Abstract
Purpose:
Study the biology and identify markers of severe cytokine release syndrome (CRS) and immune effector cell-associated neurotoxicity syndrome (ICANS) in children post chimeric antigen receptor T-cell (CART) treatment.
Experimental Design:
We use comprehensive proteomic profiling to measure over 1400 serum proteins at multiple serial timepoints in a cohort of patients with B-cell acute lymphoblastic leukemia treated with the CD19 targeted CART CTL019 on two clinical trials
Results:
Identified FLT3 and MILR1 as pre-infusion predictive biomarkers of severe CRS. Demonstrated that CRS is an interferon gamma driven process with a protein signature overlapping with hemophagocytic lymphohistiocytosis (HLH). Identified IL18 as a potentially targetable cytokine associated with the development of ICANS.
Conclusion/Discussion:
We identified pre-infusion biomarkers that can be used to predict severe CRS with a sensitivity, specificity, and accuracy superior to the current gold standard of disease burden. We demonstrated the fundamental role of the interferon gamma pathway in driving CRS which suggests that CRS and carHLH are overlapping rather than distinct phenomena which has important treatment implications. We found IL-18 as a possible targetable cytokine in ICANS, providing rationale for IL-18 blockading therapies to be translated into clinical trials in ICANS.
Statement of Translational Relevance
We identify two pre-infusion biomarkers for CRS that can predict severity. These markers can be utilized in replacement of bone marrow aspirate to prognosticate CRS. We demonstrate significant overlap between HLH and CRS which suggests the use of cytokine blockade therapies for treatment. We identify IL-18 as a novel targetable biomarker for the treatment and prevention of ICANS. IL-18 blockade is currently being evaluated in other inflammatory conditions and could rapidly be translated into clinical trials in ICANS. In conclusion, these findings are clinically relevant with important implications for the development and management of CART.
INTRODUCTION
Chimeric antigen receptor T-cells (CART) against CD19 (CART19) have revolutionized the treatment of B-cell malignancies.(1) The most common adverse events associated with CART are cytokine release syndrome (CRS) and immune effector cell associated neurotoxicity syndrome (ICANS).(2) CRS is a disorder characterized by fever and inflammation related to CART and bystander cell activation.(2) Our group and others have previously demonstrated that the pathophysiology of CRS is related to the immune dysregulation syndrome hemophagocytic lymphohistiocytosis (HLH).(3,4) ICANS is a poorly understood phenomenon characterized by the development of a spectrum of neurologic symptoms that may include encephalopathy, seizure, or cerebral edema, following CART infusion.(2) Despite their frequency, the biology of both conditions is incompletely understood.
The current best predictor of both CRS and ICANS is disease burden prior to CART infusion, with patients with higher disease burden at higher risk of severe CRS and ICANS.(2) While disease burden is somewhat sensitive for the development of severe CRS it has poor specificity. Our group and others have recently shown through prospective clinical trials that early intervention with cytokine blockade may mitigate risk for severe CRS in the appropriate context.(5,6)
Cytokines, including IFNγ, IL-6, IL-10 and GCSF have been implicated in the pathophysiology of CRS and ICANS. Cytokine blockade targeting the IL-6 pathway is the current standard of care for the treatment of CRS.(2) Corticosteroids are the current standard of care for ICANS, but their efficacy is limited and steroids may theoretically impede CART efficacy.(2) Elevations of IL-1 in the CSF have been associated with the development of ICANS, and blockade of IL-1 has been shown to ameliorate ICANS in murine models, leading to the investigation of IL-1 blockade with anakinra in early phase clinical trials (NCT04150913, NCT04359784, NCT04148430).(7,8)
To better understand the biology of CRS and ICANS, and to identify potential biomarkers of severity and targets for cytokine blockade, we performed serial comprehensive profiling to measure 1463 soluble proteins in the serum of pediatric and young adult patients prior to and following infusion with the 41BB-containing CART19 CTL019. We make the following novel observations: (i) we identify candidate pre-infusion biomarkers of CRS severity, (ii) we provide a detailed serum proteomic analysis of patients with severe CRS, showing the fundamental role of IFNγ in this pathophysiology and identifying potentially targetable pathways, (iii) we identify targetable cytokines associated with the development of ICANS including IL-18, and (iv) we demonstrate that perturbations in the complement cascade may occur in association with CRS and ICANS.
MATERIALS AND METHODS
Study Design, Population, Clinical Categorization and Sample Collection
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board (IRB) at CHOP. Written informed consent was obtained from all patients. Serial time points were analyzed on pediatric patients enrolled on clinical trials of the 41BB containing CD19 directed CART CTL019 (NCT01626495 & NCT02906371). Proteomic analysis was performed on serial time points on an initial 26 patients, 13 with minimal and 13 with severe CRS. Details on this patient cohort have been reported previously.(3) A limited proteomic analysis was performed on 17 additional patients (4 severe and 13 minimal) at the pre-infusion timepoint. Disease burden was measured at pre-infusion and defined as the highest percentage of blasts by multiparameter flow for minimal residual disease, aspirate morphology, or biopsy.
CRS was graded according to the UPENN scale as previously described (Supplemental Table S1).(3) Minimal CRS was defined as patients with no CRS, grade 1, grade 2 or Grade 3 CRS without the need for pressors or non-invasive ventilatory support. Severe CRS included patients with grade 3 CRS who required pressors or non-invasive ventilatory support, and those with grade 4 CRS. This cohort did not include any patients with Grade 5 CRS. Neurotoxicity symptoms were abstracted from severe adverse event reports on both clinical trials. Onset of neurotoxicity was defined as the first day of any neurologic symptoms, with resolution defined as the last day of any neurologic symptoms.
Samples were collected as previously described at pre-infusion, post-infusion day 7-10, day 14 and resolution.(3) Per patient timepoints are shown in Supplemental Figure S1.
Proteomic Measurement
The Olink Explore 1536/384 panel (Olink Proteomics, Upppsala, Sweden) was used to measure 1463 proteins on serial timepoints from the serum of the initial cohort of 26 patients. A subset of 732 of these proteins, measured in the Explore 384 Oncology and Inflammation panels, were measured in the expansion cohort of 17 patients at the pre-infusion timepoint. All protein data is reported in normalized expression values (NPX), on a Log2 scale.
Measurement of sC5b9
Sc5b9 levels were measured using enzyme-linked immunosorbent assays (ELISA; BD Biosciences, CA, USA, #558315) at two dilutions and in triplicate.
Flow Cytometry
Flow cytometry was performed on PDX samples created from patients included in this analysis. All PDX were assessed for surface expression of MILR1 and FLT3. Antibodies were obtained from Miltenyi (MA, USA) and R&D Systems Inc (MN, USA). Data were analyzed using FlowJo version 10.8 (BD Biosciences, NJ, USA). A cut-off of 20% marker positivity was used as a binary criterion for positivity of expression.
Protein Analysis
Analysis of proteins was performed in R (version 4.0.4) using RStudio (Rstudio, MA, USA). False discovery rates were calculated using the Benjamini-Hochberg correction. Clustering analysis was performed using tSNE for R and Factoextra was used to abstract data from PCA. PathfindR was used for pathway analysis. Details of the machine learning analysis, protein-protein clustering, RNA sequencing analysis and Cystosig analyses and other methods are presented in Supplemental Methods S1.
Data Availability
The data generated in this study are available upon request from the corresponding author. In addition, publicly available data generated by others were used by the authors and are available from the Office of Cancer Genomics (https://ocg.cancer.gov/programs/target/data-matrix).
RESULTS
Clinical Description of Patients
Our initial discovery cohort included 26 patients treated with CTL019 on the clinical trials NCT01626495 & NCT02906371: 13 patients with minimal CRS and 13 patients with severe CRS, classified using the UPENN scale (Supplemental Table S1). Two patients with minimal and 12 patients with severe CRS developed ICANS. We collected serum at timepoints from pre-infusion to 35 days post-infusion, measuring 1463 proteins at 128 unique datapoints. Clinical details of patients are presented in Supplemental Table S2. Timing of the development of CRS, ICANS, tocilizumab administration and sample collection is presented in Supplemental Figure S1.
Pre-infusion Biomarkers for CRS
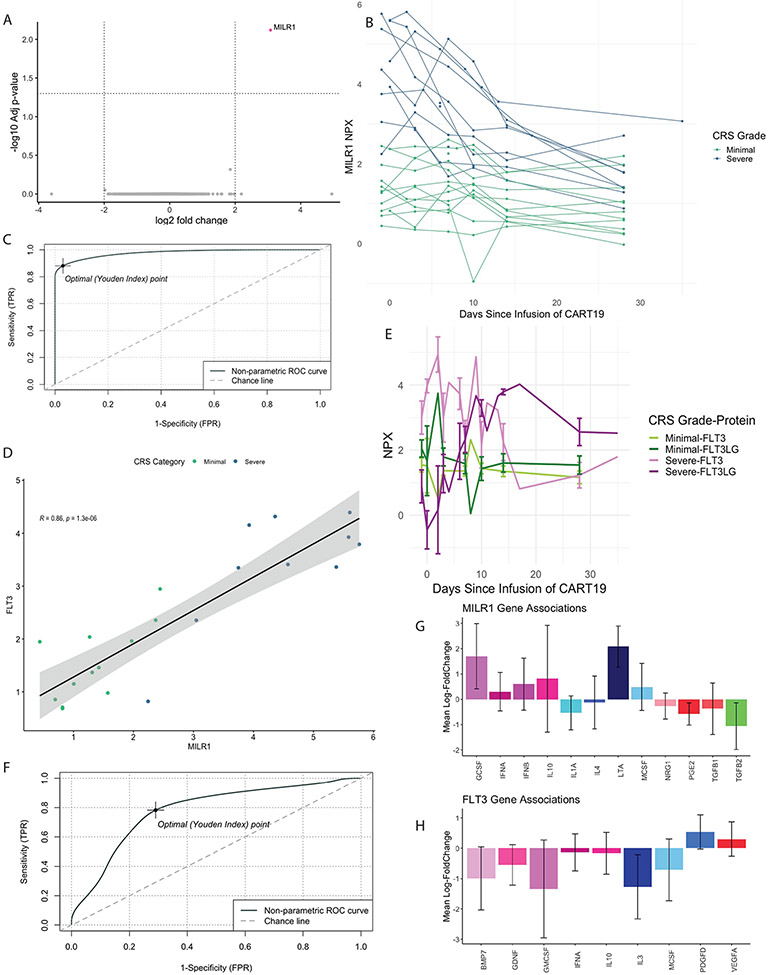
We sought to identify pre-infusion biomarkers predictive for severe CRS. In our discovery cohort, we performed a differential expression analysis (DEA) of proteins comparing severe and minimal CRS at pre-infusion (N=22; Figure 1A). A single protein, Mast Cell Immunoglobulin Like Receptor-1 (MILR1), a negative regulator of hypersensitivity reactions not previously associated with B-ALL, was identified.(9) We found that in patients who developed severe CRS (but not those with minimal CRS), MILR1 levels were initially high and declined over time (Figure 1B). Elevations in MILR1 at the pre-infusion timepoint predicted development of severe CRS with a sensitivity of 88% and a specificity of 97% (area under the curve = 0.977, N=22; Figure 1C).
Figure 1. Identification and validation of pre-infusion biomarkers for cytokine release syndrome.
(A) Differential expression analysis of proteins at the pre-infusion timepoint (N=22) comparing patients with severe and minimal cytokine release syndrome (CRS) identified a single significant marker, Mast Cell Immunoglobulin-like Receptor 1 (MILR1). An absolute fold change threshold of 2 and a false discovery rate (FDR) threshold of 0.05 were used. (B) MILR1 levels in patients with severe (N=13) and minimal CRS (N=13) over time. Patients with severe CRS had high levels of MILR1 at pre-infusion that decreased over time, where those with minimal CRS had consistent levels over time. (C) Receiver operative characteristic (ROC) curve of MILR1 as a binary predictor of severe CRS (N=22). (D) FLT3 levels and MILR1 levels at the pre-infusion timepoint are highly correlated (R=0.86, p=1.3x106, N=22). Dots are colored by CRS category with higher levels of both biomarkers in patients with severe CRS compared to those with minimal CRS. (E) Mean levels of FLT3 and FLT3-ligand (FLT3LG) in patients with severe (green lines) and minimal (purple lines) CRS. Patients with severe CRS have high levels of FLT3 at pre-infusion that decline over time in an inverse manner to levels of FLT3LG. (F) ROC curve of FLT3 and MILR1 in an expanded validation cohort (N=39) predicted the development of severe CRS with a sensitivity of 0.78 and a specificity of 0.71 (AUC=0.78). Cytosig outputs of proteins known to be associated with MILR1 (G) and FLT3 (H) from previously reported RNA sequencing data sets. Y-axis represents mean log fold change of significantly different cytokines (p ≤0.05) listed on the X-axis.
We next used linear regression to identify proteins that were highly correlated at the pre-infusion timepoint with MILR1 levels (Supplemental Data 1). Levels of the soluble form of the type III cytokine receptor fms-like tyrosine kinase 3 (FLT3) were highly correlated with MILR1 levels (Figure 1D; R=0.86, p=1.3x106). FLT3 showed a similar pattern over time to MILR1, with higher levels at pre-infusion decreasing over time in patients with severe CRS. Interestingly, FLT3’s canonical ligand, FLT3-ligand (FLT3LG) changed in an inverse pattern to FLT3 levels in patients with severe CRS (Figure 1E).
In this initial cohort both FLT3 and MILR1 were highly correlated with disease burden (Spearman R=0.85 and 0.82, respectively, Supplemental Figure S2). As disease burden is the current gold standard for predicting severe CRS, we looked at the candidate biomarkers FLT3 and MILR1 in a larger set of patients and expanded the cohort from 22 to 39 patients enrolled on the same clinical trials. To bias towards the null hypothesis that FLT3 and MILR1 were not predictive of severe CRS, we added 17 additional subjects with known low disease burden/severe CRS and high disease burden/minimal CRS (Supplemental Methods and Table S3). In these 17 additional patients we measured a smaller panel of proteins that incorporated both MILR1 and FLT3 at the pre-infusion timepoint. In the combined cohort of 39 patients, disease burden predicted severe CRS with a sensitivity 73% and specificity 52% (AUC=0.61, N=39). Both MILR1 (sensitivity = 71%, specificity = 70%, AUC = 0.73, N=39) and FLT3 (sensitivity = 68%, specificity = 62%, AUC = 0.63, N=39) were equal or better to disease burden in predicting the development of severe CRS. The combination of FLT3 and MILR1 predicted severe CRS with sensitivity 78% and specificity 71% (Figure 1F; AUC=0.78, N=39).
We next applied a machine-learning feature forward selection approach with leave-one-out-cross validation to identify pre-infusion biomarkers that are classifiers of severe CRS among the 732 proteins measured in all 39 patients. TGFA and LAIR1 were identified as the top two classifiers, with a model accuracy of 77.2% +/− 7.9 for prediction of severe CRS (Supplemental Table S4 and Figure S3). MILR1 alone in this model had an accuracy of 64.2% +/− 8.3. The combination of MILR1, TGFA and LAIR1 had an accuracy was 79.7% +/− 12 for prediction of severe CRS. Both MILR1 separately and the combination of MILR1, LAIR1 and TGFA out-performed disease burden (accuracy 54.2% +/− 14.2%).
As MILR1 and FLT3 correlated with disease burden, we hypothesized that MILR1 and FLT3 may be secreted by leukemic blasts. We examined the TARGET RNA-sequencing data set of pediatric patients with B-ALL (https://ocg.cancer.gov/programs/target/data-matrix; N=145) and found that B-ALL blasts expressed FLT3 and MILR1 (Supplemental Figure S4). We used flow cytometry to look at surface expression in patient derived xenografts (PDX) made from a subset of 9 patients included in this analysis. A representative flow plot is presented in Supplemental Figure S5. All samples were positive for surface FLT3 expression, and two of nine samples were positive for surface MILR1 expression (Supplemental Table S5).
To better understand the cytokine responses induced by MILR1 and FLT3 we queried the Cytokine Signaling Analyzer (CytoSig; Figure 1G&H).(10) Notably, MILR1 is associated with induction of the lymphocyte-produced cytotoxic protein LTA, with the T-cell inhibitory cytokine IL-10 and the neutrophil chemotactic factor G-CSF.
Cytokine Architecture at Peak CRS
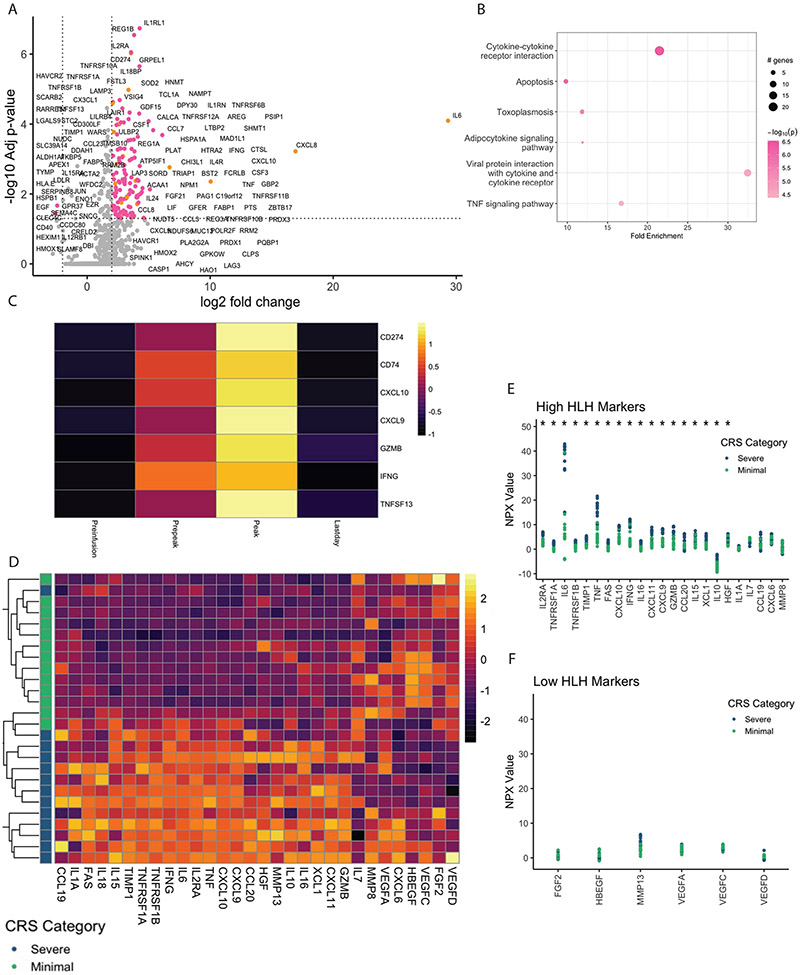
We next looked at cytokines at peak CRS, defined as the first day of measured cytokines after admission to the intensive care unit. In patients with minimal CRS, the peak timepoint was defined as day 7. The overall architecture of proteins at this timepoint visualized with principal component analysis (PCA) and t-distributed stochastic neighbor embedding (tSNE) are presented in Supplemental Figures S6-8. Patients with severe CRS cluster separately from patients with minimal CRS. Differentially expressed proteins (DEP) at peak CRS are presented in Figure 2A. Given the putative role of IFNγ in CRS pathophysiology, we classified DEP as IFNγ responsive based on a recently published comprehensive network map of IFNγ signaling.(11) Out of 131 DEP, 20 were IFNγ responsive (Figure 2A and Supplemental Data 2). We performed pathway analysis on DEPs to identify potentially targetable pathways (Figure 2B). Notably, the TNF-signaling pathway was identified in this analysis.
Figure 2. Insights into the pathophysiology of cytokine release syndrome at the peak timepoint.
(A) Differential expression analysis at the peak timepoint for patients with severe (N=13) compared to minimal (N=13) CRS. Colored dots meet the threshold of significance. Orange dots are dots known to be associated with IFNγ signaling. Pink dots are not known to be associated with IFNγ signaling. A fold change threshold of 2 and a false discovery rate (FDR) threshold of 0.05 were used to define significance. (B) Pathway analysis of significantly differentially expressed proteins is presented. Notably, the TNF signaling pathway was significantly perturbed. (C) Protein-protein clustering at pre-infusion, prior to peak, peak and last day timepoints in severe patients in whom all 4 timepoints were available (N=8). The cluster with the highest degree of correlation between the proteins and the cluster centroid (0.96) is presented. Notably IFNγ, and its canonical responders CXCL9 and CXCL10 were highly clustered together, as was the T-cell exhaustion marker CD274 (PDL1). (D) Heatmap of 29-protein signature associated with HLH clusters patients in to severe vs. minimal CRS when unsupervised clustering is applied at the peak timepoint (N=26). Data for the heatmap was mean-centered. Dot plot demonstrating per patient levels cytokines previously shown to be elevated (E) or decreased (F) in the setting of HLH. Patients with severe CRS (N=13) demonstrate a pattern very similar to that seen in patients with HLH. Y axis represents NPX log2 scale, *represents significantly different proteins with p-value ≤0.05 calculated using a Wilcoxon test. Green dots indicate patients with minimal CRS and blue dots represent patients with severe CRS.
We used unbiased, k-means protein-protein clustering to identify patterns of association over time in patients with severe CRS and identified 3 protein clusters. The cluster with proteins that have the highest correlation to the centroid is shown in Figure 2C with the others presented in Supplemental Figure S9. Notably, IFNγ, and its canonically induced chemokines CXCL9 and CXCL10, and the T-cell exhaustion associated protein CD274 (Programmed death-ligand 1) clustered together over time (Figure 2C).
We have previously reported on the similarities between HLH and severe CRS in a smaller panel of cytokines.(3) We sought to test this similarity by comparison of our Olink dataset to a recent proteomic analysis measuring 140 proteins comparing classical HLH patients to healthy controls reported by Lin et al.(12) 69 of the 140 proteins measured by Lin were also measured in our Olink dataset. 54 proteins were found to be significantly different in patients with HLH compared to healthy controls with 29 of these proteins measured by Olink. Unsupervised clustering using this 29-protein signature groups patients as having severe or minimal CRS, implying that a common pathophysiology underlies both HLH and CRS (Figure 2D).
Lin et al reported 39 proteins elevated in HLH relative to healthy controls. Twenty-three of these proteins were measured by Olink and 18 were significantly elevated in patients with severe compared to minimal CRS (Wilcoxon p-value ≤0.05; Figure 2E). They also identified 11 proteins which were lower in HLH than healthy controls. Six of these 11 proteins were measured by Olink and, as expected, all were low in all patients (Figure 2F).
IL-18 is associated with ICANS
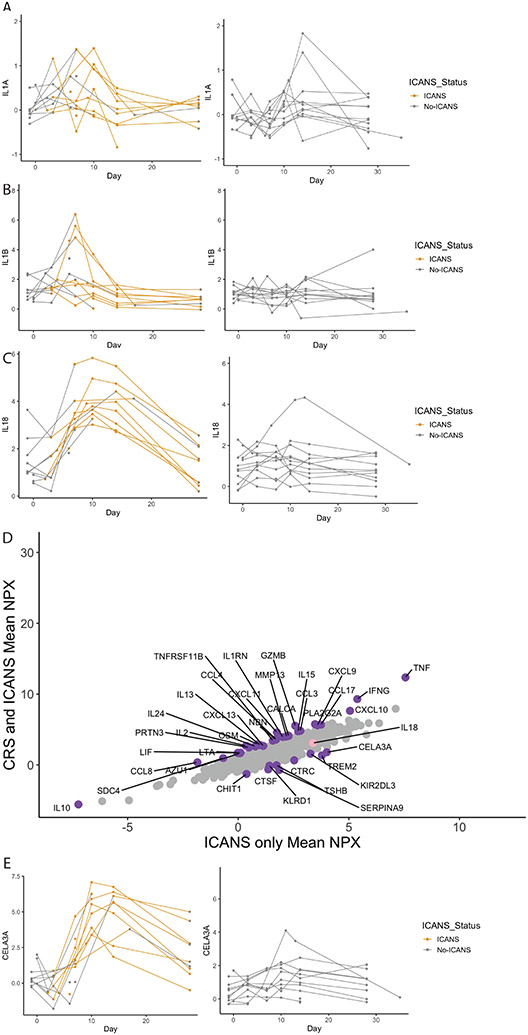
In animal models, IL-1 has been invoked as potentially related to the pathophysiology ICANS.(8) We thus looked at the association between the IL-1 family of proteins and the timing of ICANS symptom onset. Interestingly, neither IL-1A or IL-1B were associated with the onset of ICANS symptoms (Figure 3A&B). We then looked at all proteins in the IL-1 family measured by Olink (Supplemental Figure S10). We found that IL-18 best recapitulated the onset of ICANS symptoms (Figure 3C). IL-18 binding protein demonstrated a similar pattern (Supplemental Figure S11). Only one patient categorized as not having ICANS had significant elevations in IL-18. Notably, this child was infused prior to the recognition of ICANS as a clinical entity. The patient’s neurologic assessment was confounded due to intubation and sedating medications.
Figure 3. Immune effector cell associated neurotoxicity syndrome (ICANS) associated cytokines.
Longitudinal levels for patients who did (left; N=14) and did not (right; N=12) develop ICANS are demonstrated. Yellow coloration represents the onset of ICANS symptoms for each patient. The cytokines IL-1A (A) and IL-1B (B) were not associated with the development of ICANS symptoms when comparing the area under the curve (AUC) (p=0.38 and p=0.54, respectively) or peak values during ICANs (p=0.55 and p=0.8) in those who did (left) vs did not (right) develop ICANS by Wilcoxon. IL-18 levels (C) were associated with the onset of ICANS symptoms in most patients (AUC: p<0.01; peak values during ICANS: p<0.01). X-axis demonstrates day, Y-axis represents NPX for each protein. Expression of proteins associated with both CRS and ICANS (Y-axis) or with ICANS alone (X-axis) is presented in (D). Purple dots are statistically significant proteins with a nominal p-value ≤ 0.05 computed with a t-test and have an absolute fold change ≥ 1.5. IL-18 is colored in pink. (E) Longitudinal levels of CELA3A, the highest expressed protein from (D) is shown associated with onset of ICANS symptoms (AUC: p<0.01; peak values during ICANS: p<0.01). X-axis demonstrates day, Y-axis represents NPX for each protein.
We next expanded our search for ICANS biomarkers beyond the IL-1 family of cytokines. Given the significant overlap between patients who developed ICANS and those who developed CRS, we leveraged the serial nature of our data to identify timepoints when both CRS and ICANS were occurring and compared them to timepoints when only ICANS was occurring using DEA (Figure 3D). Proteins significantly more associated with both CRS and ICANS or with ICANS alone are listed in Supplemental Tables S6&7. Notable proteins associated with ICANS alone included CHIT1 and TREM2, which have been associated with neuroinflammation in other contexts.(13-17) The protein with the highest mean NPX, CELA3A (chymotrypsin like elastase 3A) increased in concordance with the onset of ICANS symptoms when visualized serially (Figure 3E). Time course for all identified proteins with onset of ICANS symptoms are shown in Supplemental Figure S12. For each cytokine identified in the previous analyses, we performed statistical comparisons between patients with and without ICANS, comparing both area under the curve and the highest value of each cytokine. Data for both analyses are presented in Supplemental Table S8.
Complement Dysregulation in CRS and ICANS
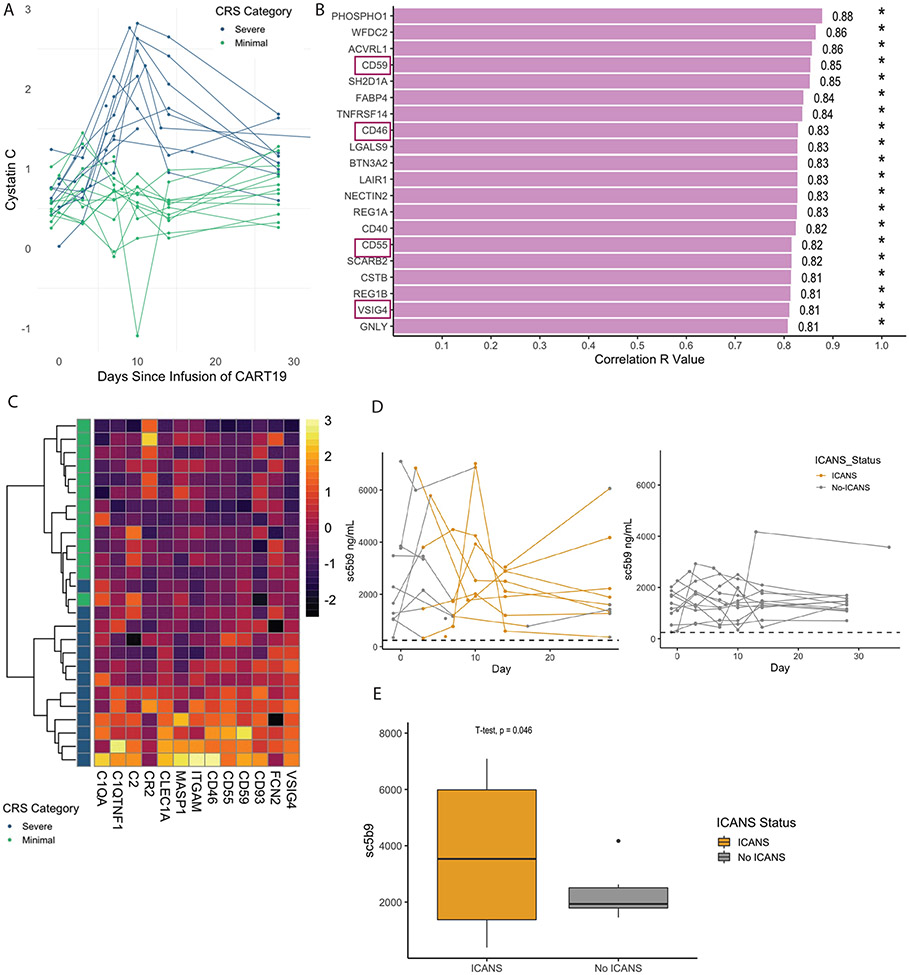
Patients with severe CRS can develop significant but typically transient renal dysfunction. It is unknown if this is merely a consequence of decreased perfusion or kidney damage from alternative pathophysiology.(18) To better understand this biology, we looked at proteins that were highly associated with renal dysfunction using cystatin C as a surrogate marker.(19) Severe CRS was associated with increases in cystatin C, especially at the peak timepoint (Figure 4A). We used linear regression to identify proteins that were associated with cystatin C at this timepoint (Figure 4B). Notably, negative regulators of complement CD46, CD59, CD55 and VSIG4 were all significantly associated with elevations in cystatin C. We looked at all complement related proteins measured and found that these proteins classified patients as having severe or minimal CRS when unsupervised clustering was used (Figure 4C).
Figure 4. Evidence of complement dysregulation related to the development of CRS and ICANS.
Severe cytokine release syndrome (CRS) is associated with renal dysfunction as demonstrated by increases in the renal dysfunction marker Cystatin C (A), particularly at the peak timepoint (p < 0.001). (B) Top twenty linear correlates of Cystatin C at the peak timepoint. Proteins related to complement are denoted with purple rectangles. (C) A heatmap of all complement related proteins measured by Olink with unsupervised hierarchical clustering applied to all patients at the peak CRS timepoint (N=26). Patients with minimal CRS are colored in green and patients with severe CRS are colored in blue. Color bar represents variable range of NPX value. (D) Soluble c5b9 (sc5b9) levels (ng/mL) in patients with (N=14) and without (N=12) ICANS symptoms measured over time. Yellow lines represent occurrence of ICANS symptoms with grey lines denoting absence of ICANS symptoms. Dashed horizontal line represents upper limit of normal (ULN) on sC5b9 assay (247 ng/mL). Area under the curve (AUC) comparison of those who did vs did not develop ICANS showed a trend toward a significant difference (p=0.07 by Wilcoxon), but there were outliers (E) Comparing the highest sC5b9 level in patients who did (N=14) and did not (N=12) develop ICANS compared with t-test, p=0.046 did demonstrate a difference between groups.
Vascular endothelial dysfunction has previously been demonstrated to be associated with the development of ICANS.(20) We hypothesized that complement activation may also be related to the development of ICANS and measured the marker of terminal complement activation sC5b9 and its association with ICANS (Figure 4D). Notably sC5b9 levels were highly elevated relative to normal in all patients. We compared the highest sC5b9 at any point in patients with and without ICANS and found elevations in sC5b9 were significantly associated with ICANS symptoms (Figure 4E; t-test p=0.046).
DISCUSSION/CONCLUSION
We performed comprehensive proteomic profiling of patients treated with CTL019 to make several novel observations about the pathophysiology of CRS and ICANS. First, we identify and validate the proteins MILR1 and FLT3 as candidate pre-infusion serum biomarkers that predict the development of CRS at least as well as the current gold standard of disease burden. Second, we demonstrate that CRS is associated with IFNγ and its related proteins, supporting the fundamental clinicopathological similarities of severe CRS to HLH. Third, we identify novel, potentially targetable biomarkers for the development of ICANS, including IL-18 and CELA3A. Finally, we show that complement dysregulation may play a role in the development of CRS and ICANS.
We identify two potential pre-infusion biomarkers of severe CRS: MILR1 and FTL3. MILR1 has not been previously reported in pediatric leukemia biology. It primarily serves as a negative regulator of IgE-mediated hypersensitivity reactions.(9) We demonstrate that MILR1 is expressed on the surface of B-ALL blasts in a subset of patients and on a transcriptomic level. FLT3 is known to be expressed on B-ALL blasts and has previously been targeted in B-ALL.(21) The remarkable inverse relationship between FLT3 and FLT3LG over time implies that FLT3 and FLT3LG may play a role in modulating CRS. FDA-approved FLT3 inhibitors have been extensively studied in the pediatric population and our data suggest they should be investigated in CRS. Importantly, serum biomarkers like FLT3 and MILR1 could obviate the need for the current practice of pre-infusion bone marrow aspirate to prognosticate CRS. Given that the relative amount of marrow disease can vary between different sites of bone marrow aspirates and biopsies, it is possible that MILR1 and FLT3 are better at defining disease burden than a single bone marrow aspirate at pre-infusion.
Next, we demonstrate the key role of IFNγ and its related proteins in the development of severe CRS. IFNγ is an effector T-cell cytokine and an in vitro marker of T-cell activation, and initially blockade of IFNγ was avoided as a treatment strategy in CRS because of concern that this would impede T-cell proliferation and anti-tumor efficacy. However, recent data demonstrates that IFNγ blockade does not impede CART cell activity in vitro or in vivo in preclinical models.(22) Our group has successfully treated multiple pediatric patients with severe CRS refractory to steroids and tocilizumab with the IFNγ-blocking medication emapalumab, which is currently FDA-approved for the treatment of pediatric HLH.(23) Based on the data we present here and our clinical success using this medication in patients, we plan to prospectively evaluate emapalumab as a treatment for severe refractory CRS.
Like most biologic phenomena, CRS and HLH exist on a gradient of severity. Our current and previous data demonstrate no appreciable difference in the immunophysiology of severe CRS and HLH. HLH is a process with diverse contributors and a spectrum of clinical severities.(24) Even in familial HLH, infants with PRF1 deficiency can present with a spectrum of mild inflammation to critical illness. Although cytotoxic therapies are the mainstay of treatment for familial HLH, most other HLH subtypes can be successfully managed with steroids, cytokine blockade, and immunomodulators. Indeed, use of chemotherapy has been shown to be harmful, worsening outcomes in some cases of infection driven HLH.(25) We caution against categorizing those with severe CRS or late CRS as uniquely having HLH. While future work may find the biology of refractory and late CRS is different and novel therapies are required, there are no data to suggest that these patients benefit from medications such as etoposide. These cases may be more likely to be successfully treated with targeted therapies, such as IFNγ inhibitors, Jak-Stat inhibitors, or IL-18 inhibitors.
Although targeting specific cytokines has proven to be a successful strategy in the treatment of CRS, effective targeted therapies for ICANS remain elusive and steroids remain the standard of care.(2) This may be in part related to poor CNS penetration of monoclonal antibodies, including tocilizumab. The IL-1RA-antagonist anakinra, which has excellent CNS penetration, is currently being evaluated as a potential therapy for ICANS in clinical trials (NCT04150913, NCT04359784, NCT04148430). Our results argue that IL-1A and IL-1B are not key drivers of ICANS, but that IL-18 may be playing an important role. Blockade of IL-18 with recombinant IL-18BP as a treatment strategy in HLH related conditions is currently being evaluated in a clinical trial (NCT03113760). Simultaneous blockade of IL-18 and IL-1 may be a tractable treatment strategy for ICANS and superior to IL-1 inhibition alone.
We show evidence of complement dysregulation being associated with renal dysfunction in patients with severe CRS, and terminal complement elevations in all patients, with higher levels in patients who develop symptoms of ICANS. Endothelial dysfunction has previously been identified as potentially contributing to the pathophysiology of ICANS.(20) Complement activation may drive this endothelial dysfunction. This is an important area for future inquiry as the complement pathway has not yet been targeted in CRS or ICANS. The availability of drugs that target complement activation, such as eculizumab, helps pave the way for future trials to prevent or treat ICANS.
In this report we employed longitudinal sample collection and comprehensive proteomic analysis to make unique observations about targetable cytokines that can be evaluated in patients with refractory CRS and ICANS. We show that CRS bears the hallmarks of an IFNγ-driven process with fundamental similarities to HLH. We identify FLT3 and MILR1 as potential candidate pre-infusion biomarkers for severity. Future work will validate these findings in other CART products and other diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Dr. Tina Glisovic-Aplenc. Additionally, this study was supported by the Leukemia and Lymphoma Society (CHJ, SG, RA, SAG, DTT), NIH grants R01CA193776 (DTT), R01CA264837 (DTT), R03CA256550 (DTT), U10CA180886 (DTT), U24CA196173 (DTT), X01HD100702 (DTT), P01CA214278 (CHJ, SG, SAG), Cookies for Kids Cancer (DTT), the Alex’s Lemonade Stand Foundation (DTT), The Children’s Hospital of Philadelphia Immune Dysregulation Frontier Program (EB, SC, HB, DTT) and many generous donations from patients and families for research on CAR-T, acute lymphoblastic leukemia, HLH, and immune dysregulation. CD was supported by the Canadian Institute for Health Research (CIHR) and an American Society of Clinical Oncology (ASCO) Conquer Cancer Young Investigator Award. RBL is supported by the Thrasher Research Fund #15351, the CHOP Research Institute, and the Endowed Chair of the Department of Anesthesiology and Critical Care. SC reports grants from Eunice K Shriver National Institute for Child Health and Human Development.
Footnotes
CONFLICTS OF INTEREST
DTT reports grants and personal fees from BEAM Therapeutics, grants from NeoImmune Tech, and personal fees from Sobi during the conduct of the study. Additionally, DTT has a patent for Biomarkers of Cytokine Release Syndrome pending and a patent for Chimeric Antigen Receptor T cells Targeting CD38 pending. SAG reports grants and personal fees from See ms- disclosure outside the submitted work; in addition, SAG has a patent for CAR toxicity patents managed by CHOP and UPenn issued, licensed, and with royalties paid from Novartis. BLL reports personal fees from Avectas, Akron Bio, Immusoft, In8bio, Immuneel, Ori biotech, Oxford Biomedica, Vycellix and other support from Tmunity Therapeutics and from Capstan Therapeutics outside the submitted work; in addition, BLL has a patent for Methods for treatment of cancer (US 8906682) (US 8916381)( US 9101584) issued, licensed, and with royalties paid from University of Pennsylvania, a patent for Compositions for treatment of cancer (US 8911993) (US 9102761) (US 9102760) issued, licensed, and with royalties paid from University of Pennsylvania, a patent for Method for treating chronic lymphocytic leukemia (CC) (US 9161971) issued, licensed, and with royalties paid from University of Pennsylvania, a patent for Compositions and methods for treatment of cancer (US 9464140 )(US 9518123) (US 9481728)(US 9540445) issued, licensed, and with royalties paid from University of Pennsylvania, a patent for Use of chimeric antigen receptor-modified T cells to treat cancer (US 9328156) (US 9499629) issued, licensed, and with royalties paid from University of Pennsylvania, a patent for Methods for assessing the suitability of transduced T cells for administration (US 9572836) issued, licensed, and with royalties paid from University of Pennsylvania, and a patent for Toxicity management for anti-tumor activity of CARs (10,603,378, 11,273,219) issued, licensed, and with royalties paid from University of Pennsylvania. MPL reports support from Octapharma, Dova, Principia, Shionogi, personal fees from Shionogi, Dova, Principia, Argenx, Rigel, and grants from Sysmex, Rigel, Principia, Argenx, Dova, Octapharma, AstraZeneca outside the submitted work. SC reports personal fees from Simcha Therapeutics, grants outside the submitted work. SLM reports grants and personal fees from Wugen outside the submitted work; in addition, SLM has a patent for PCT/US2017/044425: Combination Therapies of Car and PD-1 Inhibitors pending and licensed to Novartis Pharmaceuticals. SFL reports a patent for Kymriah and related biomarkers licensed to Novartis. JJM reports grants and personal fees from IASO Biotherapeutics, Poseida Therapeutics, Gilead, and Janssen outside the submitted work; in addition, JJM has a patent for WO2016/109410 A2 issued, a patent for WO2019/213282 issued, a patent for WO2018/175733 issued, a patent for WO2018/013918 issued, a patent 201806619 issued, and a patent for WO2017/210617 issued. CHJ reports other support from Tmunity and other support from Capstan Therapeutics during the conduct of the study; personal fees from Poseida, BluesphereBio, Cabaletta, Carisma, Cartography, Cellares, Danaher, Verismo, and Alaunos Therapeutics outside the submitted work. HN reports grants from National Institutes of Health. No other authors have potential conflicts of interest.
REFERENCES
- 1.June CH, Sadelain M. Chimeric antigen receptor therapy. N Engl J Med 2018;379(1):64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santomasso BD, Nastoupil LJ, Adkins S, Lacchetti C, Schneider BJ, Anadkat M, et al. Management of Immune-Related Adverse Events in Patients Treated With Chimeric Antigen Receptor T-Cell Therapy: ASCO Guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2021;39(35):3978–92 doi 10.1200/jco.21.01992. [DOI] [PubMed] [Google Scholar]
- 3.Teachey DT, Lacey SF, Shaw PA, Melenhorst JJ, Maude SL, Frey N, et al. Identification of Predictive Biomarkers for Cytokine Release Syndrome after Chimeric Antigen Receptor T cell Therapy for Acute Lymphoblastic Leukemia. Cancer discovery 2016;6(6):664–79 doi 10.1158/2159-8290.Cd-16-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neelapu SS, Tummala S, Kebriaei P, Wierda W, Gutierrez C, Locke FL, et al. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nat Rev Clin Oncol 2018;15(1):47–62 doi 10.1038/nrclinonc.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadauke S, Myers RM, Li Y, Aplenc R, Baniewicz D, Barrett DM, et al. Risk-Adapted Preemptive Tocilizumab to Prevent Severe Cytokine Release Syndrome After CTL019 for Pediatric B-Cell Acute Lymphoblastic Leukemia: A Prospective Clinical Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2021;39(8):920–30 doi 10.1200/jco.20.02477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner RA, Ceppi F, Rivers J, Annesley C, Summers C, Taraseviciute A, et al. Preemptive mitigation of CD19 CAR T-cell cytokine release syndrome without attenuation of antileukemic efficacy. Blood 2019;134(24):2149–58 doi 10.1182/blood.2019001463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santomasso BD, Park JH, Salloum D, Riviere I, Flynn J, Mead E, et al. Clinical and Biological Correlates of Neurotoxicity Associated with CAR T-cell Therapy in Patients with B-cell Acute Lymphoblastic Leukemia. Cancer discovery 2018;8(8):958–71 doi 10.1158/2159-8290.Cd-17-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norelli M, Camisa B, Barbiera G, Falcone L, Purevdorj A, Genua M, et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nature medicine 2018;24(6):739–48 doi 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 9.Hitomi K, Tahara-Hanaoka S, Someya S, Fujiki A, Tada H, Sugiyama T, et al. An immunoglobulin-like receptor, Allergin-1, inhibits immunoglobulin E-mediated immediate hypersensitivity reactions. Nat Immunol 2010;11(7):601–7 doi 10.1038/ni.1886. [DOI] [PubMed] [Google Scholar]
- 10.Jiang P, Zhang Y, Ru B, Yang Y, Vu T, Paul R, et al. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat Methods 2021;18(10):1181–91 doi 10.1038/s41592-021-01274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, et al. Comprehensive network map of interferon gamma signaling. Journal of cell communication and signaling 2018;12(4):745–51 doi 10.1007/s12079-018-0486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin H, Scull BP, Goldberg BR, Abhyankar HA, Eckstein OE, Zinn DJ, et al. IFN-γ signature in the plasma proteome distinguishes pediatric hemophagocytic lymphohistiocytosis from sepsis and SIRS. Blood advances 2021;5(17):3457–67 doi 10.1182/bloodadvances.2021004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Peng J, Sherchan P, Ma Y, Xiang S, Yan F, et al. TREM2 activation attenuates neuroinflammation and neuronal apoptosis via PI3K/Akt pathway after intracerebral hemorrhage in mice. J Neuroinflammation 2020;17(1):168 doi 10.1186/s12974-020-01853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Taso O, Wang R, Bayram S, Graham AC, Garcia-Reitboeck P, et al. Trem2 promotes anti-inflammatory responses in microglia and is suppressed under pro-inflammatory conditions. Hum Mol Genet 2020;29(19):3224–48 doi 10.1093/hmg/ddaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Feng S, Nie K, Li Y, Gao Y, Gan R, et al. TREM2 modulates microglia phenotypes in the neuroinflammation of Parkinson's disease. Biochem Biophys Res Commun 2018;499(4):797–802 doi 10.1016/j.bbrc.2018.03.226. [DOI] [PubMed] [Google Scholar]
- 16.Freigang M, Steinacker P, Wurster CD, Schreiber-Katz O, Osmanovic A, Petri S, et al. Increased chitotriosidase 1 concentration following nusinersen treatment in spinal muscular atrophy. Orphanet J Rare Dis 2021;16(1):330 doi 10.1186/s13023-021-01961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaur N, Huss E, Prell T, Steinbach R, Guerra J, Srivastava A, et al. Monocyte-Derived Macrophages Contribute to Chitinase Dysregulation in Amyotrophic Lateral Sclerosis: A Pilot Study. Front Neurol 2021;12:629332 doi 10.3389/fneur.2021.629332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MD, Strohbehn IA, Seethapathy HS, Rusibamayila N, Casey KS, Gupta S, et al. Acute Kidney Injury After the CAR-T Therapy Tisagenlecleucel. American journal of kidney diseases : the official journal of theNational Kidney Foundation 2021;77(6):990–2 doi 10.1053/j.ajkd.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012;367(1):20–9 doi 10.1056/NEJMoa1114248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gust J, Hay KA, Hanafi LA, Li D, Myerson D, Gonzalez-Cuyar LF, et al. Endothelial Activation and Blood-Brain Barrier Disruption in Neurotoxicity after Adoptive Immunotherapy with CD19 CAR-T Cells. Cancer discovery 2017;7(12):1404–19 doi 10.1158/2159-8290.Cd-17-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood 2005;105(2):812–20 doi 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 22.Bailey SR, Vatsa S, Larson RC, Bouffard AA, Scarfo I, Kann MC, et al. Blockade or deletion of IFNg reduces macrophage activation without compromising CAR-T function in hematologic malignancies. Blood Cancer Discov 2021. doi 10.1158/2643-3230.Bcd-21-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNerney KO, DiNofia AM, Teachey DT, Grupp SA, Maude SL. Potential Role of IFNγ Inhibition in Refractory Cytokine Release Syndrome Associated with CAR T-cell Therapy. Blood Cancer Discov 2021. doi 10.1158/2643-3230.Bcd-21-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canna SW, Marsh RA. Pediatric hemophagocytic lymphohistiocytosis. Blood 2020;135(16):1332–43 doi 10.1182/blood.2019000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabler SS, Hogan PG, Fritz SA, Bednarski JJ, Hunstad DA. Incidence and treatment of hemophagocytic lymphohistiocytosis in hospitalized children with Ehrlichia infection. Pediatr Blood Cancer 2020;67(10):e28436 doi 10.1002/pbc.28436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author. In addition, publicly available data generated by others were used by the authors and are available from the Office of Cancer Genomics (https://ocg.cancer.gov/programs/target/data-matrix).