Abstract
Escherichia coli ssrA encodes a small stable RNA molecule, tmRNA, that has many diverse functions, including tagging abnormal proteins for degradation, supporting phage growth, and modulating the activity of DNA binding proteins. Here we show that ssrA plays a role in Salmonella enterica serovar Typhimurium pathogenesis and in the expression of several genes known to be induced during infection. Moreover, the phage-like attachment site, attL, encoded within ssrA, serves as the site of integration of a region of Salmonella-specific sequence; adjacent to the 5′ end of ssrA is another region of Salmonella-specific sequence with extensive homology to predicted proteins encoded within the unlinked Salmonella pathogenicity island SPI4. S. enterica serovar Typhimurium ssrA mutants fail to support the growth of phage P22 and are delayed in their ability to form viable phage particles following induction of a phage P22 lysogen. These data indicate that ssrA plays a role in the pathogenesis of Salmonella, serves as an attachment site for Salmonella-specific sequences, and is required for the growth of phage P22.
Many virulence functions that contribute to in vivo fitness are encoded on pathogenicity islands, sequences presumed to have been acquired by horizontal transfer as evidenced by their atypical base composition and codon usage (17). We have recently used in vivo expression technology (20, 34, 35) to identify several Salmonella enterica serovar Typhimurium in vivo-induced (ivi) acquired sequences, which are distinct within and between Salmonella serovars and contribute to fitness within the host (10). One of these acquired virulence regions, iviXXII, is composed of a mosaic of Salmonella-specific sequences that is also serovar specific (present in a distinct set of serovars of Salmonella but not in E. coli or several other enteric pathogens). Removal of a 17-kb portion of the iviXXII region conferred a greater than 200-fold defect in virulence in a murine model of typhoid fever (10).
ssrA resides in or near the junction point of this mosaic of Salmonella-specific sequence (10) and is also known to serve as the insertion site for acquired sequences such as the cryptic phage CP4-57 in Escherichia coli (27) and pathogenicity islands in Vibrio cholerae (25, 30) and Dichelobacter nodosus (2), the causative agents of cholera and ovine foot rot, respectively. ssrA encodes a small, stable RNA molecule (∼350 nucleotides), tmRNA, which may serve as both a tRNA and an mRNA, tagging partially synthesized proteins for degradation (26). Additionally, tmRNA plays other diverse roles in E. coli including regulating an alternative protease activity (27), modulating DNA binding protein activity (41), and contributing to the growth of λ-P22 hybrid phage (40, 55). We investigated whether ssrA contributed to the pathogenesis of Salmonella since ssrA mapped within a 17-kb region that is required for pathogenesis (10) and RNA molecules are also known to play a key role in the virulence of many organisms (17, 39, 43).
MATERIALS AND METHODS
Bacterial strains and phage.
All S. enterica serovar Typhimurium strains used in this study were derived from strain ATCC 14028 (CDC 6516-60) and are listed in Table 1. The high-frequency generalized transducing bacteriophage P22 mutant HT105/1, int-201, was used for all transductional crosses (47), and phage-free, phage-sensitive transductants were isolated as previously described (5). The isolations of all ivi fusions used in this study were described previously (19). Bacteriophage P22-3657 and S. enterica serovar Typhimurium LT2 strain MST3357 were kindly provided by Stan Maloy. P22-3657 contains a deletion in gene 9, encoding phage P22 tail protein (15). MST3357 constitutively expresses P22 tail protein, which is necessary for P22-3657 to form plaques.
TABLE 1.
Bacterial strains and phage used in this study
| Strain or phagea | Genotype | Source or reference |
|---|---|---|
| Salmonella strains | ||
| ATCC 14028 | Wild type | CDC 6516-60 |
| MT1219 | DUP3133 [φ(iviXXII-purA+-lacZ+ lacY+)* pIVET1* (iviXXII+)] | 19 |
| MT1780 | DUP3458 [φ(vacB′-purA+-lacZ+ lacY+)* pIVET* (vacB+)] | 19 |
| MT2055 | Δzfj7503::Kn (17-kb deletion of ssrA region) | 10 |
| MT2057 | zjf7504::MudJ (33% linked to purA) | 10 |
| MT2094 | Δzfj7505::Kn (3.4-kb deletion removing ssrA, Slp, and Orf) | This work |
| MT2095 | Δzfj7506::Kn (2.0-kb deletion removing HlyB and HlyD) | This work |
| MT2096 | Δzfj7507::Kn (3.0-kb deletion removing HlyB and AggA) | This work |
| MT2121 | ssrA1::Tn10d-Tc | This work |
| MT2122 | zfj7508::Tn10d-Tc (within SlpA) | This work |
| MT2123 | zfj7509::Tn10d-Tc (intergenic between SlpA and Orf) | This work |
| MT2124 | zfj7510::Tn10d-Tc (within Orf) | This work |
| MT2194 | ΔssrA2::Kn | This work |
| MT2207 | P22-3657 lysogen in 14028 | This work |
| MT2208 | P22-3657 lysogen in ssrA1::Tn10d-Tc | This work |
| MT2218 | DUP3122 [φ(asnS′-purA+-lacZ+ lacY+)* pIVET1* (asnS+)] | 19 |
| MT2219 | DUP3229 [φ(thiI′-purA+-lacZ+ lacY+)* pIVET1* (thiI+)] | 19 |
| MT2220 | DUP3458 [φ(vacB′-purA+-lacZ+ lacY+)* pIVET1* (vacB+) ΔssrA2::Kn] | This work |
| MT2221 | DUP3122 [φ(asnS′-purA+-lacZ+ lacY+)* pIVET1* (asnS+) ΔssrA2::Kn] | This work |
| MT2222 | DUP3229 [φ(thiI′-purA+-lacZ+ lacY+)* pIVET1* (thiI+) ΔssrA2::Kn] | This work |
| MT2231 | DUP3360 [φ(f304-cat+-lacZ+ lacY+)* pIVET8* (f304+)] | 19 |
| MT2232 | DUP3136 [φ(dedE-purA+-lacZ+ lacY+)* pIVET1* (dedE+)] | 19 |
| MT2234 | DUP3448 [φ(yoaA-cat+-lacZ+ lacY+)* pIVET8* (yoaA+)] | 19 |
| MT2235 | DUP3360 [φ(f304-cat+-lacZ+ lacY+)* pIVET8* (f304+) ΔssrA2::Kn] | This work |
| MT2236 | DUP3136 [φ(dedE-purA+-lacZ+ lacY+)* pIVET1* (dedE+) ΔssrA2::Kn] | This work |
| MT2238 | DUP3448 [φ(yoaA-cat+-lacZ+ lacY+)* pIVET8* (yoaA+) ΔssrA2::Kn] | This work |
| MST3357 | LT2 leuA414(Am) supE Fels−/pJS28 (P22 gene 9/Apr) | Stan Maloy |
| E. coli strain | ||
| DH5α λpir | F−endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80dlacZM15 Δ(lacZYA-argF)U169 λpir | 18, 29 |
| Phage | ||
| P22 HT 105/1 HT 105/1 int-201 | 47 | |
| P22-3657 | Kn6 Pant↓-11c arc-amH1605 Psar↓subS lac2 9-delTB | Stan Maloy |
All bacterial strains with an MT designation are derivatives of S. enterica serovar Typhimurium ATCC 14028 (CDC 6516-60). MST3357 is a derivative of S. enterica serovar Typhimurium strain LT2.
Nomenclature.
The nomenclature is generally as described previously (4, 7, 13). Unless otherwise specified, the mutation designations used in this study are those of Sanderson et al. (45). The nomenclature z--::Tn10 refers to a Tn10 insertion in a “silent” DNA region; the z-- describes the map position of the insertion (44). The nomenclature used for chromosomal rearrangements is as described previously (8, 21, 33, 48).
Media and chemicals.
Luria broth (LB) (11) was the laboratory medium used in this study except for β-galactosidase assays, for which 2-(N-morpholino)ethanesulfonic acid (MES) buffered to pH 5.5 and supplemented with 0.05 mM Mg2+ was used. Unless otherwise specified, the final concentrations of antibiotics were as follows: ampicillin, 50 μg/ml (LB) or 16 μg/ml (MES); kanamycin, 50 μg/ml; and tetracycline, 20 μg/ml. Mitomycin C and o-nitrophenyl-β-d-galactopyranoside (ONPG) were purchased from Sigma.
Construction of ssrA mutations. (i) Knr deletions.
Deletions within the ssrA region were constructed by a PCR-based strategy as previously described (23). The resulting mutations have defined deletion endpoints that contain a Knr (kanamycin) determinant at the deletion joint point. The endpoints and primers used to generate the deletions were as follows: MT2055, 5′-TTTGATGCGCGGAAACAC-3′ and 5′-GCAATGCACCTTGGGTTT-3′; MT2094, 5′-TACTCATCTCCTGAGTGG-3′ and 5′-GCAATGCACCTTGGGTTT-3′; MT2095, 5′-GCGAGCGGTAAATCGTTG-3′ and 5′-CGGTGTATCAGGCTATCG-3′; MT2096, 5′-GAATGCGCAGTTAACGGC-3′ and 5′-CGATAGCCTGATACACCG-3′; and MT2194, 5′-TTTCGGACGCGGGTTCAA-3′ and 5′-GCAATGCACCTTGGGTTT-3′.
(ii) Tn10d-Tc insertions.
Tn10d-Tc insertions in the ssrA region were isolated using localized mutagenesis to a genetically linked ivi fusion, iviXXII (10). Briefly, MT1219 (iviXXII) was used as a recipient of phage P22 grown on a genomic pool of Tn10d-Tc insertions. Tetracycline-resistant transductants (Tcr) were selected and screened for loss of lacZ (inherent in ivi fusions) on LB plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). These Lac− Tn10d-Tc insertions were pooled, and a P22 lysate grown on these insertions was used to transduce S. enterica serovar Typhimurium harboring a recombinant plasmid containing a 3.4-kb DNA insert of the ssrA region (cloned within pWKS30 [53], Apr) to tetracycline resistance. The Tcr transductants were pooled, and plasmid DNA was isolated and used to electroporate E. coli DH5α λpir to tetracycline resistance. These Tcr Apr transformants contain Tn10d-Tc insertions in the 3.4-kb ssrA region. Transposon insertion joint points on the plasmid were determined by sequence analysis using a primer (5′-ACCTTTGGTCACCAACGC-3′) which hybridizes to the right end of Tn10d-Tc. To move these Tn10d-Tcr transposon insertions into the S. enterica serovar Typhimurium chromosome, phage P22 was grown on the strains containing the Tn10d-Tc-bearing plasmids and used to transduce wild-type S. enterica serovar Typhimurium 14028 to tetracycline resistance. Transductants containing a Tn10d-Tc insertion which had recombined into the chromosome were confirmed by screening for Aps (loss of pWKS30) and by verifying linkage to the parental ivi fusion, iviXXII.
Competitive index assay in BALB/c mice.
Mutant and wild-type cells were grown overnight in LB medium. BALB/c mice were intraperitoneally infected with a total of 103 cells of a 1:1 ratio of mutant to wild type (MT2057). The mice were sacrificed after 5 days, and the bacterial cells were recovered from the spleen; the competitive index is the ratio of the number of mutant to wild-type bacteria recovered.
Phage P22 assays. (i) EOP.
To determine the relative efficiency of plating EOP for ssrA strains, serial dilutions of phage P22 (HT105/1 int-201) were mixed with wild-type and ssrA strains and plated on LB medium containing 0.75% top agar. The number of PFU was determined on both wild-type and ssrA hosts. The EOP was calculated by dividing the number of PFU per milliliter obtained on the ssrA host strain by the number of PFU per milliliter obtained on the wild-type host.
(ii) Phage adsorption.
Phage P22 adsorption assays were performed by adding approximately 108 overnight-grown bacteria and 105 phage P22 to an Eppendorf tube containing prewarmed LB medium (37°C) in a total volume of 1 ml. A no-cell control was used to calculate the efficiency of adsorption. After a 15-min incubation, the cells were centrifuged at low speed for 30 s, and then at full speed spin 90 s, after which the supernatant was immediately transferred to a fresh tube. To determine the remaining PFU, supernatants were subjected to titer determination on wild-type S. enterica serovar Typhimurium. The percent adsorption is calculated as follows: [(number of phage PFU recovered from the supernatant of phage without cells − number of PFU recovered from the supernatant of phage with cells)/number of phage PFU recovered from the supernatant of phage without cells] × 100.
(iii) Lysogen formation.
The frequency of P22 lysogen formation was determined as follows. Overnight cultures of wild-type and ssrA S. enterica serovar Typhimurium were diluted 1:50 and incubated for 2.5 h at 37°C. At this point, the number of viable cells was determined and 4 × 107 bacteria were mixed with 1.5 × 109 phage P22-3657 (which contains a kanamycin resistance gene in place of the phage mnt gene [15]), for a multiplicity of infection (MOI) of approximately 40, in a total volume of 0.4 ml. Following a 1-h standing incubation at 37°C, the cell-phage mixture was serially diluted and plated on LB plates containing kanamycin to select for lysogens. The percent lysogeny was determined as follows: (number of lysogens/viable-cell count) × 100.
(iv) Lysogen induction.
Induction of viable phage from a lysogen was quantified by a single-burst assay in which overnight-grown wild-type and ssrA lysogens were subcultured 1:50 and incubated for 2.5 h at 37°C, at which time an aliquot was removed from the culture (time = 0). Mitomycin C (Sigma) was added to a final concentration of 2 μg/ml, and the cultures were returned to 37°C. Aliquots (1 ml) were removed at various times for the determination of phage production and viable cells. Phage production was determined by removing an aliquot, lysing the cells by vortexing them in chloroform, centrifuging them for 2 min, and transferring the supernatant to a fresh tube. Because phage P22-3657 is defective in tail protein production, P22 phage tails were added to each lysate in vitro by adding 6 μl of P22 tail stock prepared as described previously (37), after which the lysates were incubated overnight at room temperature. PFU were determined by subjecting the lysates to titer determination on strain MST3357, which constitutively expresses P22 gene 9 tail protein.
β-Galactosidase assays.
To determine β-galactosidase levels, a single colony was inoculated into 1 ml of MES medium buffered to pH 5.5 and containing 0.05 mM Mg2+ and was grown overnight at 37°C with shaking for 16 h. β-Galactosidase activities were determined by the method of Slauch and Silhavy (49). The activity of β-galactosidase is reported as 103 U per optical density at 600 nm unit per milliliter of cell suspension, where units are micromoles of o-nitrophenol (ONP) formed per minute (n = 3 trials; standard deviations were ±10% of the mean).
DNA sequencing and analysis.
DNA was sequenced by the dideoxy chain termination method (46). Homology searches were conducted using the FASTA program (Wisconsin Package version 10.0; Genetics Computer Group, Madison, Wis.) and the BLASTN and BLASTX algorithms available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
RESULTS
ssrA is required for Salmonella virulence.
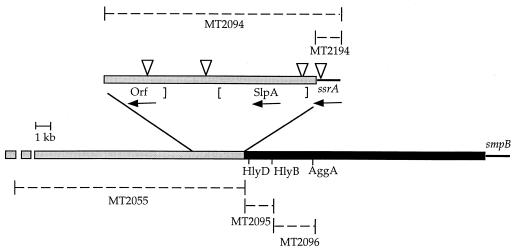
Previously, we have shown that a deletion of 17 kb of Salmonella-specific DNA at min 60 conferred a greater-than-200-fold virulence defect in BALB/c mice (10). To identify the gene(s) responsible, a series of defined deletions throughout the 17-kb region were constructed and tested for the ability to confer defects in virulence in a murine model for typhoid fever. Table 2 shows that removal of 3.4 kb of sequence (MT2094) conferred the same virulence defect as did the 17-kb deletion (MT2055). Sequence analysis of this region (Fig. 1) revealed three genes: ssrA, a predicted open reading frame with DNA homology (55%) to E. coli slpA integrase (27), and a predicted open reading frame of unknown function. Tn10d-Tc transposon insertions were isolated in each of these genes and retested for virulence. Table 2 shows that ssrA1::Tn10d-Tc (MT2121) conferred a virulence defect (70-fold) whereas all other downstream insertion (MT2122 to MT2124) and upstream deletion (MT2095 and MT2096) strains exhibited wild-type virulence. Additionally, MT2194, which contains a deletion of sequences entirely within ssrA, reproduced the 200-fold defect, suggesting that ssrA was responsible for the virulence phenotype (Table 2).
TABLE 2.
Mutations in ssrA confer a virulence defect in S. enterica serovar Typhimurium
| Strain | Relevant genotype | CI ± SD (n)a |
|---|---|---|
| MT2055 | Δzfj7503::Kn (17-kb deletion of ssrA region) | 0.0044 ± 0.0009 (4) |
| MT2094 | Δzfj7505::Kn (3.4-kb deletion of ssrA region) | 0.0042 ± 0.0020 (3) |
| MT2121 | ssrA1::Tn10d-Tc | 0.0145 ± 0.0092 (4) |
| MT2194 | ΔssrA2::Kn deletion | 0.0047 ± 0.0013 (5) |
| MT2122 | zfj7508::Tn10d-Tc (within SlpA) | 1.62 ± 0.917 (3) |
| MT2123 | zfj7509::Tn10d-Tc (intergenic between SlpA and Orf) | 1.04 ± 0.323 (5) |
| MT2124 | zfj7510::Tn10d-Tc (within Orf) | 0.836 ± 0.234 (3) |
| MT2095 | Δzfj7506::Kn (2.0-kb deletion including HlyD and HlyB) | 0.760 ± 0.507 (3) |
| MT2096 | Δzfj7507::Kn (3.0-kb deletion including HlyB and AggA) | 1.57 ± 1.51 (3) |
Competitive index (CI) assays were performed by intraperitoneal infection of BALB/c mice with 103 total bacteria in a 1:1 ratio of mutant bacteria to wild type (MT2057). At 5 days after infection, bacterial cells were recovered from the spleen. The CI is the ratio of mutant to wild-type bacteria recovered. n, number of mice tested.
FIG. 1.
Genetic organization of the S. enterica serovar Typhimurium ssrA region. Thick black and gray lines represent two distinct Salmonella-specific regions (10); thin black lines represent sequences shared between S. enterica serovar Typhimurium and E. coli (i.e., ssrA and smpB); dashed lines represent the extent of deleted DNA. The enlarged region denotes the material deleted in MT2094; Tn10d-Tc insertions in this region are designated by triangles; the arrows depict the predicted direction of transcription. Salmonella-specific sequences upstream of ssrA encode predicted proteins with similarity to sequences encoded within SPI4, an unlinked S. enterica serovar Typhimurium pathogenicity island (56). Protein similarities include HlyD and LktD, which are involved in the secretion of hemolysin in E. coli and Pasteurella haemolytica, respectively (32% identity over 176 amino acids and 26% identity over 146 amino acids to SPI4 protein D), ABC transport proteins (HlyB and LktB family, involved in the transport of hemolysin) (32% identity over 177 amino acids and 26% identity over 175 amino acids to SPI4 protein R), AggA and PrtF, which are involved in Pseudomonas putida adherence to root tip cells and protease secretion in Erwinia chrysanthemi, respectively (21% identity over 375 amino acids and 18% identical over 401 amino acids to SPI4 protein C).
ssrA is required for expression of ivi genes.
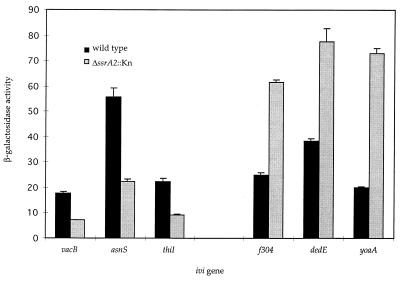
The effect of ssrA on ivi gene expression was determined by transducing an ssrA deletion (ΔssrA2::Kn) into our collection of more than 200 ivi fusion strains (19). Figure 2 shows that ssrA affected the expression of six ivi genes, three of which (vacB, asnS, and thiI) were induced by ssrA and three of which (f304, dedE, and yoaA) were repressed by ssrA. Several of these genes are required for or implicated in virulence, including vacB, which is required for the expression of the invasion plasmid antigen proteins of Shigella flexneri (28); f304, an open reading frame of unknown function which resides within a uropathogenic E. coli pathogenicity island (16); and dedE (cvpA), which is required for the production of the colicin V toxin (14). Other genes under ssrA control include asnS, which encodes asparaginyl-tRNA synthase (57); thiI, which is involved in the production of thiazole, a precursor of thiamine synthesis (54); and yoaA, a homologue of the dinG family of DNA helicases involved in the bacterial SOS response (31).
FIG. 2.
ssrA regulation of ivi genes. β-Galactosidase activities were determined following overnight incubation in MES minimal medium buffered to pH 5.5 and containing 0.05 mM Mg2+. β-Galactosidase activities are given as 103 micromoles of ONP formed per minute per optical density unit at 600 nm per milliliter of cell suspension. Values represent the average of three independent cultures, with standard deviations (±10%) shown as error bars.
Salmonella-specific sequence adjacent to ssrA.
Previously, we have shown that ssrA resides near the junction of two different regions of Salmonella-specific sequence (10). To ascertain whether ssrA was at the junction point of either of these unique DNA regions, the ssrA region was sequenced. S. typhimurium ssrA shows 96.1% homology to the E. coli ssrA gene. However, homology to E. coli is lost immediately after the 3′ end of ssrA, which contains the P4-like bacteriophage attachment site, attL (27). attL serves as the attachment site for the E. coli cryptic prophage, CP4-57 (27), and pathogenicity islands in Vibrio cholerae (30) and Dichelobacter nodosus (2), the causative agents of cholera and sheep foot rot, respectively. These data suggest that ssrA attL sequences serve as the attachment site for one of the two regions of Salmonella-specific sequences at min 60. Moreover, Salmonella-specific sequence with limited or no homology to E. coli continues for several kilobases 3′ to the ssrA attL attachment site. The first 180 nucleotides downstream of attL have no homology to the GenEMBL DNA database, after which limited homology to the E. coli DNA sequence (55%) extends for 1.2 kb (Fig. 1). This coincides with the location of E. coli slpA, which encodes the cryptic phage CP4-57 integrase SlpA.
Analysis of the sequence at the 5′ end of ssrA revealed homology to the E. coli sequence for 116 nucleotides (67%), after which no homology to the DNA database was detected for at least another 1.1 kb. No bacterial attachment sites or IS sequences were detected at or near the junction point of this region of Salmonella-specific sequence. Figure 1 shows that this region of Salmonella-specific sequence encodes predicted proteins with similarity to sequences encoded within SPI4, an unlinked S. enterica serovar Typhimurium pathogenicity island (56). Protein similarities include HlyD/LktD (SPI4 protein D), involved in the secretion of hemolysin in E. coli and Pasteurella haemolytica; HlyB/LktB family (SPI4 protein R), involved in the transport of hemolysin; and AggA/PrtF (SPI4 protein C), involved in Pseudomonas putida adherence to root tip cells and protease secretion in Erwinia chrysanthemi, respectively. Deletion of these SPI4-like sequences did not have an effect on virulence (Table 2).
ssrA is required for support of the growth of phage P22 and for the induction of P22 lysogens.
Table 3 shows that S. enterica serovar Typhimurium ssrA hosts are severely restricted (nearly 10,000-fold) in their EOP of phage P22. Moreover, the rare phage plaques produced on an ssrA host appeared to be mutant by two criteria: (i) the plaques produced on an ssrA host have a clear plaque morphology relative to the turbid plaques produced by phage P22 on wild-type S. enterica serovar Typhimurium (data not shown); and (ii) clear-plaque phage showed a wild-type EOP when plated on either ssrA or wild-type hosts (data not shown).
TABLE 3.
Effects of S. enterica serovar Typhimurium ssrA on EOP, phage P22 adsorption, and formation of P22 lysogens
| Strain | Relevant genotype | EOPa | % P22 adsorptionb (P22 HT 105/1, int-201) | % P22 lysogenyc (P22-3657) |
|---|---|---|---|---|
| 14028 | Wild type | 1 | 99.5 ± 0.17 | 41.1 ± 4.1 |
| MT2121 | ssrA1::Tn10d-Tc | 1.11 × 10−4 | 99.8 ± 0.06 | 40.9 ± 5.3 |
| MT2194 | ΔssrA2::Kn | 1.05 × 10−4 | 99.7 ± 0.06 | NDe |
| DH5α λpir | E. coli | NAd | 4.6 ± 0.70 | ND |
EOP experiments were performed by plaquing P22 HT 105/1, int-201 on the recipient strain indicated. The EOP the wild-type strain was defined as 1; the EOP of the mutant strains was calculated as follows: (number of PFU/ml obtained on the mutant strain)/(number of PFU/ml obtained on the wild-type strain). Values represent the average of three independent experiments with a standard deviation of <10% of the mean.
P22 adsorption assays were performed by incubating the generalized transducing phage mutant P22 HT105/1, int-201 with bacteria for 15 min at 37°C, at which time the cells were centrifuged and the supernatants were recovered. Values are reported as the percentage of phage recovered, according to the formula [(PFU of phage without cells − PFU of phage with cells)/PFU of phage without cells] × 100. Values represent the average of three independent experiments ± standard deviation.
P22 lysogeny experiments were performed using phage P22-3657, which contains a kanamycin resistance gene in place of the phage mnt gene (15), allowing direct selection of lysogens. The percent lysogeny was calculated as follows: (number of lysogens/viable-cell count determined at the time when cells and phage were mixed) × 100. Values represent the average of three independent experiments ± standard deviation.
NA, not applicable.
ND, not determined.
To characterize the EOP defect, we tested the effect of ssrA on phage P22 adsorption, P22 lysogen formation, and P22 lysogen induction. Table 3 shows that ssrA strains are proficient for phage adsorption, suggesting that the role of ssrA in supporting phage growth occurs at a postadsorption step. The efficiency of P22 lysogeny of ssrA strains was tested using the kanamycin-resistant phage P22-3657 (15), which allows direct selection for phage P22 lysogens. Table 3 shows that P22-3657 was able to lysogenize an ssrA strain (MT2121) with wild-type efficiency, indicating that the lack of ssrA did not impair the formation of stable phage P22 lysogens.
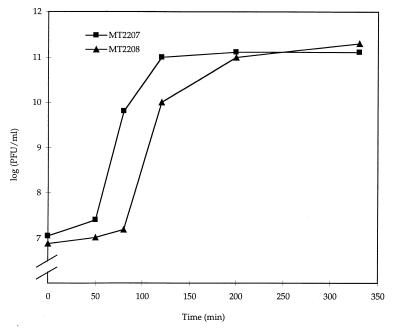
The ability to produce viable phage from P22 lysogens was tested in ssrA strains. Phage P22-3657 was also used in this analysis since it lacks the P22 tail protein (encoded by gene 9 [15]), and thus progeny phage are unable to reinfect other host cells. Single-step phage production from P22-3657 lysogens was assessed in wild-type and ssrA backgrounds. Following lysogen induction with mitomycin C, bacterial cells were lysed with chloroform at several time points to quantitate the phage production. Figure 3 shows that the ssrA1::Tn10dTc lysogen exhibited a 40-min delay in the production of phage following the addition of mitomycin C. However, despite the delay, the titer of viable phage in ssrA lysogens after 3 h of induction was indistinguishable from the titer obtained in wild-type host strains. The phage growth delay was not due to differences in host strain survival in response to phage induction, since the wild-type and ssrA lysogens displayed very similar viable-cell counts over time in response to mitomycin C treatment (data not shown). Moreover, the 40-min delay in recovery of viable phage was not due to a phage P22 lysis defect, since the host cells were lysed before viable phage were quantified. Taken together, these data suggest that ssrA is required for lytic growth of phage P22 and for the induction of a P22 lysogen in S. enterica serovar Typhimurium.
FIG. 3.
Effect of ssrA on the induction of phage P22 lysogens. Phage production in wild-type (MT2207) and ssrA (MT2208) strains [represented as the log (PFU/ml) on the y axis] is shown as a function of time after mitomycin C induction of a P22-3657 lysogen. P22-3657 contains a deletion of P22 tail gene 9, and thus single-burst progeny phage in the supernatant were subjected to titer determination on strain MST3375, which constitutively expresses the P22 tail protein. Values represent the average of three independent experiments, where the standard deviation is <20%.
DISCUSSION
ssrA encodes a multifunctional RNA molecule that plays many diverse roles in the bacterial cell, ranging from tagging abnormal proteins for degradation to supporting phage growth to modulating the activity of DNA binding proteins. Here we show that ssrA (i) is required for full virulence in S. enterica serovar Typhimurium, (ii) affects the expression of several genes induced during infection, (iii) is the integration site for a large region of Salmonella-specific sequence, and (iv) is required to support the growth of bacteriophage P22 and for the induction of P22 lysogens.
The role of ssrA in Salmonella virulence may be attributed to its known role as a tmRNA that tags aberrant polypeptides for degradation. Keiler et al. (26) proposed that tmRNA initially functions as a tRNA and then as an mRNA, resulting in the release of the parental mRNA template from the ribosome and translocation of the nascent peptide to tmRNA. The subsequent addition of an tmRNA-encoded tag marks the nascent polypeptide for degradation (26). In the macrophage phagosome, reactive oxidants are a major source of DNA and/or protein damage and play a major role in bacterial killing (1). Accordingly, the repair of damaged DNA/or protein has been implicated in Salmonella pathogenesis (3). ssrA may counter, in part, the oxidative damage by preventing the buildup of abnormal proteins which may be toxic to the cell.
Another role of ssrA in Salmonella pathogenesis may be its effect on the expression of virulence functions since RNA molecules are known to control protein production at the transcriptional (e.g., transcriptional silencing [50]) and posttranscriptional (e.g., antisense [12, 22]), anti-antisense [36], and mRNA decay [32, 43] mechanisms) levels. Moreover, small RNA molecules which control the expression of virulence genes and functions have been identified for several pathogens including (i) tRNA molecules implicated for translation of virulence genes within pathogenicity islands of uropathogenic E. coli (17, 42) and type 1 fimbriae in Salmonella spp. (9, 52); (ii) RNAIII, a global Staphylococcus aureus regulator involved in the expression of several extracellular toxins and cell surface proteins (39); and a putative Erwinia carotovora RNA molecule, AepH, an activator of extracellular proteases involved in tissue maceration in plant hosts (38).
Similarly, a subset of the ssrA-regulated genes identified in this work are required for or implicated in virulence: vacB is an RNase R which regulates the production of proteins required for invasion, intracellular dissemination, and macrophage apoptosis in Shigella flexneri (6, 28); f304 is contained within a pathogenicity island in uropathogenic E. coli strain CFT073, and its presence has been correlated with more acute clinical symptoms (16, 24); dedE is required for the production of the toxin colicin V (14); and yoaA encodes a homologue of the dinG family of DNA helicases involved in the bacterial SOS response (31). Additionally, ssrA may affect the expression of other virulence genes in Salmonella.
The integration sites for pathogenicity islands are often within genes encoding small RNA molecules, e.g., tRNA genes (17). Similarly, a portion of the bacteriophage P4 attachment site, attL, resides within the 3′ end of ssrA, which serves as the insertion site for acquired sequences in several organisms, including the cryptic phage CP4-57 in E. coli (27), and pathogenicity islands in Vibrio cholerae (25, 30) and Dichelobacter nodosus (2). Here we show that the ssrA attL is also the attachment site for a region of acquired sequence in Salmonella. Additionally, another region of acquired sequence maps 5′ to ssrA, integrating between the corresponding ssrA and smpB genes in E. coli. Sequence analysis of the ssrA-proximal junction region did not reveal bacteriophage attachment sites, IS sequences, or sequence with significant repeats, and thus the mechanism of inheritance of this region remains unclear.
The temperate bacteriophage P22 is subject to complex regulatory mechanisms (51). Here we have shown that Salmonella ssrA mutants were deficient in the ability to support the growth of phage P22, as evidenced by a severe reduction in the EOP (nearly 10,000-fold). ssrA mutants also showed a delay in the production of viable P22 phage when induced from a lysogen, which may arise from inefficient phage P22 excision or from an inability to effectively initiate and/or sustain phage replication. Retallack et al. (40) have shown that ssrA mutants impair the growth of hybrid λ/P22 phage in an E. coli host. However, phage mutants that lack the C1 gene product, which activates transcription of the P22 repressor (c2), produce phage at wild-type levels in E. coli ssrA hosts. These researchers propose a model in which tmRNA directly or indirectly affects the expression of C1-controlled genes/functions required for phage growth. Their model suggests that the lack of tmRNA alters the affinity of a phage-regulatory protein(s) for its cognate DNA binding site (40) and/or leads to a deficiency of aminoacylated tmRNA molecules required for translation of phage mRNA (55). In addition to the defects in phage P22 growth presented here for S. enterica serovar Typhimurium, tmRNA-mediated alterations in DNA-protein interactions and/or translation may contribute to the role of ssrA in bacterial gene expression and in the virulence of Salmonella and other pathogens.
ACKNOWLEDGMENTS
We thank Bob Sinsheimer for critically reading the manuscript.
This work was supported by NIH grant AI36373, Santa Barbara Cottage Hospital Research Foundation, and private donations from Jim and Deanna Dehlsen (M.J.M.).
REFERENCES
- 1.Beaman L, Beaman B L. The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 2.Billington S J, Johnston J L, Rood J I. Virulence regions and virulence factors of the ovine footrot pathogen, Dichelobacter nodosus. FEMS Microbiol Lett. 1996;145:147–156. doi: 10.1111/j.1574-6968.1996.tb08570.x. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier N A, Lipps C J, So M Y H, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 4.Campbell A, Berg D, Lederberg E, Starlinger P, Botstein D, Novick R, Szybalski W. Nomenclature of transposable elements on prokaryotes. In: Bukhari I A, Shapiro J A, Adhya S L, editors. DNA insertion elements, plasmids, and episomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1977. pp. 15–22. [Google Scholar]
- 5.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Z, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 7.Chumley F G, Menzel R, Roth J R. Hfr formation directed by Tn10. Genetics. 1979;91:639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chumley F G, Roth J R. Rearrangement of the bacterial chromosome using Tn10 as a region of homology. Genetics. 1980;94:1–14. [Google Scholar]
- 9.Clouthier S C, Collinson S K, White A P, Banser P A, Kay W W. tRNA(Arg) (fimU) and expression of SEF14 and SEF21 in Salmonella enteritidis. J Bacteriol. 1998;180:840–845. doi: 10.1128/jb.180.4.840-845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 12.Delihaus N. Regulation of gene expression by trans-encoded antisense RNAs. Mol Microbiol. 1995;15:411–414. doi: 10.1111/j.1365-2958.1995.tb02254.x. [DOI] [PubMed] [Google Scholar]
- 13.Demerec M, Adelberg A E, Clark A J, Hartman P E. A proposal for uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fath M J, Mahanty H, Kolter R. Characterization of a purF operon mutation which affects colicin V production. J Bacteriol. 1989;171:3158–3161. doi: 10.1128/jb.171.6.3158-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grana D, Gardella T, Susskind M M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyer D M, Kao J, Mobley H L. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Heithoff D M, Connor C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 21.Hughes K T, Roth J R. Directed formation of deletions and duplications using Mud (Ap, lac) Genetics. 1985;109:263–282. doi: 10.1093/genetics/109.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye M, Delihaus N. Small RNAs in the prokaryotes: a growing list of diverse roles. Cell. 1988;53:5–7. doi: 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- 23.Julio S M, Conner C P, Heithoff D M, Mahan M J. Directed formation of chromosomal deletions in Salmonella typhimurium: targeting of specific genes induced during infection. Mol Gen Genet. 1998;258:178–181. doi: 10.1007/s004380050721. [DOI] [PubMed] [Google Scholar]
- 24.Kao J, Stucker D M, Warren J W, Mobley H L T. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis D K R, Johnson J A, Bailey C C, Boedecker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keiler K C, Waller P R H, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 27.Kirby J E, Trempy J E, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobe T, Sasakawa C, Okada N, Honma Y, Yoshikawa M. vacB, a novel chromosomal gene required for the expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol. 1992;174:6359–6367. doi: 10.1128/jb.174.20.6359-6367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 30.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 31.Lewis L K, Jenkins M E, Mount D W. Isolation of DNA damage-inducible promoters in Escherichia coli: regulation of polB (dinA), dinG, and dinH by Lex-A repressor. J Bacteriol. 1992;174:3377–3385. doi: 10.1128/jb.174.10.3377-3385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M Y, Gui G, Wei B, Preston III J F, Oakford L, Yuksel U, Giedroc D P, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 33.Mahan M J, Roth J R. Ability of bacterial chromosome segment to invert is dictated by included material rather than flanking sequence. Genetics. 1991;129:1021–1032. doi: 10.1093/genetics/129.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial genes that are specifically induced in host tissues. Science. 1993;259:666–668. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 35.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier J R, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majdalani N, Cunning C, Sledjeski D, Elliot T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. p. 477. [Google Scholar]
- 38.Murata H, Chatterjee A, Liu Y, Chatterjee A K. Regulation of the production of extracellular pectinase, cellulase, and protease in the soft rot bacterium Erwinia carotovora subsp. carotovora: evidence that aepH of E. carotovora subsp. carotovora 71 activates gene expression in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Escherichia coli. Appl Environ Microbiol. 1994;60:3150–3159. doi: 10.1128/aem.60.9.3150-3159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick N P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Retallack D M, Johnson L L, Friedman D I. Role for 10Sa RNA in the growth of λ-P22 hybrid phage. J Bacteriol. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Retallack D M, Friedman D I. A role for a small stable RNA in modulating the activity of DNA binding proteins. Cell. 1995;83:227–235. doi: 10.1016/0092-8674(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 42.Ritter A, Blum G, Embody L, Kerenyi M, Bock A, Neuhierl W R, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 43.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson K E, Roth J R. Linkage map of Salmonella typhimurium, edition 7. Microbiol Rev. 1988;52:485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanderson K E, Hessel A, Liu S-L, Rudd K. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Linn E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1903–1999. [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmeiger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 48.Schmid M B, Roth J R. Circularization of transducing fragments: a mechanism for adding segments to the bacterial chromosome. Genetics. 1980;94:15–29. doi: 10.1093/genetics/94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slauch J M, Silhavy T. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Susskind M M, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swenson D L, Kim K J, Six E W, Clegg S. The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol Gen Genet. 1994;244:216–218. doi: 10.1007/BF00283525. [DOI] [PubMed] [Google Scholar]
- 53.Wang R, Kushner S. Construction of versitile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 54.Webb E, Claas K, Downs D M. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Withey J, Friedman D. Analysis of the role of trans-translation in the requirement of tmRNA for λimmP22 growth in Escherichia coli. J Bacteriol. 1999;181:2148–2157. doi: 10.1128/jb.181.7.2148-2157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong K, McClelland M, Stillwell L C, Sisk E C, Thurston S J, Saffer J D. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect Immun. 1998;66:3365–3371. doi: 10.1128/iai.66.7.3365-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto M, Nomura M, Oshawa H, Maruo B. Identification of a temperature-sensitive asparaginyl-transfer ribonucleic acid synthase mutant of Escherichia coli. J Bacteriol. 1977;132:127–131. doi: 10.1128/jb.132.1.127-131.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]