Abstract
Purpose:
Current endoscopy-based screening and surveillance programs have not been proven effective at decreasing esophageal adenocarcinoma (EAC) mortality, creating an unmet need for effective molecular tests for early detection of this highly lethal cancer. We conducted a genome-wide methylation screen to identify novel methylation markers that distinguish EAC and high grade dysplasia (HGD) from normal squamous epithelium (SQ) or non-dysplastic BE (NDBE).
Experimental Design:
DNA methylation profiling of samples from SQ, NDBE, HGD and EAC was performed using HM450 methylation arrays (Illumina) and reduced-representation bisulfite-sequencing. Ultra-sensitive methylation-specific droplet digital PCR and NGS-based bisulfite-sequencing assays were developed to detect the methylation level of candidate CpGs in independent esophageal biopsy and endoscopic brushing samples.
Results:
Five candidate methylation markers were significantly hypermethylated in HGD/EAC samples compared to SQ or NDBE (P < 0.01) in both esophageal biopsy and endoscopic brushing samples. In an independent set of brushing samples used to construct biomarker panels, a 4-marker panel (Model 1) demonstrated sensitivity of 85.0% and 90.8% for HGD and EACs respectively, with 84.2% and 97.9% specificity for NDBE and SQ respectively. In a validation set of brushing samples, the panel achieved sensitivity of 80% and 82.5% for HGD and EAC respectively, at specificity of 67.6% and 96.3% for NDBE and SQ samples.
Conclusions:
A novel DNA methylation marker panel differentiates HGD/EAC from SQ/NDBE. DNA-methylation based molecular assays holds promise for the detection of HGD/EAC using esophageal brushing samples.
Keywords: esophageal adenocarcinoma, Barrett’s esophagus, high-grade dysplasia, DNA methylation, biomarkers for detection
Introduction
Esophageal adenocarcinoma (EAC) incidence is rapidly increasing in the US and affects approximately 20,000 people each year1. It has a poor prognosis with five year survival rates of <20% 2. Survival rates dramatically improve if EAC is detected at an early stage, when it can be cured by surgical or endoscopic resection. Importantly, virtually all EAC is believed to arise from a precancerous condition called Barrett’s esophagus (BE), which can evolve into EAC over time through a BE→low grade dysplasia (LGD)→high grade dysplasia (HGD)→EAC progression sequence. Important recent developments in the treatment of HGD and early EAC are radiofrequency ablation (RFA) and endoscopic mucosal resection (EMR), which are endoscopic therapies that can remove these lesions, allowing ingrowth of neosquamous tissue, and dramatically reducing cancer risk3, 4. These endoscopic treatments demonstrate low morbidity and mortality, substantially reducing the risk for EAC therapy related death. Given these low morbidity and mortality treatment options, assays that can detect HGD and early stage EAC are of high value and have the potential to reduce EAC-related death and preserve the quality of life of BE patients when included in a surveillance program.
Aberrant DNA methylation has been shown to be a common molecular feature of BE and EAC 5–8. Although the functional significance of the majority of these DNA methylation events is unclear, they have proven to be highly promising as biomarkers 9–12. Aberrantly methylated DNA biomarkers have been used to develop a “molecular cytology” assay based on methylated VIM in DNA samples from esophageal cytology brushings obtained during endoscopies of 322 individuals, divided into training and validation cohorts 11. The assay showed 91% sensitivity and 93% specificity for detecting BE, BE with dysplasia, and EAC, with essentially identical results obtained in both the training and validation cohorts. In a subsequent study that used an assay to detect mVIM and mCCNA1 in samples collected via a non-invasive device, EsoCheck, the assay detected 90.3% of BE patients with 91.7% specificity10.
In light of these studies, we conducted a genome-wide methylation screen to identify potential biomarkers specific for HGD and EAC. We discovered and validated two models that have potential to be used as DNA-methylation based molecular assays for the minimally invasive detection of HGD and EAC. These results demonstrate a novel assay to detect non-BE related DNA methylation that is specific for HGD and EAC.
Materials and Methods
Study design
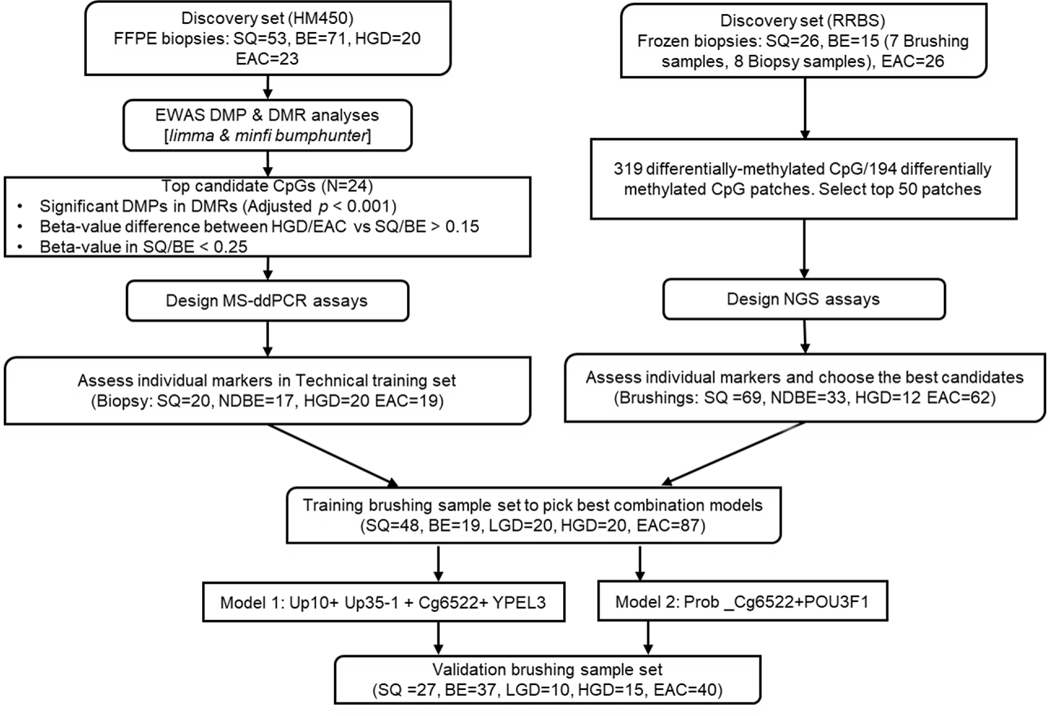
This was a non-randomized observational study. Study size was not pre-specified, and results are reported for esophageal brushing samples accrued from June 2011 to April of 2019. The primary endpoint of detection of BE and more advanced lesions were pre-specified before study initiation. All brushing samples were assayed by investigators blinded to the clinical status of the subjects from whom the samples were obtained. The clinical trial registration number at ClinicalTrials.gov is NCT00288119 for the endoscopic cytology brushings study. See Figure 1 for the overall study design and workflow.
Figure 1:
Development of DNA-methylation Biomarkers for HGD/EAC Early Detection (workflow)
Tissue Samples
For the Illumina HumanMethylation450 BeadChip arrays (HM450 arrays), the discovery set of formalin-fixed paraffin-embedded (FFPE) slides included a total of 53 normal squamous tissue (SQ), 71 BE, 20 HGD, and 23 EAC samples. They were obtained at the time of endoscopic exam or surgical resection (with no neoadjuvant therapy) from Case Western Reserve University/University Hospitals of Cleveland (Cleveland, OH), Cleveland Clinic (Cleveland, OH), University of North Carolina School of Medicine (Chapel Hill, NC), and University of Washington Medical Center (Seattle, WA), following protocols approved by the Institutional Review Board of each institution. H&E-stained slides were created for each sample and examined by an expert gastrointestinal pathologist (JEW or DR) to confirm diagnosis and identify precise areas with the histological subtypes of interest. Unstained slides were then matched with the annotated H&E slides and a sterile razor blade was used to remove the tissue for DNA extraction. In cases with mixed histology, special care was taken to separate histological subtypes before extraction.
For the Reduced-representation bisulfite sequencing study preformed as previously described10, the discovery set consisted of 26 biopsy pairs of EACs with respective matched normal squamous (SQ) samples, 8 BE biopsies, 7 BE endoscopic brushings, and 5 esophageal cancer cell lines. The biopsies and brushings were collected at Case Western Reserve University, Washington University, and John Hopkins University. Four esophageal cell lines (SKGT4, FLO1, OE19, OE33) were obtained from Sigma Aldrich. JH-Esoad1 esophageal cancer cell line was a kind gift from Drs. Anirban Maitra and James Eshleman13. Mycoplasma testing was preformed using an MycoAlert kit obtained from Lonza, on all cell lines after 10 passages. All cell lines were used at less than 20 passages and discarded at passage 20. NGS- -based methylation assays were designed for the top candidate loci identified by RRBS analysis.
The ddPCR assay for the top candidate markers identified via array analysis was tested on an independent set of endoscopic biopsies and endoscopic mucosal resection (EMR) samples obtained from University of Washington Medical Center (UWMC), with a total of 20squamous, 17 non-dysplastic BE (NDBE), 20 HGD, and 19 EAC samples. DNA from squamous samples was extracted from fresh-frozen tissue and all other tissue types were extracted from FFPE slides examined by an expert gastrointestinal pathologist DR. This sample set was used to test individual markers with MS-ddPCR assays and construct logistic regression model.
All brushing specimens were obtained at the time of the endoscopic exam using a through-the-scope cytology brush, prior to passage of the endoscope through the distal esophagus. Demographic information on the subjects in each sample set can be found in Supplementary Table 1. Control subjects had no endoscopic evidence of BE and no histological evidence of intestinal metaplasia. In the brushing sample sets, junctional cancer cases (JCA) were analyzed with EAC cases jointly.
Bisulfite DNA preparation
Genomic DNA was extracted from fresh-frozen tissues with the DNeasy Blood and Tissue Kit, following manufacturer’s instructions and eluted into a total volume of 100 uL. The QIAamp DNA FFPE Tissue Kit (Qiagen) was used to extract genomic DNA from FFPE tissues according to manufacturer’s instructions with the modification of lysing all tissues overnight. Samples were then eluted into 25–100 uL, dependent upon tissue size and kit protocol recommendations. Quant-iT PicoGreen DNA assay kit (Life Technologies), or Qubit HS DNA kit (ThermoFisher) were used to quantify genomic DNA before bisulfite conversion. The EZ DNA Methylation Kit (ZymoResearch) or the QIAGEN Epitect kit were used for bisulfite conversion.
HM450 arrays and genome-wide differential methylation analysis
The Infinium HD FFPE DNA Restore Kit (Illumina Inc.) was used to process bisulfite-converted DNA according to the manufacturer’s instructions. DNA samples were submitted to the Genomics Core at the Fred Hutchinson Cancer Research Center (FHCRC) for processing and subsequently run on the HM450 arrays according to manufacturer’s instructions (Illumina Inc.). Data acquisition, normalization, filtering, and analysis were conducted as previously 6, 9. Each CpG site was evaluated for differential methylation between SQ/BE and HGD/EAC by comparing mean ‘beta-values’ for the two groups (0.0=0% methylation, 1.0=100% methylation) using R Limma with adjustment for the sample age and gender. Differentially methylated genomic regions were also analyzed between the two groups using R minfi bumphunter14. CpG sites with statistically significant higher methylation in HGD/EAC vs. SQ/BE with the adjusted p value < 0.001, beta-value difference > 0.15, baseline beta-value < 0.25 and located in differentially methylated regions were considered for assay development.
Methylation-specific ddPCR
MethyLight PCR assays were designed for three CpGs, referred to as cg6522, YPEL3, and POU3F1 (cg4156522, cg16348385, and cg38512601, respectively) and methylation-specific ddPCR was run on esophageal biopsy and brushing DNA samples. ABI Primer Express Software version 3.0.1 primer/probe test tool was used to manually design the primer and probe sequences for each region. The C-LESS-C1 assay was used as a methylation-independent control as previously described 15–17. A list of all primer/probe sequences used can be found in Supplementary Table 2.
Methylation-specific ddPCR reactions contained 2x ddPCR Supermix for Probes (no dUTP) (BioRad), locus-specific primers (900 nmol/L), and locus-specific probes (250 nmol/L). Each reaction was done in duplex with the CpG of interest and the control assay, as previously described 16, 17. Bisulfite-converted samples were used as template DNA, 100% methylated EpiTect Methyl DNA was used as positive control, and 100% unmethylated EpiTect Unmethyl DNA (Qiagen) was used as negative control; all samples were run in duplicate. Reactions underwent droplet generation using the QX200 droplet generator (BioRad). Thermocycler conditions were performed in the T100 Thermal Cycler (BioRad): 95°C for 10 min, 45 cycles of 94°C for 30 seconds followed by 60°C for 1 min, 98°C for 10 min, and hold at 4°C indefinitely. Results were generated using the QX200 Droplet Reader (BioRad) and data were analyzed with QuantaSoft Software as previously described16, 17. Methylation status in both validation sets was reported as relative methylation percentage (RM%), calculated as a ratio percentage of the amount of target methylated alleles (cg6522, YPEL3, or POU3F1) over total DNA measured by the C-LESS-C1. RM% values were determined as means of duplicates.
Bisulfite-Seq-based methylation detection (NGS Bisulfite-Seq)
Bisulfite-specific, methylation indifferent PCR primers were constructed as a mixture of primers against converted products of fully methylated or fully unmethylated templates and were used to amplify differentially-methylated regions of GRASP and MAFB (Supplementary Table 3). Platinum Taq reaction mix (Invitrogen) was supplemented with 1mM MgCl2, 0.2mM dNTP mix (New England Biolabs), 0.5M Betaine (Sigma), and a mix of the 4 primers, each at 0.1μM final concentration. PCR was performed using a touchdown protocol as follows: the activation of Taq polymerase at 95°C for 5 min, the initial cycling conditions were: 95°C for 45 sec, 67°C for 45sec, 72°C for 45sec. The annealing temperature was decreased by 3°C every 3 cycles, to a final of 55°C. An additional 35 cycles of PCR were performed at the annealing temperature of 55°C. Successful amplification was confirmed by agarose gel electrophoresis. PCR products were purified using NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), and quantitated by Qubit. The NEXTflex Rapid DNA-seq kit (BIOO Scientific) was used to prepare indexed libraries for NGS sequencing (Illumina-compatible) (based on indexed adapters), and NGS was performed at the McGill University and Génome Québec Innovation Centre, Montréal, Canada, as previously described 10.
Classification/prediction for HGD/EAC using maker methylation cutoff values
An ROC curve analysis of BE (controls) vs EAC (cases) was performed on the training set of brushings samples. The cutoffs were chosen above the ROC-derived cutoff to maximize the specificity over sensitivity.
Classification/prediction for HGD/EAC using a logistic regression model
A logistic regression model was trained on the biopsy sample set (training set) as , where p and β stand for the probability score of the specified sample being positive and the intercept or coefficient of each marker. . The trained model was then tested on the two brushing samples sets. Different combinations of the three candidate markers were compared based on area under the ROC curve (AUC), with the largest AUC indicating the best performance. The diagnostic sensitivity, specificity, and accuracy, defined as (true positives + true negatives) / total sample size, were calculated in both the brushing training and testing sets (Supplemental Figure 2).
Statistical Analysis
Differential methylation analysis of the discovery HM450 array data was performed using R minfi and limma. Results shown here from the training and validation sample sets comparing methylation levels between different histologic groups were generated using GraphPad Prism version 7 (GraphPad Software Inc.). Chi-Square test and ANOVA were used to test difference of categoric and numeric clinical variables among different tissue samples.
Data Availability
A summarized version of the RRBS data and of the HM450 array data supporting the findings of this study are available within the article’s supplementary files ((Supplemental Table 10–11). The raw sequencing data and HM450 array data are not publicly available due to patient privacy requirements but are available upon request from the corresponding author. Other data generated in this study are available within the article and its supplementary data files.
Results:
DNA methylation array discovery of biomarkers for detection of esophageal high-grade dysplastic and malignant lesions
We conducted genome-wide DNA methylation profiling using Illumina HumanMethylation450 (HM450) Beadchip arrays on a set of DNA samples from normal esophageal squamous epithelium (SQ) (N=53), non-dysplastic BE (N=71), HGD (N=20), and EAC (N=23). After data normalization and filtering as previously described 6, 9, there were 426,464 CpGs for further evaluation. We performed differentially methylated probe and differentially methylated region (DMP/DMR) analyses using limma & minfi bumphunter (See Methods and Figure 1 for overall study design and workflow). Twenty-four CpGs were identified to be significantly hypermethylated in the HGD and EAC samples compared to the NDBE or SQ samples, using the following cutoff values: difference in mean beta-value > 0.15 (HGD and EAC vs. BE and SQ), baseline beta values in SQ or NDBE < 0.25, and false discovery rate q < 0.001. These 24 top candidate CpGs were selected for further assessment and validation (Supplementary Table 4).
We assessed the association of the methylation status of CpG sites with potential confounding factors, such as age and gender, using the univariate linear regression model (P <0.05 was considered significant). We did not find age or gender to be significant confounding variables. We did not assess race as a confounding variate because the majority of the subjects from which our samples were obtained are Caucasians (Supplementary Table 1).).
We designed droplet digital PCR assays that measure the methylation level of the ~150 bp region surrounding the target CpGs on the HM450 arrays for the top candidate CpGs (Supplementary Table 4). We determined the limit of detection and limit of quantification for each assay, selected the best performing assays for Cg6522, YPEL3 and POU3F1 and then used the assays on DNA extracted from tissue samples. The primer and probe sequences for these MS-ddPCR assays are listed in Supplementary Table 2.
Next, we used the MS-ddPCR assays for Cg6522, YPEL3 and POU3F1 to assess their methylation status in an independent set of DNA samples extracted from endoscopic esophageal biopsies (SQ=20, NDBE =17, HGD=20, EAC=19). As shown in Supplementary Figure 1, the mean methylation levels of all three genes were significantly elevated in EAC samples and for POU3F1 in HGD samples (p < 0.01, compared to the SQ or NDBE).
Reduced-representation bisulfite sequencing (RRBS) discovery of DNA methylation-based biomarkers for detection of esophageal high-grade dysplastic and malignant lesions
In a separate discovery approach, we performed RRBS analysis as previously described10, on a set of 26 esophageal adenocarcinoma (EAC) biopsies and their respective matched normal squamous biopsies (SQ), 15 biopsy or brushing samples of Barrett’s Esophagus (BE), and 5 esophageal cancer cell lines. Out of 3,091,193 analyzable CpGs, 21,911 CpGs showed a methylation level below 5% in all the informative BE samples (requiring at least 3 informative BEs each having sequencing depth of at least 20x), and additionally had >90% of the informative SQ samples at methylation level below 12% (requiring at least 4 informative SQ samples, each having sequencing depth of at least 20x). 319 of these CpGs additionally showed a level > 20% methylation in at least 5 of the informative EAC cases (all having sequencing depth of >20X).
These 319 CpGs that were differentially methylated between BE and SQ vs EAC samples were clustered into 194 differentially methylated CpG patches (defined as clusters of differentially methylated CpGs each less than 400bp apart). Fifty one of these patches were selected for further inspection (Supplementary Table 5). Of these, the top candidates for discriminating nondysplastic BE lesions from esophageal cancer were Up10, a 2-CpG patch located on chromosome 12, in the CpG island spanning the promoter and 5’ UTR of General Receptor for Phosphoinositides 1-associated Scaffold Protein (GRASP), and Up35, comprising two patches, one of 3 CpGs (up35–1), another of single CpG (up35–2), both located 844 bp apart in the CpG island of MAF bZIP transcription factor B (MFAB) gene on chromosome 20.
To further interrogate the Up10- and Up35- associated DNA methylation patches, we designed a next-generation sequencing (NGS)-based assay for targeted resequencing of these differentially-methylated regions (Supplementary Table 3) as previously described 10. We used this method to characterize a set of esophageal cytology brushings and to compare the performance of DNA methylation at the Up10 and Up35 loci. An Up10 read was considered methylated if any 26 out of 32 CpGs in the Up10 patch were methylated in a single read. For Up35, a read was methylated if methylation was observed in at least 14 out of 20 CpGs in the amplicon. The samples were classified as methylated for a given marker if more than 1% of total observed marker reads were methylated.
Assessment of top candidate methylated DNA biomarkers in a common training set of esophageal brushing samples
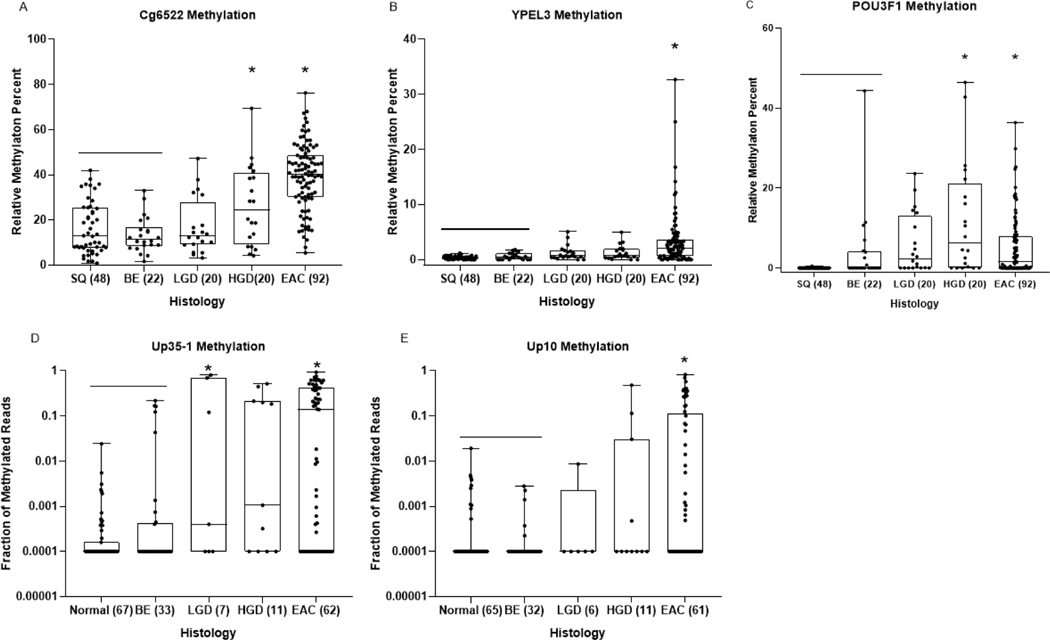
To assess the performance of individual markers and to construct the optimal marker panel for detecting HGD and/or early EAC, we examined the marker performance in a common set of cytology brushings from 202 individuals composed of 48 controls with normal esophageal endoscopic exams, 22 with NDBE, 20 with LGD, 20 with HGD and 92 cases with EAC (Figure 2, Table 1, Supplementary Table 7). The methylation level of each individual marker was significantly increased in brushings from EAC cases, and Cg6522 and POU3F1 methylation was also significantly increased in brushings from HGD, when compared to brushings obtained from normal squamous tissue (P<0.01).
Figure 2:
Dot-plot graphs of individual markers Cg6522, YPEL3, POU3F1, Up35–1 and Up10 (A, B, C, D, E, respectively) in DNA extracted from esophageal cytology brushing sample set 1 (training brushing sample set). Each sample was defined by histologic diagnosis, shown on x-axis, as determined by pathologist D.R. The y-axis shows the relative methylation percent when compared to the reference gene C-LESS for total DNA, as measured by MS-ddPCR assays for each marker in panels A,B,C. The fraction of methylated reads is plotted on the Y axis in panels D and E. One way ANOVA was used to determine statistical significance between histologic groups based on relative methylation percent. P-values < 0.01 when compared to SQ/BE tissue are considered significant (*).
Table 1:
Performance of model 1 and model 2 in Training set of Esophageal Brushings
| Performance of Individual markers | Model 1 | Model 2 | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| # samples | Up10 | Up35-1 | cg6522 | YPEL3 | POU3F1 | Up10+Up35-1 + Cg6522+ YPEL3 | Prob_Cg6522+POU3F1 | |
| Cutoff for positivity | 0.01 | 0.01 | 40 | 2.5 | 12 | Cutoffs as for Individual markers | 0 45 | |
|
| ||||||||
| %Specificity SQ’s (95% Cl) | 48 | 100.0 | 100.0 | 97.9(93.8–100) | 100.0 | 100.0 | 97.9(93.8–100.0) | 85.4(74.6–96.2) |
|
|
||||||||
| %Specificity BE (95% Cl) | 19 | 100.0 | 84.2(66.3–100.0) | 100.0 | 100.0 | 100.0 | 84.2(66.3–100.0) | 94.7(84.4–100.0) |
|
|
||||||||
| %Sens LGD (95% Cl) | 20 | 20.0(2.0–38.0) | 35.0(0–70.0) | 5.0(0–47 7) | 15.0(0–37.1) | 25.0(0–63.0) | 50.0(19.0–81.0) | 45.0(12.5–77.5) |
|
|
||||||||
| %Sens HGD (9S% Cl) | 20 | 25.0(0.63.0) | 75.0(53.1–96.9) | 25.0(0–63.0) | 15.0(0–55.4) | 35.0(0–70.0) | 85.0(68.0–100.0) | 80.0(60.4–99.6) |
|
|
||||||||
| %Sens Early EAC (95% Cl) | 32 | 42.9(18.6–67.1) | 46.4(21.1–71.7) | 50.0(23.8–76.2) | 21.4(4.3–38.6) | 25.0(6.5–43.5) | 80.8(46.2–100.0) | 89.2(54.3–100.0) |
|
|
||||||||
| %Sens EAC (95% Cl) | 87 | 34.5(17.5–51.5) | 51.7(37.1–66.3) | 50.6(35.8–65.3) | 43.7(27.9–59.5) | 16.1(0–35.3) | 90.8(84.4–97.2) | 87.4(79.9–94.8) |
Next, we determined the sensitivity and specificity of each biomarker when used singly and in combination as a panel, and constructed a model with combination of four markers, Up10, Up35–1, Cg6522 and YPEL3 (Model 1). With each marker at the cutoff values (Table 1) and calling a sample positive if any individual marker scored as methylated, Model 1 demonstrated a high sensitivity for both HGD and EAC at 85% (95% CI: 68.0–100.0) and 90.8% (95%CI 84.4–97.2), respectively, with specificity 84.2% (95% CI 66.3–100.0) for NDBE. As an alternative approach using a logistic regression model and area under the curve (AUC) for individual marker and marker combinations (Supplemental Table 8), we constructed a second model composed of Cg6522 and POU3F1, which was logit(p) = −3.75 + 0.1130×Cg6522 + 0.1926× POU3F1. When the cutoff p value was set at 0.45, the AUC in the brushing training set was 0.93 with a sensitivity of detecting HGD and EAC at 80% (95% CI: 60.4–99.6) and 87.4% (95% CI: 79.9–94.8) respectively, with specificity for SQ and NDBE at 85.4% (95% CI: 74.6–96.2) and 94.7% (95% CI: 84.4–100.0), respectively (Table 1, Supplemental Figure 2A). Both models detected roughly half of low-grade dysplasia (LDG) cases (50% (95% CI: 19.0–81.0) for Model 1, 45% (95% CI:12.5–77.5) for Model 2).
We also collected tumor stage information in this training brushing sample set to assess the accuracy of the biomarker panel for the detection of early EAC cases (tumor stage T0 and T1). In the training set of brushing samples, Model 1 achieved sensitivity of 80.8% for early EAC, with 97.9% and 84.2% specificity for SQ and NDBE; Model 2 achieved sensitivity of 89.2% for early EAC, with 85.4% and 94.7% specificity for SQ and NDBE (Table 1).
Assessment of models in an independent set of validation bushing sample set
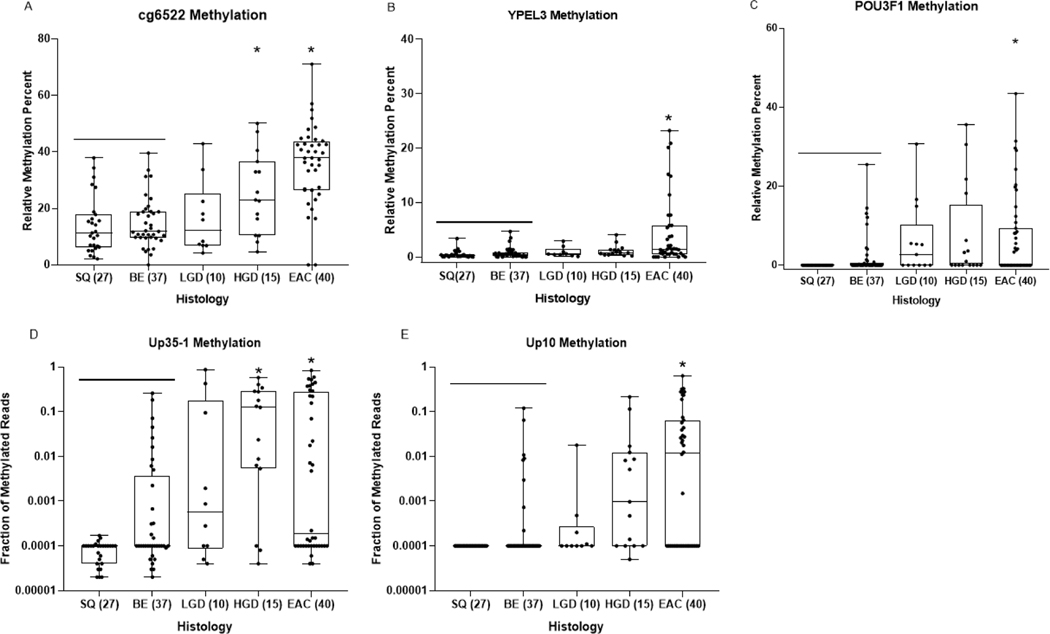
To validate the performance of the two models, we examined the methylation levels of individual markers Cg6522, YPEL3, POU3F1, Up35–1 and Up10 in a second independent set of esophageal cytology brushings from 27 control patients with normal esophageal endoscopic exams, 37 with NDBE, 10 with LGD, 15 with HGD and 40 cases with EAC (Figure 3, Supplementary Table 9). Models 1 and 2 were compared side-by-side in a core set of samples where all markers have passed QC and using the same cutoff values as in the training set. In the validation set, the 4-marker panel (Model 1) achieved sensitivity of 80% (95% CI: 57.4–100.0) and 82.5% (95% CI: 61.0–100.0) for HGD and EAC, at specificity of 67.6% (95% CI: 49.2–85.9) and 96.3% (95% CI: 89.0–100.0) for NDBE and normal squamous tissues (Table 2). In the same validation samples, the logistic regression model composed of Cg6522 and POU3F1 (Model 2) displays a higher specificity for NDBE at 83.8% (95% CI: 70.8–96.8), yet its sensitivity of detecting HGD decreased to 67% (95% CI: 42.9–90.5). The AUC in the brushing validation set was 0.90 (Supplemental Figure 2B). Both models performed equivalently for detection of EAC at sensitivity of 82.5% (Model 1, 95% CI: 61.0–100; Model 2, 95% CI: 69.5–95.5) and discriminating EAC from NDBE. For the small subset of early EAC cases (stage T0 and T1) (N=12), Model 1 demonstrated sensitivity of 70% (95% CI: 18.1–100) while Model 2 performed slightly better with sensitivity of 80% (95% CI: 24.6–100).
Figure 3:
Dot-plot graphs of individual markers Cg6522, YPEL3, POU3F1, Up35–1 and Up10 (A, B, C, D, E, respectively) in DNA extracted from esophageal cytology brushing sample set 2 (validation brushing sample set). Each sample was defined by histologic diagnosis, shown on x-axis, as determined by pathologist D.R. The y-axis shows the relative methylation percent when compared to the reference gene C-LESS for total DNA, as measured by MS-ddPCR assays for each marker in panels A,B,C. The fraction of methylated reads is plotted on the Y axis in panels D and E. One way ANOVA was used to determine statistical significance between histologic groups based on relative methylation percent. P-values < 0.01 when compared to SQ/BE tissue are considered significant (*).
Table 2:
Performarvce of model 1 and model 2 In Validation set of Esophageal brushings
| Common core set of samples | Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| # samples | Up10 | Up35-1 | Cg6522 | YPEL3 | Up10+ Up35-1 + Cg522+ YPEL3 | Prob_Cg6522+POU3F1 | |
| Cutoff | 0.01 | 0.01 | 40 | 2.5 | Cutoffs as for individual markers | 0.45 | |
|
| |||||||
| %Specificity SQ’s (95% Cl) | 27 | 100.0 | 100.0 | 100.0 | 96.3(89.0–100.6) | 96.3 (89.0–100.0) | 92.6(82.3–100.0) |
|
|
|||||||
| %Specificity BE (95% Cl) | 37 | 91 9(82.3–100.0) | 83.8(70.8–96.8) | 100 0(91.0–100.0) | 91.9(82.7–100.0) | 67.6(49.2–85.9) | 83.8(70.8–96.8) |
|
|
|||||||
| %Sens LGD (95% Cl) | 10 | 10.0 (0–70.0) | 30.0(0–80.0) | 10.0(0–68.8) | 10.0(0–68.0) | 50(6.2–93.8) | 40.(0–88.0) |
|
|
|||||||
| %Sens HGD (95% Cl) | 15 | 27.0(0–67.0) | 60.0(28.0–92.0) | 20.0(0–65.3) | 13.0(0–59.9) | 80(57.4–100.0) | 67(37.9–96.1) |
|
|
|||||||
| %Sens Early EAC (95% Cl) | 12 | 60.0(11 9–100.0) | 60.0(11.9–100.0) | 20.0(0–47.7) | 20.0(0–47.7) | 70.0(18.1–100) | 80.0(24.6–100.0) |
|
|
|||||||
| %Sens EAC (95% Cl) | 40 | 52.5(31 1–73.9) | 40.0(16.0–64.0) | 45.0(22.0–68.0) | 35.0(10.0–60.0) | 82.5(61.0–100.0) | 82.5(69.5–95.5) |
Discussion
The need to improve detection of BE progression in surveillance of patients with BE led us to determine whether DNA methylated biomarkers in cytology brushing samples have the potential to detect HGD or early EAC. In this study we first conducted a genome-wide methylation analysis by two independent methods, followed by an exhaustive candidate biomarker search. We discovered a list of candidate methylated DNA biomarkers that showed significantly higher methylation levels in HGD and EAC compared to the normal squamous tissue and BE samples. Ultra-sensitive methylation-specific droplet digital PCR assays and bisulfite-sequencing-based assays were designed for each individual marker. We determined the sensitivity and specificity of each biomarker when used singly and in combination as an assay panel and constructed two different models for discrimination of NDBE from HGD and EAC, in two independent sets of endoscopic cytology brushings. We found a 4-marker panel composed of two ddPCR assays (Cg6522 and YPEL3) and two NGS assays (Up10 and Up35–1) achieved sensitivity of 80% and 82.5% for HGD and EAC, at specificity of 67.6% and 96.3% for NDBE and normal squamous tissues in a validation set of brushing samples. These findings demonstrate the ability of DNA methylation markers to discriminate NDBE from HGD and EAC and the potential of these markers to be used in BE surveillance.
Droplet digital PCR (ddPCR) is a relatively new technology that enables the precise and sensitive detection and absolute quantification of nucleic acid targets in various clinical specimens. We and others have demonstrated its superior performance over conventional Methylight PCR for DNA methylation studies 15–17,18. In this study, we developed methylation-specific ddPCR assays for the top candidate CpG sites from the HM450 array studies. We demonstrated that MS-ddPCR assays can accurately quantify methylated Cg6522, YPEL3 and POU3F1 in as little as 4 ng of DNA from esophageal brushing samples. Since cytologic sampling yields limited amount of DNA with mixed cell types, our results support the feasibility of MS-ddPCR based assays for the development of DNA methylation-based molecular cytology assays for HGD and EAC detection. Moreover, dd-PCR has been approved by the FDA for use in clinical assays (e.g. Bio-Rad Laboratories’ QXDx BCR-ABL %IS kit and QXDX AutoDG ddPCR system), which helps minimize the barriers of our assay for adoption in the clinical setting. However, it is also important to note that a limitation of MS-ddPCR is that it can only determine the methylation status of a small number of CpGs, which can prevent the creation of technically robust assays for some promising DNA methylation-based biomarkers. The NGS approach allows for determining the methylation status for each CpGs in an amplicon across a single DNA strand, thus an evaluation of a larger stretch of CpGs within the amplicon. The patch-based algorithm suppresses background from random methylation of individual CpGs and provides enhanced discrimination of normal squamous versus diseased tissue. The NGS-based assay also allows for greater flexibility when designing the assays in the difficult to amplify genomic regions, as is often the case with GC-rich templates found in the CpG-dense areas. For example, while the MAFB region containing DNA methylation patch UP35 was identified as differentially methylated in both methylation array, and the RRBS screens, an adequately performing MS ddPCR assay was difficult to design for the specific CpGs identified in the array screen, while a NGS-based assay performed well. We propose that both assay technologies should be considered when designing methylated DNA based assays. We believe novel DNA methylation-based biomarkers combined with non-endoscopic samples devices, such as Cytosponge or Esocheck, hold promise to be a low-cost and minimally invasive surveillance method for individuals with BE for the early detection of HGD/EAC.
The strength of this best-performing biomarker panel lies in its capability to differentiate HGD and EAC from normal squamous tissue and NDBE. There are no existing assays that can accomplish this beyond tissue histology, which requires endoscopy, although NGS assays assessing TP53 mutations may have this potential 19. If combined with promising BE markers identified and validated by our group and others, we propose that these markers have potential to enable molecular enhancement of EAC screening and BE surveillance. The limitation of our studies includes lack of a pre-specified sample size at discovery and the relatively small sample size of the training and validation sets of esophageal brushing samples, thus requires further validation in larger cohorts. Furthermore, given the emerging technical advances in swallowable collection devices 20, we plan to validate the DNA-methylation based molecular assay in esophageal balloon collected samples, as this sample collection method appears to have a high likelihood of near-term adoption into clinical practice for at least BE screening. Finally, it is worth noting that the primary focus of this study is to identify and develop methylation markers that distinguish EAC/HGD from normal squamous epithelium or NDBE. Our results raise the question of whether these biomarkers can be used to identify HGD patients who will develop EAC in the future. Although intriguing, our current study design and sample collections preclude the assessment of the BE progression biomarkers, which is beyond the scope of this study.
In summary, these findings establish the proof of principal that DNA methylation can detect progression of esophageal neoplasia to cancer and lay the molecular foundation for further trials of a DNA methylation-based tests for the detection of HGD and EAC.
Supplementary Material
Statement of translational relevance.
Esophageal adenocarcinoma (EAC) is a common and lethal cancer of the GI tract which arises from Barrett’s esophagus (BE), a metaplastic alteration of the esophageal mucosa. Patients with BE are at risk for EAC and are placed in surveillance programs requiring upper endoscopy. The current screening and surveillance program for BE and EAC is suboptimal because it uses an expensive, invasive surveillance method with modest accuracy and limited efficacy. With recent advances in the management of HGD and early EAC with well-tolerated endoscopic therapies, there is a need for more convenient and sensitive non-endoscopic surveillance methods that can save costs, avoid harm, and reduce cancer incidence and related deaths. Newer methods that use swallowed esophageal cytology collection devices (Cytosponge and Esocheck) and molecular biomarkers have the potential for the non-invasive detection of HGD and early EAC and to meet these unmet needs. In our study, we have discovered and validated novel methylated DNA biomarkers for the detection of HGD and EAC.
Acknowledgments
Funding: These studies were supported by funding from the NCI:R50CA233042 to MY, NIH:UO1CA152756, RO1CA220004, P30CA015704, U54CA163060, UO1CA086402, UO1CA182940 and the Prevent Cancer Foundation to WMG, P50CA150964 and UO1CA152756 to SDM, , and U54CA163060 and P30DK097948 to AC. PGI receives funding from Exact Sciences, Pentax Medical, Cernostics. Funding is also provided by the Cottrell Family Fund, Evergreen Fund, and Listwin Foundation to WMG.
Conflicts of Interest:
W. M. Grady is an advisory board member for Freenome, Guardant Health, and SEngine and consultant for DiaCarta, Nephron, Guidepoint and GLG. He is an investigator in a clinical trial sponsored by Janssen Pharmaceuticals and receives research support from Tempus and LucidDx. S. D. Markowitz receives income related to patents licensed to Exact Sciences, has founders shares and stock options in LucidDx, serves as a consultant to LucidDx, has sponsored research with LucidDx, and has a royalty interest in patents licensed to LucidDx. S. Markowitz also is a consultant for and has royalty and milestone interests in intellectual property licensed to Amgen. S. Markowitz is also a consultant to and shareholder in Summit Biolabs. A. Chak is an equity holder and advisor for Lucid Diagnostics, consultant for Interpace Diagnostics, and receives research support from C2Therapeutics/Pentax Inc. J. Willis is an equity holder and advisor for Lucid Diagnostics. Helen R. Moinova is a consultant for Lucid diagnostics and has a royalty interest in patents licensed to LucidDx. Dr. Shaheen receives research funding from Medtronic, Steris, Pentax, CDx Diagnostics, Interpace Diagnostics, and Lucid Medical. He is a consultant for Cernostics, Phathom Pharmaceuticals, Exact Sciences, Aqua Medical, and Cook Medical. Prashanthi Thota receives research support from Interpace diagnostics. Dr. Prasad is a consultant for Exact Sciences, Pentax Medical, Cernostics, CDx Medical, Ambu, and Symple Surgical. Other authors declare no conflicts of interest.
References
- 1.Hur C, Miller M, Kong CY, et al. Trends in esophageal adenocarcinoma incidence and mortality. Cancer 2013;119:1149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers 2017;3:17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phoa KN, van Vilsteren FG, Weusten BL, et al. Radiofrequency ablation vs endoscopic surveillance for patients with Barrett esophagus and low-grade dysplasia: a randomized clinical trial. JAMA 2014;311:1209–17. [DOI] [PubMed] [Google Scholar]
- 4.Shaheen NJ, Overholt BF, Sampliner RE, et al. Durability of radiofrequency ablation in Barrett’s esophagus with dysplasia. Gastroenterology 2011;141:460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaz AM, Grady WM, Stachler MD, et al. Genetic and Epigenetic Alterations in Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterol Clin North Am 2015;44:473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, Maden SK, Stachler M, et al. Subtypes of Barrett’s oesophagus and oesophageal adenocarcinoma based on genome-wide methylation analysis. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jammula S, Katz-Summercorn AC, Li X, et al. Identification of Subtypes of Barrett’s Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuester D, El-Rifai W, Peng D, et al. Silencing of MGMT expression by promoter hypermethylation in the metaplasia-dysplasia-carcinoma sequence of Barrett’s esophagus. Cancer Lett 2009;275:117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, O’Leary RM, Kaz AM, et al. Methylated B3GAT2 and ZNF793 Are Potential Detection Biomarkers for Barrett’s Esophagus. Cancer Epidemiol Biomarkers Prev 2015;24:1890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moinova HR, LaFramboise T, Lutterbaugh JD, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moinova H, Leidner RS, Ravi L, et al. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev 2012;21:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Kambhampati S, Cheng Y, et al. Methylation Biomarker Panel Performance in EsophaCap Cytology Samples for Diagnosing Barrett’s Esophagus: A Prospective Validation Study. Clin Cancer Res 2019;25:2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez H, Koorstra JB, Hong SM, et al. Establishment and characterization of a bona fide barrett esophagus-associated adenocarcinoma cell line. Cancer Biol Ther 2008;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 2008;36:4689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu M, Heinzerling TJ, Grady WM. DNA Methylation Analysis Using Droplet Digital PCR. Methods Mol Biol 2018;1768:363–383. [DOI] [PubMed] [Google Scholar]
- 17.Yu M, Carter KT, Makar KW, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics 2015;10:803–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Wesenbeeck L, Janssens L, Meeuws H, et al. Droplet digital PCR is an accurate method to assess methylation status on FFPE samples. Epigenetics 2018;13:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver JM, Ross-Innes CS, Shannon N, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet 2014;46:837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaz AM, Grady WM. Novel Barrett’s esophagus screening assays based on swallowable devices: will they change the game? Transl Gastroenterol Hepatol 2019;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A summarized version of the RRBS data and of the HM450 array data supporting the findings of this study are available within the article’s supplementary files ((Supplemental Table 10–11). The raw sequencing data and HM450 array data are not publicly available due to patient privacy requirements but are available upon request from the corresponding author. Other data generated in this study are available within the article and its supplementary data files.