Abstract
Background and Aims
Plant tissue nitrogen (N) and phosphorus (P) and genome traits, such as genome size and guanine–cytosine (GC) content, scale with growth or metabolic rates and are linked to plant ecological strategy spectra. Tissue NP stoichiometry and genome traits are reported to affect plant growth, metabolic rates or ecological strategies in contrasting ways, although the elemental costs for building and maintaining DNA are typically overlooked.
Methods
We formulated stoichiometry- and ecology-based predictions on the relationship between genome size and GC content to tissue N, P and N : P and tested them on a set of 130 herbaceous species from a temperate grassland using ordinary, phylogenetic and quantile regression.
Key Results
Genome size was only negatively linked to plant N and N : P in species with very small genomes. We found no link between genome size and plant P. GC content was negatively linked to plant N and P but we found these significant links consistently in both GC-rich and GC-poor species. Finally, GC content correlated positively with plant N : P but only in species with GC-rich genomes.
Conclusions
Our results provide stronger support for the ecology-based predictions than the stoichiometry-based predictions, and for the links between GC content and plant N and P stoichiometry than for genome size. We argue that the theories of plant metabolic rates and ecological strategies (resource-acquisitive vs. conservative or ruderal vs. stress-tolerator spectra) better explain interspecific genome-NP stoichiometry relationships at the tissue level (although relatively weakly) than the stoichiometric theory based on the elemental costs for building and maintaining DNA.
Keywords: GC content, genome size, nitrogen, phosphorus, plant ecological strategies, stoichiogenomics, tissue stoichiometry
INTRODUCTION
Both nitrogen (N) and phosphorus (P) are vital bioelements and their levels in plant tissues can positively correlate with growth, photosynthetic and respiration rates (Sterner and Elser, 2002; Wright et al., 2004; Reich et al., 2006; Sardans et al., 2021). To increase metabolic and growth rates, plants need to produce large quantities of P-rich RNA and ribosomes to build N-rich photosynthetic proteins (Sterner and Elser, 2002; Hessen et al., 2010). Nevertheless, relatively high amounts of N and P can also be stored in nuclear DNA, but genome size typically correlates positively with minimum cell size (Beaulieu et al., 2008; Knight and Beaulieu, 2008; Šímová and Herben, 2012) that, in turn, correlates negatively with metabolic rates (Kozlowski et al., 2003). Therefore, we have two contrasting predictions for the relationship between N and P tissue content and plant metabolic rates that could not be resolved without knowing the relationship between genome size and tissue N and P content.
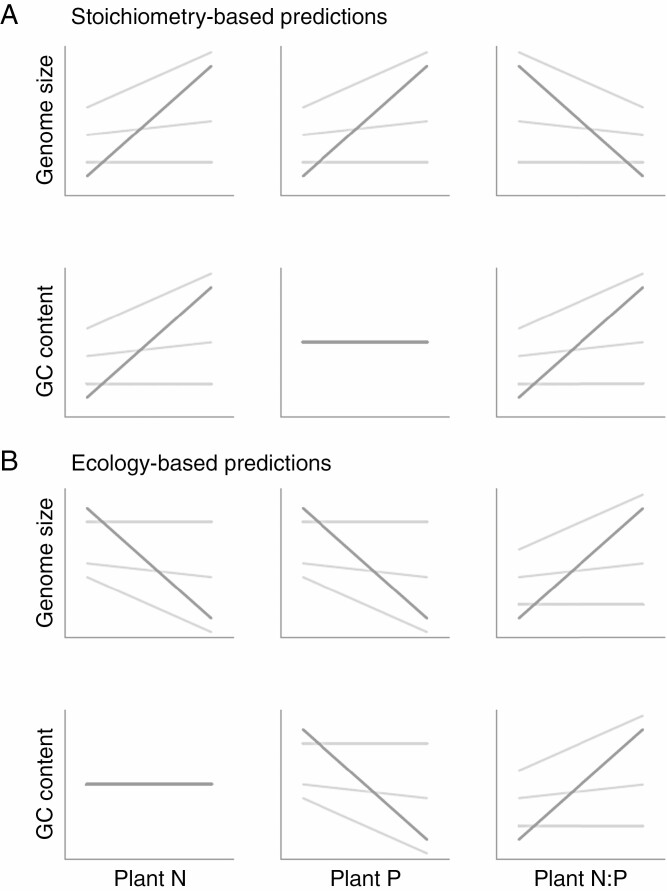
Assuming constant soil nutrient conditions, we can formulate two sets of predictions regarding the relationships between genome size and plant tissue N, P and N : P across species (Fig. 1). One follows the direct elemental costs to build and maintain DNA (the stoichiometry-based perspective, Table 1A). The second one is extrapolated from indirect links between genome traits and NP stoichiometry via ecological and metabolic frameworks, i.e. genome–cell size scaling (Beaulieu et al., 2008; Knight and Beaulieu, 2008; Šímová and Herben, 2012; Faizullah et al., 2021), the leaf economics (Wright et al., 2004; Knight and Beaulieu, 2008), photosynthetic and growth rates (Sterner and Elser, 2002; Knight and Beaulieu, 2008; Šímová and Herben, 2012; Roddy et al., 2020; Sardans et al., 2021) or stress–ruderal (Grime, 1977) spectra (the ecology-based perspective, Table 1B). The directions of the relationships between genome traits and plant tissue N, P and N : P differ for the stoichiometry- vs. ecology-based perspectives (Fig. 1).
Fig. 1.
The stoichiometry-based and ecology-based perspectives (Table 1) often suggest contradicting predictions of the relationships between plant genome size (1C or 1Cx value), guanine–cytosine (GC) content, and plant tissue nitrogen (N), phosphorus (P), and N : P ratio. Black lines indicate hypothesized trends for the central tendencies, whereas grey lines indicate hypothesized trends in lower, middle and upper quantiles. (A) DNA is an N- and P-demanding biomolecule, and thus its content (1C value) can be positively correlated with plant tissue N and P, while it can be negatively linked to plant N : P because DNA is a relatively P-rich biomolecule with low N : P. The stoichiometry-based perspective also predicts positive links of GC content to plant N and N : P, because GC pairs have more N atoms in their molecules than AT pairs. We also expected that the proposed relationships would be more pronounced in upper quantiles, i.e. in plants with large genomes and GC-rich nucleotide compositions that can contribute more to the overall plant N and P pools than small genomes or GC-poor nucleotide compositions. (B) The ecology-based perspective suggests negative links of genome size to plant N and P and a positive link to N : P. Stress-tolerant species are typically slowly growing and conservative, with larger genomes but lower N and P, and higher N : P compared to fast-growing and acquisitive ruderals. Ecological strategies are also connected to GC content as P-poor (but with high tissue N : P) stress tolerators tend to have higher GC content. The strength of the proposed relationships can vary across quantiles; for example, small genome species (lower quantiles) might allocate more P and N to RNA and proteins, respectively, to promote fast growth.
Table 1.
Possible mechanisms driving the relationships between genome size and guanine–cytosine content, and tissue N and P from (A) the stoichiometry-based and (B) ecology-based perspectives. Predictions of these perspectives are given in Fig. 1.
| Perspectives and their key ideas | Possible mechanism | Scale | References |
|---|---|---|---|
|
(A) Stoichiometry-based perspective
Predicts the links between genome traits and tissue N and P based on the N and P demands to build nucleic acids and proteins | |||
| N and P demands to build DNA | Genome size and GC content directly dictate the amount of N and P in the nucleus | Molecular | Sterner and Elser (2002), Kang et al. (2015) |
| N and P demands to maintain and repair DNA | Higher contents of histones, proteins, and ATP needed to maintain and repair large genomes | Molecular | Sterner and Elser (2002) Faizullah et al. (2021) |
| Genome → transcriptome | Large and GC-rich genomes can have large and GC-rich transcriptomes, respectively | Molecular | Elser et al. (2011), Faizullah et al. 2021 |
|
(B) Ecology-based perspective
Predicts the links between genome traits and tissue N and P through their correlations with metabolic rates and ecological strategies | |||
| Genome size → cell size | Genome size is a fundamental constraint on minimum cell size that, in turn, controls metabolic rates (correlated with tissue N and P) | Cellular | Beaulieu et al. (2008), Knight and Beaulieu (2008) |
| Genome size → stomatal guard cell size and density | Genome size positively affects guard cell size and negatively affects stomatal density. The size and density of stomata affect photosynthetic rates (correlated with tissue N and P) | Cellular, tissue, organ | Beaulieu et al. (2008), Faizullah et al. (2021) |
| Genome size → leaf cell density and mass area | Because of space constraints, genome size limits the number of cells in a tissue. Cell density in leaves affects the conductance of CO2 and water, and by extension, leaf mass per unit area and photosynthetic rate (both correlated with tissue N and P) | Organ | Wright et al. (2004), Knight and Beaulieu (2008), Roddy et al. (2020), Faizullah et al. (2021) |
| Genome size → S phase duration | DNA replication duration scales with genome size, which may affect plant growth rate (correlated with tissue N and P). | Whole plant | Sterner and Elser (2002), Šímová and Herben (2012), Sardans et al. (2021) |
| Genome traits, tissue NP → ecological strategies | The dependence of metabolic and growth rates on genome size (through its constraints on cell size), GC content (through the higher cost of synthesis for GC pairs) and NP contents (through the production of RNA and photosynthetic proteins) can determine species ecological strategy (e.g. the competitor-stress-ruderal scheme) | Whole plant | Grime (1977), Leitch et al. (2013), Guignard et al. (2019), Sardans et al. (2021) |
Considering the stoichiometry-based perspective, one can simply expect a positive relationship of genome size to plant tissue N and P across species (Fig. 1A) because large genomes store more N and P than small genomes. DNA is a biomolecule composed of 14.5 % N and 9 % P (Sterner and Elser, 2002) because nucleotides contain a phosphate group and a nitrogen base (plus a sugar group without N or P) and more N is stored in guanine–cytosine (GC) bases. Guanine–cytosine pairs in the whole genome and in all gene transcripts require one more N atom than adenine–thymine (AT) pairs (eight vs. seven, respectively), and thus relatively GC-rich plant species should need more N to build and maintain their genomes. This also implies a positive relationship of GC content to tissue N and N : P (Fig. 1A). N : P ratio and genome size should be inversely related (Fig. 1A) because DNA is relatively P-rich and has a lower N : P ratio than other biomolecules (Sterner and Elser, 2002). Assuming that large or GC-rich genomes contribute more to the overall plant N and P pools, it is possible that the proposed relationships between genome size and GC content and plant N, P and N : P might only be pronounced in species with relatively large genomes and GC-rich nucleotide compositions. Under this scenario, one could find significant links between genome traits and NP stoichiometry only in upper quantiles (Koenker and Bassett, 1978; Cade and Noon, 2003) when genome size and GC content are the response variables (Fig. 1).
The stoichiometry-based perspective builds upon the assumption that the elemental composition of nucleic acids affects the elemental composition in tissues of the entire plant. Clearly, the relationships between genome traits and N and P should be very strong at the DNA and nuclear levels. Genome size and GC content directly dictate the N and P demands of DNA, and higher amounts of N-rich histones and proteins or P-rich ATP to maintain large genomes might be needed in the nucleus as well (Faizullah et al., 2021). However, it is unclear whether these relationships scale up to tissues and organs. The mass of DNA in tissues is typically 1.5 % and rarely exceeds 3 % of the total mass (Sterner and Elser, 2002), suggesting that its contribution is marginal. However, relative to the total N and P pools in plant cells, DNA requires large amounts of N and especially P (Hessen et al., 2010) in comparison to other biomolecules, such as proteins (only for P because N contents of both proteins and DNA are relatively similar), carbohydrates or lipids (Sterner and Elser, 2002). In contrast to other biomolecules, the allocation of N and P to plant genomes remains unclear and might be cell-type-specific (Guignard et al., 2017) but can vary considerably across plant species as with genome size and GC content (Pellicer and Leitch, 2019). To our knowledge, the stoichiometry-based perspective has some direct empirical support, because Kang et al. (2015) found a positive correlation between leaf N (but not P) and genome size in a set of species from the genus Primulina.
The ecology-based perspective claims that growth and metabolic rates are negatively associated with genome size and GC content but positively associated with tissue N and P. We derived this perspective from several indirect correlations among multiple variables, which can be mechanistically linked through the effect of genome size on minimum cell size or cell density, i.e. phenotypic traits affecting species leaf economics, photosynthetic rate and, ultimately, its ecological strategy (Table 1B). Conservative and slowly growing stress-tolerators typically have larger genomes (Bennett, 1987; Hessen et al., 2010; Leitch et al., 2013; Guignard et al., 2019) and higher GC content (Šmarda et al., 2014) but lower tissue P and higher N : P (Sardans et al., 2021) than acquisitive and fast-growing ruderals. If plant growth rate is positively related to tissue P but negatively related to genome size, GC content and N : P (Bennett, 1987; Güsewell, 2004; Hessen et al., 2010; Sardans et al., 2021), we should mostly observe opposite trends in the relationships between genome traits and NP stoichiometry compared to the stoichiometry-based perspective (Fig. 1B). A negative correlation between growth rate and GC content is indirectly supported by the generally low GC content of genes that facilitate replication and growth in bacteria (Castillo and Almeida, 2021) and the higher cost of synthesis for GC pairs compared to AT pairs (Rocha and Danchin, 2002). For the leaf economics spectrum, N and P scale positively with metabolic rates and photosynthetic rates (Wright et al., 2004; Reich et al., 2006) but such processes are constrained by genome size (Roddy et al., 2020), and thus genome size should be negatively correlated to leaf N and P. Additionally, the relationship between genome traits and NP stoichiometry can be more pronounced in the lower quantiles (Fig. 1B); species with small genomes or GC-poor base compositions might disproportionally allocate more N and P to other biomolecules, such as RNA and photosynthetic proteins, to enhance growth and metabolic rates (Hessen et al., 2010; Elser et al., 2011).
To test if the proposed stoichiometry- or ecology-based perspectives best explain the relationship between genome traits and NP stoichiometry, we used the genome (1C, 1Cx values and GC content) and stoichiometric (above-ground tissue N, P and N : P) measurements of 130 herbs from a semi-natural temperate grassland. If the stoichiometry-based perspective best explains the relationship between genome traits and NP stoichiometry, we would expect mostly positive relationships between both genome traits and plant tissue N and P, a positive relationship between GC and plant N : P, and no relationship between GC and plant P (Fig. 1A). Alternatively, the ecology-based perspective would be the better explanation if there are mostly negative relationships between genome traits and NP stoichiometry, a positive relationship between genome traits and plant N : P, and no relationship between GC and plant N (Fig. 1B). In addition, we examined the central tendencies (using both non-phylogenetic and phylogenetic models) and the tendencies in lower, middle and upper quantiles (Fig. 1).
MATERIALS AND METHODS
Species sampling and plant N and P measurements
We collected plants from three localities with similar species composition (broad-leaved semi-dry grasslands with a dominance of Brachypodium pinnatum) and plant-available soil N and P levels (Supplementary Data Fig. S1) in the Beskydy and Javorníky Mountains (south-eastern part of the Czech Republic): Kýchová (49°17ʹ27.96″N, 18°7ʹ55.26″E), Losový (49°19ʹ3.82″N, 18°5ʹ39.07″E) and Půlčín (49°13ʹ38.97″N, 18°4ʹ45.95″E) in 2019 and 2020. In total, we sampled 130 species from 33 families (Bitomský et al., 2022). We collected above-ground biomass of flowering individuals from May to early July to control for seasonal changes in element contents (Zhang et al., 2013). Senescent plant tissue was removed and plant biomass was divided into stems (including flowers) and leaves, then dried at 60°C for 72 h. Because of differences in allocation of N and P to stems or leaves, we ensured that every sample had the same stem : leaf ratio (2.4 ± 0.12, most species had a stem : leaf ratio close to this value) in dry weight. Individuals for each species were pooled and species average total element concentrations of N and P was estimated in an accredited commercial laboratory then expressed as a percentage of an element in dry biomass.
Genome size and GC content measurements
In this study, the term genome size refers to holoploid genome size (1C value), i.e. the DNA content of one non-replicated chromosome set, or monoploid genome size (1Cx value), i.e. the total mass of DNA in the nucleus (2C value) divided by ploidy level. To estimate genome size and GC content (Meister and Barow, 2007), we used flow cytometry following a simplified protocol using LB01 isolation buffer (Doležel et al., 2007). Tissue samples (from the same samples used for the N and P measurements) were chopped together with the internal standard using a sharp razor blade in a Petri dish containing 2000 μL of isolation buffer supplemented with PVP-40 (Doležel and Bartoš, 2005; 20 mg mL−1). The crude suspension was filtered through a 48-μm nylon mesh and an additional 1000 μL of buffer was added. Then, the samples were split into two (1500 μL each) tubes: (1) one supplemented with RNase IIA and intercalating propidium iodide (PI, both 50 µg mL−1), and analysed using a Partec PAS instrument (Partec GmbH, Münster, Germany) equipped with a diode-pumped solid-state green laser; (2) the second stained with 4ʹ, 6-diamidino-2ʹ-phenylindole (DAPI, final concentration 4 µg mL−1) and analysed using a Partec CyFlow ML flow cytometer (Partec GmbH) equipped with a Partec UV LED kit. We used Raphanus sativus‘Saxa’(2C = 1.11 pg, GC = 40.30 %), Lycopersicum esculentum‘Stupické polní tyčkové rané’ (2C = 1.96 pg, GC = 38.72%), Glycine max‘Polanka’(2C = 2.50 pg, GC = 37.89 %), Pisum sativum‘Ctirad’(2C = 9.09 pg, GC = 41.77 %) and Vicia faba‘Inovec’(2C = 26.90 pg, GC = 41.15 %) as internal standards (Doležel et al., 1994, 1998; Šmarda et al., 2019). We recorded a fluorescence intensity of 5000 nuclei, with peaks accepted only if their coefficient of variance was below 5 %. We analysed three individuals from each species. Genome size data were compared with those of Šmarda et al. (2019) and if the differences were below 2 %, the genome size records of Šmarda et al. (2019) were accepted. Otherwise, absolute genome size was properly determined by at least three successive measurements of each plant sample following recommendations given in Doležel et al. (2007). The exact GC content values (%) were calculated according toŠmarda et al. (2008). Monoploid genome size (1Cx value) was calculated using published chromosome numbers (in most species according to Šmarda et al., 2019, but see Bitomský et al., 2022) and dividing the absolute genome size (2C) by ploidy level.
Statistical analysis
We performed all analyses in R 3.6.0 (R Core Team, 2019) using the packages geiger 2.0.6.2 (Pennell et al., 2014), phylolm 2.6 (Ho and Ané, 2014) and quantreg 5.54 (Koenker, 2019). To obtain an ultrametric phylogeny of our species, we used the package V.PhyloMaker (Jin and Qian, 2019). Genome size was ln-transformed prior to the analysis to improve its right-skewed distribution (Supplementary Data Fig. S2). Both genome size (1C and 1Cx values) and GC content were considered response variables, whereas plant N, P and N : P were considered explanatory variables. Additionally, we examined potential confounding effects of ploidy levels on plant N, P and N : P by comparing these values for diploids and polyploids.
First, we checked for phylogenetic signal in genome size and GC content and their residuals after model fitting using the fitContinuous function (Pennell et al., 2014). We fitted several macroevolutionary models, i.e. white (non-phylogenetic), Brownian motion, Ornstein–Uhlenbeck and Pagel’s transformations (lambda, kappa and delta; Pagel, 1999). Model selection was based on Akaike’s information criterion with a correction for small sample sizes (AICc). To test the relationships between genome size and GC content to plant N, P and N : P, we performed ordinary least-squares models (the lm function), phylogenetic linear models (the phylolm function, Ho and Ané, 2014) and quantile regression (the rq function; Koenker, 2019). Both ordinary and phylogenetic linear models aimed to estimate the central tendency, while for the quantile regression analysis we set five quantiles (q0.10, q0.25, q0.50, q0.75 and q0.90). Quantile regression is a useful tool to test trends across various quantiles of the response variable, which can be more informative than estimates through the centres of data distributions (Koenker and Bassett, 1978; Cade and Noon, 2003). For example, assuming that the relationships between genome size and plant tissue N and P might be more profound in species with large genomes, it is helpful to investigate upper quantiles.
Phylogenetic linear models were fitted using the macroevolutionary model that best fitted the residuals estimated by the ordinary linear models. To our knowledge, a method of quantile regression controlling for phylogeny has not been developed so far. However, note that phylogenetic relatedness is not always a bias that needs to be corrected (de Bello et al., 2015), and thus statistical methods controlling for phylogeny do not have to be inherently better or more precise than non-phylogenetic methods. Regardless, the standard errors (s.e.) of the estimated coefficients (b) of both ordinary and phylogenetic linear models overlapped with each other (see the Results section), so we assumed that quantile regression not accounting for phylogeny still had acceptable type I error rates.
RESULTS
Genome size (1C values) ranged from 0.26 to 32.80 pg, 1Cx values ranged from 0.20 to 13.12 pg, GC content ranged from 33.22 to 47.60 %, plant N ranged from 0.74 to 4.42 %, plant P ranged from 0.08 to 0.48 % and plant N : P ranged from 3.81 to 17.79 in our grassland species (Supplementary Data Fig. S2). Following the categorization of Leitch et al. (1998), 47 % of the species had very small genomes (1C < 1.4 pg), 31.5 % had small genomes (1.4 < 1C < 3.5 pg), 20 % had intermediate genomes (3.5 < 1C < 14 pg) and only 1.5 % had large genomes (1C > 14 pg). Both genome size and GC content were correlated with phylogeny because they were best fitted by the lambda and Ornstein–Uhlenbeck models, respectively (Table S1). We found no differences in plant tissue N, P and N : P between diploids (n = 69) and polyploids (n = 61, Fig. S3).
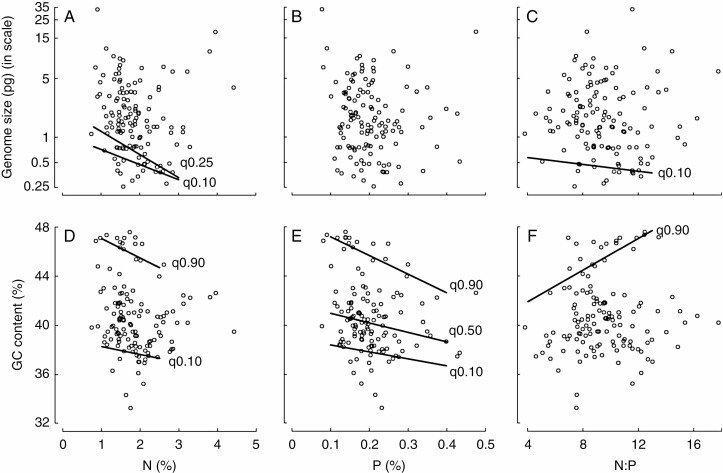
Both ordinary and phylogenetic linear models showed no central tendency for the relationship between genome size (results for 1C values in Table 2 and for 1Cx values in Supplementary Data Table S2) and plant N (Table 2A), P (Table 2B) and N : P (Table 2C). For GC content, we found no relationship to plant N (Table 2A), a weak relationship to P (Table 2B) but no relationship to P when accounting for phylogeny (Table 2B), and no relationship to N : P (Table 2C). On the other hand, quantile regression revealed significant links in some quantiles. Genome size (1C value) had a negative relationship to plant N (Table 3A) and N : P (Table 3C) in lower quantiles (q0.10 and q0.25 for N as well, Fig. 2A, C). When genome size was expressed as 1Cx values, we observed similar patterns for plant N, an additional negative link to plant P in the 0.25th quantile and no link to N : P at all (Table S3). For GC content, we observed significant relationships in upper quantiles (q0.90) where GC content was negatively related to plant N (Fig. 2D) and P (Fig. 2E), and positively related to N : P (Fig. 2F). We also observed the same negative links of GC content to plant N and P in q0.10 (both) and q0.50 (only P) (Fig. 2D, E).
Table 2.
No relationships between genome size (1C value), guanine–cytosine (GC) content and plant nitrogen (N), phosphorus (P) and N : P found for the central tendency. Coefficients (b), their standard errors (s.e.) and significance (P-value) estimated by ordinary (lm) and phylogenetic linear models (phylolm) are indicated.
| Element | 1C value (pg) [ln] | GC content (%) | ||||
|---|---|---|---|---|---|---|
| b | s.e. | P-value | b | s.e. | P-value | |
| (A) N | ||||||
| lm | −0.15 | 0.134 | 0.267 | −0.68 | 0.410 | 0.100 |
| phylolm | 0.03 | 0.133 | 0.824 | −0.04 | 0.356 | 0.920 |
| (B) P | ||||||
| lm | −1.05 | 1.198 | 0.381 | −9.18 | 3.609 | 0.012 |
| phylolm | 0.43 | 1.002 | 0.669 | −0.29 | 2.449 | 0.905 |
| (C) N : P | ||||||
| lm | −0.02 | 0.035 | 0.648 | 0.09 | 0.107 | 0.391 |
| phylolm | −0.01 | 0.033 | 0.720 | 0.00 | 0.077 | 0.964 |
Table 3.
Estimated quantile slopes (Coef.), their standard errors (s.e.) and P-values for genome size (1C value) and guanine–cytosine (GC) content regressed against plant (A) nitrogen (N), (B) phosphorus (P) and (C) N : P ratio.
| Element | 1C value (pg) [ln] | GC content (%) | ||||
|---|---|---|---|---|---|---|
| Coef. | s.e. | P-value | Coef. | s.e. | P-value | |
| (a) N | ||||||
| q0.10 | −0.41 | 0.080 | 0.001 | −0.64 | 0.308 | 0.038 |
| q0.25 | −0.52 | 0.196 | 0.009 | n.s. | ||
| q0.50 | n.s. | n.s. | ||||
| q0.75 | n.s. | n.s. | ||||
| q0.90 | n.s. | −1.59 | 0.682 | 0.021 | ||
| (B) P | ||||||
| q0.10 | n.s. | −5.69 | 2.330 | 0.016 | ||
| q0.25 | n.s. | n.s. | ||||
| q0.50 | n.s. | −7.71 | 3.374 | 0.024 | ||
| q0.75 | n.s. | n.s. | ||||
| q0.90 | n.s. | −15.2 | 7.60 | 0.047 | ||
| (C) N : P | ||||||
| q0.10 | −0.05 | 0.022 | 0.039 | n.s. | ||
| q0.25 | n.s. | n.s. | ||||
| q0.50 | n.s. | n.s. | ||||
| q0.75 | n.s. | n.s. | ||||
| q0.90 | n.s. | 0.65 | 0.299 | 0.032 | ||
Fig. 2.
Relationships of (A–C) genome size (1C value) and (D–F) guanine–cytosine (GC) content to plant tissue nitrogen (N), phosphorus (P) and N : P ratio across quantiles. Only significant quantiles (q) are shown (P < 0.05, Table 3). Genome size is ln-transformed.
DISCUSSION
In this study, the ecology-based perspective better explained interspecific patterns of genome size (both 1C and 1Cx values) and GC content in relation to tissue N and P than the stoichiometry-based perspective. However, the relationships we observed were typically weak, suggesting that the scaling of genome traits with N and P contents is not as clearly reflected at the tissue level (as found for the scaling of other traits with genome size at higher phenotypic scales; Knight and Beaulieu, 2008). For the relationship between genome size and plant N : P, our results support the stoichiometry-based perspective, while for the relationship between GC content and plant N : P, both perspectives can explain the patterns in our grassland species set. Additionally, all links were typically significant only in the lower (mostly q0.10) or upper quantiles (q0.90), while we found no links between the genome and stoichiometric traits for the central tendency.
Genome size, GC content and plant N
Genome size was negatively correlated to plant N only in lower quantiles (Fig. 1A), which does not support the prediction of the stoichiometry-based perspective. This result contradicts the findings of Kang et al. (2015), who found a positive correlation between plant N and genome size for the central tendency in a set of closely related Primulina species. However, our findings are in accordance with high N content and a small genome contributing to the resource-acquisitive plant ecological strategy. Species adopting this strategy typically have higher photosynthetic and respiration rates (Wright et al., 2004; Reich et al., 2006; Roddy et al., 2020). The diffusion of CO2 through the leaf intercellular space and into the chloroplasts lining the interior surfaces of mesophyll cells depends on their cell size (Roddy et al., 2020, Théroux-Rancourt et al., 2021). Genome size is usually positively correlated with cell size (Beaulieu et al., 2008; Šímová and Herben, 2012; Leitch et al., 2013) and large cells slow down CO2 diffusion (Roddy et al., 2020). Thus, it is probably not efficient for plants with large genomes to maintain N-rich photosynthetic tissues to boost photosynthesis, if its maximum capacity is limited by large mesophyll cells. Alternatively, cell size and density could explain the limited or non-existent relationships between genome size and tissue N and P because of interspecific differences in cell density obscuring the increased N and P costs of larger cells. Species with large genomes may reduce the total number of cells to offset the higher N and P demands of larger cells, and thus it is not clear how these genome–NP stoichiometry relationships scale up to the whole plant level and how much information can be lost when working with NP concentrations per unit dry weight of biomass (as done in this study).
By contrast, species with small genomes might be able to allocate more N to photosynthetic proteins and therefore to the overall N pool in plant leaves (Chapin, 1991; Sterner and Elser, 2002) to increase photosynthetic rates. Quantile regression revealed that genome size and plant N were negatively related only in plants with very small genomes (1C value <1.6 pg, i.e. close to the threshold of 1C value = 1.4 pg according to Leitch et al., 1998), suggesting that other factors begin to affect the relationship between genome size and plant N above this threshold. For example, plant N can reflect N availability in the soil, and thus luxury uptake or soil N deficiency might have blurred the N–genome size patterns. Plant-available soil N and P did not differ between the localities where the plant material was sampled, but the within-site variability was relatively high (Supplementary Data Fig. S1), suggesting that the co-occurring plants could have been under different nutrient regimes. Furthermore, 63 % of the species had an N : P ratio < 10, suggesting that the sampled plants were mostly N-limited (Güsewell, 2004), which is expected for the relatively young postglacial landscapes (Lambers et al., 2008) to which our localities belong.
GC content was negatively related to plant N only in the upper and lower quantiles, which contradicts the stoichiometric expectations based on the higher N costs to synthesize GC pairs. It has been hypothesized that N limitation has shaped the evolution of plant genomes or its element composition (Elser et al., 2011; Kang et al., 2015) because plants are considered to be more N-limited than animals (Elser et al., 2006). In this respect, wild plants also can have lower GC content (less N in their DNA) than their domesticated relatives that are adapted to N-rich soils as a response to N fertilization for thousands of years (Acquisti et al., 2009). However, the negative relationship between GC content and plant N detected in this study could be explained by interspecific differences in nitrogen costs for photosynthesis. Kelly (2018) has demonstrated that GC content is positively correlated with photosynthetic nitrogen use efficiency (the amount of carbon that can be fixed per unit of nitrogen invested by the plant). This suggests that species with higher GC content require less N for photosynthesis, and therefore their above-ground tissues might be generally N-poor. The slope was steeper in the 0.90th than in the 0.10th quantile, indicating that the plant N–GC content scaling is stronger in plants with richer GC base compositions (≥44 %).
Genome size, GC content and plant P
Surprisingly, we did not observe a link between genome size and plant P for the central tendency (after accounting for phylogenetic effects) or in any quantile (except for q0.25 in 1Cx values, Supplementary Data Table S3b), which does not support any of our predictions. This result could be partly explained by the overall negligibility of DNA (in terms of mass) for the whole-plant element composition. In fact, the most prominent determinant of plant P concentrations is RNA, which is far more abundant in plant cells than DNA (Sterner and Elser, 2002), and its amount could even be negatively correlated with genome size, because species with small genomes might uptake and allocate more P to RNA to boost growth rates (Sterner and Elser, 2002; Hessen et al., 2010; Sardans et al., 2021). However, the amount of RNA in cells is highly variable and depends on many factors (whereas genome size is relatively stable), so it is presumably not useful to measure RNA content for interspecific N and P stoichiometry comparisons from field data. We also did not observe any consistent correlations between genome traits and tissue NP stoichiometry, and ecological strategies predicted by the existing theories (Table S4; Grime, 1977; Bennett, 1987; Leitch et al., 2013; Šmarda et al., 2014; Sardans et al., 2021). These absent correlations of genome traits and NP stoichiometry with ecological strategies or the lack of a relationship between genome size and plant P could be an artefact of our sampling design. We exclusively collected flowering individuals that were in later life stages that might be associated with lower tissue P contents (Sardans et al., 2021). If we had collected plants during the initial stages of growth, we might have observed the relationships between genome size and plant P (positive) and N : P (negative) because plants mostly allocate P to growth in photosynthetic tissues during initial life stages and growing periods (Sardans et al., 2021).
We found a negative relationship between GC content and plant P consistently across all upper, middle and lower quantiles (q0.90, q0.50 and q0.10), supporting the ecology-based predictions. Šmarda et al. (2014) suggested that GC-rich genomes are associated with drought and cold tolerance (indicating stress tolerance). Here, we hypothesized that besides plant P (Sardans et al., 2021), GC content can also differ along the ruderal–stress tolerator spectrum (Grime, 1977), i.e. ruderals might have P-richer tissues and GC-poorer genomes than stress tolerators. This would imply a negative relationship between GC content and plant P as demonstrated here. Interestingly, we also found that the Ornstein–Uhlenbeck evolutionary model best fitted GC content in our grassland species (Supplementary Data Table S1), suggesting that GC content has been under an evolutionary pull towards an optimum value during its evolution. The same evolutionary pattern has been observed in 149 orchid species (Trávníček et al., 2019), which could be the result of physiological and environmental factors (Šmarda et al., 2014) or life-history traits, such as growth form (Trávníček et al., 2019).
Genome size, GC content and plant N : P
Genome size was also negatively correlated to plant N : P, which is in accordance with the prediction that DNA has relatively low N : P (Sterner and Elser, 2002), and thus large genomes might reduce the overall plant N : P. However, the slope was relatively small and only significant in the 0.10th quantile, i.e. only in species with very small genomes. A possible explanation of this pattern is that these species also typically had lower GC content (genome size and GC content were positively correlated, Supplementary Data Fig. S4), i.e. their DNA also contained less N and had lower N : P ratio. Finally, we found a positive link between GC content and N : P in the 0.90th quantile (see the clear threshold around a GC content of 42 %, Fig. 2F). Both stoichiometry- and ecology-based perspectives can explain our findings. More GC pairs increase the N : P ratio of nucleic acids and possibly the whole tissue N : P. Also, GC content and N : P should be positively correlated because of plant ecological strategies because species employing the stress-tolerant strategy tend to be GC-rich (Šmarda et al., 2014) and their tissue N : P relatively high (reviewed in Sardans et al., 2021). Interspecific variation in N : P is typically driven by P rather than N because P usually scales with N more rapidly (slope = 1.2; Güsewell, 2004), suggesting that investments into P-rich biomolecules and organelles, such as DNA, RNA or ribosomes, generally determine N : P differences across species. It is also notable that we found more significant links of GC content to N, P and N : P in comparison with genome size, suggesting that GC content is more closely related to tissue N than genome size. The nucleotide composition of genes also affects the nucleotide composition of RNA, which can substantially increase the whole-cell N pool, especially in highly expressed genes (e.g. those coding for photosynthetic proteins). The amount of N in genomes can also be reflected in proteomes that would also increase the overall plant N concentrations. For example, N : C ratios of prokaryotic genomes and proteomes have been positively correlated with each other (Bragg and Hyder, 2004).
CONCLUSIONS
The increasing use of genome and tissue stoichiometry traits to inform plant ecological and evolutionary models calls for better understanding of how these traits are related to each other. Here, we demonstrate that ecological and metabolic frameworks better explain the observed links between genome size, GC content and tissue NP stoichiometry in grassland herbs than the stoichiometry-based perspective. Generally, the genome–NP stoichiometry relationships were more pronounced in species with small genomes (for N and N : P) and for GC-rich (for N : P) species than in species with large genomes and GC-poor DNA. However, we typically observed weak relationships, which is not surprising because our N and P measurements were based on the whole plant level. The tightness of the relationships between genome traits and NP stoichiometry should decrease with increasing phenotypic scale (DNA → nucleus → cell → tissue → organ → whole plant) as more biomolecules containing N and P are present outside of the nucleus (e.g. chlorophyll, photosynthetic proteins or phospholipids). In summary, genome–NP stoichiometry relationships appear not to be simple and universal, and in order to fully understand them it is still necessary to evaluate them at different phenotypic scales, in different systems, plant growth forms and phylogenetic groups (e.g. non-temperate biomes, trees, aquatic plants, or ferns).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1. Phylogenetic signal of genome size, guanine–cytosine content and their residuals when regressed against plant tissue nitrogen, phosphorus and N : P ratio. Table S2. Number of relationships of genome size to plant tissue nitrogen, phosphorus and N : P found for the central tendency. Table S3. Estimated quantile slopes, their standard error, and P-values for genome size regressed against plant tissue (a) nitrogen, (b) phosphorus and (c) N : P ratio. Table S4. Expected and observed correlations between competitor-stress-ruderal scores and genome traits and tissue NP. Fig. S1. Plant-available soil nitrogen and phosphorus concentrations at the three localities where we sampled plant biomass. Fig. S2. Distributions of plant genome size, guanine–cytosine content, tissue nitrogen, phosphorus and N : P ratio. Fig. S3. Plant tissue nitrogen, phosphorus and N : P plotted separately for diploids and polyploids. Fig. S4. Positive correlation between genome size and guanine–cytosine content.
ACKNOWLEDGEMENTS
We thank F. Curtis Lubbe for correction of the English text.
Contributor Information
Martin Bitomský, Department of Ecology and Environmental Sciences, Palacký University, Šlechtitelů, Olomouc, Czech Republic; Institute of Botany of the Czech Academy of Sciences, Dukelská, Třeboň, Czech Republic.
Lucie Kobrlová, Department of Botany, Palacký University, Šlechtitelů, Olomouc, Czech Republic.
Michal Hroneš, Department of Botany, Palacký University, Šlechtitelů, Olomouc, Czech Republic.
Jitka Klimešová, Institute of Botany of the Czech Academy of Sciences, Dukelská, Třeboň, Czech Republic; Department of Botany, Charles University, Benátská, Prague, Czech Republic.
Martin Duchoslav, Department of Botany, Palacký University, Šlechtitelů, Olomouc, Czech Republic.
FUNDING
This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (INTER-EXCELLENCE, LTC18056, COST action 16212) and long-term research development project of the Czech Academy of Sciences (RVO 67985939).
CONFLICT OF INTERST
The authors declare that they have no conflicts of interest.
LITERATURE CITED
- Acquisti C, Elser JJ, Kumar S.. 2009. Ecological nitrogen limitation shapes the DNA composition of plant genomes. Molecular Biology and Evolution 26: 953–956. doi: 10.1093/molbev/msp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA.. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179: 975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- de Bello F, Berg MP, Dias ATC, et al. 2015. On the need for phylogenetic ‘corrections’ in functional trait-based approaches. Folia Geobotanica 50: 349–357. doi: 10.1007/s12224-015-9228-6. [DOI] [Google Scholar]
- Bennett MD. 1987. Variation in genome form in plants and its ecological implications. New Phytologist 196: 177–200. [Google Scholar]
- [dataset] Bitomský M, Kobrlová L, Hroneš M, Klimešová J, Duchoslav M. 2022. Stoichiometry versus ecology: Data from the relationships between genome size and guanine-cytosine content, and tissue nitrogen and phosphorus in grassland herbs. Mendeley Data. https://data.mendeley.com/datasets/7zgm6bg9y9/1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg JG, Hyder CL.. 2004. Nitrogen versus carbon use in prokaryotic genomes and proteomes. Proceedings of the Royal Society B: Biological Sciences 271: S374–S377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade BS, Noon BR.. 2003. A gentle introduction to quantile regression for ecologists. Frontiers in Ecology and the Environment 1: 412–420. doi: 10.1890/1540-9295(2003)001[0412:agitqr]2.0.co;2. [DOI] [Google Scholar]
- Castillo AI, Almeida RPP.. 2021. Evidence of gene nucleotide composition favoring replication and growth in a fastidious plant pathogen. G3 11: jkab076. doi: 10.1093/g3journal/jkab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS. 1991. Integrated responses of plants to stress. BioScience 41: 29–36. doi: 10.2307/1311538. [DOI] [Google Scholar]
- Doležel J, Bartoš J.. 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany 95: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Doležželová M, Novák FJ.. 1994. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biologia Plantarum 36: 351–357. [Google Scholar]
- Doležel J, Greilhuber J, Lucreti S, et al. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany 82: 17–26. [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2: 2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Acquisti C, Kumar S.. 2011. Stoichiogenomics: the evolutionary ecology of macromolecular elemental composition. Trends in Ecology and Evolution 26: 38–44. doi: 10.1016/j.tree.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser JJ, Fagan WF, Subramanian S, Kumar S.. 2006. Signatures of ecological resource availability in the animal and plant proteomes. Molecular Biology and Evolution 23: 1946–1951. doi: 10.1093/molbev/msl068. [DOI] [PubMed] [Google Scholar]
- Faizullah L, Morton JA, Hersch-Green EI, Walczyk AM, Leitch AR, Leitch IJ.. 2021. Exploring environmental selection on genome size in angiosperms. Trends in Plant Science 26: 1039–1049. [DOI] [PubMed] [Google Scholar]
- Grime PJ. 1977. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. The American Naturalist 111: 1169–1194. [Google Scholar]
- Guignard MS, Crawley MJ, Kovalenko D, et al. 2019. Interactions between plant genome size, nutrients and herbivory by rabbits, molluscs and insects on a temperate grassland. Proceedings of the Royal Society B: Biological Sciences 286: 20182619. doi: 10.1098/rspb.2018.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard MS, Leitch AR, Acquisti C, et al. 2017. Impacts of nitrogen and phosphorus: from genomes to natural ecosystems and agriculture. Frontiers in Ecology and Evolution 5: 70. [Google Scholar]
- Güsewell S. 2004. N:P ratios in terrestrial plants: variation and functional significance. New Phytologist 164: 243–266. doi: 10.1111/j.1469-8137.2004.01192.x. [DOI] [PubMed] [Google Scholar]
- Hessen DO, Jeyasingh PD, Neiman M, Weider LJ.. 2010. Genome streamlining and the elemental costs of growth. Trends in Ecology and Evolution 25: 75–80. doi: 10.1016/j.tree.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Ho LT, Ané C.. 2014. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Systematic Biology 63: 397–408. [DOI] [PubMed] [Google Scholar]
- Jin Y, Qian H.. 2019. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography 42: 1353–1359. doi: 10.1111/ecog.04434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Wang J, Huang H.. 2015. Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Scientific Reports 5: 11636. doi: 10.1038/srep11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. 2018. The amount of nitrogen used for photosynthesis modulates molecular evolution in plants. Molecular Biology and Evolution 35: 1616–1625. doi: 10.1093/molbev/msy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Beaulieu JM.. 2008. Genome size scaling through phenotype space. Annals of Botany 101: 759–766. doi: 10.1093/aob/mcm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenker R. 2019. quantreg: quantile regression. R package version 5.54. https://CRAN.R-project.org/package=quantreg
- Koenker R, Bassett G.. 1978. Regression quantiles. Econometrica 46: 33–50. doi: 10.2307/1913643. [DOI] [Google Scholar]
- Kozlowski J, Konarzewski M, Gawelczyk AT.. 2003. Cell size as a link between noncoding DNA and metabolic rate scaling. Proceedings of the National Academy of Sciences of the USA 100: 14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Raven JA, Shaver GR, Smith SE.. 2008. Plant nutrient-acquisition strategies change with soil age. Trends in Ecology and Evolution 23: 95–103. doi: 10.1016/j.tree.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD.. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany 82: 85–94. [Google Scholar]
- Leitch IJ, Greilhuber J, Doležel J, Wendel JF.. 2013. Plant genome diversity, Volume 2. Vienna: Springer. [Google Scholar]
- Meister A, Barow M.. 2007. DNA base composition of plant genomes. In: Doležel J, Greilhuber J, Suda J, eds. Flow cytometry with plant cells. Weinheim: John Wiley & Sons, 177–215. [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401: 877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Leitch IJ.. 2019. The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. doi: 10.1111/nph.16261. [DOI] [PubMed] [Google Scholar]
- Pennell MW, Eastman JM, Slater GJ, et al. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30: 2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. http://www.Rproject.org/ [Google Scholar]
- Reich PB, Tjoelker MG, Machado JL, Oleksyn J.. 2006. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature 439: 457–461. doi: 10.1038/nature04282. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A.. 2002. Base composition bias might result from competition for metabolic resources. Trends in Genetics 18: 291–294. [DOI] [PubMed] [Google Scholar]
- Roddy AB, Théroux-Rancourt G, Abbo T, et al. 2020. The scaling of genome size and cell size limits maximum rates of photosynthesis with implications for ecological strategies. International Journal of Plant Sciences 181: 75–87. doi: 10.1086/706186. [DOI] [Google Scholar]
- Sardans J, Janssens IA, Ciais P, Obersteiner M, Peñuelas J.. 2021. Recent advances and future research in ecological stoichiometry. Perspectives in Plant Ecology, Evolution and Systematics 50: 125611. doi: 10.1016/j.ppees.2021.125611. [DOI] [Google Scholar]
- Sterner RW, Elser JJ.. 2002. Ecological stoichiometry the biology of elements from molecules to the biosphere. Princeton: Princeton University Press. [Google Scholar]
- Šímová I, Herben T.. 2012. Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences 279: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G.. 2008. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany 101: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proceedings of the National Academy of Sciences USA 111: E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Knápek O, Březinová A, et al. 2019. Genome sizes and genomic guanine+cytosine (GC) contents of the Czech vascular flora with new estimates for 1700 species. Preslia 91: 117–142. doi: 10.23855/preslia.2019.117. [DOI] [Google Scholar]
- Théroux-Rancourt G, Roddy AB, Earles JM, et al. 2021. Maximum CO2 diffusion inside leaves is limited by the scaling of cell size and genome size. Proceedings of the Royal Society B: Biological Science 288: 20203145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trávníček P, Čertner M, Ponert J, Chumová Z, Jersáková J, Suda J.. 2019. Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytologist 224: 1642–1656. doi: 10.1111/nph.15996. [DOI] [PubMed] [Google Scholar]
- Wright IJ, Reich PB, Westoby M, et al. 2004. The worldwide leaf economics spectrum. Nature 428: 821–827. doi: 10.1038/nature02403. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wu H, Yu Q, et al. 2013. Sampling date, leaf age and root size: implications for the study of plant C:N:P stoichiometry. PLoS One 8: e60360. doi: 10.1371/journal.pone.0060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.