Abstract
Microbial activity is present at every step of the malting process. It is, therefore, critical to manage the grain-associated microbial communities for the production of high-quality malts. This study characterized barley and malt epiphytic microbiota by metabarcoding the internal transcribed spacer (ITS) 2 region and the 16S rRNA gene V1–V4 metabarcodes, respectively. We elucidated the changes in the diversity and the compositional and functional changes of the grain-associated microbiota and inferred the impact of such changes on malting efficiency and premature yeast flocculation (PYF) of the commercial malt end product. Through the malting process, the fungal diversity decreased while bacterial community diversity increased. Lactic acid bacteria (LAB) and some mycotoxin-producing fungi (e.g. Fusarium spp.) were found to be significantly enriched in malts. Most potential fungal pathogens, however, did not change in abundance through the malting process. Fungi (e.g. Aureobasidium, Candida) and bacteria (e.g. LAB, Arthrobacter, Brachybacterium) with the potential to generate organic acids or exhibit high hydrolytic enzymatic activity for degrading the endosperm cell walls and storage proteins were detected in greater abundance in kilned malt, suggesting their contribution to malting efficiency. Bacterial and fungal operational taxonomic units (OTUs) associated with PYF-positive malt were mainly identified as Aureobasidium, Candida, and Leuconostoc, while Pleosporaceae, Steptococcus, and Leucobacter were associated with PYF-negative malt. The ecological networks of the field and steeped barley samples were found to be larger and denser, while that of the malt microbiome was smaller and less connected. A decrease in the proportion of negative interactions through the malting process suggested that malting destabilized the microbial networks. In summary, this study profiled the microbiota of commercial malting barley and malt samples in western Canada; the findings expanded our knowledge in the microbiology of malting while providing potential insights regarding the management of microbial-associated problems, such as PYF, in commercial malting.
Keywords: Barley grain, Commercial malting, Microbiome, Mycotoxin-producing fungi, Premature yeast flocculation (PYF)
Graphical abstract
Highlights
-
•
Malting caused marginal decrease in fungal diversity but marked increase in bacterial diversity.
-
•
Malting enriched Lactic acid bacteria and Fusarium spp., but not most phytopathogens.
-
•
Enrichment of microbes with hydrolytic enzymatic activity may facilitate malting efficiency.
-
•
Malting destabilized cross-kingdom ecological networks.
-
•
Microbial indicators associated with PYF+ve and PYF-ve samples were identified.
1. Introduction
Malting is the first stage of beer brewing, during which barley grains undergo nutritional and functional shifts driven by the changes in temperature, humidity, and oxygen level. Malting promotes the production of hydrolytic enzymes that degrade the carbohydrate components of the barley endosperm cell walls and cell content into soluble compounds (Justé et al., 2011; Laitila et al., 2006a). The breakdown of grain into malt can be facilitated by microbial communities that reside on and in the barley seeds, which may be indigenous to the grain or originate from the field, transportation, or storage (Chen et al., 2016; Flannigan, 2003; Justé et al., 2011, 2014). Understanding the compositional and functional dynamics of the microbial communities through the malting process, therefore, is critical for producing a high-quality malt end product, which is of the utmost importance to beer brewers, and this has yet to be thoroughly investigated.
Commercial malting consists of three steps: steeping, germination, and kilning. Raw barley grains, stored at 10–14% moisture level, are first submerged in water and drained several times to raise the moisture content to 42–47% during the steeping stage (Justé et al., 2011; MacLeod and Evans, 2016). The warm and humid environment and the dissolution of nutrients in the steeping water activate dormant bacterial spores and fungal spores or hyphae, resulting in rapid microbial proliferation (Bokulich and Bamforth, 2013; Laitila et al., 2007; Noots et al., 1998). Steeped barley microbiomes were dominated by lactic acid bacteria (LAB), such as Leuconostoc spp., and basidiomycete fungi, such as Cryptococcus spp. (Justé et al., 2014; Laitila et al., 2006a). Fusarium spp. have also been found to proliferate at this stage and may inhibit yeast growth during the later fermentation stage (Bokulich and Bamforth, 2013; Justé et al., 2011; Lowe and Arendt, 2004). Steeping allows for the production of starch and sucrose degrading enzymes such as α-amylase and β-amylase within the barley grain, as well a β-glucanase which catalyzes the breakdown of cell wall polysaccharides (Aubert et al., 2018). The steeped barley then germinates under humid and aerated conditions. Previous studies suggested that the microbial count was highest at this stage, with LAB, Enterobacteria, and Pseudomonas, making up the majority of the microbiome (Justé et al., 2011; Laitila et al., 2006a; Noots et al., 1998). Certain ascomycetous fungi favored by warm germination temperatures were also found in abundance (Laitila et al., 2011). The high temperature at the kilning stage stops barley germination and reduces the moisture level to 3–4% in malt (MacLeod and Evans, 2016). Kilning restricts microbial activities and also greatly reduces the total microbial count in kilned malt (Follstad and Christensen, 1962; Noots et al., 1998). Heat-tolerant Ascomycetes spp., such as Candida spp. and Pichia spp., were commonly recovered from kilned malt (Laitila et al., 2011).
Changes in the grain-associated microbial community compositional structure during the malting process can be beneficial or detrimental to the malt end-product (Laitila, 2008). For example, the enrichment of LAB can restrict the growth of other bacteria or pathogenic fungal species, such as Fusarium spp., throughout malting and wort production (Linko et al., 1998). For the same reason, starter cultures of functional LAB (e.g. Lactobacillus spp.) or wort solution fermented by LAB strains have been used to reduce fungal load and improve malt characteristics (Laitila et al., 2006b; Oliveira et al., 2015; Peyer et al., 2017). Justé et al. (2011) showed that Candida and Arthrobacter are also beneficial to the malting process by producing hydrolytic enzymes for breaking down grain macromolecules into sugar for fermentation. Undesired fungal and bacterial growth can prolong the beer-making process and negatively affect the quality of the malt end product in terms of turbidity, off-flavors, and textures (Bokulich and Bamforth, 2013; Laitila et al., 2011). This, in turn, has detrimental consequences due to poor consumer perceptions, leading to financial loss. Of particular concern to the brewing industry are fungi, such as certain Fusarium species, that produce mycotoxins that can survive the brewing process (Gonzalez Pereyra, 2011; Wolf-Hall, 2007). In addition to inhibiting yeast growth during the fermentation process (Bokulich and Bamforth, 2013), mycotoxins can also be immunosuppressive agents and have been associated with health disorders (e.g., cancer, haematological disorders, and endocrine dysfunction, etc.) (Rodríguez-Carrasco et al., 2015). Certain species of Fusarium are also associated with hydrophobin production and proteolytic activity; the latter can increase free amino acids, reduce solubility, and lengthen the beer-making process (Geiβinger et al., 2019). An additional problem encountered is premature yeast flocculation (PYF), which occurs when cells of the brewer's yeast prematurely agglutinate and settle, resulting in incomplete fermentation and off-flavors in beer (Justé et al., 2011; Lake and Speers, 2008; van Nierop et al., 2006). Previous research suggested that PYF activities can be induced by complex polysaccharides such as arabinose and xylose degraded from arabinoxylan in barley husk. The quantity and quality of these PYF factors were found to have a strong association with high fungal load and their extracellular enzymes and possibly, high steeping pressure (Lake and Speers, 2008; Panteloglou et al., 2012; van Nierop et al., 2006). Exopolysaccharides, produced by certain Pseudomonas spp. and lactic acid bacteria, can negatively affect wort separation (Justé et al., 2011) and result in beer with an oily texture (Bokulich and Bamforth, 2013). Thus, some constituents of the grain-associated microbial community have a major impact on malt consistency and quality.
Earlier studies and reviews of the microbial members of the malting environment relied on culture-dependent and/or DNA fingerprinting methods (Bianco et al., 2018; Kaur et al., 2015; Laitila et al., 2007; Li et al., 2018). The former detects only viable and culturable community members, while the latter cannot reveal raresphere or provide taxonomic information (Kaur et al., 2015; Laitila et al., 2007). Used along or coupled with culture-dependent methods, high throughput sequencing (HTS) can comprehensively recover microbial communities of barley grains and malts (Bokulich et al., 2012; Chen et al., 2016; Justé et al., 2014; Laitila et al., 2018; Østlie et al., 2021). Fungal-bacterial interactions have been studied in the production of other fermented foods such as cheese and wine using next-generation sequencing (Wolfe and Dutton, 2015). However, the interactions between these communities at various stages of barley malting require further study.
This study aimed to comprehensively characterize the dynamics of the barley grain-associated epiphytic fungal and bacterial communities through the commercial malting process; and to obtain a better understanding of their ecological functions and associations relevant to malt quality and safety. We hypothesize that the changes in the grain-associated microbiota are suggestive to malting efficiency and end-malt quality. The barley and malt epiphytic microbial communities were profiled by metabarcoding the fungal internal transcribed spacer (ITS) 2 region and the bacterial 16S rRNA gene V1–V4 region using the 454 pyrosequencing technology. We used the 454 pyrosequencing instead of the Illumina sequencing platforms because the former was widely used when this study was carried out (2014–2016). Earlier studies also showed that several tens to several hundred putative fungal or bacterial species were recovered from individual grains (Chen et al., 2016; Links et al., 2014), suggesting the sequencing depth provided by pyrosequencing was sufficient for the microbial diversity survey of this study. By characterizing the taxonomic and functional groups associated with different malting stages, we identified microbial bioindicators potentially linked to malting efficiency and PYF.
2. Materials and methods
2.1. Sampling of malting barley and kilned malts
In 2014, barley (n = 12) of undisclosed cultivar and malt samples generated from said barley (n = 24) were sourced from a commercial malthouse located in western Canada. Barley grains were cleaned and steeped, in alternative wet and dry periods with aeration, under a controlled temperature range of 13–20 °C for a total of 36–48 h to bring the grain moisture content to 40–45%. At this stage, the barley grains were first soaked in water in conical bottomed steep to bring the moisture to 35–38%, then were sprayed with water to bring the moisture up to 40–45% in a flat bottomed steep. The steeped grain was then transferred to the germination vessels with temperature control using a constant flow of fully humidified air at 15–20 °C for 3–5 days. The germinated grain was finally kilned to reduce the grain moisture to ∼4% following a programmed heating regime with air-on temperature elevating from 50 °C to 85 °C for 20–24 h. The malting barley was germinated and dried in separate kilns (kiln1 and kiln2) under the same condition. The malt end products were collected after kilning.
Barley harvested near the malting house, but that did not undergo the malting process, was also included in this study (Table 1). The cultivar types for these samples included: AC Metcalfe (n = 3), CDC Copeland (n = 2), CDC Meredith (n = 2), and CDC Kindersley (n = 2). These samples were designated as “control” to differentiate them from barley that underwent malting (designated as “malting barley” in this study). This resulted in 9 field barley, 12 steeped barley, and 24 malt samples, for a total of 45 samples in this study.
Table 1.
Metadata and alpha-diversity indices associated with the samples used in the current study.
| Fungal ITS2 |
Bacterial 16S |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Stage | Variety | Sample | Process | Ergosterol (μg/g) | PYF | Shannon-True Diversity |
Chao1 index | Coverage | Shannon-True Diversity |
Chao1 index | Coverage |
| B245-Barley-st | Steeped | unknown | B245 | Steeping | 4.84 | 12 | 263 | 0.98 | 17 | 439 | 0.98 | |
| B245-Malt-K1 | Malt | unknown | B245 | Kiln 1 | 8.65 | 0.95 | 10 | 171 | 0.99 | 35 | 466 | 0.98 |
| B245-Malt-K2 | Malt | unknown | B245 | Kiln 2 | 7.23 | 0.97 | 13 | 167 | 0.99 | 33 | 455 | 0.98 |
| B251-Barley-st | Steeped | unknown | B251 | Steeping | 4.13 | 10 | 284 | 0.98 | 20 | 362 | 0.98 | |
| B251-Malt-K1 | Malt | unknown | B251 | Kiln 1 | 6.06 | 0.98 | 11 | 145 | 0.99 | 55 | 548 | 0.98 |
| B251-Malt-K2 | Malt | unknown | B251 | Kiln 2 | 6.45 | 0.99 | 8 | 171 | 0.99 | 70 | 585 | 0.98 |
| B254-Barley-st | Steeped | unknown | B254 | Steeping | 6.06 | 10 | 235 | 0.98 | 10 | 332 | 0.99 | |
| B254-Malt-K1 | Malt | unknown | B254 | Kiln 1 | 8.52 | 0.96 | 6 | 104 | 0.99 | 57 | 539 | 0.98 |
| B254-Malt-K2 | Malt | unknown | B254 | Kiln 2 | 8.97 | 0.96 | 7 | 112 | 0.99 | 48 | 616 | 0.98 |
| B262-Barley-st | Steeped | unknown | B262 | Steeping | 4.13 | 9 | 201 | 0.99 | 37 | 387 | 0.98 | |
| B262-Malt-K1 | Malt | unknown | B262 | Kiln 1 | 7.1 | 1.09 | 21 | 252 | 0.98 | 20 | 388 | 0.98 |
| B262-Malt-K2 | Malt | unknown | B262 | Kiln 2 | 7.61 | 1.13 | 14 | 223 | 0.99 | 48 | 649 | 0.97 |
| B265-Barley-st | Steeped | unknown | B265 | Steeping | 4.84 | 16 | 261 | 0.98 | 16 | 297 | 0.99 | |
| B265-Malt-K1 | Malt | unknown | B265 | Kiln 1 | 6.97 | 1.1 | 3 | 105 | 0.99 | 62 | 548 | 0.98 |
| B265-Malt-K2 | Malt | unknown | B265 | Kiln 2 | 6.26 | 1.07 | 5 | 143 | 0.99 | 29 | 450 | 0.98 |
| B267-Barley-st | Steeped | unknown | B267 | Steeping | 5.48 | 9 | 182 | 0.99 | 18 | 384 | 0.98 | |
| B267-Malt-K1 | Malt | unknown | B267 | Kiln 1 | 6.26 | 1.12 | 4 | 108 | 0.99 | 71 | 611 | 0.98 |
| B267-Malt-K2 | Malt | unknown | B267 | Kiln 2 | 6.18 | 1.15 | 2 | 175 | 0.99 | 75 | 516 | 0.98 |
| B270-Barley-st | Steeped | unknown | B270 | Steeping | 5.36 | 13 | 206 | 0.98 | 20 | 302 | 0.99 | |
| B270-Malt-K1 | Malt | unknown | B270 | Kiln 1 | 7.09 | 0.71 | 7 | 137 | 0.99 | 62 | 428 | 0.98 |
| B270-Malt-K2 | Malt | unknown | B270 | Kiln 2 | 8.55 | 0.7 | 7 | 68 | 1.00 | 43 | 557 | 0.98 |
| B283-Barley-st | Steeped | unknown | B283 | Steeping | 7.09 | 12 | 263 | 0.98 | 65 | 577 | 0.98 | |
| B283-Malt-K1 | Malt | unknown | B283 | Kiln 1 | 9.27 | 0.96 | 22 | 340 | 0.98 | 80 | 538 | 0.98 |
| B283-Malt-K2 | Malt | unknown | B283 | Kiln 2 | 8.18 | 0.9 | 19 | 261 | 0.98 | 42 | 614 | 0.97 |
| B285-Barley-st | Steeped | unknown | B285 | Steeping | 3.82 | 11 | 263 | 0.98 | 22 | 386 | 0.98 | |
| B285-Malt-K1 | Malt | unknown | B285 | Kiln 1 | 5.18 | 0.86 | 6 | 140 | 0.99 | 72 | 541 | 0.98 |
| B285-Malt-K2 | Malt | unknown | B285 | Kiln 2 | 6.09 | 0.87 | 7 | 126 | 0.99 | 34 | 476 | 0.98 |
| B306-Barley-st | Steeped | unknown | B306 | Steeping | 3.64 | 8 | 209 | 0.98 | 20 | 349 | 0.98 | |
| B306-Malt-K1 | Malt | unknown | B306 | Kiln 1 | 9.09 | 1.02 | 9 | 148 | 0.99 | 45 | 458 | 0.98 |
| B306-Malt-K2 | Malt | unknown | B306 | Kiln 2 | 6.27 | 1.02 | 7 | 173 | 0.99 | 49 | 371 | 0.99 |
| B317-Barley-st | Steeped | unknown | B317 | Steeping | 5 | 10 | 259 | 0.98 | 40 | 399 | 0.98 | |
| B317-Malt-K1 | Malt | unknown | B317 | Kiln 1 | 11.41 | 0.99 | 19 | 255 | 0.98 | 42 | 459 | 0.98 |
| B317-Malt-K2 | Malt | unknown | B317 | Kiln 2 | 9.64 | 1.04 | 12 | 229 | 0.98 | 48 | 475 | 0.98 |
| NC-13-04 | Barley | Kindersley | NC-13-04 | Field barley | 9.51 | 13 | 191 | 0.99 | 29 | 463 | 0.98 | |
| NC-13-05 | Barley | AC_Metcalfe | NC-13-05 | Field barley | 6.16 | 8 | 158 | 0.99 | 24 | 394 | 0.98 | |
| NC-13-16 | Barley | Copeland | NC-13-16 | Field barley | 7.67 | 9 | 222 | 0.99 | 18 | 300 | 0.99 | |
| NC-13-17 | Barley | Meredith | NC-13-17 | Field barley | 5.66 | 8 | 268 | 0.99 | 23 | 408 | 0.98 | |
| NC-13-18 | Barley | AC_Metcalfe | NC-13-18 | Field barley | 4.86 | 9 | 133 | 0.99 | 22 | 374 | 0.98 | |
| NC-13-21 | Barley | Kindersley | NC-13-21 | Field barley | 4.4 | 9 | 198 | 0.99 | 19 | 333 | 0.99 | |
| NC-13-36 | Barley | AC_Metcalfe | NC-13-36 | Field barley | 6 | 10 | 134 | 0.99 | 22 | 451 | 0.98 | |
| NC-13-37 | Barley | Copeland | NC-13-37 | Field barley | 8 | 8 | 168 | 0.99 | 19 | 414 | 0.98 | |
| NC-13-38 | Barley | Meredith | NC-13-38 | Field barley | 8.51 | 5 | 188 | 0.99 | 19 | 436 | 0.98 | |
2.2. Estimation of total fungal biomass and premature yeast flocculation (PYF)
The total fungal biomass on barley and malt samples was estimated by determining the ergosterol content using the GC–MS method as described previously (Chen et al., 2016; Dong et al., 2006).
To test the malt samples for premature yeast flocculation (PYF), we used the miniature fermentation assay (ASBC Yeast-14) recommended by the ASBC (ASBC, 2022; Kaur et al., 2014). It is recognized that there is some debate regarding PYF assays; however, for the current study the ASBC Yeast-14 recommended assay was used. Fifty grams (50.0 ± 0.05 g) of each sample were ground in duplicate using a grist mill set to the ASBC ‘fine’ standard (ASBC Malt-4). Samples were mashed according to the ASBC Congress standard mashing regime (ASBC Malt-4) using a mash bath. A total of 100 g malt yielded sufficient wort to complete a ‘miniature fermentation’ assay (30 test-tube fermenters with 15 mL of wort each). After completion of the mash cycle, samples were filtered through coarse fluted filter paper to remove solids and boiled for 10 min over high heat. The subsequent wort samples were cooled and stored overnight at 5 °C. Following a cold break, the wort samples were centrifuged at 5000 rpm for 15 min. Prior to fermentation, the wort samples were placed in sterile beakers and each wort was adjusted to a final density of 12.6 oP with sterile water and D-glucose. The adjusted wort samples were oxygenated by bubbling medical-grade compressed oxygen. Stock yeast from Wyeast (Odell, OR, USA) was washed three times with sterile water, mixed, and centrifuged. The yeast was suspended in 100 mL of sterile water and enumerated according to the methods of ASBC Yeast-4: a small aliquot of the water-washed yeast slurry was diluted with 0.1 N sodium acetate buffer and the number of cells assessed using a hemocytometer. The sterile wort samples were pitched with the water-washed yeast at a rate of 1.5 × 107 cells/mL. The pitched wort samples were aseptically distributed to the 30 sterile fermentation tubes (15 mL each) containing sterile boiling stones. Each tube was stoppered with a sponge bung and the tubes were set to ferment (in a water bath) at 21 °C until sampling. Samples were taken at 0, 1, 3, 19, 23, 26 43, 47, 51, 67, 71, 74 h, or as close to these times as was practical. At each reading, the samples were tested for density and absorbance. Absorbance was measured at 600 nm from the top 3.5 ml of each of two tubes transferred to clear-sided cuvettes. Positive (PYF+ve) and negative (PYF-ve) PYF control malts were tested with each set of samples.
To quantify the degree of PYF in malt samples, we developed a putative quantification method based on the observation that the absorbance readings for PYF-ve controls were relatively high as the yeast cells remained suspended in the fermenting wort samples almost for the entire duration of the tests. By contrast, the absorbance readings for PYF+ve controls remained high for about 30 h of the fermentation experiments and dropped drastically between 40 and 55 h of fermentation and thereafter. Therefore, the PYF value was expressed for each sample as a percentage of the average absorbance between 40 and 55 h of fermentation in relation to the values (100%) obtained for the PYF-ve controls. We acknowledge that this in-house developed PYF quantification assay requires confirmation with greater numbers of PYF-ve and PYF+ve samples from a broad range of origins and to be further validated by commercial malthouse.
2.3. Extraction of genomic DNA from seed-associated epiphytic microbiomes
All barley and malt samples were processed directly after sampling. The malting barley and kilned malt samples were washed as described by Chen et al. (2016). Briefly, 25 g of the whole grain of each sample was incubated in buffered peptone water at room temperature, shaking at 200 rpm for 1 h. The samples were then pelleted by centrifugation and the supernatant was removed. Pellets were lysed with Genomic DNA Buffer 1 (Qiagen, Venlo, Limburg, Netherlands), lysozyme, proteinase K, RNase A, and lyticase (all from Sigma, St. Louis, Missouri, USA), and subsequently in Genomic DNA Buffer 2 (Qiagen, Venlo, Limburg, Netherlands). DNA was extracted with chloroform/isoamyalcohol and then precipitated with isopropanol. The purified DNA pellet was resuspended in Tris buffer pH 8.0 and stored at −30 °C. DNA was quantified using a Quant-IT Picogreen DNA kit (Life Technologies Inc., Grand Island, New York, USA) and a fluorometer.
2.4. Amplicon library preparation and high-throughput sequencing
At the time of the study, the 454 pyrosequencing platform was widely used and was chosen for sequencing the ITS and 16S rRNA gene metabarcodes, using the following methods. The DNA libraries were prepared and sequenced as described previously in Chen et al. (2016). The fungal ITS region was amplified with ITS5 forward primer (5′- GGA AGT AAA AGT CGT AAC AAG G -3′) and ITS4 reverse primer (5′- TCC TCC GCT TAT TGA TAT GC-3′) (White et al., 1990). The forward UNBacF primer (5′- GAT CCT GGC TCA GGA TGA AC -3′) and reverse UNBacR primer (5′- GGA CTA CCA GGG TAT CTA ATC -3′) were used to amplify the bacterial 16S V1–V4 region as described previously (Chen et al., 2018b). All primers were fused with unique 24-nucleotides MID (Roche Multiplex Identifiers) barcodes. ITS regions were amplified following the PCR conditions listed in Chen et al. (2016). Amplicons were purified with the PureLink PCR Micro Purification kit (Invitrogen now Life Technologies Inc., Grand Island, New York, USA), quantified with the Nanodrop ND-1000 (Thermo Fisher Scientific Inc. Waltham, Massachusetts, USA), and normalized to 50 ng/μL. Eight replicates of 24 equimolar MID-tagged amplicon libraries were pooled and tagged with 454 pyrosequencing adaptor L. Unidirectional 454 pyrosequencing was completed at the Aquatic and Crop Resource Development Research Centre of the National Research Council of Canada in Saskatoon, SK. All samples were sequenced in one run.
2.5. Pyrosequencing data processing
The 454-pyrosequencing platform generated raw sequencing reads at different lengths, ranging from 350 to 750 bp. The ITS metabarcodes were processed using the bioinformatics pipelines described in Chen et al. (2016) with a few modifications. Demultiplexed raw sequences were trimmed using trim.seqs in MOTHUR (version 1.35.1) with a window size of 10, a qwindow average of 25, and a minimum length of 100. Chimeric sequences were removed using chimera.uchime in mothur against the UNITE fungal ITS reference database (version 8.2, released on 2020-02-20). The ITS1 and ITS2 regions were extracted using ITSx 1.1.2 (Bengtsson-Palme et al., 2013). Our previous study showed that the fungal communities recovered by the same primer pair and 454-pyrosequencing had similar diversity based on ITS1 or ITS2 regions (Chen et al., 2016), but the ITS1 region of some fungal taxa (e.g., rust fungi) contains indels that impede direct sequencing and analyses (Chen et al. 2018c, 2021). Therefore, we decided to use the ITS2 region for this study. ITS2 sequences shorter than 100 bp were removed. The quality ITS2 sequences were clustered in to Operational Taxonomic Units (OTUs) at 97% sequence identity using CD-HIT-EST (version 4.8.1) (Fu et al., 2012). The representative sequences of OTUs were assigned to taxonomic lineages using q2-feature-classifier (Bokulich et al., 2018) implemented in QIIME2 (Bolyen et al., 2019) against the UNITE ITS database (version 8.2).
The raw 16S rRNA gene V1–V4 metabarcodes were quality-trimmed using the parameters listed above. Chimeric sequences were removed using chimera.uchime in MOTHUR against the SILVA Gold database (https://drive5.com/uchime/gold.fa). The quality sequences were clustered in to OTUs at 97% similarity using CD-HIT-EST. The representative sequences were classified against the Greengenes database (version May 2019, pre-clustered at 97% sequence identity) using q2-feature-classifier implemented in QIIME2.
OTUs containing a single sequence (singletons) were removed. Functional guilds in the fungal and the bacterial communities were annotated using FUNGuilds (Nguyen et al., 2016) and FAPROTAX (Louca et al., 2016), respectively. Because the FAPROTAX database was constructed mainly for marine and freshwater microbiomes, the Phylogenetic Investigations of Communities by Reconstruction of Unobserved States 2 software (PICRUSt2, version 2.3.0-b) was also used to predict the metagenome functional content (gene families and MetaCyc pathways) from the 16S rRNA gene OTUs (Douglas et al., 2020).
2.6. Statistical analyses
Data analysis and visualization were carried out in R environment (R Core Team, 2020). After removing the samples (1 steeped and 2 kilned) with low sequencing reads, the final bacterial and fungal OTU tables contained 42 samples of 1) the raw field barley grains that were not subjected to the malting process (field barley, n = 9); 2) the malting barley grain samples that went through the steeping process (steeped barley, n = 11); and 3) kilned malts, which went through germination and were subsequently kilned in two separate chambers under the same kilning condition (kiln1, n = 11; kiln2, n = 11). The OTU tables were normalized to the same sample size (fungal ITS2 OTU n = 3829 reads; bacterial 16S rRNA gene OTU n = 6108 reads) prior to statistical analysis. Corrections were made to the ITS2 OTU tables for generic synonyms using the list provided in Supplementary Table S1.
To calculate the alpha- and beta-diversity indices, functions in vegan (Oksanen et al., 2018), entropart (Marcon and Hérault, 2015), or RAM (Chen et al., 2018a) were used. Shannon-Weiner (SW) and Gini-Simpson (GS) indices were converted to true diversity indices, which allowed for direct comparison of real diversity (Jost, 2006). To assess the differences in abundance of OTUs, taxonomic groups, or functional guilds at different malting stages, Kruskal-Wallis rank sum tests were performed, followed by post-hoc pairwise tests in R. The influence of malting stages on the divergence of the fungal or bacterial community compositional structure was visualized by the non-metric multidimensional scaling (NMDS) based on Bray-Curtis distance matrices. The differentially abundant MetaCyc pathways predicted by PICRUSt2 were determined using DESeq2 (Love et al., 2014), with a stringent threshold set at |log2 fold change| ≥ 2 and the FDR adjusted p-value ≤ 0.05.
To reconstruct the cross-kingdom ecological networks, OTUs with >0.1% abundance in any sample (1465 bacterial and fungal OTUs) were retained. The abundance table was Hellinger transformed and then uploaded to the Molecular Ecology Network Analysis Pipeline (MENAP, http://ieg4.rccc.ou.edu/mena/) (Deng et al., 2012). The pipeline computed Spearman rank correlation coefficients and reconstructed the networks using a fast-greedy modularity optimization algorithm as described previously (Chen et al., 2018b). The networks were visualized in Gephi 0.9.2 (Bastian et al., 2009). Nodes with <2° were removed.
2.7. Sequencing data accessibility
All raw sequencing data are accessible in Sequence Read Archive (SRA) under BioProject PRJNA661245 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA661245).
3. Results
After normalizing the OTU tables to equal sample size (n = 3829 reads for fungal ITS2 OTU; n = 6108 reads for bacterial 16S rRNA gene OUT), we recovered 54–192 fungal OTUs (98.68 ± 0.41% coverage), and 177–360 bacterial OTUs (98.10 ± 0.35% coverage) for each sample, respectively. The high coverage (>98%) suggested that the sequencing depth provided by the pyrosequencing technology sufficiently recovered the species diversity of the grain-associated microbiome in this study. The number of reads assigned to each taxonomic rank is shown in Supplementary Table S2. Of the 758 fungal OTUs, 569 were assigned to Ascomycota (representing 80.3% of total reads), while the rest were Basidiomycota (19.7%). The 1468 bacterial OTUs were assigned to Actinobacteria (702 OTUs, representing 66.0% of total reads), Firmicutes (397, 24.9%), Proteobacteria (165, 6.4%), and Bacteroidetes (196, 2.7%). Gemmatimonadetes, TM7, and Acidobacteria were also detected but at extremely low abundance (≤5 OTUs and <0.003% of total reads). The relative abundance of the most abundant taxa at each taxonomic rank is shown in Supplementary Figs. S1 and S2.
3.1. Diversity and compositional shift in fungal and bacterial communities through the malting process
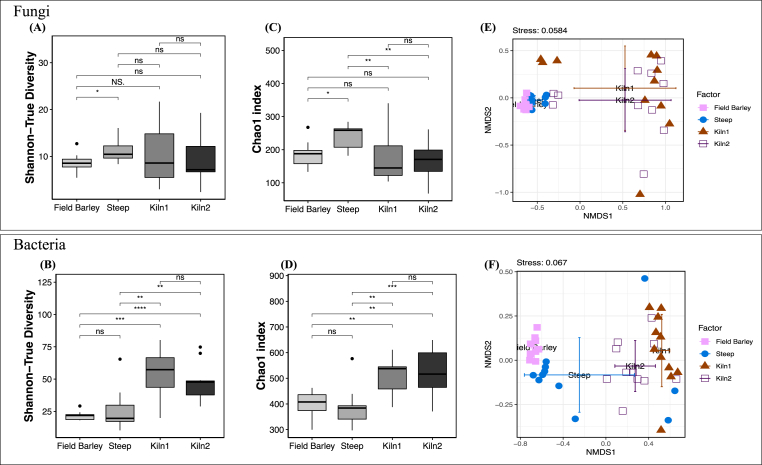
The alpha-diversity of the fungal community increased after steeping (p < 0.05) and then decreased after kilning based on the Chao1 index (p ≤ 0.05), but not the Shannon-Wiener-based true diversity index (p > 0.05) (Fig. 1A,C). An opposite trend was observed for the bacterial communities, in which the alpha-diversity increased significantly after malting (Chao1, p < 0.01; Shannon's TD, p < 0.001) (Fig. 1B,D). No significant difference in the diversity of fungal and bacterial communities was detected between the malts collected from different kilning chambers. The compositional structure of both fungal (ANOSIM R = 0.51, p ≤ 0.001) and bacterial communities (R = 0.69, p ≤ 0.01) shifted significantly through the malting process, as shown in the NMDS plots (Fig. 1E and F).
Fig. 1.
Diversity of the fungal and bacterial communities associated with the field barley grains, steeped barley grains, and kilned malts at the OTU level. Alpha-diversity indices of (A & C) fungi and (B & D) bacteria at the three malting stages (with malt samples separated depending on the kilning batch). Beta-diversity analysis using non-metric multidimensional scaling (NMDS) ordination plots based on the Bray-Curtis distances of the (E) fungi and (F) bacteria communities, calculated for hellinger transformed OTU abundance matrices. Significant differences in relative abundance between barley and malt are denoted by: ***p ≤ 0.001; ** 0.001 < p ≤ 0.01; * 0.01 < p ≤ 0.05.
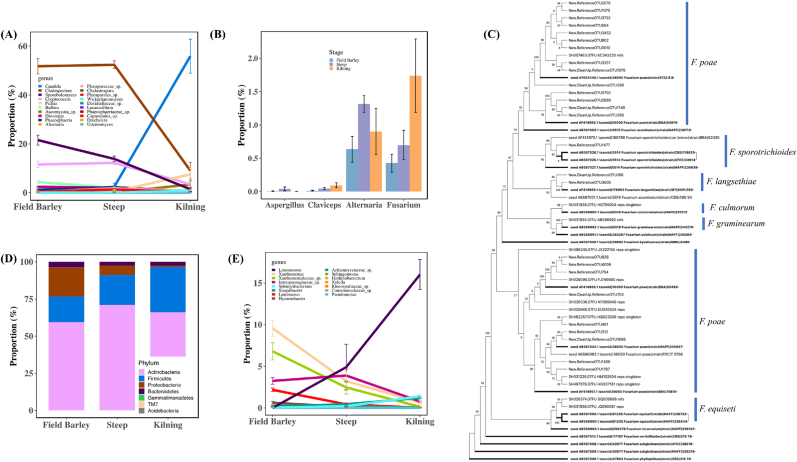
In the epiphytic mycobiota, the Ascomycota abundance increased from 45.1 ± 21.8% (MEAN ± SD) in field barley grain to 64.3 ± 7.5% in steeped grain samples (Tukey's adjusted p < 0.05), and further increased to 90.5 ± 11.0% in malt end products (p < 0.0001, Supplementary Fig. S1). By contrast, the Basidiomycota abundance decreased throughout the process, from 54.9 ± 21.8% in field grain to 30.7 ± 7.5% in steeped grain (p < 0.05), and then to 6.1 ± 11.0% in malts (p < 0.0001). Of the 48 fungal genera recovered, Cladosporium (51.7 ± 9.3% in field barley, 51.8 ± 5.2% in steeped barley), Sporobolomyces (21.4 ± 6.1%, 14.0 ± 3.8%), Cryptococcus (11.9 ± 4.1%, 12.2 ± 2.4%), and Bullera (4.1 ± 2%, 2.1 ± 1.1%) were most abundant in field barley and steeped barley samples. Candida (55.8 ± 32.5%), Cladosporium (9 ± 15.9%), and Pichia (7.6 ± 11.8%), were most abundant in malt samples (Fig. 2A, Supplementary Fig. S1). Aureobasidium, Candida, Fusarium, Pichia, and Wickerhamomyces were enriched, while Bullera, Cladosporium, Cryptococcus, Dioszegia, Phaeosphaeria, and Sporobolomyces were depleted in malt samples (absolute value of log2 Fold Change <2, FDR adjusted p < 0.0001) (Fig. 2A).
Fig. 2.
Changes in the abundance of (A) fungal genera, (B) fungal genera containing mycotoxin-producing species groups, (C) bacterial phyla, and (D) bacterial genera, through the malting process. (E) The phylogenetic tree of the 38 Fusarium ITS OTUs recovered through the malting process.
FUNGuild assigned 206 ITS OTUs as potential plant pathogens, of which none were identified as “highly probable” pathogens. The 106 OTUs annotated as probable plant pathogens (2.1% in abundance) belong to Ascochyta, Claviceps, Rhodotorula, Ustilago, Leptosphaeriaceae, Mycosphaerellaceae (including Ramularia), Phaeosphaeriaceae, and Pleosporaceae (including Bipolaris, Chalastospora, Drechslera, and Pleospora). The 100 OTUs that were considered possible plant pathogens (35.1% in abundance) belong to Acremonium, Alternaria, Aureobasidium, Camarosporium, Cladosporium, Fusarium, and Microdochium. Of these fungal genera, only Aureobasidium (p ≤ 0.05) was increased while Cladosporium (p ≤ 0.0001) was decreased significantly in abundance through the malting process. Mycotoxin-producing fungi were recovered in low abundance in both barley grains and malts. Aspergillus (3 OTUs, 0.01%) and Claviceps (6, 0.06%) did not change significantly in abundance through the malting process (FDR adjusted p > 0.05). Alternaria (16 OTUs, 1% in abundance) was highest in steeped samples and was significantly reduced in malt (p < 0.05), whereas Fusarium (34, 1.2%) was significantly increased in malt after the kilning process (p < 0.0001) (Fig. 2B). Based on BLAST results and phylogenetic analysis, Fusarium OTUs were classified to F. culmorum, F. equiseti, F. graminearum, F. langsethiae, F. poae, and F. sporotrichioides (Fig. 2C).
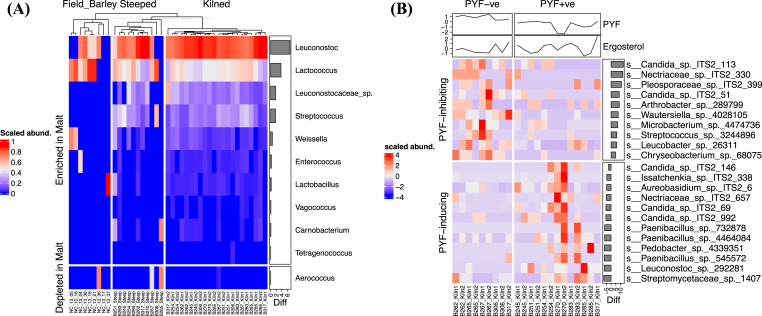
Of the eight bacterial phyla recovered from the barley grain and the malt surface, Proteobacteria (along with 9 genera in this phylum) were depleted (p < 0.0001), while members of the Firmicutes, particularly the Lactobacillales order, which includes lactic acid bacteria (LAB), were enriched (p < 0.01) through the malting process (Fig. 2D). We recovered 157 known bacteria genera from both barley and malt samples, of which, Arthrobacter (17.4 ± 2.2%), Leuconostoc (9.2 ± 1.4%), and Paenibacillus (9.0 ± 1.1%) were most abundant. Xanthomonas, Pedobacter, and Luteimonas decreased in abundance through the malting process (log2FC < −2, FDR adjusted p < 0.01). Arthrobacter, Corynebacterium, Leuconostoc, Lactococcus, Microbacterium, Sanguibacter, Sphingobacterium, and Streptococcus were enriched in malt (p ≤ 0.05) (Fig. 2E). The relative abundance of all LAB OTUs, except for those of Aerococcus spp., increased through the malting process (≤2% in barley grains but increased to 23% in the kilned malts) (Fig. 3A). Core LAB recovered include Lactobacillus, Leuconostoc, Lactococcus, and Streptococcus, while peripheral LAB recovered include Aerococcus, Carnobacterium, Enterococcus, Tetragenococcus, Vagococcus, and Weissella.
Fig. 3.
(A) Enrichment of lactic acid bacteria through the malting process. (B) Fungal and bacterial OTUs associated with PYF-positive (PYF+ve) and PYF-negative (PYF-ve) samples. The PYF value in Fig. 3B was scaled to zero mean and unit variance.
3.2. Bacteria and fungi correlated to PYF
Only the kilned malt samples were tested for PYF, which ranged from 0.70 to 1.15, with a median of 0.99 (Table 1). A PYF value > 1 was considered PYF-ve (PYF inhibiting), while a PYF value < 1 was considered PYF + ve (PYF inducing). Among the OTUs with more than 0.01% abundance (seven sequence reads), four fungal OTUs (belonging to Candida, Nectriaceae, Pleosporaceae) and six bacterial OTUs (belonging to Arthrobacter, Chryseobacterium, Leucobacter, Microbacterium, Streptococcus, and Wautersiella) were significantly correlated to PYF-negative (PYF-ve) and were considered PYF-inhibiting (qvalue ≤0.05). The OTUs associated with PYF-positive (PYF+ve), i.e. hypothesized to induce PYF, included six fungal and six bacterial OTUs (qvalue ≤0.05) (Fig. 3B). Of the potential PYF-inducing OTUs, the most abundant three were identified as Aureobasidium spp. (3.5 ± 3.6%), Candida spp. (1.0 ± 2.1%), and nectriaceous species (0.4 ± 0.9%) and were significantly enriched through the malting process. These potential indicators showed high correlation with the samples had the highest and lowest PYF values (Fig. 3B). However, the majority of the OTUs that potentially induced PYF were found in low abundance in the epiphytic microbiome of malts, which may suggest the proliferation or growth of these microbes may not be the direct cause of PYF. The total fungal biomass, represented by ergosterol content of fungal cell wall, ranged from 3.64 to 11.41 μg/g and was not correlated to PYF occurrence (p ≥ 0.05) (Fig. 3B, Table 1).
3.3. Prediction of bacterial community functionality
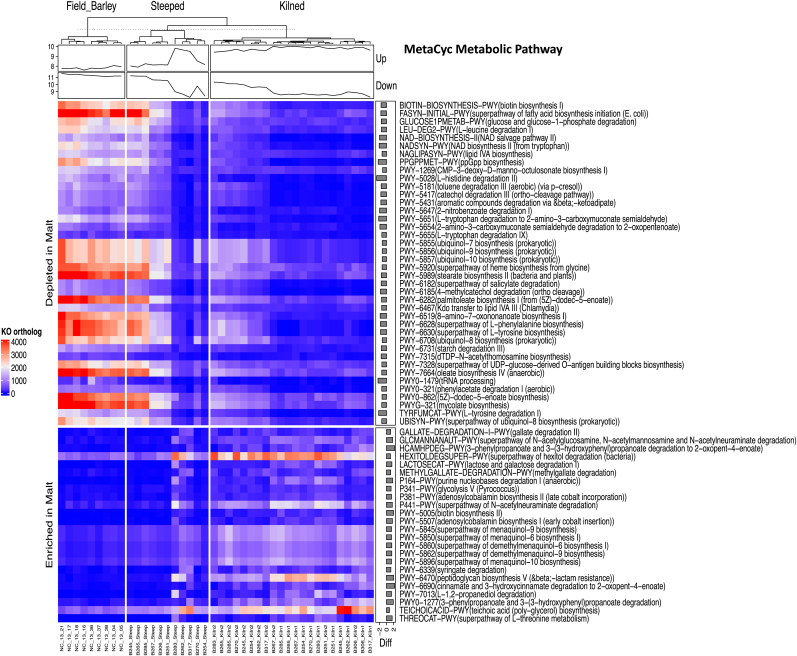
Although the seed-associated bacterial communities may have relatively smaller contribution to the degradation of starch and endosperm cell walls compared to the vast bulk of hydrolytic enzymes released by the host during malting, it is still interesting to explore the potential functions of the bacterial taxa in facilitating the malting process. The weighted nearest sequenced taxon index (weighted NSTI) score suggested that the average bacterial OTUs in our samples can be predicted from close relatives (95 ± 7% identity) by PICRUSt2. It was predicted that the steeping process potentially induced pathways (Fig. 4) affiliated with the degradation of aromatic compounds, amines and polyamines, amino acids, starch, nucleosides and nucleotides, and antibiotic resistance, as well as enzyme cofactor biosynthesis. This is attributed to the enrichment of Arthrobacter, Lactococcus, Microbacterium, Streptococcus etc. during steeping (Supplementary Table S3). The kilning process further enriched pathways affiliated with glycolysis and phenolic degradation, potentially due to the enrichment of Arthrobacter, Lactococcus, Leuconostoc, Microbacterium, Streptococcus, Sanguibacter, and Corynebacterium, etc. In contrast, the steeping and kilning processes combined were predicted to suppress glucose degradation, L-tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde, L-tyrosine degradation I, L-histidine degradation II, ppGpp biosynthesis, NAD biosynthesis II (from tryptophan) and NAD salvage pathway II, and tRNA process.
Fig. 4.
A heatmap showing the metabolic pathways that were potentially enriched or depleted through the malting process.
3.4. Co-occurrence ecological networks
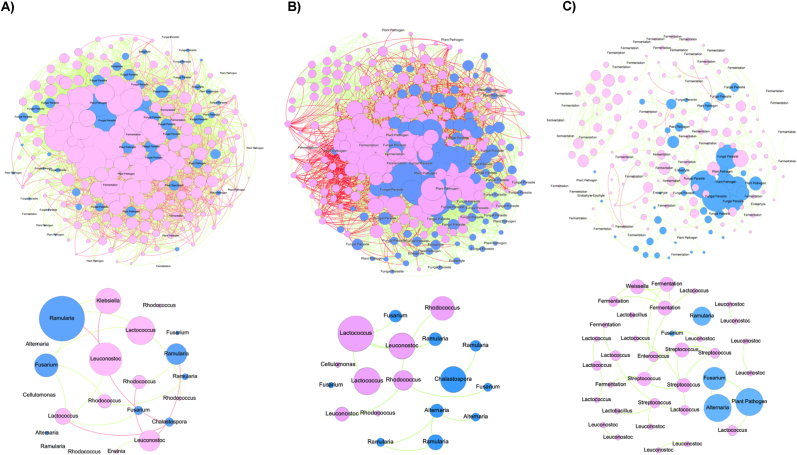
Co-occurrence network analysis showed a shift in the cross-kingdom interactions through the malting process. The inter-kingdom networks contained 380 nodes (OTUs) (3820 edges) for the field barley samples, 527 nodes (6558 edges) for the steeped barley samples, and 308 nodes (483 edges) for the kilned malt samples (Fig. 5). The steeped community, therefore, formed the largest and the most densely connected network with the highest average degree (the average number of edges per node was 14.9), while the malt network was the smallest and least connected with the lowest average degree (an average of 1.99 edges per node) (Supplementary Table S4).
Fig. 5.
Co-occurrence networks of fungal and bacterial OTUs in (A) field barley grains, (B) steeped barley grains, and (C) kilned malts. Fungal and bacterial OTU nodes are shown in blue and pink, respectively. Correlations are depicted by the green (positive correlations) or purple (negative correlations) linkages. Nodes are proportionally sized depending on their degree. Top figures display the whole ecological network, while bottom figures show the associations between fermentation bacteria and potential plant pathogens. Barley and steeped barley networks are larger, more connected, and have a higher proportion of negative interactions than those of kilned malts. The malt community has a higher proportion of bacteria with fermentation capacities that are positively correlated. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The proportion of negative interactions decreased through the malting process. The malt community had much fewer negative interactions (4.8%) than either the field barley (24.6%) or steeped barley communities (23.5%) (Fig. 5). The antagonistic relationships between the bacterial fermenters (e.g. LAB Leuconostoc spp. and Lactococcus spp.) and potential fungal pathogens (e.g.: Ramularia spp., Chalastospora spp., and some Fusarium spp.) were observed in field and steeped barley networks, but not in the malt network, despite that LAB and Fusarium were significantly enriched through the malting process (Fig. 2A, B, D, Fig. 5). By contrast, some potential plant pathogens and fermenters (e.g. Streptococcus, Lactobacillus, and Weissella) in the malt ecological network displayed positive correlations.
4. Discussion
We characterized the epiphytic fungal and bacterial communities of barley grains and malts and identified microbial indicators associated with different stages of the malting process as well as malt quality measured by PYF. The predicted functional roles of the microbial communities and the cross-kingdom co-existence patterns were also examined, suggesting the influential roles of the microbial communities during the malting process.
This study revealed yeasts, filamentous fungi, and bacteria from the epiphytic microbiome of barley and malt grains which underwent significant changes during the malting process (Fig. 1E & F). Our results support previous studies where the microbial profiles in barley and malt were vastly different (as reviewed by Noots et al. (1998), Justé et al. (2011), and Bokulich and Bamforth (2013)). However, malting affected the fungi and bacterial community differently. The diversity of the fungal community temporally increased after steeping but decreased significantly after kilning (Fig. 1A & C) as also suggested by Justé et al. (2011). A similar trend in yeasts was observed by Li et al. (2018) based on culture-dependent approaches, however, the authors reported an increase in filamentous fungi load after kilning. In contrast, our study showed that the bacterial diversity increased significantly after malting due to the enrichment of lactic acid bacteria (LAB) (Fig. 1B & D, Fig. 3A). Østlie et al. (2021) and Li et al. (2018) reported a significant increase in bacterial diversity in germinated barley grains, which may decrease after kilning (Li et al., 2018). Similarly, Petters et al. (1988) reported that kilning reduced the viable counts of the aerobic heterotrophic bacteria and filamentous fungi to the levels of the field barley, but that of the LAB can be 1000-fold higher than that in dry barley grains. Despite the observation that trends in diversity differed, the malting process increased the community compositional structure heterogeneity of both the fungal and the bacterial communities (Fig. 1E & F). Such shift in community composition, together with the fact that all bacterial and fungal OTUs were found in both barley and malt samples, suggest that the majority, if not all, taxa recovered originated from the barley grains and were proliferating and growing at different rates through malting. Considering high-throughput sequencing of environmental DNAs detects both viable and non-viable targets (Jofre and Blanch, 2010), this observation may also suggest that the DNAs of suppressed microorganisms may have not been fully degraded after the malting process and therefore detected by high throughput sequencing. No significant discrepancy in diversity or compositional structure was noted between samples from different kilning chambers.
The barley grain-associated bacterial community was dominated by Curtobacterium, Clavibacter, Leucobacter, and Salinibacterium, all of which belong to the Microbacteriaceae family. This observation indicates the transmission of microbes from the root to the seeds since the barley root microbiome is noted for the enrichment of Microbacteriaceae (Bulgarelli et al., 2015; Wang et al., 2016). Xanthomonas, commonly detected in barley grain after harvesting (Justé et al., 2011), was also found abundant in this study. The LAB (Lactobacillales) were significantly enriched after malting (Li et al., 2018) due to the nutrient-rich environment (Arimah and Ogunlowo, 2014). In particular, yeasts such as Candida spp. can stimulate LAB growth by providing essential metabolites (Wulijideligen et al., 2013), which was supported by the positive correlations between LAB and Candida spp. discovered in this study. Positive correlations between LAB and Arthrobacter spp. (53 OTUs) were also observed in this study, possibly due to the ability of Arthrobacter spp. to catabolize the lactic acid generated by LAB strains (Monnet et al., 2010). We also observe a positive correlation between the yeasts and Arthrobacter spp. A previous study showed that yeasts (e.g., Candida and Pichia) can consume the glucose in the malting environment and allow for the production of extracellular amylase enzymes capable of hydrolyzing starch in Arthrobacter (Smith and Zahnley, 2005). However, high temperature during steeping and kilning may significantly suppress such microbial activities.
LAB are known as important producers of organic acid or secondary metabolites that can inhibit the growth of other bacteria and fungi, and therefore dominate the malt microbiome (Alvarez-Sieiro et al., 2016; Lowe and Arendt, 2004; Sadiq et al., 2019; Xiraphi et al., 2008). In this study, Leuconostoc was the most abundant LAB, possibly because some Leuconostoc species can produce bacteriocins against other LAB, plant pathogens, or spoilage organisms (Hitendra et al., 2015; Xiraphi et al., 2008). This is supported by the negative correlations between Leuconstoc spp. and Xanthomonas spp. as well as Xylella spp. in the present study. Recent studies showed that LAB strains isolated from wine fermentations had the potential to promote the growth of tomato plants and to protect the plants from Fusarium oxysporum infection (López-Seijas et al., 2020).
The fungal community was composed of both Ascomycota (e.g. Cladosporium, Cryptococcus) and Basidiomycota (e.g. Sporobolomyces) in barley grains; but was dominated by Ascomycota spp. in malts, mainly Candida and Pichia, with some Pichia spp. being classified as teleomorphs of Candida spp. (Chen et al., 2016; Justé et al., 2011; Østlie et al., 2021). The high abundance of xerophilic fungi associated with barley may have been encouraged by dry storage conditions after harvesting, while the heat-tolerant ascomycetes species associated with malts were able to withstand the high kilning temperatures (Laitila et al., 2011). Candida spp. were abundant in most of the malt samples in this study, except for sample B364, where a high proportion of Wickerhamomyces was recovered. Wickerhamomyces anomalus secretes toxins that can inhibit Candida growth (Farkas et al., 2012; Li et al., 2018), possibly accounting for this observation. Furthermore, in this study, Fusarium spp. were significantly enriched in malts, which was also reported by other studies (Bokulich and Bamforth, 2013; Laitila et al., 2002). Similarly, Justé et al. (2011) observed an exponential increase in Fusarium spp. load in germinated malts, however, their abundances decreased after kilning. This discrepancy may be attributed to an inefficient drying process of the germination bed at the kilning step, therefore allowing for continued microbial proliferation, as suggested by Noots et al. (1998). A recent study has shown that Fusarium spp. located on the husk, vascular bundle, and pericarp cavities of barley grains can continue to grow their hyphal into the endosperm and embryo through the malting process (Jin et al., 2021). The presence of certain Fusarium spp. in malt has been attributed to many brewing problems, such as reducing barley germination, mycotoxin production, gushing, and degrading the colour and taste in beer (Jin et al., 2021; Salas et al., 1999; Sarlin et al., 2012; Wolf-Hall, 2007). Therefore, some bacterial or fungal taxa can inhibit the proliferation of Fusarium spp. and/or other mycotoxin-producing fungi, may help alleviate the above brewing issues.
Considering bacteria and fungi form close physical associations on barley grains, their interactions are inevitable (Frey-Klett et al., 2011) through the malting process, which can be revealed by molecular ecological network analysis as demonstrated by studies in different ecosystems (Legrand et al., 2019; Li and Wu, 2018; Zhang et al., 2020). For example, LAB and yeasts (e.g. W. anomalus, Aureobasidium pullulans, and certain Candida spp.) may inhibit mould growth by competing for space or nutrients, as well as producing exotoxins with antimicrobial activities (Lowe and Arendt, 2004; Peyer et al., 2016). Juodeikiene et al. (2018) demonstrated the selected LAB strains and their metabolites were able to inhibit the growth of Fusarium species and to reduce the production of mycotoxins, including zearalenone, deoxynivalenol, T-2, and HT-2 toxins. The enrichment of LAB and the yeasts and yeast-like fungi in malt may account for the low levels (less than 3.5% of the total relative abundance) of mycotoxin-producing fungi found in this study. However, molecular ecological network reconstruction revealed disconnected bacteria fermenters and fungal OTUs (belonging to Cryptococcus, Sporobolomyces, Hannaella, Alternaria, and Fusarium) in the malt-associated microbiome (Fig. 5C). The elevated temperature at the kilning step may have compromised the effectiveness of some antimicrobial compounds by inhibiting their production or by changing the pH (Lax et al., 2020). In addition, the decrease in competitive (negative) cross-kingdom interactions may indicate a decrease in community stability (Coyte et al., 2015; de Vries et al., 2018). Therefore, compared with the barley grain microbiome, the kilned malt microbiome may be more vulnerable to environmental perturbations, which could lead to a drastic shift in microbial diversity and compositional structure and consequently, a reduced level of consistency in malt quality. The capacity of the beneficial bacterial and yeasts to inhibit the pathogens or spoilage organisms require further investigation.
PYF causes incomplete fermentation, lowers the alcohol content, and produces off-flavoured beer (Panteloglou et al., 2012). Although the exact cause of PYF is still unknown, microbial growth is hypothesized to play a role (Kaur et al., 2012; van Nierop et al., 2008). We identified both fungal (e.g. Candida spp., Aureobasidium spp. and members of Nectriaceae) and bacterial OTUs that were correlated to PYF (Fig. 3B). This is in contrast to the findings of Kaur et al. (2012) where an association between bacteria and PYF was negligible. Kaur et al. (2012) used terminal restriction fragment length polymorphism to identify the microbial malt community, whereas high-throughput sequencing was applied in this study. The different detection methods may have allowed for the identification of additional bacterial taxa, resulting in their association to PYF. Barley exposed to Cochliobolus sativus and F. graminearum were found to display PYF characteristics after malting (MacIntosh et al., 2014). However, we did not detect an association between PYF and Cochliobolus (anamorphs Bipolaris, Curvularia) or Fusarium, which may be attributed to the low abundance of both genera recovered from this study. Panteloglou et al. (2012) found the degradation of arabinoxylans in barley husks can generate bridging polysaccharides thereby encouraging PYF. Genes encoding arabinoxylan hydrolases were found in the Lactobacillus genome (Michlmayr et al., 2013), we however did not observe an association between Lactobacillus spp. with PYF. Most recently, Shang et al. (2022) reported that PYF can be inhibited by recombinant barley xylanase inhibitor produced by transformants of Pichia pastoris during fermentation of PYF+ve worts, confirming the importance of arabinoxylan in PYF formation as also suggested by van Nierop et al. (2004). We also noticed that Candida spp., Arthrobacter spp., and Leucobacter spp. were abundant in PYF-ve malt samples, therefore our study cannot conclusively associate these microbes with PYF, and the exact nature of how these microorganisms are associated with PYF remains to be elucidated. Further investigation including sampling through the fermentation stages and monitoring the changes of these microbes and their secondary metabolites may eventually reveal the true cause of PYF to aid brewers to improve the consistency of their product.
We acknowledge that the current study focused only on the epiphytic microbiota of the grains, while the endophytic communities colonising the seeds were not explored. Nonetheless, in this study, the steeping process was predicted to promote the metabolic pathways of the bacterial communities affiliated to the degradation of carbohydrates such as aromatic compounds, polyamine and amino acids, sugars, etc., suggesting their involvement in the endosperm cell wall and starch degradation and consequently important for germination (Andriotis et al., 2016). Interestingly, it was predicted that the degradation of phenolic compounds was stimulated by the kilning process, which may result in the increase of phenolic antioxidant compounds in malts and potentially improve the health-promoting properties of the end-products of the brewing process (Carciochi et al., 2016; Xu et al., 2020). Notably, more pathways were predicted to be inhibited than promoted by the malting process. The repression of the degradation of L-tryptophan, L-tyrosine, L-histidine, NAD biosynthesis (Fig. 4), etc. through the malting process, perhaps suggested that the malt microbiome suffered from nutrients and energy starvation (Traxler et al., 2008). Our study showed that bacteria (e.g. members of Arthrobacter, Brachybacterium, Cellulomonas, Clavibacter, Paenibacillus, Pseudomonas, and Sphingobacterium) and fungi (e.g. members of Cryptococcus, Sporobolomyces, Aureobasidium, Bullera, Candida, and Cladosporium) that can produce hydrolytic enzymes (Laitila et al., 2006a; Malfliet et al., 2013) were in general enriched in the kilned malt and could have contributed to malting efficiency. We acknowledge that malting is a rather selective process, during which the high temperatures at steeping and kilning may suppress microbial activities. Yet, certain microorganisms may be able to survive, and their signals can be captured through high-throughput sequencing. Although speculative, PICRUSt provides some insights into their potential roles or even survival strategies in such unique niches.
5. Conclusions
The microbial community of field and steeped barley grains and kilned malts at a commercial malthouse in western Canada was characterized using a metabarcoding approach. The fungal diversity decreased while the bacterial diversity increased through the malting process. We also identified fungal and bacterial taxa potentially having high hydrolytic enzymatic activity and those associated with PYF+ve or PYF-ve malts. While lactic acid bacteria (LAB) and Fusarium spp. were significantly enriched in malts, these bacterial fermenters and phytopathogens did not show strong interactions in malts through ecological network analysis. Future studies should measure additional malt quality indicators and integrate proteomics and metabolomics to investigate the underlying mechanisms of grain and malt microbiomes in affecting the quality of malt end products.
Funding
This work was supported by the Agriculture Innovation Program (AIP) of the National Barley Research Cluster (projec ID: AIP-CL07). The computational infrastructure was partially supported by Agriculture & Agri-Food Canada (AAFC) funded projects (J-002216, J-002272, and J-002305), and the Genomics Research and Development Initiative (GRDI) Ecobiomics project (J-001263).
CRediT authorship contribution statement
Wen Chen: Project administration, Supervision, Data curation, Software, Validation, Writing – original draft, Writing - reviewing & editing. H.Y. Kitty Cheung: Formal analysis, Visualization, Writing – original draft, Writing - reviewing & editing. Morgan McMillan: Formal analysis, Visualization, Writing – original draft, Writing - reviewing & editing. Thomas Kelly Turkington: Conceptualization, Funding acquisition, Writing - reviewing & editing. Marta S. Izydorczyk: Methodology, Writing - reviewing & editing. Tom Gräfenhan: Conceptualization, Funding acquisition, Writing - reviewing & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The assistance of Rahr Malting Canada, Ltd., Alix, Alberta, with samples is sincerely appreciated. We thank Michael Edney (Owen Sound, ON, Canada), John O'Donovan and Neil Harker (both Lacombe, Alberta, Canada), Brian Beres (Lethbridge, AB, Canada), Eric Johnson (Scott, SK, Canada), Bill May (Indian Head, SK, Canada), Ramona Mohr (Brandon, MB, Canada) and all the field staff involved for their vital support. The technical assistance of Inge Roewer and the Aquatic and Crop Resource Development Research Centre of the National Research Council of Canada in Saskatoon, SK is graciously acknowledged for the 454-pyrosequencing service. We thank Kevin Sich (Supply Chain Director, Rahr Malting Canada Ltd., Alix, AB, Canada) and Xiang S, Yin (Director of Brewing Research and Innovation, Rahr Malting Co., Shakopee, MN, U.S.A.) for providing the technical conditions for commercial malting.
Handling editor: Dr. Siyun Wang
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2022.08.009.
Contributor Information
Wen Chen, Email: wen.chen@agr.gc.ca.
Tom Gräfenhan, Email: tom.graefenhan@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alvarez-Sieiro P., Montalbán-López M., Mu D., Kuipers O.P. Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 2016;100(7):2939–2951. doi: 10.1007/s00253-016-7343-9I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriotis V.M.E., Rejzek M., Barclay E., Rugen M.D., Field R.A., Smith A.M. Cell wall degradation is required for normal starch mobilisation in barley endosperm. Sci. Rep. 2016;6(1):33215. doi: 10.1038/srep33215I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimah B.D., Ogunlowo O.P. Identification of lactic acid bacteria isolated from Nigerian foods: medical importance and comparison of their bacteriocins activities. J. Nat. Sci. Res. 2014;4(23):76–86. [Google Scholar]
- ASBC . ASBC Headquarters; St. Paul, MN: 2022. American Society of Brewing Chemists (ASBC) Methods of Analysis: Microbiology, Yeast.https://www.asbcnet.org/Methods/MicrobiologyMethods/Pages/default.aspx Online: [Google Scholar]
- Aubert M.K., Coventry S., Shirley N.J., Betts N.S., Würschum T., Burton R.A., Tucker M.R. Differences in hydrolytic enzyme activity accompany natural variation in mature aleurone morphology in barley (Hordeum vulgare L.) Sci. Rep. 2018;8(1):1–14. doi: 10.1038/s41598-018-29068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S., Jacomy M. Vol. 3. 2009. Gephi: An Open Source Software for Exploring and Manipulating Networks; pp. 361–362.https://ojs.aaai.org/index.php/ICWSM/article/view/13937 (Proceedings of the International AAAI Conference on Web and Social Media). (1). Retrieved from. [Google Scholar]
- Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., De Wit P., Sánchez-García M., Ebersberger I., de Sousa F., Amend A., Jumpponen A., Unterseher M., Kristiansson E., Abarenkov K., Bertrand Y.J.K., Sanli K., Eriksson K.M., Vik U., Veldre V., Nilsson R.H. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013;4(10):914–919. doi: 10.1111/2041-210X.12073. [DOI] [Google Scholar]
- Bianco A., Fancello F., Balmas V., Zara G., Dettori M., Budroni M. The microbiome of Sardinian barley and malt. J. Inst. Brew. 2018;124(4):344–351. doi: 10.1002/jib.522. [DOI] [Google Scholar]
- Bokulich N.A., Bamforth C.W., Mills D.A. Brewhouse-resident microbiota are responsible for multi-stage fermentation of American coolship ale. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Bamforth Charles W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. 2013;77(2):157–172. doi: 10.1128/MMBR.00060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Kang K.B., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.-X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J.J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Garrido-Oter R., Münch P.C., Weiman A., Dröge J., Pan Y., McHardy A.C., Schulze-Lefert P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17(3):392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carciochi R.A., Dimitrov K., Galván D Alessandro L. Effect of malting conditions on phenolic content, Maillard reaction products formation, and antioxidant activity of quinoa seeds. J. Food Sci. Technol. 2016;53(11):3978–3985. doi: 10.1007/s13197-016-2393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Simpson J., Levesque C.A. 2018. RAM: R for Amplicon-Sequencing-Based Microbial-Ecology.https://CRAN.R-project.org/package=RAM R package version 1.2.1.7. [Google Scholar]
- Chen W., Radford D., Hambleton S. Towards improved detection and identification of rust fungal pathogens in environmental samples using a metabarcoding approach. Phytopathology. 2021;112(3):535–548. doi: 10.1094/phyto-01-21-0020-r. [DOI] [PubMed] [Google Scholar]
- Chen W., Wilkes G., Khan I.U.H., Pintar K.D.M., Thomas J.L., Lévesque C.A., Chapados J.T., Topp E., Lapen D.R. Aquatic Bacterial Communities Associated With Land Use and Environmental Factors in Agricultural Landscapes Using a Metabarcoding Approach. Front. Microbiol. 2018;9(2301) doi: 10.3389/fmicb.2018.02301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Radford D., Hambleton S. Towards improved detection and identification of rust fungal pathogens in environmental samples using a metabarcoding approach. Phytopathology. 2021;112(3):535–548. doi: 10.1094/phyto-01-21-0020-r. [DOI] [PubMed] [Google Scholar]
- Chen W., Turkington T.K., Lévesque C.A., Bamforth J.M., Patrick S.K., Lewis C.T., Chapados J.T., Gaba D., Tittlemier S.A., MacLeod A. Geography and agronomical practices drive diversification of the epiphytic mycoflora associated with barley and its malt end product in western Canada. Agric. Ecosyst. Environ. 2016;226:43–55. doi: 10.1016/j.agee.2016.03.030. [DOI] [Google Scholar]
- Coyte K.Z., Schluter J., Foster K.R. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350(6261):663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- de Vries F.T., Griffiths R.I., Bailey M., Craig H., Girlanda M., Gweon H.S., Hallin S., Kaisermann A., Keith A.M., Kretzschmar M., Lemanceau P., Lumini E., Mason K.E., Oliver A., Ostle N., Prosser J.I., Thion C., Thomson B., Bardgett R.D. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018;9(1) doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Jiang Y.-H., Yang Y., He Z., Luo F., Zhou J. Molecular ecological network analyses. BMC Bioinf. 2012;13(113):1–20. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Steffenson B.J., Mirocha C.J. Analysis of ergosterol in single kernel and ground grain by gas Chromatography−Mass spectrometry. J. Agric. Food Chem. 2006;54(12):4121–4125. doi: 10.1021/jf060149f. [DOI] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas Z., Márki-Zay J., Kucsera J., Vágvölgyi C., Golubev W.I., Pfeiffer I. Characterization of two different toxins of Wickerhamomyces anomalus (Pichia anomala) VKM Y-159. Acta Biol. Hung. 2012;63(2):277–287. doi: 10.1556/ABiol.63.2012.2.9. [DOI] [PubMed] [Google Scholar]
- Flannigan B.A. In: Brewing Microbiology. F. G. Priest. Campbell I., editor. Springer; Boston, MA: 2003. The microbiota of barley and malt; pp. 113–180. [Google Scholar]
- Follstad M., Christensen C. Microflora of barley kernels. Appl. Microbiol. 1962;10(4):331–336. doi: 10.1128/am.10.4.331-336.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Klett P., Burlinson P., Deveau A., Barret M., Tarkka M., Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011;75(4):583–609. doi: 10.1128/mmbr.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geißinger C., Whitehead I., Hofer K., Heß M., Habler K., Becker T., Gastl M. Influence of Fusarium avenaceum infections on barley malt: monitoring changes in the albumin fraction of barley during the malting process. Int. J. Food Microbiol. 2019;293:7–16. doi: 10.1016/j.ijfoodmicro.2018.12.026. [DOI] [PubMed] [Google Scholar]
- Gonzalez Pereyra M.L., Rosa C.A.R., Dalcero A.M., Cavaglieri L.R. Mycobiota and mycotoxins in malted barley and brewer’s spent grain from Argentinean breweries. Lett. Appl. Microbiol. 2011;53(6):649–655. doi: 10.1111/j.1472-765X.2011.03157.x. [DOI] [PubMed] [Google Scholar]
- Hitendra J., Suvarna V.C., Niveditha S.B. Antimicrobial attributes of Leuconostoc isolates against fruit and vegetable spoilage organisms. Int. J. Curr. Microbiol. App. Sci. 2015;4(11):160–166. [Google Scholar]
- Jin Z., Solanki S., Ameen G., Gross T., Poudel R.S., Borowicz P., Brueggeman R.S., Schwarz P. Expansion of internal hyphal growth in Fusarium Head blight–infected grains contributes to the elevated mycotoxin production during the malting process. Mol. Plant Microbe Interact. 2021;34(7):793–802. doi: 10.1094/MPMI-01-21-0024-R. [DOI] [PubMed] [Google Scholar]
- Jofre J., Blanch A.R. Feasibility of methods based on nucleic acid amplification techniques to fulfil the requirements for microbiological analysis of water quality. J. Appl. Microbiol. 2010;109(6):1853–1867. doi: 10.1111/j.1365-2672.2010.04830.x. [DOI] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113(2):363–375. doi: 10.1111/j.2006.0030-1299.14714.x. [DOI] [Google Scholar]
- Juodeikiene G., Bartkiene E., Cernauskas D., Cizeikiene D., Zadeike D., Lele V., Bartkevics V. Antifungal activity of lactic acid bacteria and their application for Fusarium mycotoxin reduction in malting wheat grains. LWT (Lebensm.-Wiss. & Technol.) 2018;89:307–314. doi: 10.1016/j.lwt.2017.10.061. [DOI] [Google Scholar]
- Justé A., Malfliet S., Lenaerts M., de Cooman L., Aerts G., Willems K.A., B L. Microflora during malting of barley: overview and impact on malt quality. BrSc. 2011;64(3–4):22–31. [Google Scholar]
- Justé A., Malfliet S., Waud M., Crauwels S., De Cooman L., Aerts G., Marsh T.L., Ruyters S., Willems K., Busschaert P., Lievens B. Bacterial community dynamics during industrial matling, with an emphasis on lactic acid bacteria. Food Microbiol. 2014;39:39–46. doi: 10.1016/j.fm.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Kaur M., Bowman J.P., Stewart D.C., Sheehy M., Janusz A., Speers R.A., Koutoulis A., Evans D.E. TRFLP analysis reveals that fungi rather than bacteria are associated with premature yeast flocculation in brewing. J. Ind. Microbiol. Biotechnol. 2012;39(12):1821–1832. doi: 10.1007/s10295-012-1188-8. [DOI] [PubMed] [Google Scholar]
- Kaur M., Bowman J.P., Stewart D.C., Evans D.E. The fungal community structure of barley malts from diverse geographical regions correlates with malt quality parameters. Int. J. Food Microbiol. 2015;215:71–78. doi: 10.1016/j.ijfoodmicro.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Kaur M., Evans D.E., Stewart D.C., Janusz A., Bowman J., Koutoulis A. American Society of Brewing Chemists; 2014. Malt screening for premature yeast flocculation (PYF) based on qPCR detection of the microbial genera associated with or causal of PYF. (Proceedings of the 77th American Society of Brewing Chemists convention, Chicago, USA). [Google Scholar]
- Laitila A. More good than bad: microbes in the maltings. Brewer. Distiller Int. 2008;4(8):52–54. [Google Scholar]
- Laitila A., Alakomi H.-L., Raaska L., Mattila-Sandholm T., Haikara A. Antifungal activities of two Lactobacillus plantarum strains against Fusarium moulds in vitro and in malting of barley. J. Appl. Microbiol. 2002;93(4):566–576. doi: 10.1046/j.1365-2672.2002.01731.x. [DOI] [PubMed] [Google Scholar]
- Laitila A., Kotaviita E., Peltola P., Home S., Wilhelmson A. Indigenous microbial community of barley greatly influences grain germination and malt quality. J. Inst. Brew. 2007;113(1):9–20. doi: 10.1002/j.2050-0416.2007.tb00250.x. [DOI] [Google Scholar]
- Laitila A., Wilhelmson A., Kotaviita E., Olkku J., Home S., Juvonen R. Yeasts in an industrial malting ecosystem. J. Ind. Microbiol. Biotechnol. 2006;33(11):953–966. doi: 10.1007/s10295-006-0150-z. [DOI] [PubMed] [Google Scholar]
- Laitila A., Manninen J., Priha O., Smart K., Tsitko I., James S. Characterisation of barley-associated bacteria and their impact on wort separation performance. J. Inst. Brew. 2018;124(4):314–324. doi: 10.1002/jib.509. [DOI] [Google Scholar]
- Laitila A., Sweins H., Vilpola A., Kotaviita E., Olkku J., Home S., Haikara A. Lactobacillus plantarum and Pediococcus pentosaceus Starter Cultures as a Tool for Microflora Management in Malting and for Enhancement of Malt Processability. J. Agric. Food Chem. 2006;54(11):3840–3851. doi: 10.1021/jf052979j. [DOI] [PubMed] [Google Scholar]
- Laitila A., Sarlin T., Raulio M., Wilhelson A., Kotaviita E., Huttunen T., Juvonen R. Yeasts in malting, with special emphasis on Wickerhamomyces anomalus (synonym Pichia anomala) Antonie Leeuwenhoek. 2011;99(1) doi: 10.1007/s10482-010-9511-8. [DOI] [PubMed] [Google Scholar]
- Lake J.C., Speers R.A. A discussion of malt-induced premature yeast flocculation. Tech. Q. - Master Brew. Assoc. Am. 2008;45(3):253–262. doi: 10.1094/TQ-45-3-0253. [DOI] [Google Scholar]
- Lax S., Abreu C.I., Gore J. Higher temperatures generically favour slower-growing bacterial species in multispecies communities. Nat. Ecol. Evol. 2020;4(4):560–567. doi: 10.1038/s41559-020-1126-5. [DOI] [PubMed] [Google Scholar]
- Legrand F., Chen W., Cobo-Díaz J.F., Picot A., Floch G.L. Co-occurrence analysis reveal that biotic and abiotic factors influence soil fungistasis against Fusarium graminearum. FEMS Microbiol. Ecol. 2019;95(5) doi: 10.1093/femsec/fiz056. [DOI] [PubMed] [Google Scholar]
- Li S., Wu F. Diversity and Co-occurrence patterns of soil bacterial and fungal communities in seven intercropping Systems. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cai G., Wu D., Zhang M., Lin C., Lu J. Microbial community dynamics of Dan’er barley grain during the industrial malting process. Food Microbiol. 2018;76:110–116. doi: 10.1016/j.fm.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Linko M., Haikara A., Ritala A., Penttilä M. Recent advances in the malting and brewing industry. J. Biotechnol. 1998;65(2–3):95–98. doi: 10.1016/S0168-1656(98)00135-7. [DOI] [Google Scholar]
- Links M.G., Demeke T., Gräfenhan T., Hill J.E., Hemmingsen S.M., Dumonceaux T.J. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 2014;202(2):542–553. doi: 10.1111/nph.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Seijas J., García-Fraga B., da Silva A.F., Sieiro C. Wine lactic acid bacteria with antimicrobial activity as potential. Biocontrol Agents against Fusarium oxysporum f. sp. lycopersici. Agronomy. 2020;10(1):31. doi: 10.3390/agronomy10010031. [DOI] [Google Scholar]
- Louca S., Parfrey L.W., Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D.P., Arendt E.K. The use and effects of lactic acid bacteria in malting and brewing with their relationships to antifungal activity, mycotoxins and gushing: a review. J. Inst. Brew. 2004;110(3):163–180. doi: 10.1002/j.2050-0416.2004.tb00199.x. [DOI] [Google Scholar]
- MacIntosh A.J., MacLeod A., Beattie A.D., Eck E., Edney M., Rossnagel B., Speers R.A. Assessing the effect of fungal infection of barley and malt on premature yeast flocculation. J. Am. Soc. Brew. Chem. 2014;72:66–72. doi: 10.1094/ASBCJ-2014-0204-01. [DOI] [Google Scholar]
- MacLeod L.C., Evans D.E. In: Encyclopedia of Food Grains Vol. Volume 1 - the World of Food Grains. Wrigley C., Corke H., Seetharaman K., Faubion J., editors. Oxford: Academic Press; London: 2016. Barley: malting; pp. 423–433. [Google Scholar]
- Malfliet S., Justé A., Crauwels S., Willems K., De Cooman L., Lievens B., Aerts G. Assessing the xylanolytic bacterial diversity during the malting process. Food Microbiol. 2013;36(2):406–415. doi: 10.1016/j.fm.2013.06.025. [DOI] [PubMed] [Google Scholar]
- Marcon E., Hérault B. entropart: an R package to measure and partition diversity. J. Stat. Software. 2015;67(8) doi: 10.18637/jss.v067.i08. [DOI] [Google Scholar]
- Michlmayr H., Hell J., Böhmdorfer S., Rosenau T., Kneifel W. Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl. Environ. Microbiol. 2013;79(21):6747–6754. doi: 10.1128/AEM.02130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnet C., Loux V., Gibrat J.-F., Spinnler E., Barbe V., Vacherie B., Gavory F., Gourbeyre E., Siguier P., Chandler M., Elleuch R., Irlinger F., Vallaeys T. The Arthrobacter arilaitensis Re117 genome sequence reveals its genetic adaptation to the surface of cheese. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.H., Song Z., Bates S.T., Branco S., Tedersoo L., Menke J., Schilling J.S., Kennedy P.G. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–248. doi: 10.1016/j.funeco.2015.06.006. [DOI] [Google Scholar]
- Noots I., Delcour J.A., Michiels C.W. From field barley to malt: detection and specification of microbial activity for quality aspects. Crit. Rev. Microbiol. 1998;25(2):121–153. doi: 10.1080/10408419991299257. [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. Vegan: community ecology package. 2018. https://CRAN.R-project.org/package=vegan
- Oliveira P., Brosnan B., Jacob F., Furey A., Coffey A., Zannini E., Arendt E.K. Lactic acid bacteria bioprotection applied to the malting process. Part II: substrate impact and mycotoxin reduction. Food Control. 2015;51:444–452. doi: 10.1016/j.foodcont.2014.11.011. [DOI] [Google Scholar]
- Østlie H.M., Porcellato D., Kvam G., Wicklund T. Investigation of the microbiota associated with ungerminated and germinated Norwegian barley cultivars with focus on lactic acid bacteria. Int. J. Food Microbiol. 2021;341 doi: 10.1016/j.ijfoodmicro.2021.109059. [DOI] [PubMed] [Google Scholar]
- Panteloglou A.G., Smart K.A., Cook D.J. Malt-induced premature yeast flocculation: current perspectives. Journal of Industrial Microbiology and Biotechnology. 2012;39(6):813–822. doi: 10.1007/s10295-012-1086-0. [DOI] [PubMed] [Google Scholar]
- Petters H.I., Flannigan B., Austin B. Quantitative and qualitative studies of the microflora of barley malt production. J. Appl. Bacteriol. 1988;65(4):279–297. doi: 10.1111/j.1365-2672.1988.tb01895.x. [DOI] [Google Scholar]
- Peyer L., Axel C., Lynch K.M., Zannini E., Jacob F., Arendt E.K. Inhibition of Fusarium culmorum by carboxylic acids released from lactic acid bacteria in a barley malt substrate. Food Control. 2016;69:227–236. doi: 10.1016/j.foodcont.2016.05.010. [DOI] [Google Scholar]
- Peyer L.C., De Kruijf M., O’Mahony J., De Colli L., Danaher M., Zarnkow M., Jacob F., Arendt E.K. Lactobacillus brevis R2Δ as starter culture to improve biological and technological qualities of barley malt. Eur. Food Res. Technol. 2017;243:1363–1374. doi: 10.1007/s00217-017-2847-9. [DOI] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rodríguez-Carrasco Y., Fattore M., Albrizio S., Berrada H., Mañes J. Occurrence of Fusarium mycotoxins and their dietary intake through beer consumption by the European population. Food Chem. 2015;178:149–155. doi: 10.1016/j.foodchem.2015.01.092. [DOI] [PubMed] [Google Scholar]
- Sadiq F., Yan B., Tian F., Zhao J., Zhang H., Chen W. Lactic acid bacteria as antifungal and anti‐mycotoxigenic agents: a comprehensive review. Compr. Rev. Food Sci. Food Saf. 2019;18:1403–1436. doi: 10.1111/1541-4337.12481. [DOI] [PubMed] [Google Scholar]
- Salas B., Steffenson B.J., Casper H.H., Tacke B., Prom L.K., Fetch T.G.J., Schwarz P.B. Fusarium species pathogenic to barley and their associated mycotoxins. Plant Dis. 1999;83(7):667–674. doi: 10.1094/PDIS.1999.83.7.667. [DOI] [PubMed] [Google Scholar]
- Sarlin T., Kivioja T., Kalkkinen N., Linder M.B., Nakari-Setälä T. Identification and characterization of gushing-active hydrophobins from Fusarium graminearum and related species. J. Basic Microbiol. 2012;52(2):184–194. doi: 10.1002/jobm.201100053. [DOI] [PubMed] [Google Scholar]
- Shang Y., Li X., Cai G., Wang D., Li F., Lu J., Yu X. Production of recombinant barley xylanase inhibitor in Pichia pastoris and its inhibitory effect on premature yeast flocculation. J. Inst. Brew. 2022;128:59–65. doi: 10.1002/jib.688. [DOI] [Google Scholar]
- Smith M.R., Zahnley J.C. Production of amylase by Arthrobacter psychrolactophilus. J. Ind. Microbiol. Biotechnol. 2005;32(7):277–283. doi: 10.1007/s10295-005-0240-3. [DOI] [PubMed] [Google Scholar]
- Traxler M.F., Summers S.M., Nguyen H.T., Zacharia V.M., Hightower G.A., Smith J.T., Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 2008;68(5):1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nierop S.N.E., Cameron-Clarke A., Axcell B.C. Enzymatic generation of factors from malt responsible for premature yeast flocculation. J. Am. Soc. Brew. Chem. 2004;62(3):108–116. doi: 10.1094/ASBCJ-62-0108. [DOI] [Google Scholar]
- van Nierop S.N.E., Rautenbach M., Axcell B.C., Cantrell I.C. The impact of microorganisms on barley and malt quality—a Review. J. Am. Soc. Brew. Chem. 2006;64(2):69–78. doi: 10.1094/ASBCJ-64-0069. [DOI] [Google Scholar]
- van Nierop S.N.E., Axell B.C., Cantrell I.C., Rautenbach M. Optimised quantification of the antiyeast activity of different barley malts towards a lager brewing yeast strain. Food Microbiol. 2008;25(7):895–901. doi: 10.1016/j.fm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wang W., Zhai Y., Cao L., Tan H., Zhang R. Illumina-based analysis of core actinobacteriome in roots, stems, and grains of rice. Microbiol. Res. 2016;190:12–18. doi: 10.1016/j.micres.2016.05.003. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: Guide. Method. Appl. 1990;18:315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- Wolf-Hall C.E. Mold and mycotoxin problems encountered during malting and brewing. Int. J. Food Microbiol. 2007;119(1–2):89–94. doi: 10.1016/j.ijfoodmicro.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Wolfe B.E., Dutton R.J. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015;161(1):49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- Wulijideligen S., Arakawa K., Miyamoto M., Miyamoto T. Interaction between lactic acid bacteria and yeasts in airag, an alcoholic fermented milk. Anim. Sci. J. 2013;84(1):66–74. doi: 10.1111/j.1740-0929.2012.01035.x. [DOI] [PubMed] [Google Scholar]
- Xiraphi N., Georgalaki M., Rantsiou K., Cocolin L., Tsakalidou E., Drosinos E.H. Purification and characterization of a bacteriocin produced by Leuconostoc mesenteroides E131. Meat Sci. 2008;80(2):194–203. doi: 10.1016/j.meatsci.2007.11.020. [DOI] [PubMed] [Google Scholar]
- Xu M., Rao J., Chen B. Phenolic compounds in germinated cereal and pulse seeds: classification, transformation, and metabolic process. Crit. Rev. Food. 2020;60(5):740–759. doi: 10.1080/10408398.2018.1550051. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chen W., Zhao B., Phillips L.A., Zhou Y., Lapen D.R., Liu J. Sandy soils amended with bentonite induced changes in soil microbiota and fungistasis in maize fields. Appl. Soil Ecol. 2020;146 doi: 10.1016/j.apsoil.2019.103378. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data are accessible in Sequence Read Archive (SRA) under BioProject PRJNA661245 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA661245).