Abstract
Chimeric-antigen receptor (CAR) T-cell therapy has led to tremendous successes in the treatment of B-cell malignancies. However, a large fraction of treated patients relapse, often with disease expressing reduced levels of the target antigen. Here, we report that exposing CD19+ B-cell acute lymphoblastic leukemia (B-ALL) cells to CD19 CAR T cells reduced CD19 expression within hours. Initially, CD19 CAR T cells caused clustering of CD19 at the T-cell – leukemia cell interface, followed by CD19 internalization and decreased CD19 surface expression on the B-ALL cells. CD19 expression was then repressed by transcriptional rewiring. Using single-cell RNA-sequencing and single-cell ATAC-sequencing, we demonstrated that a subset of refractory CD19low cells sustained decreased CD19 expression through transcriptional programs of physiological B-cell activation and germinal center reaction. Inhibiting B-cell activation programs with the BTK inhibitor ibrutinib increased the cytotoxicity of CD19 CAR T cells without affecting CAR T-cell viability. These results demonstrate transcriptional plasticity as an underlying mechanism of escape from CAR T cells and highlight the importance of combining CAR T-cell therapy with targeted therapies that aim to overcome this plasticity.
Keywords: B-ALL, CAR T cells, B-cell activation, germinal center reaction, single-cell sequencing
Introduction
The application of CD19 chimeric-antigen receptor (CAR) T–cell therapies has shown remarkable success in treating relapsed CD19+ B-cell acute lymphoblastic leukemia (B-ALL) [1,2]. Up to 90% of the heavily pretreated patients who receive this therapy enter into remission [2–6]. However, a large fraction of patients relapse [3–6]. Loss of CD19 expression via mutations [7,8], lineage switch [9,10], and loss of the targeted epitope through alternative splicing [11] are causes of resistance to CD19 CAR T-cell therapy and relapse. Additional studies have shown that intercellular processes, such as trogocytosis whereby membrane fragments transfer between cells, can result in emergence of CD19low B-ALL cells [12] and low expression of the target antigen can lead to resistance [13] and less effective killing [12,14]. Antigen density thresholds, however, are difficult to define and depend on CAR constructs [14], tumor type [15], and target antigens [13,16]. Other resistance mechanisms that have been described include CD19+ resistance through antigen masking [17] and impaired death receptor signaling [18].
CD19 is a coreceptor of the B-cell receptor (BCR) complex and is expressed throughout the B-cell lineage until the plasma-cell stage. CD19 clusters upon antigen binding [19] and forms BCR–antigen complexes, which are internalized and processed thereafter, thus contributing to B-cell activation [19]. Although CD19 targeting by and resistance to CAR T cells has been well established [7,8,11], the dynamics of CD19 antigen expression and its regulation during the early phases of interactions between CAR T cells and CD19+ target cells remain unclear. These early dynamics may have important implications for initial treatment failures.
In this study, we hypothesized that the early interactions of CAR T cells and CD19+ target cells trigger discrete regulatory programs that allow for escape of some target cells. By combining live single-cell microscopy with single-cell RNA-sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq), we show that the interaction between CD19 CAR T cells and CD19-expressing B-ALL cells caused clustering and internalization of CD19 in all leukemia cells that resulted in reduced surface expression of CD19. We found that a subpopulation of target cells employed physiologic B-cell activation programs reminiscent of germinal center reactions in normal B cells that caused sustained transcriptional downregulation of CD19. Initiation of these programs was inhibited by pretreating leukemic cells with the Brutońs tyrosine kinase (BTK) inhibitor ibrutinib, providing a mechanistic explanation for how ibrutinib increases effectiveness of CD19 CAR T–cell cytotoxicity.
Materials and methods
Cell lines
293T and Jurkat cell lines were obtained from ATCC in 2016 and 2015, respectively, NALM-6, REH and KOPN-8 cell lines were obtained from the Broad Institute’s CCLE in 2016. ALL cell lines were cultured in RPMI-1640 (Thermo Fisher Scientific, 11875119) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gemini, 900–208) and 100 μM penicillin/streptomycin (Invitrogen, 15140122). 293T cells were cultured in DMEM (Life Technologies, 11995073) supplemented with 10% FBS and 100 μM penicillin/streptomycin. Cell line authentication was performed by short tandem repeat profiling and re-authentication was performed within the past year. Cultures were tested for mycoplasma contamination and found to be negative. Cells were used for experiments before 20 passages.
Ibrutinib (Selleckchem, S2680) was dissolved in DMSO (ThermoFisher, BP231100) and titrated. IC10 concentration for NALM-6 cells was determined after 72h using Cell Titer Glo (Promega, G1781) on a Spectramax M5 microplate reader (Molecular devices).
CD19 CAR T-Cell Production
Vector and lentivirus production
Two third generation CD19 CAR constructs containing a single chain variable fragment targeting the FMC63 locus of the CD19 antigen with an intracellular CD3ζ as well as 4–1BB and CD28 costimulatory domains were used. Constructs contained a truncated epidermal growth factor receptor (tEGFR), separated from the CAR by a T2A sequence. Construct 1 was obtained from Creative Biolabs (pCAR-T-h; anti-CD19 scFv–CD28–4–1BB–CD3ζ) and consisted of an anti-CD19 single-chain variable fragment, CD8 hinge, CD28 transmembrane domain, 4–1BB/CD3 zeta signaling module, T2A sequence and a tEGFR. Construct 2 was designed in-house, custom-ordered at Genewiz and cloned in a pLVX-CMV100 vector backbone (Addgene, 110718). The components of construct 2 are displayed in Supplementary Fig. S1a. Construct 1 was used in the experiments unless stated differently. 293T cells were co-transfected with the CAR lentiviral construct, psPAX2 (Addgene, 12260) and pCMV-VSV-G (Addgene, 8454) packaging vectors using Lipofectamine 3000 (Thermo Fisher Scientific, L3000015) according to manufacturer’s protocol. Lentivirus-containing medium was collected, and fresh medium added after 12h, 24h, and 36h. Virus-containing media was filtered and concentrated by ultracentrifugation for 2h at 49,000g at 4°C.
T-cell isolation and transduction
Human blood from healthy donors was obtained from Research Blood Components, LLC, or the Crimson Core of the Brigham and Women’s Hospital. Mononuclear cells (PBMCs) were isolated by Ficoll-Paque PLUS (Global Life Sciences Solutions USA LLC, 45001750). PBMCs were further processed by isolating CD3+ T cells with the EasySep™ Human T Cell Enrichment Kit (STEMCELL Technologies, 19051) with an EasySep™ magnet (STEMCELL Technologies, 18000) according to the manufacturer’s protocol. For CD4+ T-cell purification, selection was performed using EasySep™ Release Human CD4 Positive Selection Kit (STEMCELL Technologies, 17752) according to the manufacturer’s protocol.
Isolated T cells were activated by Dynabeads™ Human T-Activator CD3/CD28 (Thermo Fisher Scientific, 11132D) and cultured in X-VIVO 15 Media (Lonza, 04–418Q) supplemented with 5% Human Serum (Sigma-Aldrich, H4522). 50 IU/ml IL2 (Miltenyi Biotec, 130–097-743) was added every other day. One day after isolation, T cells or Jurkat cells were infected by spinoculation at multiplicity of infection (MOI)=5. After 7 days, infection efficiencies were determined by flow cytometry as described below using a hEGFR-specific antibody (Biotinylated, Cetuximab; R&D Systems, FAB9577B-100) and a secondary APC streptavidin-conjugated antibody (BD Biosciences, 554067). CAR-expressing cells were isolated by magnetic isolation using the EasySep™ Release Human Biotin Positive Selection Kit (STEMCELL Technologies, 17653) according to the manufacturer’s protocol. Persistent expression of the CAR construct (detected using biotinylated anti-hEGFR – Cetuximab, R&D Systems; and secondary APC Streptavidin-conjugated antibody; BD Biosciences) and CD8 percentage (HIT8a, Biolegend, 300912) was assessed by flow cytometry prior to co-culture (Supplementary Fig. S1b). Activation beads were removed after 10 days with restimulations according to manufacturer’s protocol. Uninfected T cells from the same donor or uninfected Jurkat cells were maintained in parallel and used as controls.
Flow cytometry and single-cell sorting
CAR T cells were stained for CD3 PE (HIT3a, Biolegend, 300308), CD8 APC (HIT8a, Biolegend, 300912) and hEGFR (Biotinylated, Cetuximab; R&D Systems, FAB9577B-100) using a secondary APC Streptavidin-conjugated antibody (BD Biosciences, 554067). B-ALL target cells were stained for CD19 PE (HIB19, BioLegend, 302208), CD40 PE (5C3, Biolegend, 334307) or, where applicable, a primary unconjugated CD19 (HIB19, BioLegend, 302202) and a secondary PE-conjugated goat anti-mouse IgG (Poly4053, BioLegend, 405307). Fixation and permeabilization for intracellular staining were performed using the BD Cytofix/Cytoperm™ kit (BD Biosciences, 555028) according to manufacturer’s protocol. Cells were stained with 7-AAD (Thermo Fisher Scientific, A1310) to exclude dead cells. Cell nuclei were stained with DAPI (1 μg/mL, Sigma Aldrich, D9542). B-ALL target cells were labeled with CFSE (Thermo Fisher Scientific, C34554), as indicated.
Samples were analyzed on a BD Accuri C6 Flow Cytometer (BD Biosciences) or BD FACSMelody (BD Biosciences) with compensation being performed by AbC™ Total Antibody Compensation Bead Kit (Thermo Fisher Scientific, A10497) or single-stained controls. Where applicable, absolute cell counts were measured with Precision Count beads (BioLegend, 424902).
Flow cytometric analyses were performed with FlowJo V10 (BD Biosciences). CD19 fluorescence signal was normalized to the mean fluorescence intensity of CD19 in control leukemia cells that were not co-cultured with any effector cells.
For scRNA-seq and scATAC-seq experiments, NALM-6 cells were stained for CD19 and the 30% highest CD19-expressing cells (CD19high cells) were sorted and rested overnight. The next day, sorted CD19high cells were labeled with CFSE (Thermo Fisher Scientific, C34554), counted and co-cultured at 1×105 target cells with CAR T cells or uninfected T cells in a 5E:1T ratio for 24h in 96-well round bottom plates. After co-culture, cells were stained with 7-AAD (Thermo Fisher Scientific, A1310) and anti-CD19 PE (HIB19, BioLegend, 302208) antibody. Subsequently, 7-AAD negative and CFSE+ NALM-6 cells were sorted into 96-well plates (ThermoFisher, E0030129504) containing TCL buffer (Qiagen, 1031576) and 1% 2-Mercaptoethanol (Life Technologies, 21985023) based on CD19 expression (Supplementary Table S1) using a Sony SH800 sorter. Plates were spun down immediately, snap frozen on dry ice and stored at −80°C until further processing.
Co-culture experiments
NALM-6, REH and KOPN-8 cells were labeled with CFSE (Thermo Fisher Scientific, C34554) and used as target cells (Target, T). 1×105 target cells were co-cultured with CAR T cells or CAR Jurkat cells (Effector, E) at an E:T ratio of 1:1, 2:1 and 5:1 in a 96-well round bottom plate for 1, 2, 4, 6, 12, and 24h in X-VIVO 15 Media supplemented with 5% Human Serum. After co-culture, cells were stained with 7-AAD (Thermo Fisher Scientific, A1310) and an anti-CD19 PE (HIB19, BioLegend, 302208) and analyzed on a BD Accuri C6 Flow Cytometer (BD Biosciences) or BD FACSMelody (BD Biosciences) with compensation being performed by AbC™ Total Antibody Compensation Bead Kit (Thermo Fisher Scientific, A10497) or single-stained controls. Cytotoxicity was calculated as number of 7-AAD positive (dead) target cells divided by total number of target cells.
Where applicable, 7-AAD–negative target cells were isolated from effector cells after 24h of co-culture using a Sony SH800 sorter. Sorted target cells were taken back into culture and viability was assessed by trypan blue staining using a Neubauer cell counting chamber (Fisher Scientific, 026716) after 24h, 48h and 72h.
For re-exposure experiments, NALM-6 cells were co-cultured with CAR T cells or uninfected T cells at an E:T ratio of 2:1 in a 96-well round bottom plate overnight. Cells were then pooled and NALM-6 cells were isolated from effector cells using the EasySep™ Human CD3 Positive Selection Kit II (STEMCELL Technologies, 17851) according to the manufacturer’s protocol. Successful isolation was assessed by positivity of CFSE-labeling via flow cytometry. NALM-6 cells were subsequently re-exposed to fresh effector cells at an E:T ratio of 2:1 in a 96-well round bottom plate for 6h. After co-culture, cells were stained with 7-AAD and analyzed as described above.
For transwell experiments, cytotoxicity assays were performed in a Corning HTS Transwell-96 system with 0.4um pore size (Sigma-Aldrich, CLS3381). For ibrutinib experiments, NALM-6 cells were pre-treated with ibrutinib for 72h at IC10 dosing. To exclude dead cells, NALM-6 cells were then ficolled, washed, counted and co-cultured with effector cells either in presence of DMSO (ThermoFisher, BP231100) or ibrutinib at IC10 dosing for 6h. After co-culture, cells were stained with 7-AAD and analyzed as described above.
Microscopy
Live cell imaging was performed in X-VIVO 15 media without phenol red (Lonza, 04–744Q) supplemented with 5% Human Serum. NALM-6 cells were labeled with CFSE or an AF647-conjugated anti-CD19 (HIB19, BioLegend, 302220). A stage top incubator was used to maintain constant humidified O2 and CO2 flow at 37°C (Okolab). 1×106 cells were seeded on a petri dish and allowed to settle for at least 30min before starting the timelapse. CAR T cells were carefully added at a 1:1 ratio. Where applicable, SYTOX Blue Dead Cell Stain (Thermo Fisher Scientific, S11348) was added to the media at 1mM.
For holotomography-based three-dimensional live microscopy, interactions were recorded by measurement of refractive index and CFSE fluorescence signal using a 3D Cell Explorer microscopy system with a 60x magnifying objective at 512×512 resolution (Nanolive). Images were further processed with Nanolive’s software STEVE v1.6.3496 to display three dimensional timelapses.
For CD19 antigen movement tracking, a Nikon Eclipse Ti microscope system recorded interaction sites every 10min for 6h with a 20x magnifying objective at 2048×2048 resolution. Z-stack images were recorded focusing on the middle layer of the cells as well as 2μm above and below using Nikon’s Perfect Focus System. Four dimensional hyperstacks (x-, y-, z- and time axis) were assembled using the Fiji distribution of ImageJ 1.52p. For representative still images, effector cells were pseudo-colored in orange. Cells were first analyzed qualitatively with respect to CD19 clustering and internalization. Interactions were characterized as “cluster formation”, when CD19 clustering was observed in at least two sequential frames or if CD19 clustering was followed by CD19 internalization in two sequential frames. Interactions that did not meet these criteria but lasted for at least two sequential frames were characterized as “no cluster”. Dead cells were determined by positivity of SYTOX Blue Stain. Analysis only included NALM-6 cells that were SYTOX Blue–negative at time of analysis unless stated differently. Mean fluorescence intensity of the three Z-stack images was then quantified in the Fiji distribution of ImageJ 1.52p. Photobleaching was accounted for by normalizing to the mean loss of fluorescence after 6h in NALM-6 cells that did not interact with any effector cells in the recorded frames during the same experiment. Analysis was performed in a blinded manner.
scRNA-seq library preparation
Full-length scRNA-seq libraries were prepared using the SMART-seq2 protocol [20]. As described above, cells were sorted into 96-well plates containing 10μl TCL buffer (Qiagen, 1031576) and 1% 2-mercaptoethanol. RNA was purified using RNAClean XP beads (Beckman Coulter, A63987) according to the manufacturer’s protocol and reverse transcribed using Maxima RNAse H-minus (Thermo Fisher Scientific, FEREP0752) in the presence of oligo-dT30VN (Integrated DNA Technologies, (/5BiosG/AAGCAGTGGTATCAACGCAGAGTACT(30)VN), template-switching oligonucleotides (Qiagen, /5BiosG/AAGCAGTGGTATCAACGCAGAGTACATrGrG+G), and betaine (Sigma Aldrich, B0300–5VL). cDNA was then amplified using the KAPA Hifi Hotstart ReadyMix (Kapa Biosystems, KK2602) with ISPCR primers (Integrated DNA Technologies, (/5BiosG/AAGCAGTGGTATCAACGCAGAGT). After purification with Agencourt Ampure XP beads (Beckmann Coulter, A63881), product size distribution was assessed on a Bioanalyzer using a High Sensitivity DNA Kit (Agilent Technologies, 5067–4626) and quantified by Qubit (Thermo Fisher Scientific, Q32854), followed by fragmentation and indexing using Nextera XT and Nextera PCR primers (Illumina, FC-131–1096). Libraries were pooled and paired-end sequencing was performed on a NextSeq 500 (Illumina) aiming for sequencing depth of 1 million reads per single cell.
scATAC-seq library preparation
Single-cell ATAC-seq libraries were prepared following the protocol by Chen et al. [21]. NALM-6 cells were pre-sorted, co-cultured and processed as described above (see Flow cytometry and single-cell sorting), followed by DAPI-staining (1 ug/mL, Sigma-Aldrich, D9542) and sorting into 96-well plates containing lysis buffer (recipe as described by Chen et. al [21]: 2X lysis buffer - 100 mM Tris.HCl, pH 8.0 (Boston BioProducts, BM-320); 100mM NaCl (Sigma-Aldrich, 71386); 40 μg/ml Proteinase K (Ambion, AM2546); 0.4% SDS (Boston BioProducts, BM-230A)) and Nextera XT indexes (Illumina, FC-131–1096). Libraries were pooled and purified using a Qiagen MinElute PCR purification kit (Qiagen, 28004) followed by size selection using Agencourt Ampure XP beads, (Beckman Coulter, A63881). Paired-end sequencing was performed on a NextSeq 500 aiming for sequencing depth of 1 million reads per single cell.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9. Normal distribution was tested using the D’Agostino and Pearson test or the Shapiro-Wilk-Test unless otherwise indicated. For parametric data, the two-tailed unpaired Student’s t-test was used, or an ordinary one-way ANOVA with Tukey’s multiple comparisons in data with more than two groups. For non-parametric data, the two-tailed Mann Whitney test was used, or Kruskal-Wallis test with Dunn’s multiple comparisons testing in data with more than two groups.
Processing of scRNA-seq data and quality filtering
Raw sequencing reads were trimmed using trimmomatic and aligned to the hg19 genome using STAR [22] with the following parameters ‘-- twopassMode Basic --alignIntronMax 100000 --alignMatesGapMax 100000 --alignSJDBoverhangMin 10 --alignSJstitchMismatchNmax 5 −1 5 5. HTSeq [23] and RSEM [24] were used to calculate raw counts and normalized transcript per million (TPM) values from the aligned bam files.
Four different parameters were used to filter out low quality cells – library size, number of detected genes, percentage of reads mapping to mitochondrial genes, and percentage of reads mapping to house-keeping genes. Cells were distributed according to these parameters and determined as low quality when falling beyond three median absolute deviations (MAD).
Analysis of scRNA-seq profiles
Clustering of high-quality cells was performed using PAGODA2 [25] (https://github.com/kharchenkolab/pagoda2). Due to batch effects one cluster was uniquely formed by cells from one well plate, and thus excluded from further analysis. 7 clusters were identified using multilevel graph-based clustering within PAGODA2.
Each individual cell was analyzed for expression of G1, G2M and S phase markers to predict the cell-cycle phase using Seurat2 [26] (https://github.com/satijalab/seurat). The findMarkers function in the scran package (https://github.com/elswob/SCRAN) was used to identify differentially regulated genes in each of these clusters. The mean log2FC of differentially regulated genes in each cluster compared to all other clusters was used to perform gene-set enrichment analysis (GSEA) on all hallmark gene sets. Monocle2 [27,28] (http://cole-trapnell-lab.github.io/monocle-release/) was employed to discover distinct transcriptional states within the NALM-6 cells cocultured with CAR T cells. Transcriptional regulons associated with each Monocle2 state were determined using SCENIC [29] (https://github.com/aertslab/SCENIC).
Signatures for hematopoietic cell types were derived from previously published literature comprising bulk RNA-seq studies on purely sorted immune populations [30,31]. The preB signature was taken from the Human Cell Atlas [32]. The germinal center signatures were derived from Holmes et al. [33]. The genes in each of these signatures were scored using the AUCell package (https://github.com/aertslab/AUCell). The mean Z-scores of a cell type of each state were chosen for further visualization.
Analysis of published RNA-seq datasets
Gene expression data from bulk-sorted populations of human naïve B cells and germinal center B cells from Holmes et al. [33] were analyzed. All three datasets for the naïve B cells and for the germinal center B cells from three independent donors were used. The gene-expression count matrix (Transcripts Per Kilobase Million (TPM)) was analyzed using R version 4.0.1 and the ggplot2 package (https://github.com/tidyverse/ggplot2).
Processing of scATAC-seq data
The reads from scATAC-seq samples were aggregated by each 96-well plate prior to peak calling and analyzed using the nf-core ATAC pipeline [34] with the ‘--narrowPeak’ parameter, employing MACS2 for peak calling and HOMER for annotation of peaks. In total, the union of all peak calls resulted in 33,125 consensus peaks. For gene-enhancer relationships, annotations were derived from GREAT [35]. Raw fastq files from individual scATAC-seq samples were processed using the snapATAC pipeline (https://github.com/r3fang/SnapATAC). Briefly, single-cell bam files were processed to obtain read counts in the consensus peaks. Peaks were resized to a fixed width of 500 bp, centered at the summit and overlapping peaks were removed. Filtering was performed using the parameters ‘min_depth=1500, min_in_peaks=0.15’. Clustering of cells was performed using cisTopic [36] (https://github.com/aertslab/cisTopic) and densityClust [37] and visualization of clusters using uniform manifold approximation and projection (UMAP). Differential motif accessibility was investigated using ‘chromVAR’ [38] (https://github.com/GreenleafLab/chromVAR)). Motifs were imported from the JASPAR database and differential accessibility was determined using the ‘differentialDeviations’ function. Bias-corrected deviation and mean Z-scores were calculated and used for visualization.
Data availability:
Sequencing data are deposited in GEO under accession number GSE176418. All other data are available within the manuscript and its accompanying supplementary data files or from the corresponding author upon reasonable request.
Code availability:
For processing of sequencing data, we used trimmomatic, STAR aligner, HTSeq, RSEM and the nf-core ATAC pipeline, and for downstream analysis R version 4.0.1, python 2.7.2 and the following packages: PAGODA2, MONOCLE2, SingleR, scran, SCENIC, Seurat, chromVAR, cisTopic, all of which are available on github.
Results
CD19 CAR T cells cause CD19 internalization, reducing surface expression on CD19+ B-ALL cells
To investigate possible early escape of CD19-expressing B-ALL leukemic cells from CD19 CAR T cells, we first analyzed interactions between CD19 CAR T cells and NALM-6, a CD19+ B-ALL cell line. For these studies, we used commercially available or in-house designed third generation CD19 CAR constructs containing 4–1BB and CD28 costimulatory domains, which we expressed in T cells isolated from blood of normal donors. Using holotomography-based three-dimensional live microscopy, CAR T-cell engagement with the NALM-6 cells was identified by cell–cell contacts suggesting immunological synapse formation, as has previously been reported [39]. We identified three major types of interactions between CAR T cells and B-ALL cells (Figure 1a, Supplementary Movies S1–S3). 1) Effective cytotoxic killing was shown by the leukemia cell blebbing after approximately 20min and subsequent nuclear condensation within 1h (Supplementary Movie S1). 2) “Scanning interactions” were characterized by CAR T-cell – leukemia cell interactions in which contacts were sustained over at least two hours without subsequent cytotoxic activity (Supplementary Movie S2). 3) “Escape interactions” occurred when leukemia cells moved away within an hour of initial contact with a CAR T cell (Supplementary Movie S3).
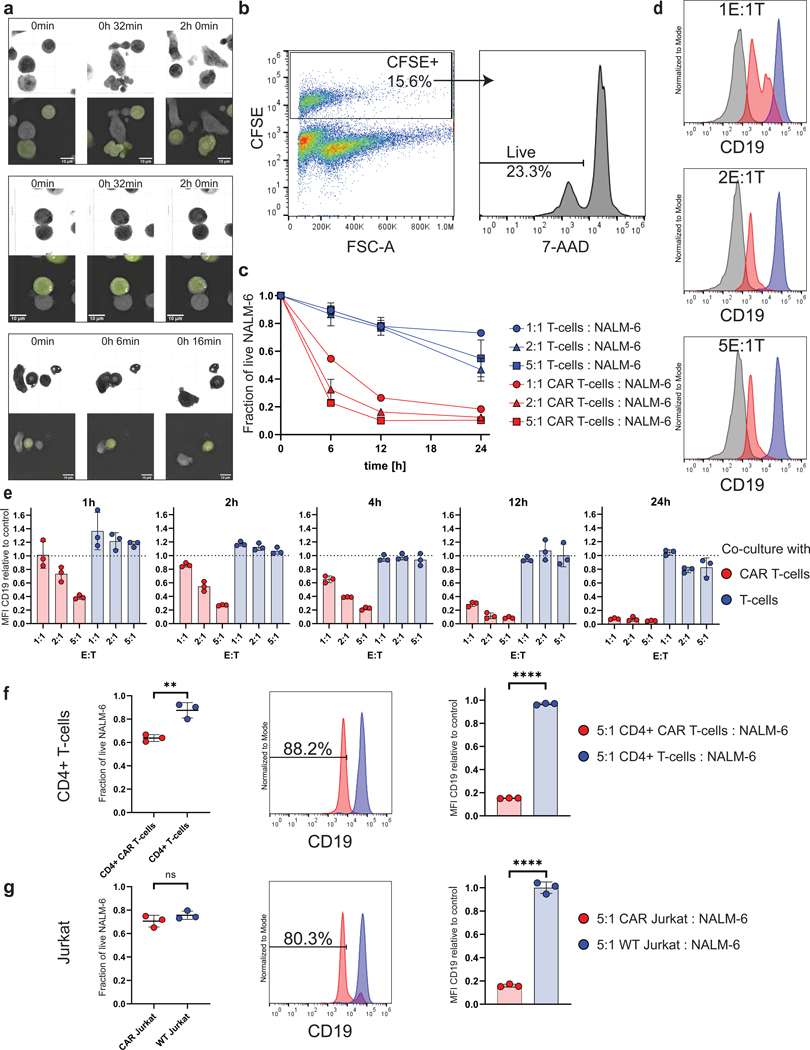
Figure 1. CD19 CAR T-cell treatment induces rapid reduction of CD19 surface protein on B-ALL cells.
a, Three-dimensional holotomography-based live cell microscopy reveals different types of interactions between CD19 CAR T cells and NALM-6 B-ALL cells (60x magnification). Upper sections show refractive index, lower sections show selected two-dimensional Z-stack slice (slice 0, 0 and 15 respectively) for identification of CFSE-labeled NALM-6 target cells. Top, productive interaction with cell–cell contacts suggesting immunological synapse formation, target-cell blebbing and nuclear condensation (Supplementary Movie S1). Middle, sustained immunological synapse without cytotoxicity (Supplementary Movie S2). Bottom, leukemia cell escape after interaction (Supplementary Movie S3). b-e, CFSE-labeled B-ALL NALM-6 cells (Target, T) were co-cultured with either CD19 CAR T cells or uninfected T cells (Effector, E). Quantification of 7-AAD negative live cells and CD19 expression performed by flow cytometry. Data are representative of at least two independent experiments from two different T-cell donors, n=3. b, Gating strategy to identify CFSE+ (left) and 7-AAD– live (right) target cells in a representative 5E:1T CAR T cell co-culture sample. c, Cell viability of NALM-6 B-ALL cells after 6, 12, and 24h co-culture with CD19 CAR T cells (red) compared to the viability of NALM-6 B-ALL cells co-cultured with uninfected T cells (blue). d, Representative histograms demonstrating CD19 expression on live NALM-6 cells (target cell population) with increasing E:T ratios after 6h of co-culture with CAR T cells (red histogram) or uninfected control T cells (blue histogram). Gray histograms show CD19negative T cells from the same experiment. e, Normalized mean fluorescence intensity (MFI) for CD19 expression on live NALM-6 B-ALL cells co-cultured at the indicated E:T ratios for 1, 2, 4, 12, and 24h (data normalized to triple-stained (CD19-PE, CFSE, 7-AAD) control cells that have not been co-cultured with effector cells). f-g, Reduced CD19 expression on B-ALL cells after exposure to CD4+ CAR T (f) or CAR Jurkat (g) cells is independent of CAR effector killing efficacy. CD4+ CAR T-cells or CAR Jurkat cells co-cultured with NALM-6 B-ALL target cells at an E:T ratio of 5:1 for 24 hours. Left, remaining fraction of live NALM-6 cells; middle, CD19 expression (MFI) on live NALM-6 cells; right, relative mean CD19 surface expression. Data representative of two independent experiments with n=3 each (red=NALM-6 cells from CAR T cell co-culture, blue=NALM-6 cells from uninfected T cell co-culture). P values determined by unpaired two-tailed Student’s t-test. ns=p>0.05 **=p<0.01, ****=p<0.0001. Data are mean ± s.d.
We hypothesized that these differences in interactions may be related to heterogenous expression of the target antigen CD19. To address this question, we used flow cytometry to measure CD19 expression by NALM-6 cells after co-culture with CD19 CAR T cells and compared CD19 expression levels with cytotoxicity. Increased time of co-culture and increased E:T ratio led to more killing of leukemia cells (Figure 1b, c). However, among the remaining live leukemia cells in these conditions, we detected progressively reduced levels of CD19 expression with higher E:T ratios and longer periods of co-culture, resulting in more than 95% downregulation of CD19 expression compared with CD19 expression of control NALM-6 cells that had not been co-cultured with any effector cells after 24h (Figure 1d, e). To confirm that the remaining leukemia cells were viable, we sorted live NALM-6 cells and monitored cell viability for 24h, 48h and 72h after removal from co-culture. Viability of CAR T-cell exposed leukemia cells remained relatively stable over the course of 72 hours (50% after 48h, 58% after 72h), and the expression of CD19 recovered, underscoring that a significant fraction of the leukemia cells that survive CAR T-cell cytotoxicity are viable (Supplementary Fig. S2a–b). We next assessed whether these CD19low expressing leukemic cells were refractory to CAR T-cell killing. Indeed, NALM-6 cells that had been co-cultured with CD19 CAR T cells overnight and were re-exposed to fresh CD19 CAR T-cells escaped CAR T cell–mediated killing (viability 98.85% vs 63.91%; Supplementary Fig. 2c).
To ensure that CD19 downregulation was induced by CAR T-cell – leukemia cell interactions and not due to selection of pre-existing CD19low leukemia cells, we performed co-culture experiments using either CD4+ CAR T cells or Jurkat cells transfected with the CAR construct, both of which are less cytotoxic than CD8+ CAR T cells. Using a mixture of CD8+ and CD4+ CAR T cells at a 5:1 E:T ratio, only 10% of NALM-6 cells survived after 24h of co-culture (Figure 1c). Using the same co-culture conditions, 64% and 71% of leukemia cells survived in the CD4+ CAR T cell and CAR Jurkat cell co-cultures, respectively (Figure 1f, g). Although the majority of leukemia cells survived in these co-cultures, the degree of CD19 downregulation was similar to that observed among leukemia cells co-cultured with a mixture of CD8+ and CD4+ CAR T cells (Figure 1f, g compared to Figure 1e) and leukemia cells exhibited an 85% and 84% downregulation of CD19 expression when exposed to CD4+ CAR T cells or CAR Jurkat cells, respectively, (Figure 1f, g right) compared to CD19 expression of NALM-6 cells without co-culture of any effector cells (control). Experiments using two other B-ALL cell lines, REH and KOPN-8, provided similar results (Supplementary Fig. S3). These data suggest that the occurrence of CD19low expressing cells is mediated by active processes induced by CAR T cells causing CD19 downregulation rather than by selection of pre-existing CD19low cells.
Reduced CD19 surface expression is accompanied by CD19 internalization
To analyze the kinetics of decreased CD19 expression, we recorded time-lapse images of CAR T-cell – leukemia cell interactions using live microscopy tracking the CD19 protein in the leukemia cells. When interacting with the CAR T cells, CD19 first clustered at the interface of the interaction with CAR T cells (Figure 2a, Supplementary Movie S4, Supplementary Image File S1). These clusters were primarily seen in leukemia cells interacting with CAR T cells; in co-cultures with uninfected T cells, there was no change in CD19 antigen distribution (Figure 2b, Supplementary Movie S5).
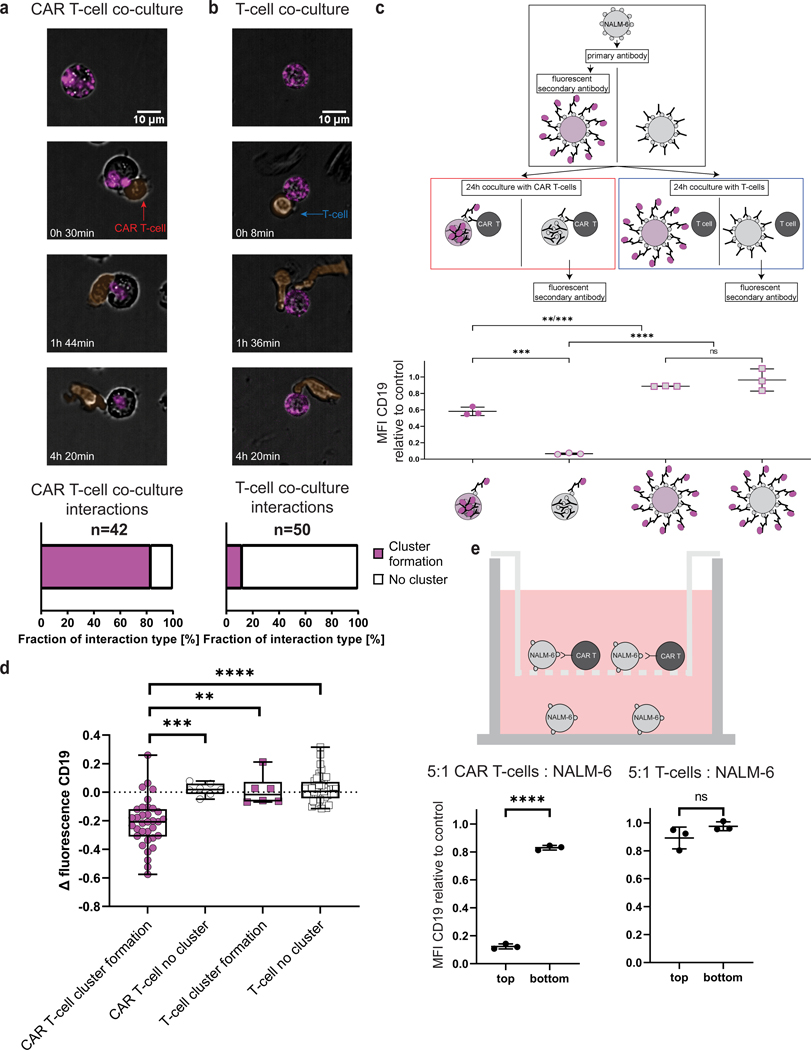
Figure 2. CD19 CAR T cells promote target receptor clustering and internalization in B-ALL cells.
a-b, Time lapse of CD19 distribution in NALM-6 B-ALL cells interacting with CAR T cells (a) or uninfected control T cells (b). Effector and target cells were co-cultured at a 1:1 ratio for 6h (end point), at 20x magnification. Scale bar length 10μm. Effector cells pseudo-colored in orange, CD19 labeled with an AF647-conjugated anti-CD19. a, Representative interaction of a CAR T cell and a NALM-6 B-ALL cell with CD19 clustering and internalization characterized as “cluster formation” (Supplementary Movie S4) with quantification below (83.33% of CAR T – NALM-6 interactions showed CD19 cluster formations during interactions). b, Representative interaction of a control uninfected T cell and a NALM-6 B-ALL cell with no change in CD19 distribution characterized as “no cluster” (Supplementary Movie S5) with quantification below (88% of T-cell – NALM-6 interactions do not show a change of CD19 distribution). c, Experimental work-flow to quantify CD19 internalization. Target cells stained with primary unconjugated anti-CD19 and secondary PE-conjugated antibody either before or after 24h of co-culture. PE-conjugated antibody staining after 24h of co-culture only stains CD19 molecules on the cell surface that were not internalized (data normalized to triple-stained (CD19 PE, CFSE, 7-AAD) control cells that have not been co-cultured with effector cells, representative of two independent experiments, n=3. P values determined by Tukey’s multiple comparisons test). d, Fluorescence intensity decreases in CAR T–cell exposed NALM-6 cells with CD19 “clustering” over 6h compared to uninfected control T cell–exposed NALM-6 cells. Boxplots show minimum, 25th percentile, median, 75th percentile and maximum values. P values determined by two-tailed Mann Whitney test. e, Top, schematic of transwell assay to determine influence of soluble mediators for CD19 internalization. Bottom, mean fluorescence intensity of CD19 expression by live target cells in bottom well after 24h of co-culture (data normalized to triple-stained (CD19 PE, CFSE, 7-AAD) control cells that have not been co-cultured with target cells). P values determined by unpaired t-test, n=3. **=p<0.01, ***=p<0.001, ****=p<0.0001, ns=not significant. Data are mean ± s.d).
During and after interaction with CAR T cells, CD19-specific antibody fluorescence was detectable inside the leukemia cell, indicating that CD19 is internalized after cluster formation (Figure 2a). To further evaluate this hypothesis, we stained NALM-6 cells with an anti-CD19 prior to co-culture with subsequent staining using a fluorescently tagged secondary antibody either before or after the 24h co-culture (Figure 2c). Staining with the secondary antibody after co-culture led to significantly decreased surface fluorescence intensity compared to staining before co-culture (58.16% vs 6.61%, p-value = 0.0001), consistent with CD19 internalization (Figure 2c), which we confirmed by intracellular flow cytometry for CD19 (Supplementary Fig. S4). Cytotoxicity of the CAR T cells was not affected by the presence of the anti-CD19 during co-culture, thus excluding interference of the CD19-specific antibody with target epitope recognition by the CAR T cells (Supplementary Fig. S5).
When accounting for antibody bleaching, we observed a significant reduction of CD19 signal within the cells that formed clusters when interacting with CAR T cells (Figure 2d), suggesting that these leukemia cells may degrade CD19 after internalization. In contrast, leukemia cells that interacted with CAR T cells without cluster formation or leukemia cells that interacted with uninfected T cells did not show changes in fluorescence intensity on either the cell surface or intracellularly.
Leukemia cells that were killed by CAR T cells demonstrated reduced CD19 expression after six hours of co-culture (Supplementary Fig. S6), implying that internalization of CD19 is triggered by CD19–CAR interactions independent of cytotoxic or apoptotic outcomes.
To exclude the possibility that soluble mediators such as cytokines were causing CD19 internalization, we performed co-culture assays in transwells, where leukemia cells that did not interact with CAR T cells shared the same media with leukemia cells co-cultured with CAR T cells (Figure 2e). In this scenario, CD19 expression by the leukemia cells not able to contact CAR T cells did not change.
Taken together, these data demonstrate that direct interaction between the CAR and CD19 is necessary for CD19 clustering, internalization, and downregulation.
scRNA-seq reveals B cell–activation signatures in a subset of B-ALL cells interacting with CAR T cells
CD19 cluster formation is a well-described phenomenon of B-cell activation in response to BCR stimulation by ligand [19]. We hypothesized that CD19 clustering and internalization may lead to transcriptional programs of B-cell activation, similar to what has been described upon BCR stimulation–induced CD19 clustering in normal B cells. To address this question, we performed scRNA-seq of surviving leukemic cells after co-culture with CAR T cells for 24 hours and compared these data with scRNA-seq of leukemic cells co-cultured with uninfected T cells. For these experiments, leukemic cells were presorted for high CD19 expression to exclude pre-existing cells expressing low CD19 prior to co-culture.
A total of 1,039 cells were sorted and sequenced based on CD19 expression after co-culture, with 831 cells remaining after quality filtering steps (Supplementary Fig. S7a–b). Pagoda2 clustering and visualization by t-Stochastic Neighbor Embedding (t-SNE) revealed a total of 7 clusters. Clusters 2–7 contained a mixture of leukemic cells co-cultured with CAR T cells and uninfected T cells (Figure 3a, Supplementary Table S2). In contrast, cluster 1 solely consisted of leukemic cells that had been co-cultured with CAR T cells (Figure 3a).
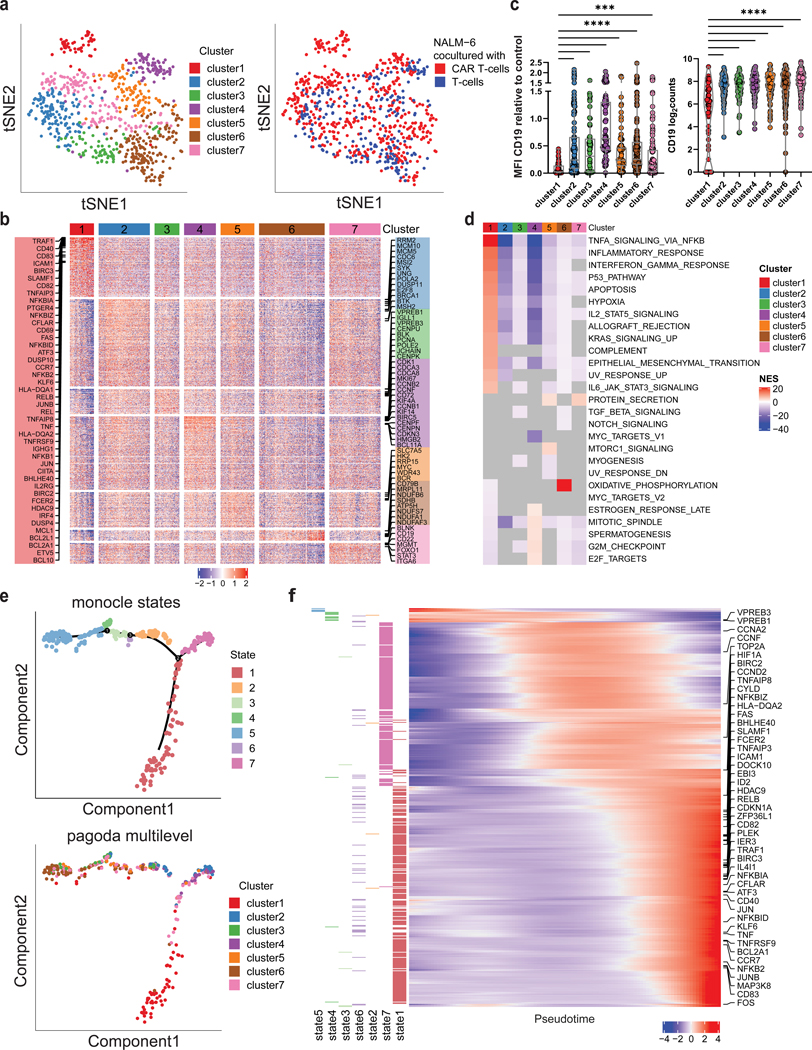
Figure 3. CD19 CAR T cells trigger activation of B-ALL cells with subsequent transcriptional rewiring.
a, ScRNA sequencing of NALM-6 B-ALL cells exposed to CD19 CAR T cells or uninfected control T cells at 5:1 ratio (E:T) for 24h. tSNE plot of B-ALL target cells showing PAGODA2 clusters (left) and type of effector cells (right). Only pagoda cluster 1 consists exclusively of B-ALL target cells exposed to CAR T cells. b, Heatmap of marker genes expressed in each PAGODA2 cluster with a mean log2FC > 1. c, CD19 surface protein (left) and mRNA (right) expression in cluster 1 cells compared to clusters 2–7 (P values determined by Dunn’s multiple comparison test. **=p<0.01, ****=p<0.0001, log2counts calculated as log2(Counts Per Million (CPM)+1)). Boxplots show minimum, 25th percentile, median, 75th percentile and maximum values. d, Heatmap of normalized enrichment scores (NES) of significantly enriched hallmark gene sets in individual clusters. Grey values depict no significance. All gene sets with a significant NES in at least one cluster are shown. e, Top, Monocle2 pseudotime analysis of B-ALL cells co-cultured with CD19 CAR T cells identifying 7 states. Bottom, projection of pagoda clusters over monocle2 pseudotime distribution. f, Heatmap of genes that informed the pseudotemporal distribution. Increasing expression of NFkB- and CD40-signaling pathway genes over pseudotime (TNFAIP8, NFKBIZ, FAS, SLAMF1, TNFAIP3, RELB, TRAF1, NFKBIA, CFLAR, CD40, NFKBID, KLF6, TNF, BCL2A1, CCR7, NFKB2) with highest enrichment in state 1. Left annotation depicts state, in which each gene is expressed at highest.
To understand how the clusters differed from each other, we first assessed differences in cell cycle, a known driver of cluster formation in single cell analysis [40] using Seurat2 [26]. We found that clusters 2 and 3 were enriched for cells in S-phase and clusters 4 and 5 for cells in G2M phase (Supplementary Fig. S8). Cluster 7 was enriched for cells in G1, whereas clusters 1 and 6 showed a mixture of cell cycle phases. Accordingly, marker gene analysis of the respective clusters revealed expression of genes involved in cell-cycle regulation, such as CDK1, CDC20, CCNB1/B2, CCNF, CDCA2/3/5/8, MKI67, HMGB2, TOP2A in cluster 4, and MYC and RRP15 in cluster 5 (Figure 3b, Supplementary Table S3). Consistent with cells in S-phase, clusters 2 and 3 showed high expression of DNA synthesis and repair genes such as RRM2, MCM5/10, PCNA and POLE2, BRCA1, MSH2. Cells in cluster 6 expressed various genes of the NADH:ubiquinone oxidoreductase and ATP synthase subunits, both of which are involved in the mitochondrial respiratory chain. Additionally, clusters 2–6 expressed genes that are part of several known signaling pathways in B cells – SYK and BTK expression were highest in cluster 2; VPREB1/3, IGLL1, BLK and JCHAIN highest in cluster 3; CD72 highest in cluster 4; BCR highest in cluster 5; CD79B highest in cluster 6; and BLNK, CD19 and CD22 highest in cluster 7 (Supplementary Table S3).
Cluster 1, which was the only cluster consisting exclusively of leukemic cells exposed to CD19 CAR T cells, demonstrated the lowest CD19 expression of all clusters, both at the protein (Figure 3c, left panel, Supplementary Table S4) and transcript level (Figure 3c, right panel). We therefore hypothesized that this cluster might represent a unique population of leukemic cells refractory to CAR T cells. This cluster demonstrated strong enrichment of B-cell activation signatures with expression of CD69, CD40 and CCR7. To test whether these transcriptional changes reflected protein expression, we assessed CD40 protein expression by flow cytometry. We observed increased CD40 expression in NALM-6 cells that survived CAR T-cell co-culture (Supplementary Fig. S9). Cluster 1 also expressed genes involved in the NFkB- and CD40-signaling pathway (NFKB2, NFKBIA/D/Z, REL, RELB, TRAF1, TNF, TNFAIP3/8, BCL2A1; Figure 3b, d). GSEA revealed enrichment for NFkB signatures, several other immune response signatures related to cytokines (interferon gamma response, IL2–STAT5 signaling and IL6–JAK–STAT3 signaling) and inflammatory response signatures, indicating the activation of immunomodulatory pathways in cluster1 cells (Figure 3d, Supplementary Table S5). These data suggest that CAR-mediated CD19 crosslinking causes transcriptional programs of B-cell activation in a subset of B-ALL cells.
To gain further insights into the transcriptional dynamics of CAR T cell–exposed B-ALL cells, we employed pseudotime analysis by Monocle2. This analysis revealed seven distinct states distributed along a single common trajectory (Figure 3e, top). State 5 cells at one end of the trajectory showed high expression of two genes, VPREB1 and VPREB3, that are normally expressed in preB cells [41] (Figure 3f). Genes informing the pseudotemporal distribution were also involved in cell-cycle regulation (CCNA2, CCNF, TOP2A, CDK1). State 1 cells, which consisted of Pagoda2 cluster 1 cells, were located at the opposite end of the trajectory (Figure 3e, bottom). Cells towards this end of the pseudotime distribution exhibited increased expression of genes involved in NFkB signaling (NFKBIZ, NFKBIA, TNFAIP3, NFKBID, NFKB2, RELB, NFKB1, TNFAIP9, FCER2) and CD40 signaling (CD40, CD80, TRAF1, ICAM1, CFLAR, BCL2A1, EBI3), consistent with the identified marker genes of cluster 1 (Figure 3f, Supplementary Table S6). These analyses suggest a continuous differentiation process of leukemic cells with state 1 cells representing the most differentiated state, exclusively containing CAR T cell–exposed CD19+ B-ALL cells.
A subset of CAR T cell–exposed B-ALL cells activates germinal center–reaction regulatory programs
B-cell activation normally leads to B-cell differentiation and initiation of the germinal center reaction, processes that are tightly regulated by master transcription factors [42,43]. Given their distinct transcriptional programs, we postulated that the subset of leukemic cells that clustered in state 1 were employing physiological programs of B-cell differentiation and activation. To assess a possible involvement of B-cell activation programs, we analyzed transcriptional regulon activity using SCENIC. We found that pseudotime states 2, 4, and 7 cells were enriched for transcriptional regulons involved in cell-cycle regulation such as E2F1/2/4/8, TP53, MYC, BRCA1, STAT3. States 2–5 demonstrated enrichment of the early B-cell regulators PAX5, POU2F1, BACH1, BACH2 and KLF3, consistent with their developmental state. State 1 cells showed high activity of IRF4, BCL6 and ZBTB7A regulons, which are known master transcription factors of the germinal center reaction (Figure 4a, left, Supplementary Table S7).
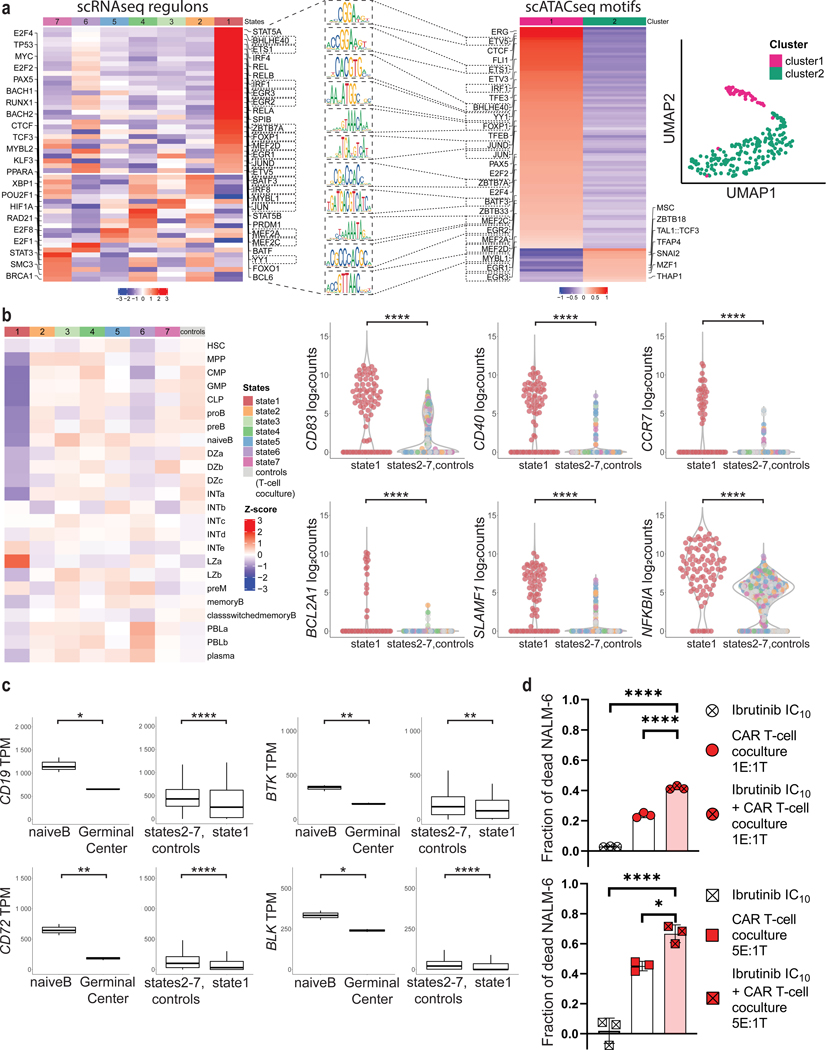
Figure 4. Interactions with CD19 CAR T cells induce germinal center reaction regulatory programs in B-ALL cells.
a, Left, activity of selected transcriptional regulons involved in B-cell lineage in NALM-6 cells, as predicted by SCENIC. Right, UMAP projection and cluster identification of scATAC-seq peaks from B-ALL cells that have been co-cultured with CAR T cells at 5:1 (E:T) ratio for 24h. Rectangular markings depict regulons and motifs that overlap in state 1 (scRNA-seq) and cluster 1 (scATAC-seq). Middle, visualization of selected motifs from overlap (significant differential accessibility of germinal center motifs in cluster 1 was tested by Fisher’s exact test (p=0.0003)). b, Left, heatmap demonstrating expression of HSC, MPP, CLP, proB, preB, naïveB, Germinal Center (GC) Dark Zone, GC Intermediate Zone, GC Light Zone, preMemory, Memory, class-switched Memory, Plasmablast, Plasma cell - signatures in monocle2 states of NALM-6 cells exposed to CAR T cells and uninfected T cells (controls). Right, violin plots of selected germinal center light zone gene expressions CD83, CD40, CCR7, BCL2A1, SLAMF1 and NFKBIA from monocle state 1 compared with states 2–7 and NALM-6 exposed to uninfected T cells (controls). Log2counts calculated as log2 (Counts Per Million (CPM)+1). c, Violin plots displaying lower CD19, BTK, BLK and CD72 expression (Transcripts Per Kilobase Million (TPM)) in germinal center B cells compared to naïve B cells in previously published dataset from Holmes et al. [33] (left panels), and B-ALL cells from monocle state 1 compared with states 2–7 and NALM-6 exposed to uninfected T cells (right panels). Boxplots show minimum, 25th percentile, median, 75th percentile and maximum values. d, Cytotoxicity of CD19 CAR T cells in the presence of ibrutinib at IC10 concentration of 0.963μM after 6h of co-culture with NALM-6 cells (percentages of dead cells normalized to baseline cytotoxicity of uninfected T-cells. P values determined by Tukey’s multiple comparisons test, n=3, representative of two independent experiments using either CAR constructs 1 or 2, respectively. *=p<0.05, ****=p<0.0001. Data are mean ± s.d.).
As further proof of transcriptional reprogramming we assessed transcription factor–motif enrichment by scATAC-seq in NALM-6 cells after 24h of co-culture with CD19 CAR T cells. From 288 cells sorted and sequenced, 245 passed quality control (Supplementary Fig. S10a–b). Clustering of ATAC-peaks using cisTopic and visualization using UMAP identified 2 major clusters (Figure 4a, right) with 99 differentially regulated motifs (adjusted p-value < 0.05, Supplementary Table S8) that were chosen for further analysis. Differential motif–enrichment analysis showed that germinal center motifs such as ETV5, CTCF, ETS1, BHLHE40, YY1, FOXP1, TFEB, JUND, ZBTB7A, MEF2A/C/D, EGR1/2/3, MYBL1 were more accessible in peaks in cluster 1 (Figure 4a, right). Additionally, we found that motifs of other regulators of B-cell function and differentiation were enriched in peaks in cluster 1, including ERG, PAX5, FLI1, NFYA, NFYB and transcription factors of the ETS and Sp/KLF family. In contrast, cluster 2 peaks were enriched for motifs of SNAI2, a transcription factor active in early lymphoid cell development [44], and MSC, an inhibitor of the critical B-cell development regulator E47 [45,46]. We also detected motifs of the nuclear proapoptotic factor THAP1 [47] and the tumor/growth suppressor MZF1 [48], which were enriched in cluster 2 peaks. Additionally, motifs of the known cell cycle regulators E2F2 and E2F4 were enriched in cluster 1 peaks, corresponding to the G2M-phase signatures identified in our RNA-seq data in clusters 4 and 5 (Figure 3b, d and Figure 4a, left).
To further define which B-cell differentiation programs leukemic cells employed in response to interaction with CD19 CAR T cells, we assessed expression of validated B cell–differentiation markers, including hematopoietic stem cell (HSC), multipotent progenitor (MPP), common lymphoid progenitor (CLP), pro-B, pre-B, naïve B, germinal center dark zone (GC DZ), germinal center intermediate (GC INT), germinal center light zone (GC LZ), pre-memory, memory, class-switched memory, plasmablast and plasma cell signatures. We compiled these signatures from several datasets (BLUEPRINT [30], Human Cell Atlas [32]) and recently published signatures of the germinal center reaction from Holmes et al. (2020) ([33]; Figure 4b, left, Supplementary Table S9). Most states showed enrichment for several immature signatures of the B-cell lineage, consistent with their B-ALL developmental state. For example, NALM-6 cells co-cultured with uninfected T cells were enriched for the proB signature, which is consistent with NALM-6 cells arrested at this developmental stage. In contrast, immature signatures were downregulated in state 1 cells and these cells demonstrated enrichment of mature B–cell signatures including the light zone a signature (Figure 4b, left). Furthermore, state 1 cells expressed the light zone marker CD83 and other germinal center markers like CD40, BCL2A1, SLAMF1, NFKBIA (Figure 4b, right). As the physiologic B-cell germinal center reaction is accompanied by decreased CD19 expression (Figure 4c, analyzing RNA-seq data from Holmes et al. [33]), these data suggest that state 1 leukemia cells employ germinal center reaction signatures to transcriptionally downregulate CD19 and maintain lower CD19 expression for escape from CD19 CAR T cells.
As the tyrosine kinase BTK is required for normal B-cell activation [49,50], we next tested whether inhibiting BTK prevented B-cell activation induced by CD19 CAR T cells and inhibited escape from CAR T cell–mediated killing. To this end, we pretreated leukemic cells with the BTK inhibitor ibrutinib for 72h at IC10 dosing (Supplementary Fig. S11a) prior to co-culture with CAR T cells. Ibrutinib-treated leukemic cells exposed to CAR T cells were killed at higher rates compared to non-ibrutinib–treated leukemic cells (43.17% vs 25.45% at E:T ratio of 1:1, p-value = 6E-08; 66.62% vs 45.11% at E:T ratio of 5:1, p-value = 0.0131; Figure 4d). Increased cytotoxicity was also observed after ibrutinib wash-out before co-culture and we did not observe any reduced viability of either CAR T cells or leukemic cells with the ibrutinib dosing used in these assays (Figure 4d, Supplementary Fig. S11b, c). Thus, these data demonstrate that leukemic cells may employ B-cell activation and germinal center reaction programs to escape from CAR T cell–mediated killing, a process that can be suppressed by inhibiting B cell–activation programs.
Discussion
It is well established that target antigen recognition by CAR T cells is dependent on antigen density and that efficacy of CAR T cell–mediated cytotoxicity decreases with lower antigen expression on target cells [12,14,16,51]. Therefore, low expression or nearly complete loss of target antigen is a challenge for CAR T-cell treatments and other immunotherapies [13,51,52]. Trogocytosis, a process by which membrane fragments are transferred from target to effector cells, has been identified as a possible mechanism for lowering target epitope expression [12]. Enhancing CAR signaling has been proposed to tune CAR T-cells’ antigen recognition and counteract low target epitope expression [14,53]. However, little is known about the dynamic regulation of CD19 expression by target cells during their early interactions with CAR T cells and how these interactions may affect cytotoxic efficacy.
Here, we show that CD19 CAR T-cell therapy induces rapid clustering of CD19 on the leukemia cell – T-cell interface and internalization of CD19 by B-ALL leukemic cells, resulting in decreased CD19 surface antigen expression among CAR T cell–exposed B-ALL cells. Among the initially CD19+ leukemic cells that survived CAR T-cell killing, we identified a distinct subpopulation in which low CD19 expression was sustained through transcriptional rewiring and engagement of unique transcriptional regulators associated with B-cell differentiation. Specifically, we show that these cells engage regulatory programs of normal B-cell activation such as the germinal center reaction, and the NFkB- and CD40-signaling pathways.
Our studies raise important issues about CAR-mediated receptor engagement on cancer cells and their consequences. This is particularly important as CAR T cells may lose efficacy over time, such as through T-cell exhaustion [54]. As their killing efficacy decreases, those CAR T cells may still maintain their state change–inducing effects on the cancer cells, which may propagate a greater number of surviving cancer cells over time. Further work is needed to characterize the functional effects of CAR-mediated receptor engagement in relevant preclinical disease models in vivo and how they translate to treating relapsed disease in patients.
Our findings also raise questions about the effects of T cell–targeting immunotherapies other than CAR T cells on their cancer cell targets. In this context, treatment with the CD3/CD19 bispecific antibody blinatumomab has been associated with CD19low/negative relapses [55,56]. Further studies are needed to understand the relative impact of bispecific immunotherapy agents and CAR T cells on CD19low/negative relapses and the optimal sequence of administering different immunotherapies.
Cellular plasticity is a basis for treatment failure in several cancers [57,58]. Cancer cells may engage transcriptional programs that allow for de-differentiation or cross-differentiation into other lineages [9,10]. Our data demonstrate that rather than de-differentiating into more stem-like states, engagement of lineage-appropriate differentiation and activation programs may allow for less effective recognition by CD19 CAR T cells. This is reminiscent of what has been shown for key signaling pathways that operate in normal B cells and remain essential for leukemia cell survival in B cell receptor signaling–dependent precursor B-ALL, thus providing a rationale for targeting these pathways [59–61].
Our studies highlight the importance of combining CAR T-cell therapy with other targeted therapies to overcome the plasticity of leukemia cells. More work is needed to identify the exact epigenetic mediators that orchestrate the engagement of programs of normal B-cell activation and differentiation in B-ALL cells in response to CD19 CAR T cell interactions, as well as their dependencies on B cell–signaling pathways. While other groups have shown favorable combination of CAR T cells and ibrutinib in other B-cell malignancies, such as chronic lymphocytic leukemia [62] and mantle cell lymphoma [63], previous studies of combination treatment in B-ALL have been inconclusive [64,65]. Our data show that inhibiting BTK, an essential mediator of B-cell activation [49,50], increases CD19 CAR T cell–mediated B-ALL cytotoxicity, presumably through inhibiting the engagement of normal B-cell differentiation and activation programs.
In summary, our studies highlight how CAR T cells might trigger engagement of physiological regulatory programs in their target cells, and how CAR T cells’ failure to kill their targets at first interaction may promote target-cell survival.
Supplementary Material
Synopsis.
The authors show CD19-expressing B-ALL cells can employ regulatory programs of normal B-cell activation and germinal center reaction to transcriptionally downregulate and maintain lower CD19 expression, allowing for enhanced survival in the early phases of CAR T-cell exposure.
Acknowledgments
We thank the Knoechel and Lohr lab members for discussions and helpful comments on the manuscript. We thank the Jimmy Fund DFCI Flow Cytometry core for their support with flow sorting. We thank Min Gyu Im for his help with editing the movies. N.G.I is supported by the Kind-Philipp-Stiftung für pädiatrisch-onkologische Forschung. This research was supported by the NCI (K08CA191091 to B.K. and K08CA191026 to J.G.L.) and the Ambrose Monell Foundation (to B.K.).
Funding information:
This research was supported by the NCI (K08CA191091 to B.K. and K08CA191026 to J.G.L.), and the Ambrose Monell Foundation (B.K.). N.G.I was supported by the Kind-Philipp-Stiftung für pädiatrisch-onkologische Forschung.
COI disclosure statement:
J.G.L. receives research funding from Celgene for an unrelated research project. All other authors declare no conflicts of interest.
References
- [1].Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. New England Journal of Medicine 2013;368:1509–18. Available from: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. New England Journal of Medicine 2014;371:1507–17. Available from: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. New England Journal of Medicine 2018;378:439–48. Available from: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, et al. Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. New England Journal of Medicine 2018;378:449–59. Available from: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gardner RA, Finney O, Annesley C, Brakke H, Summers C, Leger K, et al. Intent to treat leukemia remission by CD19CAR T cells of defined formulation and dose in children and young adults. Blood 2017:blood‐2017‐02‐769208. Available from: 10.1182/blood-2017-02-769208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maude SL, Teachey DT, Rheingold SR, Shaw PA, Aplenc R, Barrett DM, et al. Sustained remissions with CD19-specific chimeric antigen receptor (CAR)-modified T cells in children with relapsed/refractory ALL. Journal of Clinical Oncology 2016;34:3011. Available from: 10.1200/JCO.2016.34.15_suppl.3011. [DOI] [Google Scholar]
- [7].Orlando EJ, Han X, Tribouley C, Wood PA, Leary RJ, Riester M, et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nature medicine 2018;24:1504–06. [DOI] [PubMed] [Google Scholar]
- [8].Asnani M, Hayer KE, Naqvi AS, Zheng S, Yang SY, Oldridge D, et al. Retention of CD19 intron 2 contributes to CART-19 resistance in leukemias with subclonal frameshift mutations in CD19. Leukemia 2019. Available from: http://www.nature.com/articles/s41375-019-0580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gardner R, Wu D, Cherian S, Fang M, Hanafi L-A, Finney O, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 2016;127:2406–10. Available from: 10.1182/blood-2015-08-665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nature communications 2016;7. Available from: http://www.nature.com/articles/ncomms12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, et al. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer discovery 2015;5:1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J, et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 2019;568:112–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nature medicine 2018;24:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, et al. Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer discovery 2020;10:702–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nerreter T, Letschert S, Götz R, Doose S, Danhof S, Einsele H, et al. Super-resolution microscopy reveals ultra-low CD19 expression on myeloma cells that triggers elimination by CD19 CAR-T. Nature communications 2019;10:3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ramakrishna S, Highfill SL, Walsh Z, Nguyen SM, Lei H, Shern JF, et al. Modulation of Target Antigen Density Improves CAR T-cell Functionality and Persistence. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25:5329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ruella M, Xu J, Barrett DM, Fraietta JA, Reich TJ, Ambrose DE, et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nature medicine 2018;24:1499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Singh N, Lee YG, Shestova O, Ravikumar P, Hayer KE, Hong SJ, et al. Impaired Death Receptor Signaling in Leukemia Causes Antigen-Independent Resistance by Inducing CAR T-cell Dysfunction. Cancer discovery 2020;10:552–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Depoil D, Fleire S, Treanor BL, Weber M, Harwood NE, Marchbank KL, et al. CD19 is essential for B cell activation by promoting B cell receptor–antigen microcluster formation in response to membrane-bound ligand. Nature Immunology 2008;9:63–72. Available from: http://www.nature.com/articles/ni1547. [DOI] [PubMed] [Google Scholar]
- [20].Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols 2014;9:171–81. Available from: http://www.nature.com/articles/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- [21].Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. A rapid and robust method for single cell chromatin accessibility profiling. Nature communications 2018;9:5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nature methods 2016;13:241–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14:979–82. Available from: https://www.nature.com/articles/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 2014;32:381–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nature methods 2017;14:1083–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fernández JM, La de Torre V, Richardson D, Royo R, Puiggròs M, Moncunill V, et al. The BLUEPRINT Data Analysis Portal. Cell systems 2016;3:491–495.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome biology 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Experimental hematology 2018;68:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Holmes AB, Corinaldesi C, Shen Q, Kumar R, Compagno N, Wang Z, et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. The Journal of experimental medicine 2020;217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nature biotechnology 2020;38:276–78. [DOI] [PubMed] [Google Scholar]
- [35].McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nature methods 2019;16:397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rodriguez A, Laio A. Machine learning. Clustering by fast search and find of density peaks. Science (New York, N.Y.) 2014;344:1492–96. [DOI] [PubMed] [Google Scholar]
- [38].Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature methods 2017;14:975–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xiong W, Chen Y, Kang X, Chen Z, Zheng P, Hsu Y-H, et al. Immunological Synapse Predicts Effectiveness of Chimeric Antigen Receptor Cells. Molecular therapy : the journal of the American Society of Gene Therapy 2018;26:963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nature reviews. Genetics 2015;16:133–45. [DOI] [PubMed] [Google Scholar]
- [41].Rodig SJ, Kutok JL, Paterson JC, Nitta H, Zhang W, Chapuy B, et al. The pre-B-cell receptor associated protein VpreB3 is a useful diagnostic marker for identifying c-MYC translocated lymphomas. Haematologica 2010;95:2056–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cyster JG, Allen CDC. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 2019;177:524–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Silva de NS, Klein U.Dynamics of B cells in germinal centres. Nature reviews. Immunology 2015:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pioli PD, Weis JH. Snail transcription factors in hematopoietic cell development: a model of functional redundancy. Experimental hematology 2014;42:425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Massari ME, Rivera RR, Voland JR, Quong MW, Breit TM, van Dongen JJ, et al. Characterization of ABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Molecular and Cellular Biology 1998;18:3130–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. The Journal of experimental medicine 2009;206:2271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard J-P. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene 2003;22:2432–42. [DOI] [PubMed] [Google Scholar]
- [48].Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, et al. Mzf1 controls cell proliferation and tumorigenesis. Genes & Development 2001;15:1625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pal Singh S, Dammeijer F, Hendriks RW. Role of Bruton’s tyrosine kinase in B cells and malignancies. Molecular cancer 2018;17:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Middendorp S, Dingjan GM, Maas A, Dahlenborg K, Hendriks RW. Function of Bruton’s tyrosine kinase during B cell development is partially independent of its catalytic activity. Journal of immunology (Baltimore, Md. : 1950) 2003;171:5988–96. [DOI] [PubMed] [Google Scholar]
- [51].Watanabe K, Terakura S, Martens AC, van Meerten T, Uchiyama S, Imai M, et al. Target Antigen Density Governs the Efficacy of Anti–CD20-CD28-CD3 ζ Chimeric Antigen Receptor–Modified Effector CD8 ^+ T Cells. The Journal of Immunology 2015;194:911–20. Available from: 10.4049/jimmunol.1402346. [DOI] [PubMed] [Google Scholar]
- [52].Walker AJ, Majzner RG, Zhang L, Wanhainen K, Long AH, Nguyen SM, et al. Tumor Antigen and Receptor Densities Regulate Efficacy of a Chimeric Antigen Receptor Targeting Anaplastic Lymphoma Kinase. Molecular therapy : the journal of the American Society of Gene Therapy 2017;25:2189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gudipati V, Rydzek J, Doel-Perez I, Gonçalves VDR, Scharf L, Königsberger S, et al. Inefficient CAR-proximal signaling blunts antigen sensitivity. Nature Immunology 2020;21:848–56. [DOI] [PubMed] [Google Scholar]
- [54].Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019;576:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Aldoss I, Song J, Stiller T, Nguyen T, Palmer J, O’Donnell M, et al. Correlates of resistance and relapse during blinatumomab therapy for relapsed/refractory acute lymphoblastic leukemia. American Journal of Hematology 2017;92:858–65. [DOI] [PubMed] [Google Scholar]
- [56].Libert D, Yuan CM, Masih KE, Galera P, Salem D, Shalabi H, et al. Serial evaluation of CD19 surface expression in pediatric B-cell malignancies following CD19-targeted therapy. Leukemia 2020. Available from: http://www.nature.com/articles/s41375-020-0760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Quintanal-Villalonga Á, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nature reviews. Clinical oncology 2020;17:360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Biehs B, Dijkgraaf GJP, Piskol R, Alicke B, Boumahdi S, Peale F, et al. A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition. Nature 2018;562:429–33. [DOI] [PubMed] [Google Scholar]
- [59].Köhrer S, Havranek O, Seyfried F, Hurtz C, Coffey GP, Kim E, et al. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/AKT, FOXO1 and MYC, and can be targeted by SYK inhibition. Leukemia 2016;30:1246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, et al. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 2015;27:409–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kim E, Hurtz C, Koehrer S, Wang Z, Balasubramanian S, Chang BY, et al. Ibrutinib inhibits pre-BCR+ B-cell acute lymphoblastic leukemia progression by targeting BTK and BLK. Blood 2017;129:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fraietta JA, Beckwith KA, Patel PR, Ruella M, Zheng Z, Barrett DM, et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood 2016;127:1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ruella M, Kenderian SS, Shestova O, Fraietta JA, Qayyum S, Zhang Q, et al. The Addition of the BTK Inhibitor Ibrutinib to Anti-CD19 Chimeric Antigen Receptor T Cells (CART19) Improves Responses against Mantle Cell Lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22:2684–96. [DOI] [PubMed] [Google Scholar]
- [64].Qin JS, Johnstone TG, Baturevych A, Hause RJ, Ragan SP, Clouser CR, et al. Antitumor Potency of an Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy, Lisocabtagene Maraleucel in Combination With Ibrutinib or Acalabrutinib. Journal of Immunotherapy 2020:1. Available from: http://Insights.ovid.com/crossref?an=00002371-900000000-99404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dufva O, Koski J, Maliniemi P, Ianevski A, Klievink J, Leitner J, et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T cell cytotoxicity. Blood 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kochenderfer JN, Feldman SA, Zhao Y, Xu H, Black MA, Morgan RA, et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. Journal of immunotherapy (Hagerstown, Md. : 1997) 2009;32:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are deposited in GEO under accession number GSE176418. All other data are available within the manuscript and its accompanying supplementary data files or from the corresponding author upon reasonable request.