Highlights
-
•
We evaluated antibacterial and antibiofilm activity of Lactobacillus strains against E. coli isolated from women with UTI.
-
•
L. acidophilus and L. casei were able to tolerate pH 3, bile salts, and pancreatic enzymes and adhere to intestinal epithelial cells.
-
•
L. acidophilus and L. casei strains showed a good antibacterial and antibiofilm against E. coli isolates with resistant to antibiotics.
Keywords: Anti-bacterial agents, Probiotics, Lactobacillus, Urinary tract infection
Abstract
The purpose of this study was to antibacterial, and antibiofilm activity of two Lactobacillus strains secretome and extraction against E. coli isolated from women with urinary tract infection (UTI). We isolated 100 E. coli samples from women with UTI. Lactobacillus acidophilus and Lactobacillus casei characteristics were evaluated, and their secretome and extraction were prepared. The antibacterial and antibiofilm activity of secretome and extraction of both Lactobacillus strains were evaluated against isolated E. coli samples. L. acidophilus and L. casei were able to tolerate pH 3, bile salts, and pancreatic enzymes. Both probiotics were not resistant to antibiotics and demonstrated an appropriate ability to adhere to the intestinal epithelial cells. Secretome and extraction of L. acidophilus and L. casei strains showed a good antibacterial and antibiofilm against E. coli isolates. Generally, present study suggested that the secretome and extraction of L. acidophilus and L. casei strains exhibits a good antimicrobial activity.
1. Introduction
Urinary tract infection (UTI) is one of the important health problem in women, which usually is occurred due to E. coli. Approximately, 150,000,000 individuals are diagnosed with UTI per year worldwide [1]. UTI is frequently observed with urological problems, which can lead to hypertension of renal failure continued [2]. In fact, a limited serotypes of E. coli can cause UTI; these serotypes have been observed with a high tissue invasion, colonization, and adhesion characteristics as compared to other non-pathogenic microorganisms [3, 4].
The major problem in treatment of UTI is related to acquire resistance to common antibiotics through various mechanisms such as changes in efflux pump, outer membrane permeability, target modification, and antibiotic inactivating [5, 6]. Previous studies have reported a high antibiotic resistance (especially to fluoroquinolones and beta-lactam antibiotics) in E. coli isolated from patients with UTI [7, 8]. Due to increased antibiotic resistance worldwide and emergence of multi-drug resistant strains, many studies have been conducted to introduce natural compounds as antimicrobial agents [8].
Lactic acid bacteria (LAB) are non-spore forming gram positive and non-respiratory probiotics which produce lactic acid by carbohydrates fermentation. Lactobacillus are an important group of microorganisms that known as probiotic, and recently considered as an antibacterial agent for treatment of human infection [9]. Due to antibacterial effects on pathogenic microorganisms, probiotics are well-known. The antibacterial effects of probiotics are due to production of organic acids (antagonize pathogens), adherence to pathogens, and reduction of bacterial adherents [10, 11]. Moreover, previous studies reported that Lactobacilli bacteria produce several bactericidal compounds [12, 13]. The evidence suggested that production of organic acids and bacteriocin are the major cause of antibacterial activity by Lactobacillus strains [14], [15], [16].
The purpose of this study was to assess antibacterial activity of Lactobacillus acidophilus and Lactobacillus casei secretome and extractions as probiotic, against E. coli isolated from women with UTI.
2. Materials and methods
2.1. Sample collection
In the present study, 100 E. coli isolated from patients with UTI were evaluated, which were identified using the biochemical analyzes. The isolates obtained from women with UTI referred to Asadabadi hospital, Tabriz, Iran. The urine samples of studied patients were cultured in eosin methylene blue (EMB) agar medium. To identification of E. coli isolates, the bacterial colonies were evaluated using biochemical analysis, including Voges-Proskauer, methyl red, triple sugar iron (TSI) agar, indole, and citrate. The isolated E. coli were cultured in tryptic soy broth (TSB) medium supplemented by glycerol (40%), and then were stored at −20 °C.
2.2. Samples isolation and identification
Genomic DNA was extracted using the salting-out method, and amplification of 16sRNA gene was conducted using polymerase chain reaction (PCR) to molecular identification of isolated E. coli. The used primers: F: 5`-ACTCTGTTATTAGGGAAGAA-3` and R: 5`-AACGCTTGCCACCTACGTAT-3`. Amplification was conducted in 25 μL total volume: 2.5 μL PCR buffer, 1 μL extracted DNA (50 ng), 1 μL primers (25 pmol), Taq DNA polymerase (1.5 unit), Mgcl2 (1.5 mmol/L), and dNTP (0.1 mmol), in the following condition: initial denaturation: 1 cycle (4 min at 95 °C), denaturation: 35 cycles (1 min at 94 °C), annealing: 35 cycles (1 min at 53 °C), extension: 35 cycles (1 min at 72 °C), and final extension: 1 cycle (10 min at 72 °C). The products of PCR were separated on 2% agarose gel and evaluated by gel document.
2.3. Preparation of Lactobacillus strains
Two Lactobacillus strains, include L. casei (ATCC 393) and L. acidophilus (ATCC 4356) were purchased strains from Persian Type Culture Collection (PTCC). The suspensions of Lactobacillus strains were prepared as follows: lyophilized culture (5 μL) added to 5 mL tryptic soy broth (TSB) and 5 mL de Mann Rogosa and Sharpe broth (MRS), and then standardized using visible-ultraviolet spectrophotometer (600 nm).
2.4. Preparation of Lactobacillus secretome and extraction
The Lactobacillus strains was cultured on the de Man, Rogosa and Sharpe (MRS) agar medium. The obtained colonies were inoculated into liquid MRS medium and incubated for 24 h. The bacterial culture was sub-cultured in fresh MRS medium and its absorbance was adjusted on 1 at 600 nm. The obtained bacterial culture was centrifuged and the supernatant was sterilized using a 0.22 μm syringe filter. The different concentrations of supernatant were prepared using MRS broth. Also, the same concentrations of MRS medium were prepared and considered as negative controls.
Moreover, the bacterial plate was resuspended by phosphate buffered saline (PBS) and lysized using ultrasonic bath. The obtained bacterial lysates were sterilized using a 0.22 μm syringe filter. The different concentrations of bacterial extract were prepared using PBS.
2.5. Evaluation of acid tolerance of probiotic
The Lactobacillus strains (7–8 log CFU/mL PBS) were inoculated into PBS (pH 3) and PBS (pH 7.2), and incubated for 3 h anaerobically at 37 °C. Next, serial dilutions of each strains were prepared by PBS. Then, 100 µL from bacterial suspension was spread plated on MRS agar and incubated anaerobically for 24 h at 37 °C. The obtained bacterial colonies on MRS agar were enumerated as CFU/mL. The acidic tolerance was evaluated by viability of Lactobacillus strains counts after exposure to normal condition and acidic condition (pH 3). This assay was in triplicate repeat.
2.6. Evaluation of bile tolerance of probiotic
Overnight culture of the Lactobacillus strains (adjusted to a 7–8 log CFU/mL) was cultured in MRS broth (10 mL) in presence or absence of oxgall (0.3%), and then incubated anaerobically for 4 h at 37 °C. Next, we prepared serial dilutions tenfold (up to 10−7) by PBS. The diluted sample (100 µL) was cultured on MRS agar medium, and then incubated as previous condition. Next, viability of Lactobacillus strains was evaluated by colony counts (CFU/mL). The bile tolerance was evaluated by viability of Lactobacillus strains counts in presence or absent of bile (oxgall).
2.7. Evaluation of pancreatic enzyme tolerance of probiotic
Harvested cell pellet of overnight culture of the Lactobacillus strains were resuspended by PBS (7–8 log CFU/mL), and the resuspended cells (1%) was cultured in 10 mL prepared solution (1.9 mg/mL pancreatin and 150 mM NaHCO3 were diluted in PBS, pH 8) and control solution (PBS, pH 7.2), and then incubated anaerobically for 3 h at 37 °C. Next, we prepared serial dilutions tenfold (up to 10−7) by PBS. The diluted sample (100 µL) was cultured on MRS agar medium, and then incubated anaerobically for 24 h at 37 °C. The viability of Lactobacillus strains was evaluated by enumeration of colonies (CFU/mL). The pancreatic enzymes tolerance was evaluated by viability of Lactobacillus strains counts in presence or absent of pancreatin (prepared solution).
2.8. Evaluation of adherence of probiotic
Human intestinal epithelial cell line (HT-29) was used to probiotic adherence investigation. The cancer cells were cultured using RPMI-1640 medium contains 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (5000 units/mL–5000 mg/mL) antibiotics. Next, the cancer cells (1 × 105/well) were seeded in 6-well plate containing fresh culture medium and incubated at 37 °C in 5% CO2. In addition, the overnight culture of Lactobacillus strains (10 mL) were harvested, and then resuspended in sterile PBS at 8 log CFU/mL concentration. The bacterial suspension (100 µL) and fresh culture medium (2 mL) was added to the all wells and then incubated for 1 h. Then, the cells monolayer was washed with PBS, and fixed with methanol (3 mL), and placed at room temperature for 10 min. The cells monolayer was Gram stained and evaluated using a light microscope at 20 random microscopic fields. This assay was in triplicate repeat.
2.9. Evaluation of antibiotic susceptibility
The antibiotic susceptibility of Lactobacillus strains was by disc diffusion method. The used antibiotics included: streptomycin (10 μg/ml), ampicillin (10 μg/ml), tetracycline (30 μg/ml), kanamycin (25 μg/ ml), and erythromycin (15 μg/ml). The cultures of Lactobacillus strains (100 μl) was swabbed on surface of nutrient agar medium. Then, the antibiotic discs were placed on the plates. The plates were anaerobically incubated at 37 °C for 24 to 48 h. The diameters of inhibition zones were investigated using calipers and considered as susceptible, S (≥21 mm), resistance, R (≤15 mm), intermediate, I (16–20 mm).
2.10. Evaluation of antibacterial activity by microdilution method
Antibacterial activity of the Lactobacillus strains was assessed by a broth microdilution susceptibility test. Briefly, 100 μL of the diluted (1:2 through 1:512) Lactobacillus strains was transferred to a 96-well plates in presence of LB broth medium. The prepared suspension (108 CFU/ml) was then cultured in a 96-well plate, and then incubated for 24 h at 37 °C. Next, the optical density each well (OD) was measured at 620 nm. Finally, samples were serially diluted by PBS and cultured on LB agar medium (in triplicate repeat).
2.11. Evaluation of antibacterial activity by disk-diffusion method
Antibacterial activity of the Lactobacillus strains was assessed by disk diffusion method in Mueller Hinton agar medium. Bacterial inoculums were spread plated on Mueller-Hinton agar. Next, empty Whatman discs were impregnated with different concentration of both Lactobacillus strains secretome and extraction and were placed on the plates. The plates were anaerobically incubated at 37 °C for 24 to 48 h. The diameters of inhibition zones were investigated using calipers.
2.12. Evaluation of antibiofilm activity by microtiter plate-crystal violet method
Antibiofilm effects of the both Lactobacillus strains were investigated using microtiter plate-crystal violet assay. For this purpose, serially diluted (1:2 through 1:512) strains were cultured in 96-well plates containing LB broth and sucrose. In addition, the E. coli strain suspension (108 CFU/ml) was added to 96-well plates and incubated for 24 h at 37 °C. Next, all wells were stained with crystal violet, and de-stained with 95% ethanol. Finally, optical density (OD) of biofilm-related crystal violet was investigated at 570 nm wavelength.
2.13. Statistical analysis
In the present study, all experiments were performed in triplicate, and results were presented as mean ± standard deviation (SD). We used SPSS (ver. 21.0) and GraphPad Prism (ver. 6) softwares to analyze of the obtained data. The statistical analysis was performed using Tukey's multiple comparison tests and one-way ANOVA analysis. The p < 0.05 was considered as statistically significant.
3. Results and discussion
3.1. Acid tolerance of probiotic
Both studied Lactobacillus strains showed a high acid tolerance, but the level of tolerance varied among two strains. In present study, L. acidophilus showed high acid tolerance (viability loss: 0.16 log units). The acid tolerance of L. acidophilus strain was significantly higher than L. casei strain (viability loss: 0.30 log unit) (p < 0.05). The viability of the Lactobacillus strains at pH 3 and pH 7.2 presented in Table 1. In a similar study, Mourad and Nour-Eddine have indicated that L. plantarum isolated from fermented olives showed survival percentages of 55–65%, when exposed to pH 3 for 3 h [17]. Moreover, our results are in agreement with study of Rajoka et al. and Akalu et al., which reported that the isolated Lactobacillus strains from various sources presents above 80% survival rate at pH 3 for several hours [18, 19]. Other previous studies reported that Lactobacillus strains showed a high survival rate (more than 89%) at pH 3 for several hours [20]. However, survival rate of both Lactobacillus strains were significantly decreased at low acidity.
Table 1.
The acid tolerance of Lactobacillus strains.
| Strains | Cell viability (log CFU/mL) ± SD | Viability loss (log units) | |
|---|---|---|---|
| pH 7.2 | pH 3.0 | ||
| L. acidophilus | 7.15 ± 0.10 | 6.99 ± 0.07 | 0.16 a |
| L. casei | 7.03 ± 0.03 | 6.73 ± 0.11 | 0.30 b |
Standard Deviation (SD); The values with different superscript letters are significantly different (p < 0.05).
3.2. Bile tolerance of probiotic
Both studied Lactobacillus strains showed a high bile tolerance, but the level of tolerance varied among two strains. The L. acidophilus showed the highest tolerance to bile salt (viability loss: 0.12 log units). Moreover, we observed a slight reduction in cell viability of L. casei strain (viability loss: 0.44 log units). However, the bile salt tolerance levels of L. acidophilus strain were significantly higher than L. casei strain (p < 0.05). The bile tolerance of the Lactobacillus strains presented in Table 2. Similar to our study, other studies have reported that the isolated Lactobacillus strains are indicate high bile salt tolerance with 88–92% survival rates [21, 22]. In a study by Akalu et al. reported that the isolated Lactobacillus strains are with high tolerance in presence of 0.3% bile salt [18]. In contrast, Boke et al. demonstrated that Lactobacillus strains presents a low levels of bile salts tolerance with decreased survival rates [21]. In addition, Rajoka et al. demonstrated that the Lactobacillus isolates presents a low levels of bile salts tolerance with less than 50% survival rate in presence of bile salts [23].
Table 2.
The bile tolerance of Lactobacillus strains.
| Strains | Cell viability (log CFU/mL) ± SD | Viability loss (log units) | |
|---|---|---|---|
| MRS | MRS + bile salt | ||
| L. acidophilus | 7.71 ± 0.13 | 7.59 ± 0.01 | 0.12 a |
| L. casei | 7.93 ± 0.01 | 7.49 ± 0.08 | 0.44 b |
Standard Deviation (SD); The values with different superscript letters are significantly different (p < 0.05).
3.3. Pancreatic enzyme tolerance of probiotic
Both studied Lactobacillus strains exhibited an appropriate tolerance to pancreatic enzymes. The L. acidophilus showed highest tolerance to the pancreatic enzymes (viability loss: 0.29 log units). The pancreatic enzymes tolerance of L. acidophilus strain was higher than L. casei strain (viability loss: 0.35 log unit), but this difference was not statistical significant (p > 0.05). The pancreatic enzyme tolerance of the Lactobacillus strains presented in Table 3. Pancreatic enzymes in the small intestine are involved in digestion of carbohydrates, fats, and proteins of foods. Tolerate to pancreatic enzymes is another selection criterion to use probiotics [24, 25]. In a similar study by Rönkä et al. reported that the survival rate of L. brevis strain was decreased slightly in presence of pancreatic enzymes [24]. Moreover, Ruiz-Moyano et al. also reported that more than 90% tested Lactobacillus strains survived after 3 h of treating with pancreatic enzymes [26].
Table 3.
The pancreatic enzyme tolerance of Lactobacillus strains.
| Strains | Cell viability (log CFU/mL) ± SD | Viability loss (log units) | |
|---|---|---|---|
| Control | Pancreatic enzymes | ||
| L. acidophilus | 7.88 ± 0.21 | 7.59 ± 0.01 | 0.29 a |
| L. casei | 7.71 ± 0.11 | 7.36 ± 0.12 | 0.35 a |
Standard Deviation (SD); The values with different superscript letters are significantly different (p < 0.05).
3.4. Adherence assay of probiotic
Both studied Lactobacillus strains were adhered to the HT-29 intestinal epithelial cell line, but adherence ability was different in two Lactobacillus strains. The highest adherence ability was exhibited by L. casei (51.8 attached cells per HT-29 cell), and the lowest adherence ability was exhibited by L. acidophilus (29.3 attached cells per HT-29 cell). However, this difference was not statistical significant (p > 0.05). Ability to attach to intestine epithelial cells and colonize is an important criterion for selection of probiotic isolates which can be established in the intestine [23]. In addition, adherence to intestine epithelial cells is essential probiotics activity, such as antimicrobial activities, cholesterol lowering activity, and immune-modulation. In this study, we used HT-29 cell line (human intestinal cell line) for attachment of two Lactobacillus strains, due to its similar physiological and morphological characteristics to normal human intestine epithelial cells [27]. In a related study by Jacobsen et al. reported variable adhere ability (from strong to low adhesion) by several Lactobacillus strains to HT-29 cell line [28]. Gopal et al. also found that L. acidophilus exhibited strong ability to adhere to the HT-29 human epithelial cell lines [25]. Previous studies demonstrated that the adhesion molecules, exopolysaccharides, on the cell walls were involved in adherence ability of Lactobacillus strains [29, 30]. Moreover, various adhesion factors of Lactobacillus strains are loosely bound to the epithelial cells surface by noncovalent interaction [31].
3.5. Antibiotic susceptibility
The antibiotic susceptibility of Lactobacillus strains demonstrated sensitivity to, erythromycin, and tetracycline, ampicillin. However, both Lactobacillus strains showed a resistance to streptomycin and kanamycin. The antibiotic susceptibility of the Lactobacillus strains presented in Table 4. Resistant to antibiotics is a main characteristic of probiotic bacteria. In another study by Tigu et al. reported that isolated Lactobacillus strains from fermented condiments were sensitive to tetracycline, ampicillin, and erythromycin [32]. On the contrary, Sukmarini et al. reported that isolated Lactobacillus strains from Indonesian fermented foods were resistant to erythromycin [30]. In addition, Pan et al. reported that isolated Lactobacillus strains from Chinese fermented foods were resistant to erythromycin, ampicillin, and tetracycline [31].
Table 4.
The antibiotic susceptibility profile of Lactobacillus strains.
| Antibiotics | Dose (μg/ml) | Lactobacillus strains | |
|---|---|---|---|
| L. acidophilus | L. casei | ||
| Streptomycin | 10 | R | R |
| Ampicillin | 10 | R | R |
| Tetracycline | 30 | S | S |
| Kanamycin | 25 | R | R |
| Erythromycin | 15 | S | S |
Susceptible: S (≥21 mm); Intermediate: I (16–20 mm); Resistance: R (≤15 mm).
3.6. Antibacterial activity of probiotic
The secretome and extraction of Lactobacillus strains was shown to inhibit the growth of the E. coli isolates. According to the obtained results, the extraction of L. acidophilus showed a high antibacterial activity. The extraction of L. casei presented a high antibacterial activity against the E. coli isolates. Moreover, the extraction of both Lactobacillus strains showed a high antibacterial activity than secretome.
The obtained results showed that the largest growth inhibition zone was related to the extraction of L. acidophilus (16 mm) and L. casei (15 mm) stains. Moreover, the secretome of both Lactobacillus strains showed a larger growth inhibition zone. However, the enrofloxacin created the largest inhibition zone in the E. coli isolates (Table 5).
Table 5.
Inhibition zone diameter of secretome and extraction of Lactobacillus strains on E. coli isolates.
| Bacterial isolate | L. acidophilus | L. casei | Enrofloxacin | ||
|---|---|---|---|---|---|
| Extraction | Secretome | Extraction | Secretome | ||
| E. coli | 16 mm | 9 mm | 15 mm | 6 mm | 23 mm |
| E. coli (PTCC 43,889) | 19 mm | 12 mm | 17 mm | 9 mm | 26 mm |
In a similar study by Bassyouni et al. reported that isolated Lactobacillus strains from fermented condiments presents an antibacterial activity against E. coli [33]. Similarly, our study showed a 16 mm inhibition zone diameter in the UTI isolated E. coli by extraction of L. acidophilus. In agreement to the present study, Tadesse et al. reported that the Lactobacillus isolates can inhibit the E. coli strain with 15–17 mm inhibition zone diameters [34]. Tigu et al. have also revealed that Lactobacillus isolates inhibited the growth of E. coli with inhibition zones ranging from 10 to 14 mm in diameters [32]. In line with this, Haghshenas et al. have reported that Lactobacillus isolated from Iranian fermented dairy products, Lactobacillus species showed the most efficient antibacterial activity against E. coli with inhibition zones of 12.3 mm diameters [22].
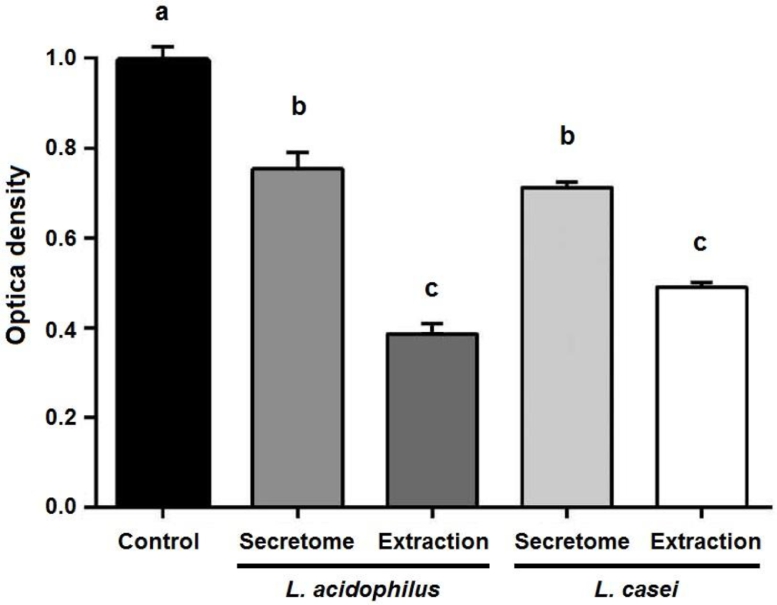
3.7. Antibiofilm activity of probiotic
The secretome and extraction of Lactobacillus strains was shown to inhibit biofilm formation of the E. coli isolates. The extraction of L. acidophilus and L. casei strains showed a high antibiofilm activity than their secretome (Fig. 1). This finding indicated that, the L. acidophilus and L. casei probiotics used in the present study had ability to inhibit biofilm formation by E. coli isolates. In parallel with our findings, Rao et al. have declared that cell free supernatant of Lactobacillus strains showed good antibiofilm activity [35]. Moreover, Khiralla et al. reported that three Lactobacillus strains isolated from traditional products are strongly recommended as biocontrol agents by inhibition of pathogens ability to form biofilm [36].
Fig. 1.
The antibiofilm activity of secretome and extraction of Lactobacillus strains on E. coli isolates. The values with different superscript letters are significantly different (p < 0.05).
4. Conclusion
In conclusion, we suggested that the Lactobacillus strains in the present study displayed potential probiotic properties. These strains had significant antimicrobial effect against E. coli isolated from patients with UTI. Moreover, we showed the antibiofilm effect of Lactobacillus strains against E. coli isolates. The effects of L. acidophilus and L. casei probiotics are not limited only to promote of human healthy, it also provides antibacterial effect against pathogenic bacteria. The results of this study indicated that L. acidophilus and L. casei probiotics directly interact with cancer cells and indirectly inhibit growth of E. coli isolate by release various bacitracin and metabolites. However, further studies are needed to investigate probiotic characteristics of various Lactobacillus strains.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
None applicable.
References
- 1.Mahdavi S., Tanhaeivash E., Isazadeh A. Investigating the presence and expression of stx1 gene in Escherichia coli isolated from women with urinary tract infection using real-time PCR in Tabriz, Iran. Int. J. Enteric Pathog. 2018;6(4):104–107. [Google Scholar]
- 2.Stamm W.E., Norrby S.R. Urinary tract infections: disease panorama and challenges. J. Infect. Dis. 2001;183(1):1–4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 3.Terlizzi M.E., Gribaudo G., Maffei M.E. UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017;8:1566. doi: 10.3389/fmicb.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yari Z., Mahdavi S., Khayati S., Ghorbani R., Isazadeh A. Evaluation of antibiotic resistance patterns in Staphylococcus aureus isolates collected from urinary tract infections in women referred to Shahid Beheshti educational and therapeutic center in Maragheh city. Med. J. Tabriz Univ. Med. Sci. Health Serv. 2020;41(6):106–112. [Google Scholar]
- 5.Mahdavi S., Isazadeh A.R. Investigation of contamination rate and determination of pattern of antibiotic resistance in coagulase positive staphylococcus aureus isolated from domestic cheeses in Maragheh. Iran. Pathobiol. Res. 2019;22(2):85–89. [Google Scholar]
- 6.Wright G.D. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug. Deliv. Rev. 2005;57(10):1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Glasner C., Albiger B., Buist G., Tambić Andrašević A., Canton R., Carmeli Y., et al. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill. 2013;18(28):20525. doi: 10.2807/1560-7917.es2013.18.28.20525. [DOI] [PubMed] [Google Scholar]
- 8.Mahdavi S., Kheyrollahi M., Sheikhloei H., Isazadeh A. Antibacterial and antioxidant activities of essential oil on food borne bacteria. Open Microbiol. J. 2019;13(1):81–85. [Google Scholar]
- 9.Tagg J.R., Dierksen K.P. Bacterial replacement therapy: adapting ‘germ warfare'to infection prevention. Trends Biotechnol. 2003;21(5):217–223. doi: 10.1016/S0167-7799(03)00085-4. [DOI] [PubMed] [Google Scholar]
- 10.Mahdavi S., Chalabi P., Zomorodi S., Isazadeh A. Effect of bananas puree on survival of Lactobacillus casei in coktel apple and banana juice during storage. Pharm. Biomed. Res. 2018;4(2):23–27. [Google Scholar]
- 11.Anas M., Eddine H.J., Mebrouk K. Antimicrobial activity of Lactobacillus species isolated from Algerian raw goat's milk against Staphylococcus aureus. World J. Dairy Food Sci. 2008;3(2):39–49. [Google Scholar]
- 12.Ennahar S., Sashihara T., Sonomoto K., Ishizaki A. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 2000;24(1):85–106. doi: 10.1111/j.1574-6976.2000.tb00534.x. [DOI] [PubMed] [Google Scholar]
- 13.McAuliffe O., Ross R.P., Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001;25(3):285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 14.Mahdavi S., Isazadeh A. Lactobacillus casei suppresses hfq gene expression in Escherichia coli O157:H7. Br. J. Biomed. Sci. 2019;76(2):92–94. doi: 10.1080/09674845.2019.1567903. [DOI] [PubMed] [Google Scholar]
- 15.Simova E.D., Beshkova D.B., Dimitrov Z.P. Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian dairy products. J. Appl. Microbiol. 2009;106(2):692–701. doi: 10.1111/j.1365-2672.2008.04052.x. [DOI] [PubMed] [Google Scholar]
- 16.Rather I.A., Choi K.H., Bajpai V.K., Park Y.H. Antiviral mode of action of Lactobacillus plantarum YML009 on Influenza virus H1N1. Bangladesh J. Pharmacol. 2015;10(2):475–482. [Google Scholar]
- 17.Mourad K., Nour-Eddine K. In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. Int. J. Probiotics Prebiotics. 2006;1(1):27. [Google Scholar]
- 18.Akalu N., Assefa F., Dessalegn A. In vitro evaluation of lactic acid bacteria isolated from traditional fermented Shamita and Kocho for their desirable characteristics as probiotics. Afr. J. Biotechnol. 2017;16(12):594–606. [Google Scholar]
- 19.Rajoka M.S., Mehwish H.M., Siddiq M., Haobin Z., Zhu J., Yan L., et al. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT-food Sci. Technol. 2017;84:271–280. [Google Scholar]
- 20.Oh Y.J., Jung D.S. Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT-food Sci. Technol. 2015;63(1):437–444. [Google Scholar]
- 21.Boke H., Aslim B., Alp G. The role of resistance to bile salts and acid tolerance of exopolysaccharides (EPSS) produced by yogurt starter bacteria. Arch. Biol. Sci. 2010;62(2):323–328. [Google Scholar]
- 22.Haghshenas B., Nami Y., Almasi A., Abdullah N., Radiah D., Rosli R., et al. Isolation and characterization of probiotics from dairies. Iran. J. Microbiol. 2017;9(4):234. [PMC free article] [PubMed] [Google Scholar]
- 23.Rajoka M.S., Hayat H.F., Sarwar S., Mehwish H.M., Ahmad F., Hussain N., et al. Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology. 2018;87(1):116–126. [Google Scholar]
- 24.Rönkä E., Malinen E., Saarela M., Rinta-Koski M., Aarnikunnas J., Palva A. Probiotic and milk technological properties of Lactobacillus brevis. Int. J. Food Microbiol. 2003;83(1):63–74. doi: 10.1016/s0168-1605(02)00315-x. [DOI] [PubMed] [Google Scholar]
- 25.Gopal P.K., Prasad J., Smart J., Gill H.S. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int. J. Food Microbiol. 2001;67(3):207–216. doi: 10.1016/s0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz-Moyano S., Martín A., Benito M.J., Nevado F.P., de Guía Córdoba M. Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Sci. 2008;80(3):715–721. doi: 10.1016/j.meatsci.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Shokryazdan P., Sieo C.C., Kalavathy R., Liang J.B., Alitheen N.B., Faseleh Jahromi M., et al. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobsen C.N., Nielsen V.R., Hayford A.E., Møller P.L., Michaelsen K.F., Paerregaard A., et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 1999;65(11):4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tallon R., Bressollier P., Urdaci M.C. Isolation and characterization of two exopolysaccharides produced by Lactobacillus plantarum EP56. Res. Microbiol. 2003;154(10):705–712. doi: 10.1016/j.resmic.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Sukmarini L., Mustopa A.Z., Normawati M., Muzdalifah I. Identification of antibiotic-resistance genes from lactic acid bacteria in Indonesian fermented foods. HAYATI J. Biosci. 2014;21(3):144–150. [Google Scholar]
- 31.Pan L., Hu X., Wang X. Assessment of antibiotic resistance of lactic acid bacteria in Chinese fermented foods. Food Control. 2011;22(8):1316–1321. [Google Scholar]
- 32.Tigu F., Assefa F., Mehari T., Ashenafi M. Probiotic property of lactic acid bacteria from traditional fermented condiments: datta and awaze. Int. Food Res. J. 2016;23(2):770. [Google Scholar]
- 33.Bassyouni R.H., Abdel-all W.S., Abdel-all M.G., Kamel Z. Characterization of lactic acid bacteria isolated from dairy products in Egypt as a probiotic. Life Sci. 2012;9:4–9. [Google Scholar]
- 34.Tadesse G., Ephraim E., Ashenafi M. Assessment of the antimicrobial activity of lactic acid bacteria isolated from Borde and Shamita, traditional Ethiopian fermented beverages, on some foodborne pathogens and effect of growth medium on the inhibitory activity. Int. J. Food Saf. 2005;5:13–20. [Google Scholar]
- 35.Rao K.P., Chennappa G., Suraj U., Nagaraja H., Raj A.C., Sreenivasa M.Y. Probiotic potential of Lactobacillus strains isolated from sorghum-based traditional fermented food. Probiotics Antimicrob. Proteins. 2015;7(2):146–156. doi: 10.1007/s12602-015-9186-6. [DOI] [PubMed] [Google Scholar]
- 36.Khiralla G.M., Mohamed E.A., Farag A.G., Elhariry H. Antibiofilm effect of Lactobacillus pentosus and Lactobacillus plantarum cell-free supernatants against some bacterial pathogens. J. Biotech. Res. 2015;6:86. [Google Scholar]