Abstract
During bacterial conjugation, the single-stranded DNA molecule is transferred through the cell envelopes of the donor and the recipient cell. A membrane-spanning transfer apparatus encoded by conjugative plasmids has been proposed to facilitate protein and DNA transport. For the IncPα plasmid RP4, a thorough sequence analysis of the gene products of the transfer regions Tra1 and Tra2 revealed typical features of mainly inner membrane proteins. We localized essential RP4 transfer functions to Escherichia coli cell fractions by immunological detection with specific polyclonal antisera. Each of the gene products of the RP4 mating pair formation (Mpf) system, specified by the Tra2 core region and by traF of the Tra1 region, was found in the outer membrane fraction with one exception, the TrbB protein, which behaved like a soluble protein. The membrane preparation from Mpf-containing cells had an additional membrane fraction whose density was intermediate between those of the cytoplasmic and outer membranes, suggesting the presence of attachment zones between the two E. coli membranes. The Tra1 region is known to encode the components of the RP4 relaxosome. Several gene products of this transfer region, including the relaxase TraI, were detected in the soluble fraction, but also in the inner membrane fraction. This indicates that the nucleoprotein complex is associated with and/or assembled facing the cytoplasmic site of the E. coli cell envelope. The Tra1 protein TraG was predominantly localized to the cytoplasmic membrane, supporting its potential role as an interface between the RP4 Mpf system and the relaxosome.
The self-transmissible IncPα plasmid RP4 (60,099 bp) (44) is known for its extremely broad host range; i.e., it is stably maintained in all of the tested gram-negative bacteria. Furthermore, the RP4 transfer system can mediate DNA transmission by conjugation to a variety of different organisms, such as gram-positive bacteria and even in yeasts (5, 24, 63). Therefore, RP4 serves as a widely used model system for the study of plasmid biology in general and conjugative DNA transfer in particular. In addition, several findings provide evidence for a close relationship of RP4-mediated bacterial conjugation and T-DNA transfer from agrobacteria to plant cells (58). It has been shown that the gene organizations and amino acid sequences of the gene products of the RP4 Tra regions, the Vir regions of Ti plasmids of Agrobacterium tumefaciens, and other transfer systems possess a remarkable degree of similarity (37, 39, 42). This family of related export systems for macromolecules (for reviews, see references 30, 39 and 59) includes, in addition to the VirB operon of Ti plasmids (32, 66), conjugative transfer systems of IncN plasmid pKM101 and IncW plasmid R388 (46), as well as that of the Tra3 region of the Ti plasmid (1, 14). Most remarkably, the RP4 transfer system is also related to the pertussis toxin export system, the Ptl operon, of Bordetella pertussis (69). In none of these systems have the transport structures and the mechanism of the macromolecule export process been studied extensively.
Two distinct regions of RP4, Tra1 and Tra2, encode essential transfer functions (for a review, see reference 42). The Tra1 region mainly encodes DNA processing functions for the generation of the single-stranded DNA molecule which is transferred to the recipient cell. The Tra2 core region (11 genes) and traF of Tra1 belong to the so-called mating pair formation (Mpf) system (22, 38). On the basis of our previous studies, we postulate that three major components are required for conjugative transfer of RP4 into the recipient cell: (i) the relaxosome, which is formed by gene products of the Tra1 region, namely, TraH, TraI, TraJ, and TraK, at the transfer origin (oriT) (17, 42); (ii) the Mpf system; and (iii) TraG, which is encoded by Tra1 and is thought to connect the relaxosome and the Mpf complex (9).
The essential components of the Mpf system were determined by constructing defined, mostly nonpolar knockout mutations in each gene of the Tra2 core region. Ten Tra2 genes, trbB, trbC, trbD, trbE, trbF, trbG, trbH, trbI, trbJ, and trbL, but not trbK, were found to be essential for RP4-specific transfer. Several criteria served to further characterize the RP4 Mpf system: (i) mobilization of the non-self-transmissible IncQ plasmid RSF1010 (RSF1010 encodes its own relaxosomal components, but its transfer to a recipient cell relies on the Mpf systems of conjugative helper plasmids), (ii) propagation of donor-specific phages (PRD1, Pf3, and PRR1), and (iii) pilus production. The Mpf components which are essential for RP4 transfer are also required for RSF1010 mobilization, for pilus assembly, and for the formation of the phage receptor (22). Selection of resistant cells via donor-specific phages led to the mapping of mutations in each of the essential genes specifying the Mpf components (19). The only gene of the Tra2 core region which does not encode an essential RP4 Mpf component is trbK (19, 22). TrbK functions in entry exclusion (20).
The RP4 Mpf system is thought to encode a membrane-associated structure that is responsible for establishment of the intimate cell-to-cell contact between the donor and the recipient and also serves as a receptor complex for donor-specific phages like PRD1. Mpf components were proposed to form a pore or channel, thereby facilitating the transfer of DNA through the membranes into the recipient cell (39). The Mpf gene products exhibit typical features of bacterial membrane proteins, such as signal sequences and membrane-spanning regions, suggesting that most, if not all, of these proteins are membrane associated (37). Several proteins of the Ti VirB operon and the B. pertussis Ptl operon, homologues of the RP4 Tra2 region, were localized to the respective cell envelopes (7, 15, 29, 56, 57, 60, 61, 67). However, little is known about the architecture of the RP4-encoded or other, related transfer complexes. In addition, the release of the single-stranded DNA molecule from the relaxosome, initiating the DNA transport through the membrane, seems to require a special trigger signal raised upon contact with a potential recipient cell (22, 42). Therefore, a close interaction of the nucleoprotein complex with the Mpf system and/or the host cell membrane can be assumed. Here we describe the localization of essential RP4 transfer gene products to subcellular fractions of Escherichia coli. Our data contribute to a better understanding of the structure and function of the RP4 DNA transfer apparatus and the interplay of the individual components of this protein complex.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strain SCS1 (recA1 endA1 gyrA96 thi-1 hsdR17[rk− mk+] supE44 relA1), a DH1 derivative (23), was used as the host for plasmids. Cells were grown in YT medium (40) buffered with 25 mM 3-(N-morpholino)propanesulfonic acid (sodium salt; pH 8.0) and supplemented with 0.1% glucose and 25 μg of thiamine hydrochloride/ml. When appropriate, antibiotics were added as follows: ampicillin (sodium salt), 100 μg/ml; chloramphenicol, 10 μg/ml; and/or kanamycin sulfate, 10 μg/ml. The plasmids for expression of RP4 transfer proteins are listed in Table 1. Strains used in cell fractionation experiments are listed in Table 2.
TABLE 1.
Plasmids used for expression and purification of RP4 transfer gene products
| Gene product | Plasmid | Descriptiona | Reference |
|---|---|---|---|
| TrbF | pDB220 | pMS470Δ8Δ[NdeI-HindIII]Ω[RP4 23,100–23,861]bc | 22 |
| TrbG | pJH23 | pMS470Δ8Δ[NdeI-HindIII]Ω[RP4 23,870–24,774]bc | 22 |
| TrbH | pJH24 | pMS470Δ8Δ[NdeI-HindIII]Ω[RP4 24,767–25,285]bc | 22 |
| Trx-TrbL | pJH54 | pMS125Ω[pTrxFus, 2.724–3.080 kb, RsrII 6×-His tag]b | This work |
| TraF | pWP471 | pJF119EHΩ[RP4 NspI-HaeII, 45,909–46,577]bc | 68 |
All expression plasmids contain the pMB1 replicon and encode resistance to ampicillin as the selective marker.
Cloning and expression vector plasmids pTrxFus (35), pGZ161-3 (42, 71), pMS470Δ8 (3), and pJF119EH (16) were described previously. Plasmids pDB20 and pMS125 were described elsewhere (22).
RP4 sequence coordinates of the inserted fragment are according to published sequence data (44).
TABLE 2.
Strains used for cell fractionation experiments
| Plasmid(s) in host strain SCS1 | Description | Reference(s) |
|---|---|---|
| None | Control | |
| pML123 | Tra2 regiona | 38 |
| pML123, pWP471 | Mpf systemb | 38, 68 |
| pML123, pML100 | Tra2, traF+ traG+ | 38 |
| pML123, pVWDG23110Δ0.2 | Tra2 and Tra1 regions | 22, 38 |
| pVWDG23110Δ0.2 | Tra1 regionc | 22 |
| pMS54 | trbB+ | 42 |
| pDB179 | trbB+ trbC+ | 2, 22 |
| pKW21Δ1 | trbE+ | 22 |
| pDB220 | trbF+ | 22 |
| pJH23 | trbG+ | 22 |
| pJH24 | trbH+ | 22 |
| pDB25 | trbI+ | 22 |
| pDB260 | trbJ+ | 22 |
| pDB27 | trbK+ | 20, 22 |
| pMS125 | trbL+ | 22 |
| pWP471 | traF+ | 68 |
DNA techniques.
Standard molecular cloning techniques were performed in accordance with procedures of Sambrook et al. (52).
Protein techniques.
Isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent expression of proteins was performed as described previously (38). Proteins were analyzed by electrophoresis, using standard sodium dodecyl sulfate (SDS)-polyacrylamide gels (34) or Tricine-SDS polyacrylamide gels (53). Solid-phase immunoassays were carried out as described before (36, 62), except that proteins were transferred to polyvinylidine difluoride (PVDF) membranes (Millipore or Amersham). Cross-reactions were visualized and quantified by using a FluorImager 575 and the software package ImageQuaNT (version 4.1b) (both from Molecular Dynamics).
Protein purification for preparation of specific antisera.
In this study, proteins TrbF, TrbG, TrbH, and TraF and fusion proteins TraL-TrbD, Trx-TrbD, and Trx-TrbL were purified in a denatured state and used to raise specific antibodies in rabbits. Cultures (1.2 liters each) of SCS1 cells carrying the appropriate expression plasmids (for descriptions of plasmids, see Table 1) were grown at 37°C to an A600 of 0.5 to 0.8. IPTG was added to a final concentration of 1 mM, and shaking was continued for 4 to 5 h. Cells were harvested by centrifugation and subsequently used to partially purify their respective proteins as follows. TraL-TrbD was solubilized and denatured by lysing the cells with SDS cracking buffer (15% [wt/vol] glycerol, 5% [wt/vol] SDS, 1 M 2-mercaptoethanol, 100 mM Tris-HCl [pH 6.9]). The fusion protein was purified by preparative SDS-polyacrylamide gel electrophoresis (PAGE) as described below. For purification of Trx-TrbD, TrbF, TrbG, TrbH, Trx-TrbL, and TraF, the corresponding cells were resuspended in 25 ml of spermidine solution (100 mM spermidine, 200 mM NaCl, 2 mM EDTA) and frozen in liquid nitrogen. The cells were thawed, and 15 ml of 10% (wt/vol) sucrose (in 100 mM Tris-HCl, pH 7.6), 0.5 ml of lysozyme (50 mg/ml), 1 ml of 10% (wt/vol) Brij 58, 20 ml of 5% (wt/vol) sucrose (in 50 mM Tris-HCl [pH 7.6]–100 mM NaCl), and 15 ml of 5 M NaCl were added; the resultant suspension was kept on ice for 60 min and then centrifuged at 100,000 × g for 90 min. With the exception of Trx-TrbL, which was partially present in the resulting supernatant, the proteins were found in the sediment. Urea buffer (20 mM Tris-HCl [pH 7.6], 6 M urea, 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA) was added to these sediments, the samples were centrifuged as described above, and the respective proteins were subsequently solubilized and purified by the use of individual protocols for Trx-TrbD, TrbF, TrbG, TrbH, and TraF as described below. (i) The TrbG-containing urea supernatant was saturated with (NH4)2SO4 to 60%, stirred for 2 h, and then kept overnight at 4°C. The precipitated protein was collected by centrifugation and resuspended in urea buffer. (ii) Following two urea extraction steps, proteins TrbF and TrbH remained insoluble in the sediment and were solubilized by resuspending the pellet in SDS cracking buffer. (iii) The urea-solubilized (His)6-tag-containing Trx fusion protein Trx-TrbD and solubilized Trx-TrbL from the initial lysis step (see above) were enriched by chromatography on an Ni2+-nitrilotriacetate–agarose column (Qiagen) (25, 26) in accordance with the manufacturer's instructions. Trx-TrbD was further subjected to hydroxylapatite chromatography and eluted from the column with 100 to 150 mM sodium phosphate buffer. (iv) After the urea extraction step, TraF in the sediment was solubilized in guanidine buffer (20 mM Tris-HCl [pH 7.6], 3 M guanidine hydrochloride, 100 mM NaCl, 1 mM EDTA). Following dialysis against urea buffer containing 10% glycerol, TraF remained partially soluble and was subsequently separated from most contaminating proteins by hydroxylapatite chromatography. Preparative SDS-PAGE was used as the final step for purification of TraL-TrbD, TrbF, TrbG, TrbH, and Trx-TrbL. Protein solutions from previous purification steps (see above) were applied to polyacrylamide gels (12 to 17% acrylamide), and their respective protein bands were recovered by using a model 491 Prep Cell apparatus (Bio-Rad) in accordance with the manufacturer's instruction. Antiserum against maltose binding protein (MBP) was purchased from Sigma. Antisera against leader peptidase (LPase) and OmpA were a generous gift of Bruce L. Geller.
Cell fractionation. (i) Isolation of membranes.
The separation of cytoplasmic (CM) and outer (OM) membrane fractions was based on isopycnic sucrose density gradient centrifugation of the total membrane fraction (41). A 0.5-liter culture of SCS1 cells containing the appropriate plasmid was grown to an A600 of 0.8 to 1.0. Cells were harvested by centrifugation, resuspended in 20 ml of 10 mM Tris-HCl (pH 7.4)–1 mM MgCl2, and kept on ice during further treatments. RNase A and micrococcal nuclease were added, and the cells were broken by three passages through a precooled French pressure cell at 8,000 lb/in2 (55). The disrupted cells were treated with lysozyme (0.1 mg per ml of cell suspension; Boehringer Mannheim) for 30 min on ice. After the intact cells were removed by centrifugation, KCl was added to the supernatant to a final concentration of 0.2 M (11) and the membranes were separated from the soluble fraction by centrifugation at 100,000 × g for 2 h. The supernatant, representing the soluble fraction (cytoplasm and periplasm), was stored at −20°C. The membrane-containing pellet was resuspended in 2.5 ml of 10 mM Tris-HCl, pH 7.4. A 1-ml aliquot of this suspension was layered on top of a discontinuous sucrose density gradient consisting of 1.5 ml of 55% (wt/wt) sucrose and 2 ml each of 50, 45, 40, 35, and 30% (wt/wt) sucrose in 10 mM Tris-HCl, pH 7.4. The gradients were centrifuged for 16 h at 35,000 rpm and 4°C in a Beckman SW41 rotor.
For density flotation of the membranes, cells were grown to an A600 of 0.5 as described above, harvested by centrifugation, and resuspended in 10 mM Tris-HCl, pH 7.4. The cell suspension was disrupted by two passages through a French pressure cell. Unbroken cells were removed by centrifugation, and the total membrane fraction was collected from the supernatant by centrifugation at 100,000 × g for 2 or 10 h. The membrane pellet was resuspended in 55% (wt/wt) sucrose in 10 mM Tris-HCl (pH 7.4)–5 mM EDTA. Two density flotation gradients were used. A 2-ml sample was layered on a sucrose cushion (0.5 ml of 60% [wt/wt] sucrose) and overlaid either with 3 ml of 50%, 3 ml of 45%, 2 ml of 40%, 1 ml of 35%, and 0.5 ml of 30% (wt/wt) sucrose or with 1.5 ml of 55%, 3 ml of 50%, 2 ml of 45%, 2 ml of 40%, and 1 ml of 35% (wt/wt) sucrose in 10 mM Tris-HCl (pH 7.4)–5 mM EDTA. Centrifugation was performed in a Beckman SW41 rotor at 35,000 rpm and 4°C for 72 h. Fractions (0.5 ml) were collected starting from the top by using a Piston Gradient Fractionator (BioComp Instruments, Inc., Fredericton, New Brunswick, Canada). The refractive index and A280 nm of the gradient fractions were measured.
(ii) Release of periplasmic proteins.
SCS1 cells containing appropriate plasmids were grown as described above. Cells were collected by centrifugation and resuspended in cold 10 mM Tris-HCl (pH 7.8)–0.75 M sucrose. The cell suspension was stirred slowly at 0°C for 10 min, after which lysozyme was added to the final concentration of 160 μg/ml. Spheroplast formation was achieved by addition of cold EDTA to a final concentration of 1 mM. A sample was taken from which to recover the released periplasmic proteins and the detached OM. Spheroplasts and intact cells were removed by centrifugation, and the OM was separated from the periplasmic proteins by ultracentrifugation (100,000 × g for 3 h). The membrane pellet was resuspended in 10 mM Tris-HCl, pH 7.4.
An aliquot of the spheroplast preparation was subjected to sonication to release the cytoplasmic proteins. Unbroken cells were removed by centrifugation. The total membrane fraction (CM and OM) was collected from the supernatant by centrifugation as described above, and the membrane pellet was resuspended in 10 mM Tris-HCl, pH 7.4. The supernatant contained both periplasmic and cytoplasmic proteins. Cell fractions were analyzed by SDS-PAGE and solid-phase immunoassay as described above.
Electron microscopy.
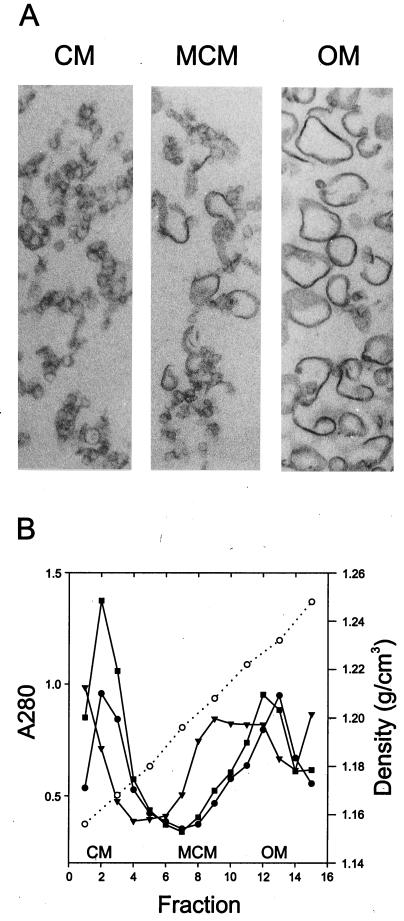
Isolation of membrane vesicles from appropriate cells was attained by performing a flotation experiment as described above. Light-scattering fractions from the sucrose gradients (representing CM, OM, and an additional membrane fraction, termed MCM [for Mpf-containing membranes] [see Fig. 3]) were collected, and membrane vesicles were fixed with 3% glutaraldehyde at 24°C for 20 min. The diluted and fixed membranes were collected by centrifugation at 60,000 × g for 15 h and processed for thin-section transmission electron microscopy as previously described (4). Micrographs were taken with a Jeol 1200 EX electron microscope operating at 60 kV.
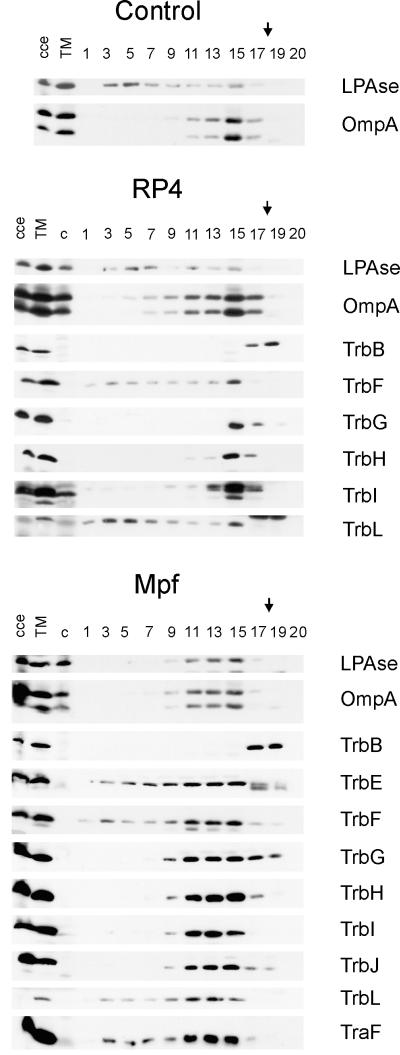
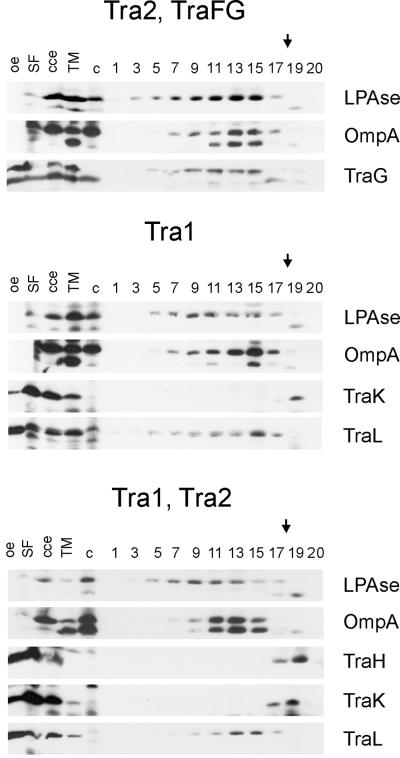
FIG. 3.
Immunological detection of RP4 Mpf components in the membrane fractions. Flotation of the total membranes in density gradients resulted in the separation of the CM and the OM of the control SCS1 strain. An additional, an intermediate light-scattering zone (MCM) was detected in cells containing either RP4 or the Mpf genes (plasmids pML123 and pWP471). Aliquots of the fractions were analyzed in standard SDS-polyacrylamide gels, and the proteins were transferred to a PVDF membrane. Incubation with specific antisera identified the Mpf components (listed on the right) in different cell fractions. cce, crude cell extract; TM, total membranes; c, crude cell extract of the control SCS1 strain; 1 to 20, fractions collected starting from the top of the gradient. The arrows indicate the sample application position.
RESULTS
RP4 transfer proteins contain potential membrane-spanning and membrane-associating structural domains.
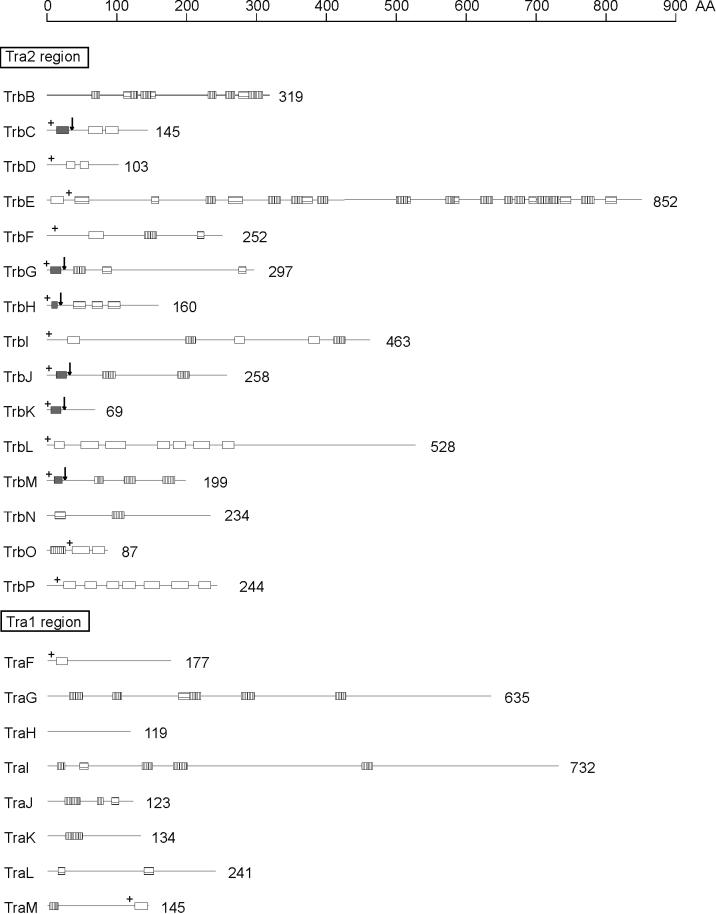
To improve our understanding of the RP4 transfer apparatus, we performed a systematic sequence search of RP4 proteins for potential secondary-structure elements, which are typical components of membrane proteins. Signal sequences for Sec-dependent protein translocation across the cytoplasmic membrane have been previously predicted for six gene products of the Tra2 region, TrbC, TrbG, TrbH, TrbJ, TrbK, and TrbM (Fig. 1) (37). Two of these proteins, TrbH and TrbK, contain signal peptides that are typical components of bacterial lipoproteins. The lipid modification of TrbK was verified in a previous study (20). In addition, the signal peptidase cleavage sites within TrbC (21), TrbG, and TrbJ (22) were determined experimentally. TrbC has been identified as the precursor of pilin. Pilin is a cyclic polypeptide (13).
FIG. 1.
Schematic representation of potential membrane-spanning or membrane-associating segments within RP4 transfer proteins. Polypeptide chains of proteins encoded by RP4 transfer regions are shown as lines corresponding to the scale at the top of the figure. The number of amino acid (AA) residues deduced from the nucleotide sequence (44) is given for each polypeptide. Signal sequences were predicted according to the method of von Heijne (64) and are shown as dark-gray bars. Arrows indicate the predicted cleavage sites. Potential transmembrane helices (white bars) were found by the PHDhtm and the PHDtopology programs (European Molecular Biology Laboratory [EMBL], Heidelberg, Germany [47, 48]). Additional stretches of at least 10 consecutive hydrophobic amino acid residues (horizontally striped bars) were identified by the hydropathy plot method of Kyte and Doolittle (33). Potential amphiphilic helical segments (vertically striped bars) were determined by using the secondary-structure prediction program PHDsec (EMBL) (49–51) in combination with the helical-wheel projection method of Schiffer and Edmundson (54). A surplus of positively charged amino acid residues, which is important for the topology of membrane proteins according to the positive-inside rule (65), is indicated by a plus sign.
Integral membrane proteins often insert into the lipid bilayer by means of one (bitopic) or more (polytopic) membrane-spanning α-helices (classification is according to the system described in reference 8). These normally consist of 18 to 24 predominantly hydrophobic amino acid residues. Potential transmembrane helices (TMH) were identified in several of the Tra2, but also in Tra1, gene products (Fig. 1). TrbE, TrbF, TraF, and TraM each contain only one TMH, which probably serves as a membrane anchor. According to the positive-inside rule of von Heijne (65), the topology of TrbF, TraF, and TraM is such that their N termini face the cytoplasm. Interestingly, in contrast to the other two proteins, the potential membrane anchor of TraM is at the C terminus; thus, the N-terminal major part of the protein would remain in the cytoplasm. Due to the presence of a number of positively charged amino acid residues at the C-terminal side of the single TMH of TrbE, the polypeptide probably inserts into the membrane in a head-first orientation, leaving the bulk of the protein in the cytoplasm (see also Fig. 7A). For TrbC, TrbD, TrbI, TrbL, TrbO, and TrbP, two or more transmembrane helices were predicted (Fig. 1), suggesting that the polypeptide chains of these gene products span the membrane multiple times.
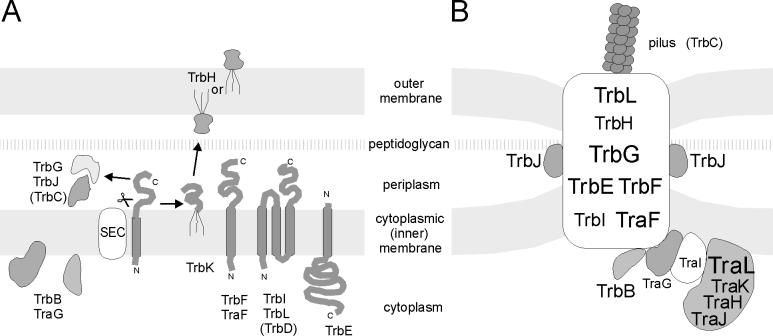
FIG. 7.
RP4 DNA transfer apparatus assembly. (A) Schematic representation of individual RP4 transfer protein insertion into, association with, or translocation across the E. coli membrane. N, N terminus; C, C terminus, SEC, general secretion pathway. (B) Model of the membrane-spanning and membrane-associated DNA transfer apparatus. The size of the lettering provides a rough estimate of the relative number of molecules per Mpf complex. The spatial arrangement of TrbE, -F, -G, -I, and -L, and TraF within the complex is hypothetical. For details, see the text.
Additional hydrophobic stretches of at least 10 amino acid residues, which may contribute to the interaction of the protein with the cell membranes, are present in a large number of RP4 transfer gene products (Fig. 1). Association of proteins with the membranes can also be facilitated directly by amphiphilic helical domains (28, 45) or indirectly by binding to integral membrane proteins. Potential α-helices of amphiphilic character were found in several RP4 transfer proteins (Fig. 1). Some of these segments may play a role in membrane association of the respective protein.
Localization of RP4 transfer proteins in E. coli cell fractions.
To obtain data supporting the hypothesis of an envelope-traversing DNA transfer apparatus, we determined the membrane localization of RP4 gene products, which are involved in conjugation. Because these proteins are minor components of a cell, the initial method chosen was the immunological identification of RP4 proteins in E. coli cell fractions. The generation and characterization of specific antisera against a number of RP4 proteins have been described previously (Table 3). Additional antisera were raised against two TrbD fusions, TrbF, TrbG, TrbH, a TrbL fusion, and TraF, which were purified as described in Materials and Methods. The antisera obtained specifically recognized the expected gene product(s) in extracts of cells that expressed the protein in the original gene arrangement (data not shown but summarized in Table 3). The two antisera against TrbD (TraL-TrbD and Trx-TrbD fusions) both recognized the respective antigen. The antiserum against Trx-TrbD also reacted with the TraL-TrbD fusion protein, suggesting specificity for the TrbD moiety. However, no specific cross-reaction was observed with extracts from trbD-containing SCS1 cells (Table 3), suggesting that the specificity and/or sensitivity of the antisera was insufficient.
TABLE 3.
Antisera used for detection of RP4 transfer gene products
| Antigena | Reactivity of antiserum | Reference(s)b |
|---|---|---|
| TrbB (n) | TrbB+ | 2 |
| TraL-TrbC (d) | TraL-TrbC+, TraL+, TrbC+ | 2, 22 |
| TrbE (d) | TrbE+ | 22 |
| TrbF (d) | TrbF+ | This work |
| TrbG/TrbGc (d) | TrbG+, TrbG*+ | This work |
| TrbH (d) | TrbH+, TrbH*+ | This work |
| TrbI (d) | TrbI+ | 22 |
| TrbJc (n) | TrbJ+, TrbJ*+ | 38 |
| TrbKc (d) | TrbK+, TrbK*+ | 20 |
| Trx-TrbL (d) | Trx-TrbL+, TrbL+ | This work |
| TraF (d) | TraF+ | This work |
| TraG (n) | TraG+ | 42 |
| TraH (n) | TraH+ | 42 |
| TraL-TraI (n) | TraL-TraI+, TraI+ | 42 |
| TraJ (n) | TraJ+ | 42 |
| TraK (n) | TraK+ | 42 |
| TraL (n) | TraL+ | 42 |
Purified proteins were used as antigens in their native (n) or denatured (d) state.
For a description of strains used for expression and purification of respective proteins in this work, see Table 1.
N-terminally processed gene product.
Essential transfer components of the Mpf and DNA transfer and replication (Dtr) system, have been previously determined, and the two separate plasmid regions, Tra1 and Tra2, encoding these functions have been cloned into compatible vector plasmids (36, 38). Also, the individual genes have been cloned separately in expression vectors. In these constructs, expression is controlled by the IPTG-inducible Ptac promoter. Because the RP4-encoded Tra components constitute only a minority of the cellular proteins, all of our studies were carried out with uninduced cells. We also approximated the amounts of proteins produced, in comparison to RP4-containing cells, by Western blotting. In the case of Mpf complex proteins produced from plasmids pML123 (TrbB, -C, -D, -E, -F, -G, -H, -I, -J, and -L) and pWP471 (TraF), 5 to 10 times more Mpf proteins were produced than for RP4 cells. This led to an approximately fourfold increase in the number of active Mpf complexes (activity was determined by plasmid transfer [36], phage propagation [37], and phage receptor saturation experiments [10, 31]) with no detectable loss in cell viability. A comparable increase was detected when individual Tra proteins were produced. Again, no adverse effects on cell viability were detected. This indicates that the results obtained were not the result of a major blockage of membrane protein translocation pathways. To determine whether the localization of certain RP4 transfer proteins requires the presence of other RP4-encoded gene products, we analyzed cells expressing different portions of the RP4 Tra system. Thus, the host strain SCS1 contained the Tra1 or the Tra2 region alone; the entire Mpf system, consisting of the Tra2 region and TraF; or all of the essential transfer functions, i.e., the Tra1 region plus the Tra2 region (Table 2). In addition, the localization of the Mpf components was determined in strains in which the respective gene products were expressed independently of any other RP4 protein (Table 2). Strain SCS1, containing no plasmid, served as a control.
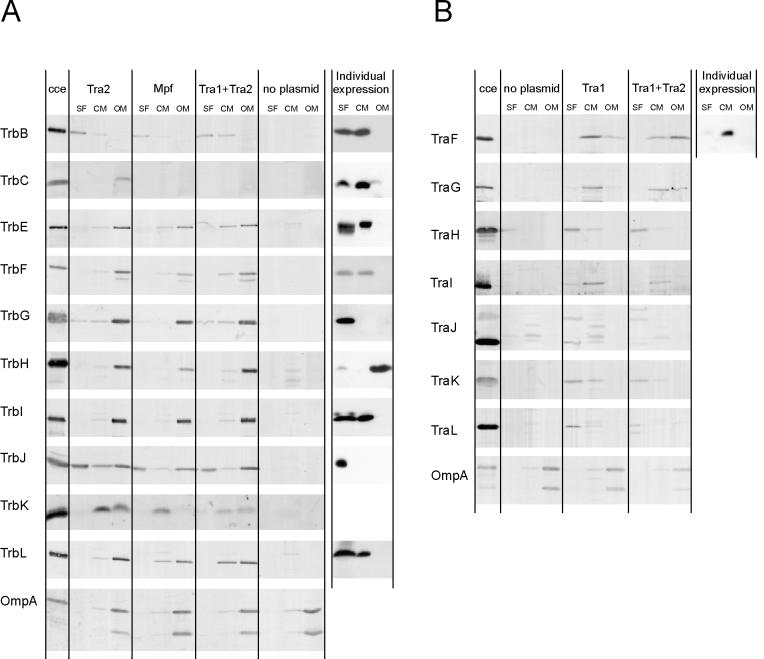
Disrupted cells were separated into three compartments, the soluble fraction, CM (inner membrane), and OM, as described in Materials and Methods. Isopycnic sucrose gradient centrifugation of the total membrane fraction resulted in two major peaks, with buoyant densities of 1.15 and 1.22 g/cm3, typical values for CM and OM, respectively. Sufficient separation of the CM and OM fractions was demonstrated by determining the SDS-PAGE protein pattern of the collected fractions (data not shown) and by immunological detection of the OM reference protein, OmpA (Fig. 2). The presence of the RP4 transfer proteins in the soluble, CM, and OM fractions was determined by solid-phase immunoassay with specific antisera.
FIG. 2.
Immunological detection of RP4 transfer proteins in E. coli cell fractions. SCS1 cells containing the appropriate plasmids were fractionated into soluble (SF), CM, and OM fractions as described in Materials and Methods. Plasmids in the host strain encoding the indicated transfer functions or individual gene products are listed in Table 2. Proteins in the soluble and the membrane fractions were separated on standard SDS-polyacrylamide gels and were transferred to PVDF membranes, which were subsequently incubated with the appropriate antiserum specific for Tra2 (A) and Tra1 (B) region gene products (Table 3). To the lanes on the left side of each panel were applied appropriate amounts of crude cell extract (cce) from the respective expression strains.
The RP4 Tra2 proteins are thought to be involved in the formation of a structure for transport through the E. coli cell envelope and to facilitate contact between the donor and recipient cells. As expected, most of the 11 essential Mpf proteins were found in the membrane fractions (Fig. 2A). In the expression strains producing individual proteins, all transfer components containing predicted α-helical transmembrane segments (TrbC, -E, -F, -I, and -L) were located in the soluble and CM fractions. The presence of these proteins in the soluble fraction was due to cross-contamination of the soluble fraction with small CM vesicles (data not shown), which are difficult to remove from that fraction under the centrifugation conditions used. Three proteins, TrbB, -G, and -J, were found in the soluble fraction (containing soluble cytoplasmic and periplasmic proteins) (Fig. 2A). TrbB was also detected in the CM fraction. The only protein associated with OM alone was TrbH. When the entire Tra2 region, the Mpf system, or all essential RP4 transfer functions were present in the cell, all Mpf components except TrbB were found in the OM fraction (Fig. 2A). TrbB was found in the soluble and CM fractions. TraF, the only Tra1 region gene product belonging to the Mpf system, was found associated with the CM fraction in cells deficient for other transfer genes but was shifted to the OM fraction in the presence of the Tra2 region (Fig. 2B).
RP4 Mpf complex forms an envelope structure connecting the CM and OM.
In equilibrium centrifugation gradients, all Tra2 proteins were found to be associated with the OM zone when Mpf-containing cells were used, even though most of these proteins were predicted to localize to the CM on the basis of their sequence characteristics and they were shown to localize to soluble and CM fractions when individual proteins were studied. This suggests that some Mpf components affect the localization of others and that this multicomponent structure links the CM and the OM. It has been reported that when cell envelopes of E. coli and Salmonella typhimurium are fractionated by flotation density gradient centrifugation, membrane fractions in addition to the major inner membrane and OM can be isolated (27). A fraction of somewhat lower density than the OM is likely to include the CM- and OM-derived adhesion zones, originally described by Bayer (6). To determine whether an additional membranous fraction in cells containing either RP4 or plasmids encoding the Mpf system could be separated, we subjected the total membrane preparation to further analysis by density flotation. In flotation gradients, membrane components float to reach their isopycnic positions while denser components remain at the site of sample application. The shape of the gradient in the flotation experiment was optimized to separate the CM and the OM or the OM and the sample application position.
In the SCS1 control strain, two major light-scattering zones were detected, at sucrose densities of 1.16 and 1.23 g/cm3 (see Fig. 4). The identities of the membranous structures in the separated fractions were determined by immunological detection of the inner membrane and OM reference proteins LPase (signal peptidase I) and OmpA, respectively (Fig. 3). Separation of the cytoplasmic and OM fractions was also verified by thin-section electron microscopy (Fig. 4A) and by determining the protein content of the gradient fractions by SDS-PAGE (data not shown). In the control (no plasmid) cells, the cytoplasmic and OM reference proteins were found in vesicles floating at densities of about 1.16 and 1.23 g/cm3, respectively (Fig. 3 and 4B). Also, the protein concentration pattern across the gradient fractions showed two major peaks, with buoyant densities of 1.16 and 1.23 g/cm3, respectively (Fig. 4B).
FIG. 4.
(A) Thin-section electron microscopy of membrane vesicles. Separation of the CM and the OM was attained by flotation gradient centrifugation. The appearance of the CM and OM preparations was the same in all analyzed strains—SCS1, SCS1 (RP4), and SCS1 (pML123, pWP471)—and that of the SCS1 control strain is shown. The intermediate band (marked MCM), shown for RP4 cells, contains material of both CM and OM origin. (B) The density profile (open circles) and the protein content (closed symbols) of the gradient fractions are shown. Closed circles, SCS1 closed squares, SCS1 (RP4); closed triangles, SCS1(pML123, pWP471).
In RP4- and Mpf-containing cells, in addition to the major CM and OM zones, a third light-scattering zone, designated the MCM, was detected at a density of about 1.21 g/cm3. There was a shift of the membranous material from CM to this density, as supported by the following observations: (i) the intensity of the CM zone decreased concomitantly with the increase in intensity of the MCM zone; (ii) the CM marker protein, LPase, moves to the MCM fraction (Fig. 3); and (iii) the MCM fraction contains morphologically CM-type vesicles (Fig. 4A). This effect was more substantial in gradients in which Mpf-containing cells were analyzed than in the corresponding fraction in experiments in which RP4-containing cells were used. This fraction also contained OM material, as shown by the presence of the OM marker protein, OmpA (Fig. 3), and by the morphology of the material in this fraction (Fig. 4A).
Since the light-scattering zone at a density of 1.21 g/cm3 was present only in cells containing RP4 transfer proteins, and not in control cells, we next determined whether the Mpf components were associated with vesicles floating at this density. In Mpf-containing cells, all components except TrbB localized in the MCM fraction but were also found in the OM fraction (Fig. 3). TrbB behaved like a soluble protein and remained at the site of sample application. In RP4-containing cells, in which the number of transfer complexes is smaller than that of Mpf cells (10), Mpf components were detected mainly in the OM fraction.
TrbJ is a periplasmic component of the transmembrane Mpf complex.
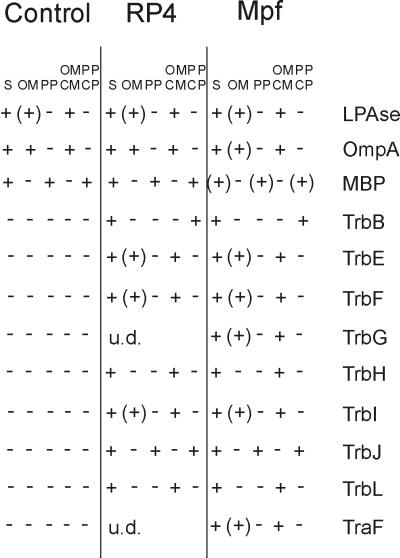
For specific release of periplasmic proteins, the OM was made leaky by EDTA-lysozyme treatment. By phase-contrast microscopy, the degree of spheroplast formation was determined to be over 95%. Detached OM was separated from the periplasmic proteins by differential ultracentrifugation. An aliquot of the spheroplast preparation was also disrupted by sonication, and fractions containing the cellular membranes (CM and OM) as well as soluble cytoplasmic and periplasmic proteins were collected. The presence of Mpf proteins was determined by solid-phase immunoassay (data not shown but summarized in Fig. 5). Efficient separation of periplasmic proteins and OM was verified by immunodetection of the OM and periplasmic reference proteins, OmpA and MBP, respectively. LPase served as a CM marker.
FIG. 5.
Specific release of periplasmic proteins. Control and RP4- and Mpf-containing cells were transformed to spheroplasts (S) and subsequently separated into fractions containing periplasmic proteins (PP), OM, soluble cytoplasmic (CP) and periplasmic (PP) proteins, and total membranes (OM and CM) as described in Materials and Methods. +, Mpf components immunologically identified in the periplasmic, total membrane, and soluble fractions; (+), Mpf proteins detected in small amounts in the OM. Some of the Mpf proteins were under the level of detection (u.d.) in RP4-containing cells.
Localization of the Mpf proteins in cells containing RP4 or the Mpf system is summarized in Fig. 5. Only one of the Mpf proteins, TrbJ, was released from the cells under the conditions used and hence was detected in the periplasmic fraction and the fraction containing both soluble cytoplasmic and periplasmic proteins. TrbB was also found in the total soluble fraction, but not in the periplasmic fraction. All of the other Mpf proteins were localized in fractions containing cell membranes. Immunodetection was strongest in the fraction containing both the CM and the OM. However, small amounts were also found in the OM detached from the spheroplasts. Mpf proteins in RP4- and Mpf-containing cells localized identically. It is intriguing, though, that MBP is found in only minute amounts in the Mpf-containing-cell fractions studied.
RP4 DNA transfer and replication functions are specified by soluble and membrane-associated components.
Most of the RP4 Tra1 region gene products specify DNA transfer and replication functions and are thus suspected to be soluble cytoplasmic proteins, some of which might be associated with the cell envelope. Immunological identification of all essential Tra1-encoded transfer components in E. coli cell fractions is shown in Fig. 2B. Tra1 gene products TraG and TraI were clearly localized to the CM fraction (Fig. 2B). When all RP4 transfer functions are expressed in the cell, TraG can also be found in the OM fraction. Relaxosomal components TraH, TraJ, and TraK were predominantly found in the soluble fraction. However, minor portions of TraH and TraK were also detectable in the CM fraction (Fig. 2B). In addition, TraL was detected predominantly in the soluble fraction but, to a small degree, also in the CM and, in the presence of the Tra2 region, even in the OM fraction (Fig. 2B).
The membrane association of TraG, TraH, TraK, and TraL in SCS1 cells containing Tra1 or both the Tra1 and Tra2 regions cloned in separate replicons was further analyzed by flotation density gradient centrifugation (Fig. 6). TraG was clearly detected in the OM fraction as well as in the intermediate band (MCM) (Fig. 3) of cells containing all Mpf components and TraG. Also, TraL associated with the OM fraction irregardless of the presence of the Tra2 region in the cells. TraH and TraK behaved as soluble proteins; TraH was detected only in the soluble fraction (as shown for cells containing Tra1 and Tra2). A fraction of TraK sedimented with total membranes but remained at the position of sample application.
FIG. 6.
Immunological detection of RP4 DNA transfer and replication components in the membrane fractions. Plasmids in the SCS1 host strain encoding the indicated transfer functions or individual gene products are listed in Table 2. Flotation of the total membranes of cells containing the Tra2 region in density gradients resulted in the separation of the CM and the OM and, in addition, an intermediate band (MCM). Aliquots of the fractions, collected starting from the top of the gradient (fraction numbering is identical to that of Fig. 3), were analyzed in standard SDS-polyacrylamide gels, and proteins were transferred to PVDF membrane. Incubation with specific antisera identified the Tra1 region components (listed on the right) in different cell fractions. oe, corresponding overexpression strain; cce, crude cell extract; TM, total membranes; c, crude cell extract of the control SCS1 strain; 1 to 20, fractions collected. The arrows indicate the sample application position.
DISCUSSION
The hypothesis that a membrane-spanning channel or pore is formed by plasmid-encoded transfer functions of the donor cell mediating protein and DNA transport is popular. Until now, the hypothesis of channel-mediated DNA transfer of IncP plasmids has rested mainly on genetic data. Some evidence that the RP4 Mpf system indeed serves as a path through the cell envelope came from studies of the electrochemical properties of Mpf-containing cells (10). In this investigation, we showed that individual RP4 transfer proteins are translocated across, inserted into, or associated with the membranes, depending on or independent of host-encoded secretion factors, as illustrated in Fig. 7A, and that the entire Mpf complex is associated with cell membranes.
Disruption of the Mpf-containing cells and subsequent separation of cell membranes in density flotation gradients led to the discovery of an additional membrane fraction, MCM, with a density close to that of the OM fraction. The flotation technique is a powerful means of separating soluble proteins from those associated with the membranes (as shown also for TrbB in this investigation). The MCM fraction contained all Mpf components except TrbB. Interestingly, both cytoplasmic (LPase) and OM (OmpA) marker proteins were also found in this fraction. This indicates that a major rearrangement of the membrane architecture has taken place. In the control strain, LPase and OmpA were found in the cytoplasmic and OM fractions, respectively. This strongly suggests that (i) the Mpf proteins form a complex and (ii) the complex connects the CM and OM, binding them together. Association of the CM marker with membranes of high density can also be seen in RP4-containing cell fractions. The number of transfer complexes in Mpf-containing cells is approximately fourfold higher than that in RP4-containing cells (10, 31). This is in accordance with the more distinct intermediate membrane zone observed in Mpf-containing cells.
The data from flotation experiments suggest that the Mpf complex is anchored to the CM, as discussed above. This is also supported by the finding that five Mpf components (TrbE, -F, -I, -L, and TraF) that were predicted to contain α-helical membrane-spanning regions localize to the cytoplasmic membrane (Fig. 2 and 3). The association of these transfer proteins with the CM is in accordance with the localization of corresponding proteins in related DNA and protein export systems (7, 15, 29, 57, 60, 67). Two of the Mpf components, TrbG and TrbJ, were suspected to be periplasmic proteins due to the presence of a signal peptide cleavage site (22). Also, TrbJ seems to be more loosely connected to other Mpf complex proteins, since it is released from cells during spheroplast formation. TrbG was instead tightly associated with the cell fraction containing all other Mpf components. Both proteins were soluble when expressed in the absence of other Mpf components. One of the Mpf proteins, TrbH, was shown to be associated with the OM. The TrbH protein contains a signature sequence common in lipoproteins targeted to the OM (18, 37, 70). The only transfer protein which could not be extracted from Mpf-containing cells along with other Tra2 region gene products was TrbB. Results from density flotation and spheroplast experiments suggest that it has a cytoplasmic localization. However, this does not exclude the possibility of an association of TrbB with the cytoplasmic portion of the Mpf complex.
Conjugative transfer of plasmid DNA probably begins with the triggered release of single-stranded DNA resulting from the site- and strand-specific cleavage of the plasmid DNA by relaxase within a high-precision nucleoprotein complex, the relaxosome (43). The results obtained from the localization experiments in this study clearly indicate that the RP4 relaxosome is located in the cytoplasm, in association with the CM (Fig. 7B). This association is independent of the membrane-spanning Mpf complex, since TraI and TraL localized, at least partially, to the CM in the absence of the Tra2 region proteins. In the presence of Mpf components, TraL was found even in the OM fraction. Relaxosome components TraH and TraK are soluble cytoplasmic proteins, since the portion of these proteins that sedimented with total membranes remained at the bottom of the flotation gradients. The binding of the components of the RP4 relaxosome to the CM could be direct, via the hydrophobic or amphiphilic segments which are predicted to lie within TraI and TraL (Fig. 1), or indirect, by interactions with integral or peripheral membrane proteins. A possible anchor protein is TraG, whose proposed function is to connect the two protein complexes, the Mpf system and the relaxosome (Fig. 7B). Preliminary results suggest that TraI and TraG indeed bind to each other (43). In the localization experiments, TraG was found to be associated with the CM irregardless of the presence of the Tra2 region. More remarkably, in Mpf-containing cells, TraG was found in the intermediate band, suggesting a connection with the Mpf complex.
Our attempt to estimate the number of molecules of the different RP4 transfer components per cell led to the observation that the number ranged from less than five (TraI relaxase) to several thousand (Fig. 7). The number of Mpf complexes which serve as a receptor for the donor-specific phage PRD1 was calculated to be about 25 per RP4-containing cell (10, 31). It is obvious that the pool of Mpf proteins exceeds the amount needed to form active complexes. In many biological systems, the expression level of the proteins is in line with the amount of proteins needed. In this light, it can be assumed that the major Mpf proteins (TrbB, -E, -F, -G, -H, -I, -J, and -L, and TraF) are present in multiple copies in each of the active complexes. It is also possible that there is a dynamic assembly and disassembly of the entire complex, which could partly explain the large pool of individual transfer proteins. It should be noted, however, that the number of Mpf proteins in a cell is small compared to that of major protein complexes such as OmpA (approximately 105 molecules per cell).
The central question in bacterial conjugation is how the DNA traverses the cell envelopes of the donor and recipient cells. The current model proposes that two major complexes, the relaxosome and the Mpf protein complex, interact via the TraG-like proteins, which serve as an interface between the two complexes. Which of these components is involved in DNA transport?
The observation that the Mpf proteins form a complex, together with genetic evidence and evidence from electron microscopy, demonstrate that (i) 11 Mpf components are essential for IncP pilus production in the absence of any DNA-processing function (22), (ii) the same 11 Mpf components are essential for extrusion of the receptor(s) for donor-specific phages on the cell surface (19, 22), and (iii) the same 11 plasmid-encoded components are sufficient to form conjugative junctions (A. L. Samuels, E. Lanka, and J. Davies, unpublished data). Conjugative junctions consist of electron-dense material of unknown nature between the cell envelopes of intimately closely positioned donor and recipient cells, generally called mating aggregates (12). The properties mentioned are certainly consistent with role for the Mpf complex in protein transport and protein-protein interaction(s). However, whether the proposed multifunctional Mpf protein complex in addition directly interacts with DNA, providing the pathway for DNA from the donor to enter the recipient cell, remains an open and tempting question. So far there has been no experimental evidence supporting the hypothesis that Mpf interacts with DNA.
Other transport systems which are phylogenetically related to conjugative Mpf systems, like the pertussis toxin secretion system, appear to function in protein transport only (42, 59). Since these systems lack TraG-like proteins, the TraG analogs of conjugative systems might play a key role in DNA transport.
ACKNOWLEDGEMENTS
We are grateful to Hans Lehrach for generous support. The expert technical assistance of Marianne Schlicht is greatly appreciated. The electron microscopy was performed at the EM Unit, Institute of Biotechnology, University of Helsinki, and Arja Strandell is acknowledged for assistance. We are indepted to Beth Traxler and Tony Pugsley for useful discussions and critical readings of the manuscript.
This work was financially supported by Sonderforschungsbereich grant 344/A8 of the Deutsche Forschungsgemeinschaft to E.L. and by the Academy of Finland (grants 41400 [to A.M.G.] and 37725 [to D.H.B.]).
REFERENCES
- 1.Alt-Mörbe J, Stryker J L, Fuqua C, Li P-L, Farrand S K, Winans S C. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmid is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzer D. Lokalisierung, Nukleotidsequenz und Genorganisation von IncP-Transferregionen. Berlin, Germany: Freie Universität Berlin; 1993. [Google Scholar]
- 3.Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992;20:1851–C858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford D H, Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi 6. Virology. 1980;107:222–228. doi: 10.1016/0042-6822(80)90287-1. [DOI] [PubMed] [Google Scholar]
- 5.Bates S, Cashmore A M, Wilkins B M. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the Tra2 mating system. J Bacteriol. 1998;180:6538–6543. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer M E. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- 7.Beijersbergen A, Smith S J, Hooykaas P J J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994;32:212–218. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- 8.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabezón E, Sastre J I, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 10.Daugelavičius R, Bamford J K H, Grahn A M, Lanka E, Bamford D H. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Maagd R A, Lugtenberg B. Fractionation of Rhizobium leguminosarum cells into outer membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J Bacteriol. 1986;167:1083–1085. doi: 10.1128/jb.167.3.1083-1085.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dürrenberger M B, Villiger W, Bächi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 13.Eisenbrandt R, Kalkum M, Lai E-M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 14.Farrand S K, Hwang I, Cook D M. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J Bacteriol. 1996;178:4233–4247. doi: 10.1128/jb.178.14.4233-4247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finberg K E, Muth T R, Young S P, Maken J B, Heitritter S M, Binns A N, Banta L M. Interactions of VirB9, -10, and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J Bacteriol. 1995;177:4881–4889. doi: 10.1128/jb.177.17.4881-4889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;43:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 17.Fürste J P, Pansegrau W, Ziegelin G, Kröger M, Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci USA. 1989;86:1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gennity J M, Inouye M. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J Biol Chem. 1991;266:16458–16464. [PubMed] [Google Scholar]
- 19.Grahn A M, Haase J, Lanka E, Bamford D H. Assembly of a functional phage PRD1 receptor depends on 11 genes of the IncP plasmid mating pair formation complex. J Bacteriol. 1997;179:4733–4740. doi: 10.1128/jb.179.15.4733-4740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase J, Kalkum M, Lanka E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncPα plasmid RP4. J Bacteriol. 1996;178:6720–6729. doi: 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Heinemann J A, Sprague G F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 25.Hochuli E, Bannwarth W, Döbeli H, Gentz R, Stüber D. Genetic approach to facilitate purification of recombinant protein with a novel metal chelate adsorbent. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 26.Hochuli E, Döbeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 27.Ishidate K, Creeger E S, Zrike J, Deb S, Glauner B, MacAlister T J, Rothfield L I. Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J Biol Chem. 1986;261:428–443. [PubMed] [Google Scholar]
- 28.Jackson M E, Pratt J M. An 18 amino acid amphiphilic helix forms the membrane-anchoring domain of the Escherichia coli penicillin-binding protein 5. Mol Microbiol. 1987;1:23–28. doi: 10.1111/j.1365-2958.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnson D J, Burns D L. Detection and subcellular localization of three Ptl proteins involved in the secretion of pertussis toxin from Bordetella pertussis. J Bacteriol. 1994;176:5350–5356. doi: 10.1128/jb.176.17.5350-5356.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 31.Kotilainen M M, Grahn A M, Bamford J K H, Bamford D H. Binding of an Escherichia coli double-stranded DNA virus PRD1 to a receptor coded by an IncP-type plasmid. J Bacteriol. 1993;175:3089–3095. doi: 10.1128/jb.175.10.3089-3095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuldau G A, De Vos G, Owen J, McCaffrey G, Zambryski P. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol Gen Genet. 1990;221:256–266. doi: 10.1007/BF00261729. [DOI] [PubMed] [Google Scholar]
- 33.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.LaVallie E R, DiBlasio E A, Kovacic S, Grant K L, Schendel P F, McCoy J M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 36.Lessl M, Balzer D, Lurz R, Waters V L, Guiney D G, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 38.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 40.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–433. [Google Scholar]
- 41.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplsmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 42.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 43.Pansegrau W, Lanka E. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPα plasmid RP4. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 44.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 45.Picot D, Patrick J L, Garavito R M. The X-ray crystal structure of membrane protein prostaglandin H2 synthase-1. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 46.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 47.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 51.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 54.Schiffer M, Edmundson A B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnaitman C A. Examination of the protein composition of the cell envelope of Escherichia coli by polyacrylamide gel electrophoresis. J Bacteriol. 1970;104:882–889. doi: 10.1128/jb.104.2.882-889.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shirasu K, Kado C I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 57.Shirasu K, Koukolikova-Nicola Z, Hohn B, Kado C I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol Microbiol. 1994;11:581–588. doi: 10.1111/j.1365-2958.1994.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 58.Stachel S E, Zambryski P C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986;47:155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- 59.Thorsted P B, Macartney D, Akhtar P, Haines A S, Ali N, Davidson P, Stafford T, Pocklington M, Pansegrau W, Wilkins B M, Lanka E, Thomas C M. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 60.Thorstenson Y R, Kuldau G A, Zambryski P C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993;175:5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorstenson Y R, Zambryski P C. The essential virulence protein VirB8 localizes to the inner membrane of Agrobacterium tumefaciens. J Bacteriol. 1994;176:1711–1717. doi: 10.1128/jb.176.6.1711-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trieu-Cuot P, Carlier C, Martin P, Courvalin P. Plasmid transfer by conjugation from Escherichia coli to gram-positive bacteria. FEMS Microbiol Lett. 1987;48:289–294. [Google Scholar]
- 64.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward J E, Akiyoshi D E, Regier D, Datta A, Gordon M P, Nester E W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988;263:5804–5814. [PubMed] [Google Scholar]
- 67.Ward J E, Jr, Dale E M, Nester E W, Binns A N. Identification of a VirB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J Bacteriol. 1990;172:5200–5210. doi: 10.1128/jb.172.9.5200-5210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waters V L, Strack B, Pansegrau W, Lanka E, Guiney D G. Mutational analysis of essential IncPα plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J Bacteriol. 1992;174:6666–6673. doi: 10.1128/jb.174.20.6666-6673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 71.Ziegelin G. Konjugativer Transfer des IncP-Plasmids RP4: die Spezifitätsdeterminanten codieren oriT-Bindungsproteine. Berlin, Germany: Freie Universität Berlin; 1989. [Google Scholar]