Abstract
There are three members of the HtrA family of serine proteases, YkdA, YvtA, and YyxA, encoded in the chromosome of Bacillus subtilis. In this study, we report on the promoter structure and regulation of ykdA expression. The ykdA gene is heat inducible, exhibiting a biphasic pattern of expression during a 60-min interval after heat shock. Increased expression after heat shock occurs at the transcriptional level. The heat-shock-inducible promoter has a single mismatch with a SigA-type −10 motif, but does not exhibit similarity to a SigA −35 region. There are six octamer repeats with a consensus TTTTCACA positioned at, and upstream of, the normal position of a −35 region. While repeats V and VI appear dispensable, repeat IV is essential for normal thermoinducible expression. This promoter structure is also found in the control region of yvtA, encoding a second member of this family of proteases. Expression of ykdA is negatively autoregulated both during the growth cycle and during heat shock. Our evidence suggests that YkdA protease activity is not required for this form of regulation. Null mutants of ykdA display increased tolerance to heat and are 80-fold more resistant to 10 mM hydrogen peroxide than wild-type cells. However, ykdA expression is not induced by hydrogen peroxide. These results indicate that the regulon to which YkdA belongs is linked to the oxidative stress response in B. subtilis.
Members of the HtrA family of serine proteases are widely distributed among bacteria and have also been found in yeast, Arabidopsis, and humans (for review, see reference 26). While some bacterial genomes encode more than one HtrA-like protease (e.g., Escherichia coli and Bacillus subtilis each contain three genes encoding HtrA-like proteases), no representative of this family has been identified in the completely sequenced genomes of Mycoplasma genitalium and the archaebacteria Methanococcus janaschii, Pyrococcus horikoshii OT3, Archaeoglobus fulgidis, and Methanobacterium thermoautotrophicum. The three HtrA-like serine proteases in E. coli, HtrA (DegP), HhoA (DegQ), and HhoB (DegS), can be divided into three structural regions: (i) an amino-terminal region that determines subcellular localization; (ii) a core domain containing the catalytic triad of amino acids (H, D, and S) required for enzymatic function, and (iii) one or more PDZ domains positioned in the carboxyl terminus. All three proteins in E. coli have signal sequences, with HtrA itself being localized to the periplasmic face of the cytoplasmic membrane (36). PDZ domains function to recognize peptide motifs usually located at the carboxyl terminus of proteins. The PDZ domain of HtrA functions to assemble protein monomers into the functional hexamic complex (33). It may also function in targeting the protease to its natural substrate in vivo (14, 26, 27). Recent work has shown that HtrA can function both as a molecular chaperone and as a protease (38). The switch between these activities is temperature dependent, with the chaperone activity predominating at lower temperatures and the protease activity predominating at high temperature (38). htrA-null mutants of E. coli are thermosensitive and are deficient in degrading abnormal periplasmic proteins (22, 39). An additional interesting feature of HtrA proteases is that they appear to play an important, but as-yet-uncharacterized role in the pathogenesis of some bacteria (26). Strains of Salmonella enterica serovar Typhimurium, Brucella abortus, and Yersinia enterocolitica with null mutations in htrA genes show attenuated virulence (11, 18, 21).
The htrA gene of E. coli is a member of the SigE stress regulon (12). This regulon functions to maintain the extracytoplasmic space free of misfolded proteins. The induction signals of this regulon include increased levels of nonnative periplasmic proteins and unbalanced levels of periplasmic proteins (6, 23, 31). Induction can also be effected by mutation of genes encoding enzymes that participate in periplasmic protein folding (for example, DsbA, DsbB, DsbC, and FkpA). Constituent genes of the SigE regulon include sigE, rpoH, fkpA (encoding a peptidyl-prolyl isomerase), and ompK (25, 26). The activity of SigE is controlled by a sigma factor/anti-sigma factor partner switching mechanism. RseA is an anti-sigma factor located in the inner membrane that can signal stress in the cell envelope and whose activity can be modulated by RseB and RseC (for reviews, see references 25 and 26). The SigE/RseA ratio determines the level of SigE activity, with the level of RseA in the cell being regulated by DegS, a HtrA paralogue (1). The htrA gene is also a member of the CpxR/CpxA regulon (6, 7). CpxR/CpxA is a two-component system that participates in the response to cell envelope stresses. It regulates expression of genes that have Sig70 (e.g., ppiA), Sig32 (e.g., ppiD), and SigE (e.g., htrA) promoters (8, 28, 29). Constituent genes of the CpxR/CpxA regulon include a periplasmically located disulfide oxidoreductase (DsbA), two peptidyl-prolyl-isomerases (PpiA and PpiD), and HtrA, indicating that one role of this regulon is to maintain protein folding homeostasis within the cell envelope. However the functional repertoire of the CpxR/CpxA regulon is likely to be more extensive, with recent reports showing that positive autoregulation is effected in conjunction with RpoS and that expression of some chemotaxis and motility genes is also CpxRA dependent (10, 30).
The B. subtilis heat shock response can be resolved into four classes of genes. There are three well-characterized regulons: HrcA/CIRCE (for review, see reference 15), SigB (for review, see reference 16), and CtsR (9, 19). The fourth class comprises a grouping of genes whose expression is responsive to heat stress, but the mechanism of induction is not effected by HrcA, SigB, or CtsR. It is likely that there are heterogeneous heat shock induction mechanisms within this group. Examination of the constituent genes indicates that none of these four gene classes corresponds to the SigE or CpxRA regulons identified in E. coli. We therefore chose to analyze how expression of ykdA (a htrA homologue) is regulated in order to ascertain if B. subtilis has an extracytoplasmic heat shock response similar to that of E. coli. In this study, we show that ykdA expression is heat inducible, that increased expression occurs at the transcriptional level, and that YkdA negatively regulates its own expression during exponential growth and during heat shock.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli and B. subtilis were routinely maintained and propagated on Luria-Bertani (LB) or Schaeffers medium (SM) supplemented with agar (Becton Dickinson, Cockeysville, Md.) (1.5% [wt/vol]) as appropriate and grown at 37°C with aeration (24, 34). E. coli and B. subtilis transformations were performed as described previously (2, 32). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added to the media at a concentration of 40 μg/ml, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added at the concentrations indicated in the text. Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 3 μg/ml; and erythromycin, 0.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli TG-1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | 32 |
| B. subtilis | ||
| 168 | trpC2 | |
| DN2 | trpC2 amyE::PykdA-bgaB Cmr | pDN2→168 |
| DN3 | trpC2 ykdA::pDN3 (PykdA-lacZ Pspac-ykdA) Ermr | pDN3→168 |
| DN8 to -11 | trpC2 amyE::pDN8 to 11(5′ΔPykdA-bgaB) Cmr | pDN8–11→168 |
| DN12 to -14 | trpC2 amyE::pDN12 to 14(3′ΔPykdA-bgaB) Cmr | pDN12–14→168 |
| DN15 to -18 | trpC2 ykdA::pMOR60 amyE::pDN8 to 11(5′ΔPykdA-bgaB) Ermr Cmr | pDN8–11→DN25 |
| DN19 to -21 | trpC2 ykdAΔ439 amyE::pDN12 to 14(3′ΔPykdA-bgaB) Cmr | pDN12–14→DN26 |
| DN25 | trpC2 ykdA::pMOR60 Cmr | pDN25→168 |
| DN26 | trpC2 ykdAΔ439 | pDN26→168 |
| DN27 | trpC2 ykdAΔ439 amyE::PykdA-bgaB Cmr | pDN2→DN26 |
| Plasmids | ||
| pDL | Integration vector for the introduction of single-copy transcriptional fusions to bgaB by double crossover at the amyE locus (Apr Cmr) | 45 |
| pMUTin4 | Integration vector containing the promoterless E. coli lacZ gene and the inducible Pspac promoter (Apr Ermr) | 40 |
| pMOR60 | pBluescript SK with the erythromycin cassette from pE194 inserted into SacI-SmaI allowing Campbell-type integration into the B. subtilis chromosome (Apr Ermr) | M. O'Reilly, unpublished data |
| pGhost4+ | Vector containing a Ts replicon for conditional integration and excision allowing construction of markerless deletions (Apr Ermr) | 3 |
| pUC19 | Cloning vector (Apr) | 43 |
| pDN2 | pDL containing the full ykdA control region on a PCR-amplified fragment (Apr Cmr) | This work |
| pDN3 | pMUTin4 with a PCR-amplified fragment containing an inactive part of the ykdA promoter, ribosome binding site, and the first 112 bp of the ykdA structural gene cloned into EcoRI-BamHI (Apr Ermr) | This work |
| pDN8 | pDL containing the ykdA control region with 47 bp deleted from the 5′ end (5′Δ47) (Apr Cmr) | This work |
| pDN9 | pDL containing the ykdA control region with 55 bp deleted from the 5′ end (5′Δ55) (Apr Cmr) | This work |
| pDN10 | pDL containing the ykdA control region with 79 bp deleted from the 5′ end (5′Δ79) (Apr Cmr) | This work |
| pDN11 | pDL containing the ykdA control region with 103 bp deleted from the 5′ end (5′Δ103) (Apr Cmr) | This work |
| pDN12 | pDL containing the ykdA control region with 33 bp deleted from the 3′ end (3′Δ33) (Apr Cmr) | This work |
| pDN13 | pDL containing the ykdA control region with 61 bp deleted from the 3′ end (3′Δ61) (Apr Cmr) | This work |
| pDN14 | pDL containing the ykdA control region with 74 bp deleted from the 3′ end (3′Δ74) (Apr Cmr) | This work |
| pDN25 | pMOR60 containing a PCR-amplified internal fragment from ykdA cloned into EcoRV (Apr Ermr) | This work |
| pDN26 | pGhost4+ containing juxtaposed 5′ flanking and internal fragments from ykdA cloned into EcoRI-HindIII (Apr Ermr) | This work |
| pDN27 | pUC19 containing the ykdA control region cloned into the EcoRI-BamHI sites (Apr) | This work |
pDN2→168 denotes plasmid pDN2 transformed into strain 168.
Strain construction.
All transcriptional fusions to the bgaB reporter gene were generated with plasmid pDL (45). PCR-generated fragments were cloned into pDL; the resultant plasmids were linearized and transformed into appropriate B. subtilis strains. Strain DN2 was constructed by cloning a 341-bp PCR-generated fragment (synthesized with primers YKDAP1 [5′-GCGGATCCGATGATGAATGACATTGC-3′] and YKDAP2 [5′-GCGAATTCAGCTGTCTAGCGATCATATC-3′]; the underlined sequences represent restriction sites introduced to facilitate cloning) into pDL to generate plasmid pDN2. Plasmid pDN2 was linearized and transformed into B. subtilis strain 168, yielding strain DN2. Deletion derivatives of the ykdA promoter region were generated by Bal31 deletion of the insert in pDN2 from the 5′ and 3′ directions. Deleted promoter fragments were recloned into pDL, the deletion end points were determined by sequencing, and appropriate deletions were then integrated into the amylase locus of selected B. subtilis strains. Plasmids containing 5′- and 3′-deleted promoter fragments were transformed into strain 168, generating strains DN8 to -14. To examine expression in a ykdA mutant background, plasmids pDN8 to -11 were transformed into strain DN25, generating strains DN15 to -18, and plasmids pDN12 to -14 were transformed into strain DN26, generating strains DN19 to -21. Strain DN3 was constructed by cloning a ykdA fragment (synthesized with primers YKDA6 [5′-GCGAATTCTAAACTCAAGTCATAAACCT-3′] and YKDAP1 [described above]) into pMUTin4 to generate plasmid pDN3. Plasmid pDN3 was then integrated into the chromosome of B. subtilis strain 168 by a Campbell-type event, to yield strain DN3. In this strain, the ykdA promoter directs expression of the lacZ reporter gene and the ykdA structural gene is under the control of the Pspac inducible promoter. Strain DN25 was constructed by cloning the 265-bp internal ykdA fragment amplified with primers YKDADEL2F (AGAAGGGGCATCATCAC) and YKDADEL2R (TTGAAACCGTTCTGTCCAC) (described above) into the EcoRV site of pMOR60 to generate plasmid pDN25. Plasmid pDN25 was then transformed into B. subtilis strain 168 to generate strain DN25. Strain DN26 has a deletion in the ykdA gene that was generated by the method of Biwas et al. (3). Plasmid pDN26 was constructed by sequential cloning of two ykdA fragments synthesized with the primers YKDA6 and YKDAP1 (fragment 1) and primers YKDADEL2R and YKDADEL2F (fragment 2) into pGhost4+. The fragments were juxtaposed in this plasmid and in the same orientation as they existed on the B. subtilis chromosome. B. subtilis strain 168 was transformed with pDN26, and erythromycin-resistant transformants were selected. Excision of the chromosomal DNA between the two homologous fragments resulted in strain DN26 (which has a 439-bp deletion within the ykdA gene). The chromosomal rearrangement was confirmed by PCR and Southern analysis. Strain DN27 was made by transforming strain DN26 with plasmid pDN2. Plasmid pDN27 was constructed by cloning the insert containing the ykdA control region from pDN2 into EcoRI-BamHI-digested pUC19. This plasmid was used to generate the sequencing ladder for the primer extensions.
Conditions for stress induction.
Cultures were heat shocked as follows. An overnight culture grown in LB broth was diluted 100-fold in fresh LB broth and grown to an optical density at 550 nm (OD550) of 0.3. This culture was then divided, and half was maintained at 37°C, while the other half was transferred to a prewarmed flask at 48°C. To measure β-galactosidase levels, culture aliquots were removed at the time points indicated in the text and centrifuged, and cell pellets were stored at −20°C. This procedure was employed for the other stressors, except that both halves of the culture were maintained at 37°C and the stressor was added to one-half of the culture. Stressors were added to the following final concentrations: ethanol, 4% (vol/vol); NaCl, 0.3 M; H2O2, 0.1 mM; and puromycin, 10 μg/ml. Buffered LB medium was used when testing for salt stress (5).
Phenotypic analysis.
Thermosensitivity was tested according to the procedure of Volker et al. (41). To test for hydrogen peroxide sensitivity, an exponentially growing culture was split, and one-half of the culture was exposed to hydrogen peroxide at a final concentration of 10 mM. The percentage of survival was determined from the number of cells in the stressed and unstressed cultures at each time point.
DNA manipulations.
All routine molecular biological procedures were performed according to the protocols described by Sambrook et al. (32). Restriction enzymes and Bal31 nuclease were purchased from New England Biolabs (Beverly, Mass.), and T4 DNA ligase was purchased from Boehringer (Mannheim, Germany). The sequences of all promoter fragments amplified by PCR were verified by sequencing. Sequencing reactions were performed with the Prism DyeTerminator kit from Perkin-Elmer (Foster City, Calif.) by electrophoresis through a 6% denaturing polyacrylamide gel (Seqagel; National Diagnostics, Atlanta, Ga.) on an ABI 373A automated sequencer.
Transcriptional analysis.
Total RNA was prepared from B. subtilis cells during normal growth and during heat shock as follows: 10-ml aliquots were harvested at designated times and centrifuged for 1 min at 4°C, and cell pellets were snap frozen in a dry ice-ethanol bath. Pellets were either stored at −80°C or processed immediately. The cell pellet was resuspended in 0.2 ml of sterile water and transferred to a 2-ml screw-cap tube containing 1 ml of a guanidine thiocyanate-acid phenol mixture (TRI reagent; Sigma, St. Louis, Mo.), 80 μl of hexa-decyltrimethylammonium bromide (CTAB), and 0.5 g of glass beads (0.1 mm in diameter; Biospec, Bartlesville, Okla.). The contents were shaken with three 1-min pulses in a mini Bead Beater (Biospec). The tube was cooled on ice for 1 min between each pulse. Chloroform (0.2 ml) was then added, the solution was mixed, and the tubes were centrifuged. The upper aqueous layer was removed and extracted with 0.5 ml of chloroform followed by a second extraction with an equal volume of acid phenol-chloroform (Sigma). The RNA was then precipitated with an equal volume of isopropanol and 1/10 volume of 3 M sodium acetate (pH 4.7). The RNA concentration was determined by measuring the OD260. Twenty-five-microgram aliquots of total RNA were electrophoresed through an agarose gel (1.2% [wt/vol]) containing 2.2 M formaldehyde according to standard methods (32). Separated RNA was transferred to a positively charged nylon membrane (Pall Gelman, Ann Arbor, Mich.) by capillary blotting. The membrane was then hybridized with a 1,046-bp digoxigenin-labelled ykdA probe generated with the synthetic primers YKDA6 (5′-CGGAATTCTAAACTCAAGTCATAAACCT-3′) and RE1 (5′-CCCAAGCTTTTTGACGTCATTGCTTGG-3′) by using a PCR Dig labelling mix (Boehringer) according to the manufacturer's instructions. Transcripts were visualized with the digoxigenin detection kit (Boehringer). Primer extension analysis was performed with 25 μg of total RNA isolated from cells harvested at appropriate times. The RNA was annealed to radioactively labelled primer YKDART3 (5′-CCTTTCGTTCTGTTTTCATCACG-3′) for 30 min at 55°C in 15 μl of 1× Superscript II buffer (Life Technologies). The reaction was then cooled to 48°C, and 5 μl of a prewarmed solution containing 2 mM deoxynucleoside triphosphates (dNTPs), 4× Superscript II buffer, 20 mM dithiothreitol, and 200 U of Superscript II was added. This reaction mixture was incubated at 48°C for 45 min. The reaction mixture was ethanol precipitated overnight at −20°C, and the pellet was resuspended in 50% 10 mM Tris-HCl–1 mM EDTA (pH 8.0)–50% stop solution (U.S. Biochemicals), denatured, and electrophoresed through a 6% denaturing polyacrylamide gel containing a sequencing reaction mixture generated with the same primer by using plasmid pDN27 as a template.
Measurement of β-galactosidase activity.
Thermostable β-galactosidase (BgaB) activity was measured as previously described (17) with the following modifications. Cells were lysed for 25 min at 37°C in Z buffer (24) containing 25 mM β-mercaptoethanol, 100 μg of lysozyme per ml, and 10 μg of DNase per ml. Lysates were heat treated at 70°C for 15 min and spun at 12,500 rpm for 5 min. Aliquots were added to reaction mixtures containing o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, and incubation was carried out at 55°C. The reaction was stopped by addition of 1.2 M NaCO3. The OD420 of the reaction was read. Measurement of LacZ activity was performed as outlined above, except that dithiothreitol (1 mM) replaced β-mercaptoethanol in the Z buffer, and the incubation at 70°C was omitted. The incubation temperature for the reaction was 28°C. The protein concentration was determined by using the Bio-Rad microassay (Bio-Rad, Hercules, Calif.) according to the instructions of the manufacturer. One activity unit is defined as 1 nanomol of ONPG hydrolyzed per min per μg of protein.
RESULTS
Sequence analysis of ykdA.
There are three HtrA-like proteases encoded in the B. subtilis chromosome and listed in SubtiList as htrA, yvtB, and yyxA (20). Because a functional correlation between the three B. subtilis members of this family and the three E. coli members (HtrA, HhoA, and HhoB) has not yet been established, we give the B. subtilis htrA gene its original designation of ykdA in this paper. The yvtB gene listed in SubtiList is a truncated HtrA-like protease. We resequenced this region of the chromosome and have shown that the full-length serine protease-encoding gene (designated yvtA [GenBank accession no. AF188296]) comprises both the yvtA and yvtB open reading frames listed in SubtiList. The yyxA gene has been renamed yycK (13). YkdA contains the catalytically important triad of histidine, aspartate, and serine residues found in this family of proteases and one PDZ domain. The E. coli and B. subtilis members of the family are approximately 40% identical (60% similar) within a core region comprising the catalytic and PDZ domains. However, there is little similarity at the amino-terminal regions. The amino-terminus region of YkdA is longer than the E. coli proteases and does not have a recognizable signal peptide. Instead the amino-terminal 50 hydrophilic amino acids are followed by a 22-amino-acid segment with the potential to form a transmembrane helix, suggesting that the protein has a membrane association.
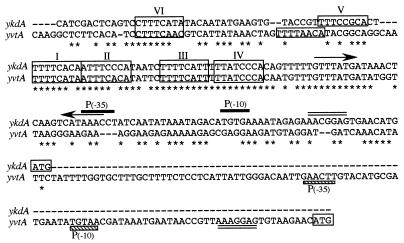
A comparison of the promoter regions of ykdA and yvtA is shown in Fig. 1. There is a 56-bp sequence found in both control regions that displays 91% identity (51 of 56 bases). This level of identity is higher than that observed between the two structural genes, confirming the importance of this sequence. Within this region are four copies of an octameric repeat similarly arranged (consensus TTTTCACA). There is a conserved 13-bp motif (TTTGTTTATGATA) positioned a half-turn of the helix downstream of octamer repeats III and IV and a putative ECF-type promoter located in the control region of ykdA that is very similar to the SigE promoter of htrA from E. coli (Fig. 1).
FIG. 1.
Comparison of the promoter regions of ykdA (top sequence) and yvtA (bottom sequence). Nucleotide identity is indicated by asterisks below the sequences, and gaps introduced to maximize the alignment are signified by dashes. The octameric repeats are boxed (labelled I through VI). The ECF-type promoters are indicated by solid bars over the sequence (ykdA) and hatched bars under the sequence (yvtA) signifying the −35 and −10 regions [P(−35) and P(−10), respectively]. The arrows represent inverted repeats, the putative ribosome binding sites have double lines over (ykdA) or under (yvtA) the sequence, and the boxed ATG is the putative start codon of each gene.
Induction of ykdA expression.
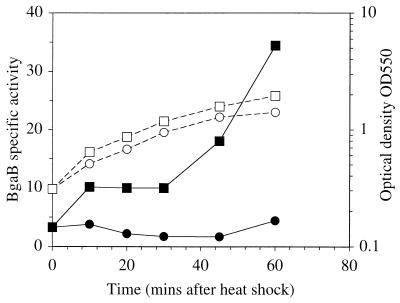
To investigate ykdA expression in B. subtilis, a 341-bp fragment containing the entire intergenic region 5′ to the gene and 112 bp of the ykdA coding sequence was directionally cloned into the integrating plasmid pDL, generating a transcriptional fusion with the bgaB (thermostable β-galactosidase) reporter gene. This fusion was then positioned in single copy at the amyE locus of the B. subtilis chromosome, generating strain DN2. The expression profile of this fusion was examined for 60 min at 37°C and after temperature upshift to 48°C (Fig. 2). Only a very low (approximately 2 U) and constant level of activity is observed when cells are grown at 37°C. However the β-galactosidase activity level increases biphasically when cells are shifted to 48°C, with an accumulation of approximately 35 U of activity after 60 min of growth at this temperature. The biphasic nature of activity accumulation is very reproducible. These data show that expression of ykdA is thermoinducible in B. subtilis.
FIG. 2.
Profile of growth and β-galactosidase (BgaB) accumulation in strain DN2 during growth at 37°C and after heat shock at 48°C. Growth (open circles) and β-galactosidase accumulation (solid circles) at 37°C: growth and β-galactosidase accumulation at 48°C are indicated by open and solid squares, respectively.
To investigate whether thermoinduction was mediated through one of the three established heat-shock regulons in B. subtilis, the kinetics of ykdA thermoinduction was investigated in hrcA-, sigB-, and ctsR-null mutant strains. The profile and level of ykdA-bgaB expression after thermoinduction in these three strains are similar to those observed in the wild-type strain (data not shown). To establish if ykdA expression is induced by stressors other than heat, strain DN2 was grown in media containing ethanol (4% [vol/vol]), NaCl (0.3 M), H2O2 (0.1 mM), and puromycin (up to 10 μg/ml). There was no increase in BgaB activity under any of these conditions (data not shown). Therefore, thermoinduction of ykdA is neither directly nor indirectly controlled by HrcA, SigB, or CtsR and must be part of a separate heat shock regulon in B. subtilis.
Control sequences involved in thermoinduction of ykdA.
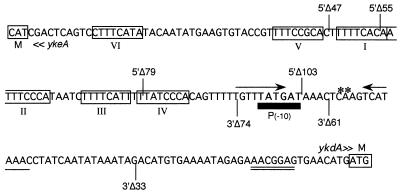
To delineate the promoter elements involved in thermoinduction, the complete ykdA control region (cloned in pDN2) was sequentially deleted from the 5′ and 3′ ends; the deletion end points are shown in Fig. 3. The promoter activity of each fragment was established by generating a bgaB transcriptional fusion, which was then placed in single copy at the amyE locus. The level of β-galactosidase activity in each strain at 30 and 60 min post-thermoinduction in wild-type (strains DN8 to -14) cells is shown in Table 2. The biphasic profile of β-galactosidase activity after thermoinduction of strain DN8 (5′Δ47) is similar to that of strain DN2 harboring the full-length control region. However, there is a twofold increase in the activity level after thermoinduction at 30 min after splitting of the culture (T30), whereas the level at T60 is approximately the same as that observed for the full-length promoter. There is also a twofold difference with this promoter deletion in a ykdA mutant background (compare levels in strains DN15 and DN27), which was observed at both time points. These data suggest the existence of a negative regulatory element positioned upstream of deletion end point 5′Δ47. Deletion of a further 8 bp (strain DN9, 5′Δ55), where most of octamer repeat I is removed, leads to attenuated thermoinduction of bgaB, especially at 30 min. Thermoinduction is further reduced in strains DN10 (5′Δ79) and DN11 (5′Δ103), with activity levels being virtually undetectable after 60 min of exposure to heat. Results from the 3′ deletions show that ykdA expression is heat inducible until a sequence between the 3′Δ61 (DN13) and 3′Δ74 (DN14) is deleted, which results in very low levels of ykdA-bgaB expression. A further nuance of the regulation of ykdA expression is revealed by strain DN13 (3′Δ61). Deletion of one arm of the putative stem-loop structure is accompanied by a twofold increase in expression after thermoinduction, suggesting the existence of a second negative regulatory element between deletion end points 3′Δ61 and 3′Δ33. These data show that the sequences required for ykdA thermoinduction are located on a 60-bp fragment (between deletion end points 5′Δ47 and 3′Δ61) containing octamer repeats I to IV and the 13-bp conserved motif and suggest the existence of two negative regulatory elements within the promoter region.
FIG. 3.
End points of the 5′ and 3′ deletions of the ykdA promoter region generated by Bal31 exonuclease. The nomenclature for each deletion indicates the end from which the deletion was made (5′ and 3′ ends above and below the sequence, respectively) and the number of bases deleted from the intergenic region. Some features of the promoter region are indicated. The octameric repeats are boxed and numbered I through VI, the initiation points of transcription are indicated by asterisks over the sequence, the −10 region of the active SigA-type promoter [P(−10)] is indicated by a box under the sequence, the inverted repeats are indicated by arrows, and the putative ribosome binding site is indicated by double lines. The start codons of ykdA and the divergently transcribed ykeA are boxed.
TABLE 2.
Expression of bgaB fused to deleted derivatives of the ykdA promoter in ykdA+ and ykdA mutant strains
| Strain | Expression (sp act units)a in ykdA+ strains
|
Strain | Expression (sp act units)a in ykdA mutant strains
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 37°C
|
48°C
|
37°C
|
48°C
|
||||||
| T30 | T60 | T30 | T60 | T30 | T60 | T30 | T60 | ||
| DN2 (full) | 2 | 4 | 10 | 34 | DN27 (full) | 6 | 12 | 230 | 723 |
| DN8 (5′Δ47) | 3 | 4 | 17 | 39 | DN15 (5′Δ47) | 12 | 25 | 275 | 825 |
| DN9 (5′Δ55) | 2 | 1 | 4 | 24 | DN16 (5′Δ55) | 1 | 1 | 11 | 55 |
| DN10 (5′Δ79) | 0 | 0 | 1 | 9 | DN17 (5′Δ79) | 0 | 0 | 0 | 5 |
| DN11 (5′Δ103) | 0 | 0 | 0 | 2 | DN18 (5′Δ103) | 0 | 0 | 0 | 3 |
| DN12 (3′Δ33) | 2 | 3 | 18 | 27 | DN19 (3′Δ33) | 16 | 24 | 308 | 729 |
| DN13 (3′Δ61) | 5 | 7 | 40 | 58 | DN20 (3′Δ61) | 78 | 108 | 479 | 940 |
| DN14 (3′Δ74) | 2 | 2 | 5 | 5 | DN21 (3′Δ74) | 0 | 0 | 0 | 0 |
Specific activity units are defined in Materials and Methods. Note that the background activity level found with a promoterless bgaB fusion has been subtracted from each value.
YkdA negatively autoregulates its own expression.
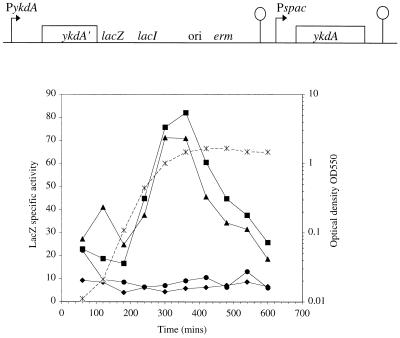
To investigate the possibility that YkdA participates in the regulation of its own expression, strain DN3 was constructed with the following salient features (Fig. 4A). (i) The full ykdA promoter region is fused to the lacZ reporter gene. (ii) An intact copy of ykdA is placed under the control of the IPTG-inducible Pspac promoter (44). The profiles of β-galactosidase accumulation during the growth cycle in the presence of different levels of IPTG inducer are presented in Fig. 4B. In cultures exposed to either 0.1 or 1 mM IPTG, β-galactosidase activity accumulates to less than 10 U, the level normally observed during growth at 37°C. However, when cells are grown either in the absence of IPTG or in the presence of 10 μM IPTG, β-galactosidase activity increases during exponential growth and reaches a maximum level of approximately 80 U at the transition phase of the growth cycle. This is followed by a sharp decrease in activity levels during the stationary phase of the growth cycle. This eightfold increase in β-galactosidase expression levels when YkdA levels are low or absent implies that the basal level of ykdA expression at 37°C during the growth cycle is negatively regulated by YkdA.
FIG. 4.
Negative autoregulation of ykdA. (A) A schematic of the construct at the ykdA locus in strain DN3. Promoters (PykdA and Pspac) are indicated by bent arrows. In this strain, the full intergenic ykdA control region is transcriptionally fused to the lacZ gene and the intact ykdA structural gene is under the control of the inducible Pspac promoter. (B) Expression of the PykdA-lacZ transcriptional fusion during the growth cycle in SM containing no IPTG (triangles), 10 μM IPTG (squares), 100 μM IPTG (diamonds), and 1 mM IPTG (circles). The growth cycle is indicated by dashed lines and is representative for the four growth curves.
To further investigate negative autoregulation of ykdA expression during the heat shock response, the full-length promoter was transferred into a ykdA mutant background, generating strain DN27. The strain was heat shocked as previously described, and the results are shown in Table 2. The level of expression is approximately twofold higher in the ykdA mutant strain than in the ykdA+ strain when cells are grown at 37°C. However, after heat shock at 48°C, expression levels are up to 20-fold higher in the ykdA mutant strain than in the ykdA+ strain at both time points. To delineate the promoter elements responsible for the elevated levels of expression during heat shock, the 5′- and 3′-deleted promoter constructs fused to bgaB were transferred into a ykdA mutant background, generating strains DN15 to -21. Expression levels are tabulated in Table 2. The profiles of expression after successive promoter deletions are similar to those obtained in the ykdA+ background, indicating that the same elements are involved in the elevated expression levels observed in the ykdA mutant strains. However, it is apparent that in cells grown at 48°C, the level of expression observed for each deletion construct is up to 20-fold higher (depending on the deleted construct) than in cells grown at 37°C.
To investigate whether the protease activity of YkdA was required for negative autoregulation, a strain was constructed in which the active-site serine was replaced by a methionine and the adjacent asparagine was replaced by a histidine (N289H, S290M). Mutation of the serine has been shown to inactivate the activity of this protease family (35). The profile and levels of ykdA expression in this strain were similar to those observed in the wild-type strain (data not shown). These data show that YkdA negatively regulates its own expression throughout the growth cycle at 37°C and also during heat shock at 48°C and that YkdA protease activity is not required for negative autoregulation.
Transcriptional analysis of ykdA expression.
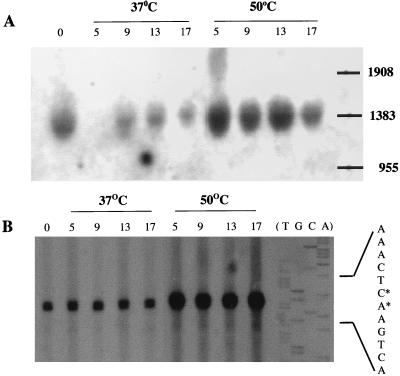
Expression of ykdA was analyzed by Northern (Fig. 5A) and by primer extension (Fig. 5B) analysis to establish transcript size and transcription initiation point. Total RNA samples were prepared from exponentially growing B. subtilis 168 cells before (0 min) and at 5, 9, 13, and 17 min after heat shock. The results of the Northern blotting show that there is a single transcript in all samples that migrates at approximately the same position as the 1,383-base RNA marker. The level of this transcript is significantly increased in the 50°C samples at 5, 9, and 13 min, but decreases to non-heat-shocked levels at 17 min after heat shock. Primer extension analysis was performed with RNA samples prepared from strain DN26 (ykdAΔ439), since expression levels are higher in this strain (see previous section). A doublet of reverse transcripts was observed in all samples that mapped to the adjacent CA bases marked by an asterisk in Fig. 3. This demonstrates that thermoinduction is effected from a single promoter. While there is a good match to the −10 consensus (TATGAT) of a SigA promoter corresponding to this initiation point of transcription, there is no apparent SigA −35 region. The level of reverse transcript in the 50°C samples was significantly higher than that observed in the 37°C samples, consistent with the observation that YkdA is a negative regulator of its own expression. These results indicate that heat shock induction of ykdA occurs at the transcriptional level from a single promoter that has a good match to the −10 region of a SigA promoter, but no apparent match to the −35 region. The size of the single transcript is consistent with the monocistronic operon predicted from the sequence.
FIG. 5.
Transcriptional analysis of ykdA. (A) Northern analysis. RNA was prepared from strain 168 cells either before (time 0) or at 5, 9, 13 or 17 min after splitting of the culture. Twenty-five micrograms of total RNA was loaded onto each lane (the sample in the 5-min lane at 37°C was lost). The positions to which the RNA size markers (1,908, 1,383, and 955 bases) migrated are indicated. (B) Primer extension analysis. RNA was prepared from strain DN26 (ykdAΔ439) before (0 min) and at 5, 9, 13, and 17 min after splitting of the culture. One-half of the culture was maintained at 37°C, while the other was heat shocked at 50°C. The amount of transcript in each lane is that obtained from 12.5 μg of total RNA. A sequencing ladder to size the reverse transcript is shown to the right of the figure, and the complement of a portion of this sequence is indicated beside it. The two bases at which transcription initiates are indicated by an asterisk.
Phenotype of ykdA mutants.
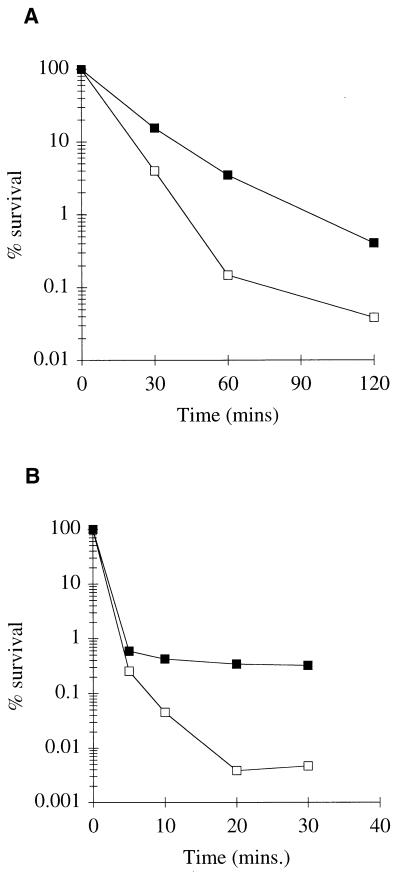
To establish if mutation of ykdA leads to altered sensitivity to stressors, strain DN26 (ykdAΔ439) was exposed to increased temperature (54°C) and hydrogen peroxide (10 mM), and cell survival profiles were compared with those of the parental B. subtilis strain, 168. Cell survival profiles (the values are the average of at least three independent experiments) after heat (Fig. 6A) and hydrogen peroxide (Fig. 6B) exposure are shown. It is evident that inactivation of ykdA leads to increased thermotolerance at all of the times sampled after temperature upshift. Similarly, inactivation of ykdA leads to an increased tolerance to hydrogen peroxide. It is evident the survival levels of wild-type and mutant cells are similar (2 to 3 logs of killing) after 5 min of exposure to hydrogen peroxide. However, whereas exposure of wild-type cells for a further 30 min results in an additional 2 logs of killing, the survival of the ykdA mutant is unaffected by this extended treatment. These experiments show that mutation of ykdA leads to increased tolerance to heat and hydrogen peroxide, indicating that YkdA expression and the peroxide stress response are somehow linked in B. subtilis.
FIG. 6.
Survival of wild-type and DN26 (Δ439) strains after exposure to heat (A) and hydrogen peroxide (B). Exponential-phase cells were exposed to heat at 54°C and 10 mM hydrogen peroxide for the times indicated. Wild-type cells are indicated by open squares, and ykdA mutant cells are indicated by solid squares.
DISCUSSION
There are three members of the HtrA serine protease family encoded in the B. subtilis genome (20). An analysis of one of these genes, ykdA, is presented in this paper. The core domain (catalytic region and one PDZ domain) of YkdA is 38% identical (61% similar) to the other B. subtilis members of this family. This core region was used to generate a phylogenetic tree of each of the three HtrA members from B. subtilis and E. coli with HtrA from Synechocystis as an outgroup. The tree shows that the three B. subtilis members are more closely related to each other than to any of the E. coli members and that ykdA is more closely related to yvtA than to yyxA (data not shown). This grouping is supported by the conservation of a regulatory motif in the promoter regions of ykdA and yvtA. Therefore, duplication of the genes encoding HtrA-like serine proteases most likely occurred after the divergence of E. coli and B. subtilis.
Expression of ykdA is thermoinducible, which is effected at the transcriptional level. Primer extension analysis shows that thermoinducible expression is driven by a promoter with a SigA-type −10 region, but the −35 region shows no similarity to SigA-type promoters. There are a series of octamer repeats positioned in this region of the promoter, at least one of which is necessary for normal thermoinduction. The importance of these repeats is indicated by the fact that both their number and positioning are conserved in the promoter region of yvtA, encoding a second HtrA-like serine protease in B. subtilis. Therefore, we predict that yvtA is also thermoinducible by a similar mechanism to ykdA. Our data also suggest the existence of two negative regulatory elements within the promoter region. These elements are worthy of further investigation, with the caveat that they may be due to novel juxtaposition of promoter and plasmid sequences generated through the deletion process. However, each region has a particular feature (octamer repeats and a stem-loop in the upstream and downstream regions, respectively) suggesting that the negative regulatory elements are physiologically relevant. Although both ykdA and yvtA have potential ECF-type promoter sequences (showing high homology to SigE-type promoters from E. coli [indicated in Fig. 1]) positioned in their control regions, we were unable to activate this putative ykdA promoter by a variety of stressors, including heat. Therefore, it is likely that ykdA expression responds to additional stimuli that can induce an ECF-type sigma factor-controlled regulon.
Expression of ykdA is also negatively autoregulated both during exponential growth at 37°C and during heat shock at 48°C. The level of β-galactosidase steadily increases in ykdA mutant cells throughout exponential growth, in contrast to ykdA+ cells, where expression levels are low and constant. This negative regulation is manifested most clearly, however, in heat-shocked cells: at 60 min post-thermoinduction, the level of β-galactosidase accumulation is 20-fold higher in ykdA mutant cells than in ykdA+ cells. Primer extension and Northern analysis show that the increased expression occurs at the level of transcription. We have evidence to show that the YkdA protease activity is not required to mediate negative autoregulation. The observed profiles of thermoinduction suggest that negative autoregulation operates by YkdA controlling the level of the inducing signal. Therefore, in ykdA-null mutants grown at 37°C, loss of YkdA results in only a small increase in the inducing signal. However, growth at 48°C results in a high level of inducing signal that remains high and persistent due to the absence of YkdA.
Counterintuitively, ykdA-null mutants are more resistant than wild-type cells to heat and to hydrogen peroxide exposure. Mutant ykdA cells are approximately 10-fold more resistant to heat exposure at 54°C than wild-type cells. This result differs from those obtained with some other bacteria in which mutation of htrA leads to a thermosensitive phenotype (26). In addition, whereas ykdA-null mutants of S. enterica serovar Typhimurium (18), Y. enterocolitica (42), and Pseudomonas aeruginosa (4) are more sensitive than wild-type strains to oxidative stress, the B. subtilis ykdA-null mutant is up to 80-fold more resistant to hydrogen peroxide than wild-type cells. It has been established in E. coli that the relationship between HtrA and oxidative stress involves the cell envelope. Cumene hydroperoxide (which partitions to the membrane) induces htrA, and htrA mutants are more sensitive to this oxidizing agent than are wild-type cells. However, mutant htrA cells are not more sensitive to hydrogen peroxide than wild-type cells, nor does hydrogen peroxide induce htrA expression (37). In addition, ferrous ions lead to oxidative damage of membrane proteins that can be alleviated by membrane-associated antioxidants, but not by cytosolic antioxidants (37). Mutation of ykdA in B. subtilis obviously results in a stimulus that leads to increased expression of a gene or genes that protect cells against both heat and hydrogen peroxide. One such candidate gene might be yvtA, encoding the closely related YkdA paralogue. Our evidence shows that the yvtA expression level increases when ykdA is mutated (D. Noone and K. M. Devine, unpublished data). Therefore, the increased thermo- and perhaps oxidative tolerance of ykdA mutants may be due to the compensatory patterns of expression of these two proteases.
ACKNOWLEDGMENTS
This work was supported by EU grants BIO2-CT93-0272, BIO2-CT95-0278, and BIO4-CT96-0655 (to K.M.D.) and by BioResearch Ireland (to D.N.) through the National Pharmaceutical Biotechnology at Trinity College, Dublin.
We thank S. L. Wong, W. Schumann, and C. Price for gifts of plasmids and strains.
REFERENCES
- 1.Ades S E, Connolly L E, Alba B M, Gross C A. The Escherichia coli SigE-dependent extracytoplasmic response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopolous C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologues of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly L, De Las Penas A, Alba B M, Gross C A. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese P N, Snyder W B, Cosma C L, Davis L J, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 8.Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derre I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 10.De Wolfe P, Kwon O, Lin E C. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol. 1999;181:6772–6778. doi: 10.1128/jb.181.21.6772-6778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elzer P H, Phillips R W, Kovach M E, Peterson K M, Roop R M., II Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect Immun. 1994;62:4135–4139. doi: 10.1128/iai.62.10.4135-4139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson J W, Gross C A. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 13.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning A S, Anderson J M. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 15.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 16.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 17.Hirata H, Negoro S, Okada H. High production of thermostable β-galactosidase of Bacillus stearothermophilus in Bacillus subtilis. Appl Environ Microbiol. 1995;49:1547–1549. doi: 10.1128/aem.49.6.1547-1549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson K, Charles I, Dougan G, Pickard D, O'Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 19.Krüger E, Hecker M. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J Bacteriol. 1998;180:6681–6688. doi: 10.1128/jb.180.24.6681-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Li S-R, Dorrell N, Everest P H, Dougan G, Wren B W. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect Immun. 1996;64:2088–2094. doi: 10.1128/iai.64.6.2088-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mecsas J, Rouviere P E, Erickson J W, Donohue T J, Gross C A. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 431–432. [Google Scholar]
- 25.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 26.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 27.Pallen M J, Ponting C R. PDZ domains in bacterial proteins. Mol Microbiol. 1997;26:411–413. doi: 10.1046/j.1365-2958.1997.5591911.x. [DOI] [PubMed] [Google Scholar]
- 28.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 29.Raina S, Missiakias D, Georgopoulos C. The rpoE gene encoding the SigE (Sig24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raivio T L, Popkin D L, Silhavy T J. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 1999;181:5263–5272. doi: 10.1128/jb.181.17.5263-5272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouviere P E, De Las Penas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sassoon N, Arie J P, Betton J M. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol Microbiol. 1999;33:583–589. doi: 10.1046/j.1365-2958.1999.01505.x. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer P, Miller J, Aubert J. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:701–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skorko-Glonek J, Wawrznow A, Krzewski K, Kurpierz K, Lipinska B. Site directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene. 1995;163:47–52. doi: 10.1016/0378-1119(95)00406-v. [DOI] [PubMed] [Google Scholar]
- 36.Skorko-Glonek J, Lipinska B, Krzewski K, Zolese G, Bertoli E, Tanfani F. HtrA heat shock protease interacts with phospholipid membranes and undergoes conformational changes. J Biol Chem. 1997;272:8974–8982. doi: 10.1074/jbc.272.14.8974. [DOI] [PubMed] [Google Scholar]
- 37.Skorko-Glonek J, Zurawa D, Kuczwara E, Wozniak M, Wypych Z, Lipinska B. The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol Gen Genet. 1999;262:342–350. doi: 10.1007/s004380051092. [DOI] [PubMed] [Google Scholar]
- 38.Spiess C, Biel A, Ehrmann M. A temperature dependent switch from chaperone to protease activity in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 39.Strauch K L, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 41.Völker U, Maul B, Hecker M. Expression of the ςB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto T, Hanawa T, Ogata S, Kamiya S. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect Immun. 1996;64:2980–2987. doi: 10.1128/iai.64.8.2980-2987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan G, Wong S-L. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]