Skotheim RI, Koster R, Chung CC, Hildebrandt MA, et al. Testicular germ cell tumor susceptibility associated with the UCK2 locus on chromosome 1q23. Hum Mol Genet 2013;22:2748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litchfield K, Holroyd A, Lloyd A, Broderick P, Nsengimana J, Eeles R, et al. Identification of four new susceptibilityloci for testicular germ cell tumour. Nat Commun 2015;6:8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristiansen W, Karlsson R, Rounge TB, Whitington T, Andreassen BK, Magnusson PK, et al. Two new loci and gene sets related to sex determination and cancer progression are associated with susceptibility to testicular germ cell tumor. Hum Mol Genet 2015;24:4138–46. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, et al. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat Genet 2017;49:1141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 2015;47:435–44. [DOI] [PubMed] [Google Scholar]
- 35.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc 2009;2009:pdb.ip71. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang C, Smith DD, Krontiris TG. Evolutionary signatures of common human cis-regulatory haplotypes. PLoS One 2008;3:e3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- 39.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PloS Comput Biol 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res 2018;46:W109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barenboim M, Zoltick BJ, Guo Y, Weinberger DR. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat 2010;31:1223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu CJ, Fu X, Xia M, Zhang Q, Gu Z, Guo AY. miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res 2021;49:D1276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 2015;31:1334–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pontén F, Jirström K, Uhlen M. The Human Protein Atlas-a tool for pathology. J Pathol 2008;216:387–93. [DOI] [PubMed] [Google Scholar]
- 50.Garner JL, Niles KM, McGraw S, Yeh JR, Cushnie DW, Hermo L, et al. Stability of DNA methylation patterns in mouse spermatogonia under conditions of MTHFR deficiency and methionine supplementation. Biol Reprod 2013;89:125. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, et al. Single-cell RNA-Seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 2017;20:858–73. [DOI] [PubMed] [Google Scholar]
- 52.Levin BL, Varga E. MTHFR: addressing genetic counseling dilemmas using evidence-based literature. J Genet Couns 2016;25:901–11. [DOI] [PubMed] [Google Scholar]
- 53.Trabert B, Zugna D, Richiardi L, McGlynn KA, Akre O. Congenital malformations and testicular germ cell tumors. Int J Cancer 2013;133:1900–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colson NJ, Naug HL, Nikbakht E, Zhang P, McCormack J. The impact of MTHFR 677 C/T genotypes on folate status markers: a meta-analysis of folic acid intervention studies. Eur J Nutr 2017;56:247–60. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D, Wen X, Wu W, Guo Y, Cui W. Elevated homocysteine level and folate deficiency associated with increased overall risk of carcinogenesis: meta-analysis of 83 case-control studies involving 35,758 individuals. PLoS One 2015;10:e0123423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro R, Rivera I, Ravasco P, Camilo ME, Jakobs C, Blom HJ, et al. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C-->T and 1298A-->C mutations are associated with DNA hypomethylation. J Med Genet 2004;41:454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ezeonwuka CD, Rastegar M. MeCP2-related diseases and animal models. Diseases 2014;2:45–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data analysed in this study have been generated in a previous work (8), and are fully available in the supporting information of the current article.