Summary
Background
Lower respiratory tract infections (LRTI) are a leading cause of critical illness and mortality in mechanically ventilated children; however, the pathogenic microbes frequently remain unknown. We combined traditional diagnostics with metagenomic next generation sequencing (mNGS) to evaluate the cause of LRTI in critically ill children.
Methods
We conducted a prospective, multicentre cohort study of critically ill children aged 31 days to 17 years with respiratory failure requiring mechanical ventilation (>72 h) in the USA. By combining bacterial culture and upper respiratory viral PCR testing with mNGS of tracheal aspirate collected from all patients within 24 h of intubation, we determined the prevalence, age distribution, and seasonal variation of viral and bacterial respiratory pathogens detected by either method in children with or without LRTI.
Findings
Between Feb 26, 2015, and Dec 31, 2017, of the 514 enrolled patients, 397 were eligible and included in the study (276 children with LRTI and 121 with no evidence of LRTI). A presumptive microbiological cause was identified in 255 (92%) children with LRTI, with respiratory syncytial virus (127 [46%]), Haemophilus influenzae (70 [25%]), and Moraxella catarrhalis (65 [24%]) being most prevalent. mNGS identified uncommon pathogens including Ureaplasma parvum and Bocavirus. Co-detection of viral and bacterial pathogens occurred in 144 (52%) patients. Incidental carriage of potentially pathogenic microbes occurred in 82 (68%) children without LRTI, with rhinovirus (30 [25%]) being most prevalent. Respiratory syncytial virus (p<0·0001), H influenzae (p=0·0006), and M catarrhalis (p=0·0002) were most common in children younger than 5 years. Viral and bacterial LRTI occurred predominantly during winter months.
Interpretation
These findings demonstrate that respiratory syncytial virus, H influenzae, and M catarrhalis contribute disproportionately to severe paediatric LRTI, co-infections are common, and incidental carriage of potentially pathogenic microbes occurs frequently. Further, we provide a framework for future epidemiological and emerging pathogen surveillance studies, highlighting the potential for metagenomics to enhance clinical diagnosis.
Funding
US National Institutes of Health and CZ Biohub
Introduction
Lower respiratory tract infections (LRTI) are a leading cause of morbidity and mortality worldwide, and their burden disproportionately affects children.1–4 Several landmark surveillance studies have evaluated the cause of LRTI in hospitalised children, but few studies have focused exclusively on those most critically ill who require mechanical ventilation support.3,5 Evaluation of the microbiological cause of LRTI in this population is challenging using conventional microbiological diagnostics given that antibiotics are frequently administered before intubation and might impede lower respiratory bacterial cultures. Additionally, specimens from the upper airways might not effectively reflect disease in the lungs.
Pathogen surveillance using metagenomic next generation sequencing (mNGS) offers an opportunity to overcome the limitations of using cultures, comprehensively assess the landscape of pathogens in the lower airways, and provides epidemiological information. Unlike traditional approaches to infectious disease diagnosis, mNGS enables a broad screen for a wide range of potential pathogens including bacteria, viruses, and fungi in a single test. Advances in mNGS bioinformatics methods in the past 5 years have enabled accurate detection of respiratory pathogens by differentiating them from the ubiquitous background, including commensal and environmental microbiota that previously obscured interpretation of results from sensitive molecular assays.6,7
Using a combination of clinically ordered testing and culture-independent mNGS diagnostics, we determined the prevalence, age distribution, and seasonal variation of lower respiratory pathogens in a multicentre cohort of critically ill children with acute respiratory failure requiring mechanical ventilation, including both patients with and without LRTI.
Methods
Study design and participants
We conducted a pre-planned secondary analysis of a prospective cohort of critically ill children with acute respiratory illnesses admitted to eight paediatric intensive care units (ICUs) participating in the National Institute of Child Health and Human Development’s Collaborative Pediatric Critical Care Research Network in the USA from Feb 26, 2015, to Dec 31, 2017.8 ICUs are listed in the appendix (p 2) and referred to by letters (A–H) in the results to avoid linking data to specific sites. The study was approved by a central institutional review board at the University of Utah. Written informed consent for study participation was obtained from legal guardians. Children aged from 31 days to 17 years who required invasive mechanical ventilation via endotracheal tube and who were suspected to need this support for more than 72 h were enrolled. Patients were excluded if there was the inability to obtain a tracheal aspirate sample within 24 h of intubation; presence of a tracheostomy tube or plans to place one; any condition in which deep tracheal suctioning was contraindicated; previous episode of mechanical ventilation during the hospitalisation; or no commitment from family or team to aggressive intensive care as indicated by do not resuscitate orders or other limitation of support. Full inclusion and exclusion criteria are shown in the appendix (p 2). Patients were followed up until hospital discharge. Prospectively collected clinical data were recorded in a research database (appendix p 3).
Case definitions and LRTI adjudication
Patients with LRTI were identified if they received a clinical diagnosis of LRTI by their treating physicians within 48 h of admission. Patients were assigned to one of four groups consistent with a recently described approach:6,9 (1) definite LRTI in which LRTI was clinically diagnosed and positive for a respiratory pathogen by clinical microbiology testing during admission; (2) suspected LRTI in which LRTI was clinically diagnosed but negative clinical microbiology testing; (3) no evidence of LRTI in which a clear alternative explanation for respiratory failure based on admission diagnosis; and (4) indeterminate LRTI in which equivocal evidence for respiratory infection. Children with indeterminate LRTI were excluded from the analyses.
Procedures
Enrolled patients received standard of care clinical respiratory microbiological diagnostics (semi-quantitative bacterial and fungal cultures from tracheal aspirate and blood, and nasopharyngeal swab respiratory viral multiplex PCR) ordered by treating clinicians at each study site. All children in the definite and suspected LRTI groups had tracheal aspirate tested by culture and nasopharyngeal specimens tested by PCR (appendix pp 3, 8).
Lower respiratory fluids sampled for research purposes by tracheal aspirate were collected within 24 h of intubation on ice and frozen in RNA stabilising agent within 30 min.8 RNA was reverse transcribed into complementary DNA and library construction was done using the NEBNext Ultra II Library Prep Kit (New England Biolabs, Ipswich, MA, USA). Human rRNA were depleted from the sample and the RNA sequencing libraries underwent paired-end sequencing on an Illumina Novaseq 6000 (Illumina, 97 San Diego, CA, USA). Detailed mNGS methods are provided in the appendix (pp 3–4).
Identities of the microbial reads were determined by querying the NCBI nucleotide and non-redundant protein databases. All sequences received species-level annotation by the IDseq pipeline.10 We performed background correction to remove taxa that did not differ in abundance compared to negative water control samples to mitigate environmental contaminants. To differentiate probable pathogens from commensal microbiota, we used a previously validated rules-based computational model6 that detects bacterial and fungal taxa with the greatest abundance in each sample relative to the rest of the lower respiratory microbiota and identifies all human respiratory viruses (appendix p 17). Additional details on these bioinformatics methods can be found in the appendix (pp 4–5). Microbial mass and lung microbiome diversity were calculated and compared between LRTI groups (appendix pp 6–7, 9). Phylogenetic analysis of respiratory syncytial virus was performed to characterise the genomic diversity of isolates (appendix p 6).
Outcomes
Primary outcomes measured were the presence or absence of a microbe by the combination of clinical diagnostics and mNGS. Secondary outcomes were length of stay in a paediatric ICU, hospital length of stay, the number of ventilator free days in 28 days, mortality, development of new morbidities, and ventilator associated pneumonia.
Statistical analysis
To identify significant co-occurrences of pathogens, we evaluated the ten most commonly detected pathogens using the Jaccard–Tanimoto coefficient.11 Co-occurrence of two pathogens was considered significant with a q value <0·05.
To determine whether pathogen distribution differed between age groups of children with definite or suspected LRTI, we performed a χ2 test for the ten most frequently detected pathogens across four age groups (31 days to 6 months, 6 months to 2 years, 2–5 years, and 5–18 years). Statistical significance was a p value <0·05.
To determine whether certain microbes were associated with worse clinical outcomes, we compared patients with definite or suspected LRTI that had a particular microbe detected (including co-detections) to patients without the microbe. Clinical outcomes are listed in the appendix (p 7). A secondary analysis compared patients with respiratory syncytial virus-A and respiratory syncytial virus-B. Statistical significance was determined by Wilcoxon rank sum (continuous variables) or Fisher’s exact test (binary variables). Next, univariate associations between clinical outcomes and age, sex, immunosuppression, functional status score12 at baseline, and presence of comorbidity13 were assessed using linear regression models for continuous outcome variables or logistic regression models for binary outcome variables. If univariate associations were significant at an α of 0·1, they were included in multivariable models.
Logistic regression was performed to determine association between presence of pathogen and LRTI diagnosis. To compare viral loads in patients between groups, we compared mNGS reads per million for both respiratory syncytial virus and rhinovirus in patients that had each virus detected by mNGS. Statistical significance was determined by Mann-Whitney tests.
We calculated the percentage of viral or bacterial cases in autumn and winter starting from January, 2016, when all sites were enrolling patients. To determine whether proportions were significantly greater than 0·5, we used a one-proportion Z test at an α of 0·05.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Of 457 children determined eligible, we included 397 in our study: 276 with clinically diagnosed or suspected LRTI and 121 with no evidence of LRTI (figure 1). The median age of children with LRTI was 11·5 months (IQR 3·2–46·7); 57% were male; and 42% had a chronic comorbid condition (table 1). In the children with no evidence of LRTI, the median age was 29·6 months (IQR 9·9–143·6); 55% were male; and 41% had a chronic comorbid condition.
Figure 1: Eligibility of children with lower respiratory tract infection.
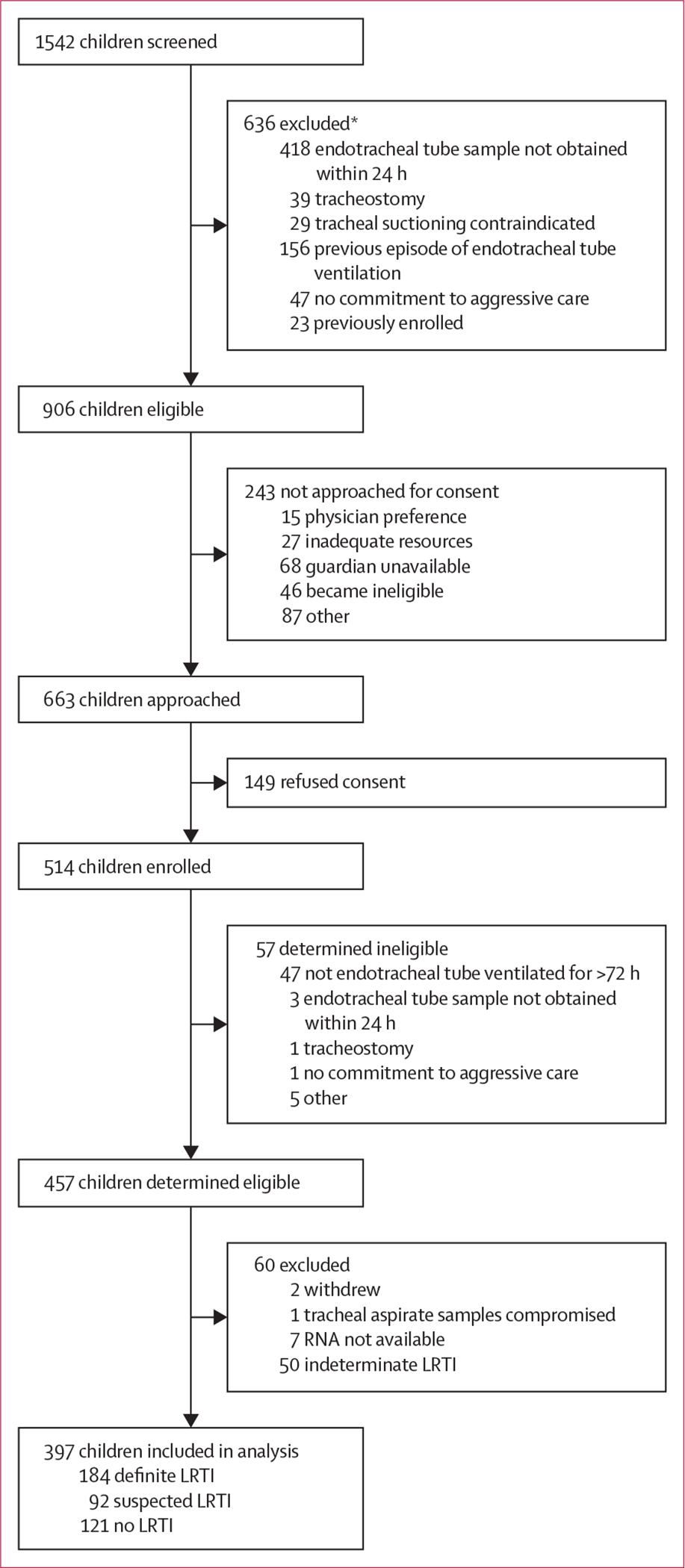
LRTI=lower respiratory tract infections. *A patient could have more than one reason for exclusion.
Table 1:
Demographics and clinical characteristics
| Definite or suspected LRTI (n=276) | No evidence of LRTI (n=121) | p value* | |
|---|---|---|---|
|
| |||
| Sex | ·· | ·· | 0·66 |
| Male | 158 (57%) | 66 (55%) | ·· |
| Female | 118 (43%) | 55 (45%) | ·· |
| Age | ·· | · | <0·0001 |
| <6 months | 97 (35%) | 20 (17%) | ·· |
| 6 months to <2 years | 88 (32%) | 36 (30%) | ·· |
| 2–5 years | 38 (14%) | 14 (12%) | ·· |
| >5 years | 53 (19%) | 51 (42%) | ·· |
| Race or ethnicity | ·· | ·· | 0·52 |
| White | 168 (61%) | 73 (60%) | ·· |
| Black or African American | 56 (20%) | 17 (14%) | ·· |
| Asian | 11 (4%) | 7 (6%) | ·· |
| American Indian or Alaskan Native | 3 (1%) | 1 (1%) | ·· |
| Native Hawaiian or other Pacific Islander | 1 (<1%) | 1 (1%) | ·· |
| More than one race | 5 (2%) | 4 (3%) | ·· |
| Unknown | 32 (12%) | 18 (15%) | ·· |
| Hispanic or Latino | 54 (20%) | 19 (16%) | 0·40 |
| Intensive care unit† | |||
| A | 17 (6%) | 10 (8%) | 0·0005 |
| B | 64 (23%) | 43 (36%) | ·· |
| C | 12 (4%) | 15 (12%) | ·· |
| D | 31 (11%) | 18 (15%) | ·· |
| E | 100 (36%) | 21 (17%) | ·· |
| F | 47 (17%) | 5 (4%) | ·· |
| G | 3 (1%) | 5 (4%) | ·· |
| H | 2 (1%) | 4 (3%) | ·· |
| Comorbidities‡ | 117 (42%) | 49 (40%) | 0·74 |
| Top five most frequent comorbidities§ | |||
| Neurological disease | 45 (16%) | 25 (21%) | 0·32 |
| Chronic airway or lung disease | 51 (18%) | 15 (12%) | 0·15 |
| Congenital anomaly or chromosomal defect | 36 (13%) | 14 (12%) | 0·74 |
| Cardiovascular disease | 29 (11%) | 10 (8%) | 0·58 |
| Cancer | 9 (3%) | 6 (5%) | 0·40 |
| Immunosuppressed | 10 (4%) | 12 (10%) | 0·016 |
| Admission category | ·· | ·· | <0.0001 |
| Medical | 275 (100%) | 75 (62%) | ·· |
| Surgical | 1 (<1%) | 20 (17%) | ·· |
| Trauma | 0 | 26 (21%) | ·· |
| Time from hospital admission to intubation, h | 5·45 (0·00–19·97) | 2·52 (0·00–20·65) | 0·19 |
| Paediatric risk of mortality III score | 4 (0–8) | 8 (3–12) | <0·0001 |
| Antibiotics in previous 7 days before intubation | 100 (36%) | 34 (28%) | 0·13 |
| Antibiotics on the day of intubation | 244 (88%) | 88 (73%) | 0·0002 |
| Hospital length of stay, days | 16 (3, 378) | 19 (5, 116) | 0·034 |
| Intensive care unit length of stay, days | 11 (3, 216) | 9 (3, 76) | 0·0041 |
| Paediatric acute respiratory distress syndrom¶ | 94 (34%) | 26 (21%) | 0·013 |
Data are n (%), median (IQR), or median (min, max). LRTI=lower respiratory tract infections.
Intensive care units are referred to by letters to avoid linking data to specific sites. Intensive care units are listed in the appendix (p 2)
Wilcoxon rank sum test used for all continuous variables; Fisher’s exact test used for all categorical variables.
Complex chronic conditions.13
Patients might have more than one comorbidity.
According to the Pediatric Acute Lung Injury Consensus Conference definition.
Of the 276 children clinically diagnosed with LRTI during admission, 184 had a microbiological cause identified by clinical testing (definite LRTI) and 92 did not (suspected LRTI). Patients with no evidence of LRTI (n=121) included 35 patients with positive clinical microbiological tests (appendix p 10).
mNGS was performed on tracheal aspirate samples from all patients. After background correction, which removed 69·9% of taxa (16·4% of sequencing reads), a computational model was implemented to distinguish probable pathogens from commensal microbiota (appendix p 17). Airway microbiome alpha diversity was lower in patients with LRTI than in patients without (appendix p 18). With respect to the 276 children with definite or suspected LRTI, we identified at least one pathogen in 184 (67%) children by PCR or culture alone, in 249 (90%) by mNGS alone, and in 255 (92%) by combining clinical and mNGS diagnostic methods (table 2; appendix p 10). The rules-based computational model detected 62% of the microbes identified by culture plus upper respiratory viral PCR testing in the definite LRTI group. Conversely, bacterial culture and viral PCR identified 66% of microbes detected by the rules-based computational model (appendix p 13). With the combination of clinical testing and mNGS, we detected more than one pathogen in 183 children (66%).
Table 2:
Established respiratory pathogens detected by culture, PCR, or mNGS.
| Upper respiratory PCR or lower respiratory culture | Lower respiratory mNGS | PCR or culture, or mNGS, or both | Co-detected (% of cases)* | |
|---|---|---|---|---|
|
| ||||
| Patients | 276 | 276 | 276 | ·· |
| Any positive | 184 (67%) | 249 (90%) | 255 (92%) | ·· |
| Respiratory syncytial virus | 113 (41%) | 116 (42%) | 127 (46%) | 108/127 (85%) |
| Haemophilus influenzae | 29 (1‘%) | 65 (24%) | 70 (25%) | 67/70 (96%) |
| Moraxella catarrhalis | 36 (13%) | 48 (17%) | 65 (24%) | 57/65 (88%) |
| Rhinovirus (A, B, C) | 12 (4%) | 57 (21%) | 61 (22%) | 41/61 (67%) |
| Streptococcus pneumoniae | 19 (7%) | 20 (7%) | 32 (12%) | 30/32 (94%) |
| Staphylococcus aureus | 21 (8%) | 7 (3%) | 25 (9%) | 22/25 (88%) |
| Parainfluenza virus (1–4) | 15 (5%) | 16 (6%) | 22 (8%) | 19/22 (86%) |
| Adenovirus | 19 (7%) | 3 (1%) | 20 (7%) | 20/20 (100%) |
| Coronavirus (HCoV-229E, HCoV-NL63, HCoV-OC4) | 16 (6%) | 9 (3%) | 18 (7%) | 18/18 (100%) |
| Human metapneumovirus | 10 (4%) | 17 (6%) | 17 (6%) | 15/17 (88%) |
| Influenza virus (A, B, C) | 8 (3%) | 9 (3%) | 11 (4%) | 9/11 (82%) |
| Escherichia coli | 6 (2%) | 3 (1%) | 9 (3%) | 8/9 (89%) |
| Human bocavirus | 0 | 9 (3%) | 9 (3%) | 4/9 (44%) |
| Pseudomonas aeruginosa | 5 (2%) | 7 (3%) | 9 (3%) | 8/9 (89%) |
| Viridans streptococci | 6 (2%) | 1 (<1%) | 7 (3%) | 7/7 (100%) |
| Human herpesvirus 6 or 7 | 0 | 5 (1·8%) | 5 (2%) | 5/5 (100%) |
| β-haemolytic streptococcus | 3 (1%) | 2 (1%) | 5 (2%) | 5/5 (100%) |
| Klebsiella pneumoniae | 3 (1%) | 0 | 3 (1%) | 3/3 (100%) |
| Pneumocystis jirovecii | 0 | 3 (1%) | 3 (1%) | 2/3 (67%) |
| Serratia marcescens | 3 (1%) | 0 | 3 (1%) | 3/3 (100%) |
| Enterobacter sp | 1 (<1%) | 1 (<1%) | 2 (1%) | 2/2 (100%) |
| Fusobacterium nucleatum | 0 | 2 (1%) | 2 (1%) | 2/2 (100%) |
| Mycoplasma pneumoniae | 0 | 2 (1%) | 2 (1%) | 0/2 (0%) |
| Parechovirus | 0 | 2 (1%) | 2 (1%) | 2/2 (100%) |
| Bordetella pertussis | 1 (<1%) | 1 (<1%) | 1 (<1%) | 1/1 (100%) |
| Citrobacter freundii | 1 (<1%) | 0 | 1 (<1%) | 1/1 (100%) |
| Morganella morganii | 0 | 1 (<1%) | 1 (<1%) | 1/1 (100%) |
| Stenotrophomonas maltophilia | 1 (<1%) | 1 (<1%) | 1 (<1%) | 1/1 (100%) |
Data are n, n (%), or n/N (%). Patients with definite LRTI had both a clinical diagnosis of LRTI or pneumonia and a microbiological diagnosis made using clinician-ordered culture and PCR-based diagnostic testing. Patients with suspected LRTI had only a clinical diagnosis of LRTI or pneumonia with negative clinician-ordered microbiologic testing. LRTI=lower respiratory tract infection. mNGS=metagenomic next generation sequencing.
Co-detected refers to a microbe that was detected along with other microbes in a patient, including multiple viruses or multiple bacteria.
Viruses were more commonly detected than bacteria (p<0·0001), with one or more viral pathogens detected in 223 (81%) children and one or more bacterial pathogens detected in 175 (63%) children. A viral pathogen was exclusively identified in 77 (28%) children and a bacterial pathogen in 31 (11%) children. Bacterial pathogens were identified by mNGS in 82 (46%) children with negative cultures. Fungal pathogens were detected by mNGS in three children (<1%). At least one bacterial and one viral pathogen were detected together in 144 (52%) children. We were unable to detect a pathogen in 21 (8%) children with suspected LRTI.
The most common pathogen overall was respiratory syncytial virus, which was detected in 127 (46%) children by the combination of clinical testing, followed by Haemophilus influenzae (70 [25%]), Moraxella catarrhalis (65 [24%]), rhinovirus A, B, C (61 [22%]), Streptococcus pneumoniae (32 [12%]), Staphylococcus aureus (25 [9%]), parainfluenza virus 1–4 (22 [8%]), adenovirus (20 [7%]), coronavirus (HCoV-229E, HCoV-NL63, or HCoV-OC43; 18 [7%]), and human metapneumovirus (17 [6%]; table 2, figure 2). Respiratory syncytial virus and H influenzae were co-detected in 47 (17%) patients and were significantly co-occurring (q=0·010). H influenzae and human metapneumovirus were co-detected in nine (3%) patients and were also significantly co-occurring (q=0·020). Co-detection of respiratory syncytial virus and rhinovirus (q=0·010) and respiratory syncytial virus and human metapneumovirus (q=0·014) were significantly unlikely to be detected together.
Figure 2: Pathogens detected in children with definite or suspected lower respiratory tract infections.
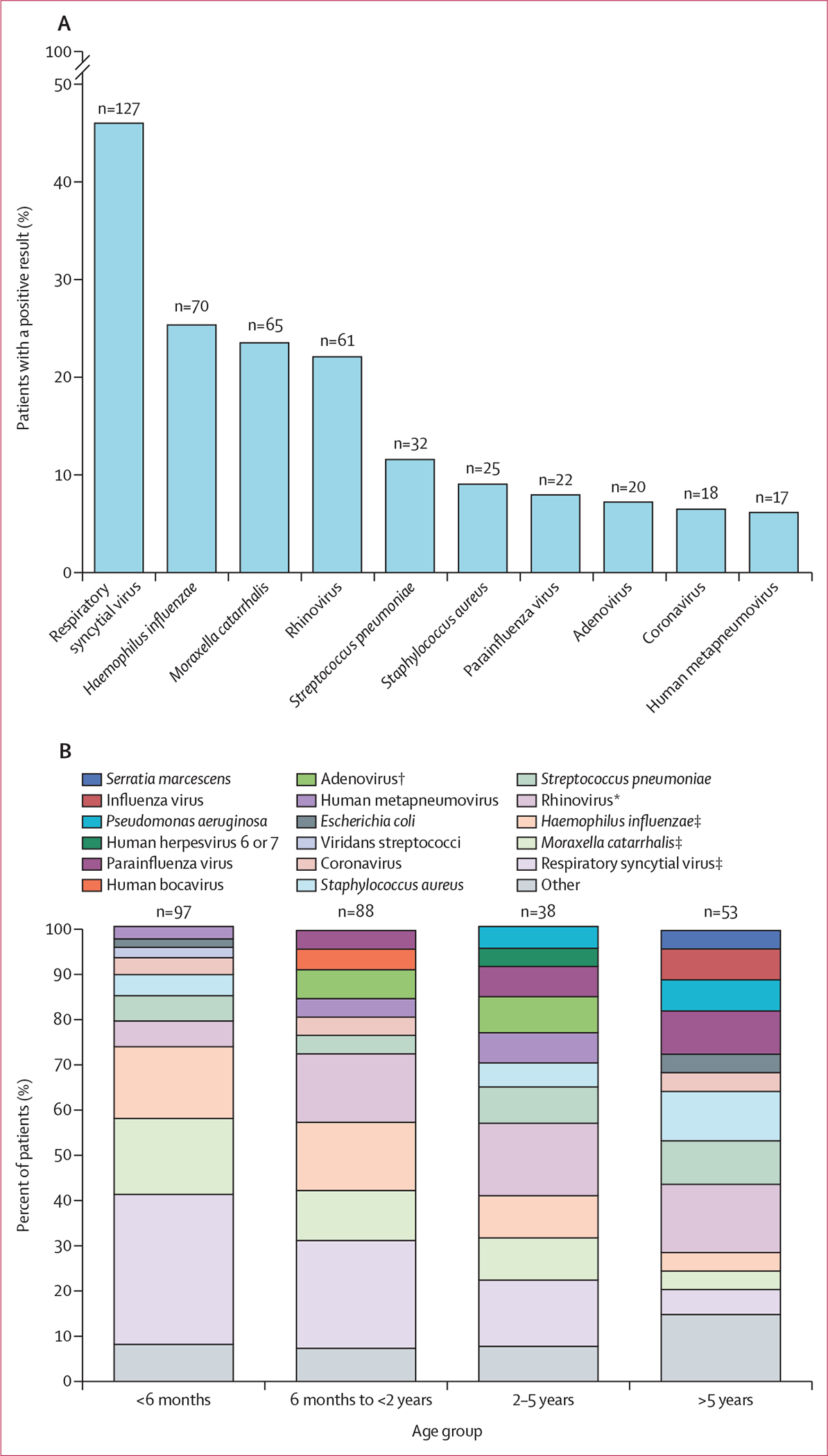
(A) Numbers (above bars) and percentages (bars) of patients with a positive result for each indicated pathogen by any method. Multiple pathogens from the same patient are included. (B) Proportions of the ten pathogens detected most frequently in each age group, with remaining pathogens grouped as other. In age groups in which there were multiple pathogens with equal numbers detected as the tenth most common pathogen, more than ten pathogens were included. Proportions are calculated as the number of cases of a given pathogen out of the total number of cases in the age group (children might have been counted more than once if they had multiple pathogens present). Statistical significance was determined by χ2 test. *p<0·05. †p<0·01. ‡p<0·001.
We also identified uncommon but putative LRTI pathogens by mNGS that were not detected by clinician-ordered testing, including human bocavirus (n=9), parechovirus (n=2), Ureaplasma parvum (n=1), Neisseria meningitidis (n=1), and Pseudomonas putida (n=1) (appendix p 10). Additionally, viruses of unclear clinical significance (Annelloviridae species and alphapapillomavirus 7) were identified in a subset of children and were always present in the context of an established LRTI pathogen (appendix p 10).
Detection of microbial pathogens in the lower airway can occur both in the context of infection as well as in the setting of asymptomatic carriage.14 To assess for this, we evaluated children with acute respiratory failure but no clinical evidence of LRTI, and a clear alternative explanation for intubation (eg, trauma, seizure, or surgery; n=121). Within this group, we detected a pathogen in 35 (29%) children by clinical diagnostics, 74 (61%) children by mNGS, and 82 (68%) by either method (appendix p 10). The most commonly detected pathogens were rhinovirus (30 [25%]) and S aureus (14 [12%]).
To determine whether certain pathogens were associated with LRTI, we compared the number of cases of each pathogen in patients with or without LRTI. Four pathogens were significantly more likely to be found in patients with LRTI: respiratory syncytial virus, human metapneumovirus, H influenzae, and M catarrhalis (figure 3; appendix p 19). Compared with patients with respiratory syncytial virus-related LRTI, patients without LRTI who had respiratory syncytial virus detected had lower viral loads (p=0·018); however, no difference was observed for rhinovirus (p=0·081; appendix p 21).
Figure 3: Microbes with established LRTI pathogenicity detected in patients with definite or suspected LRTI versus no evidence of LRTI.
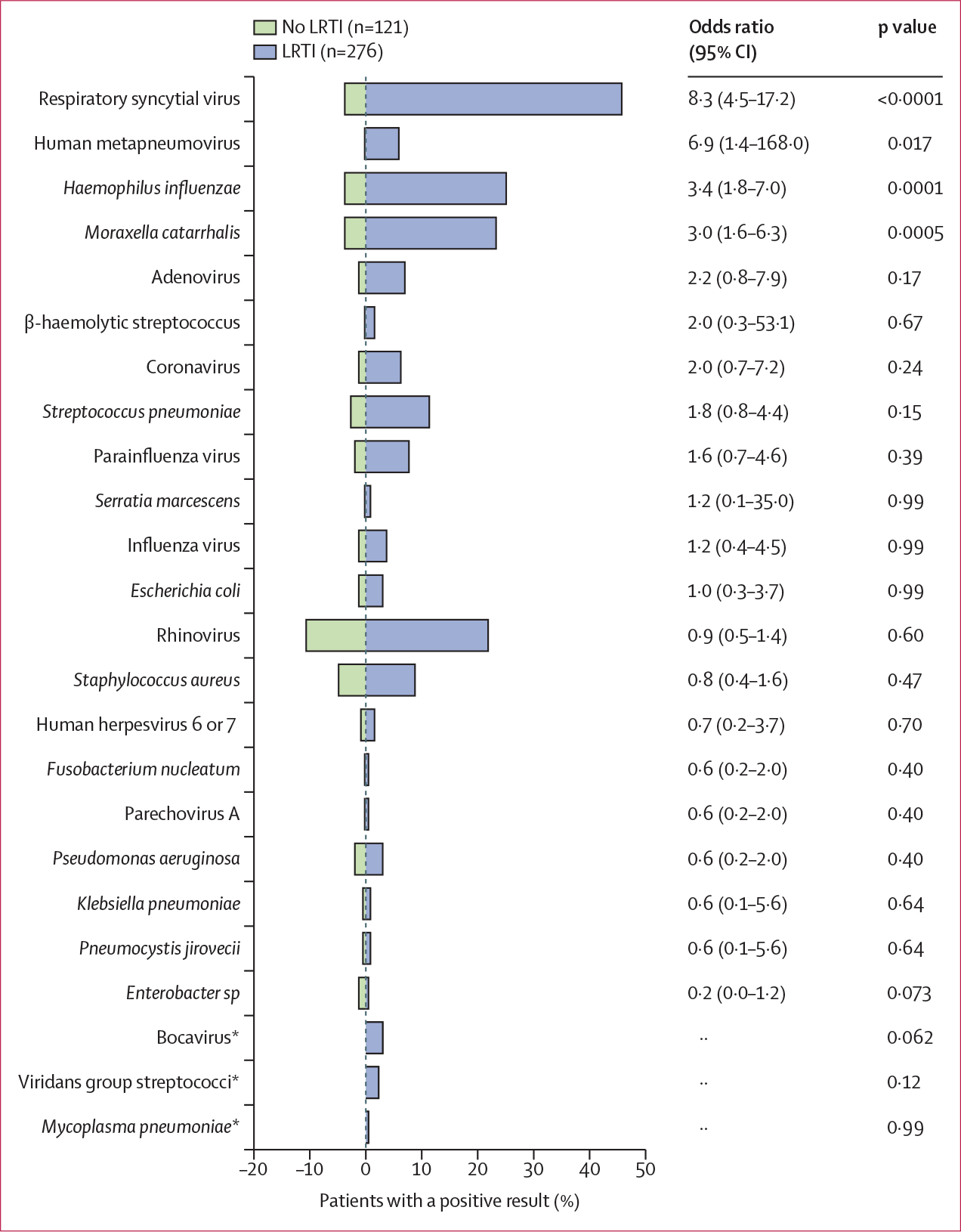
Microbes detected only once in the cohort are excluded from the graph. Bars represent the percent of patients in each group with a positive result by any method. p values were determined by Fisher’s exact test. LRTI=lower respiratory tract infections. *Cases occurred in only one group so odds ratios for these pathogens could not be calculated.
In patients with clinically adjudicated LRTI, rhinovirus was associated with a shorter length of stay in the hospital (p=0·012) and the paediatric ICU (p=0·046) after adjusting for clinical predictors (appendix pp 14–15). No other pathogens were associated with clinical outcomes after adjusting for predictors. Additionally, we did not find significant differences in clinical outcomes between patients with respiratory syncytial virus-A and respiratory syncytial virus-B (appendix p 16).
The prevalence of five pathogens was statistically different across age groups in patients with LRTI: respiratory syncytial virus (p<0·0001), H influenzae (p=0·0006), M catarrhalis (p=0·0002), adenovirus (p=0·0040), and rhinovirus (p=0·017; figure 2B). The most frequently detected pathogens in children younger than 6 months of age were respiratory syncytial virus (73%), M catarrhalis (37%), and H influenzae (35%) and prevalence of these pathogens markedly decreased with age across all groups, together accounting for only 14% of cases in the oldest group. Rhinovirus was most prevalent in children aged 6 months to 2 years (30%) and 2–5 years (32%), and adenovirus was most prevalent in children aged 2–5 years (16%).
To investigate differences in the seasonal prevalence of pathogens, we determined the distribution of cases for each of the ten most common pathogens over the course of the study. Both viral and bacterial LRTI pathogens were most prevalent in the autumn and winter months, with 72·5% of viral cases (p<0·0001) and 69·9% of bacterial cases (p<0·0001) detected between September and February (appendix p 22). We also observed differences in pathogen distribution by study site (appendix p 23).
Due to the disproportionate burden of respiratory syncytial virus in our study population we characterised the genomic diversity of respiratory syncytial virus. Both respiratory syncytial virus-A and respiratory syncytial virus-B were identified across study sites, with seasonal variation between prevalence of related isolates (appendix pp 24–25). Additionally, we observed geographical clustering by site, with one site (E) demonstrating a disproportionate number of related isolates suggesting community acquisition (appendix pp 24–25).
Discussion
We identified a putative microbiological cause in 92% of LRTI cases using a combination of clinician-ordered diagnostics and lower respiratory mNGS, a technology not previously used for LRTI surveillance in mechanically ventilated children. mNGS complemented clinical testing and enabled a microbiological diagnosis in a greater proportion of cases (90%) than clinician-ordered testing alone (67%).
Respiratory syncytial virus was the most common pathogen detected in children with LRTI (127 [46%]), as previously reported.3,5,15 Viruses were more commonly detected than bacteria (81% vs 63% of LRTI cases, p<0·0001), and 52% of children had co-detection of both a viral and bacterial pathogen, in line with previous reports.3,5,16 H influenzae and M catarrhalis were the most common bacterial pathogens detected (25% and 24% of cases, respectively), and were found at a prevalence higher than reported in previous studies of mostly noncritically ill patients.3,5 Given the reduction in invasive H influenzae serotype b disease by 99% in the USA since vaccine introduction in 1987, these findings highlight an additional opportunity for disease prevention through vaccination against non-serotype b strains. Although Mycoplasma pneumoniae was the most prevalent bacterial pathogen detected in the EPIC study of paediatric community-acquired pneumonia,5 we detected this microbe in less than 1% of children, suggesting a lower burden of disease in the most critically ill children.
In addition to established pathogens, mNGS also identified uncommon, potential LRTI pathogens. These included N meningitidis and other Neisseria species most commonly believed to be commensals but occasionally implicated in pneumonia,17 as well as U parvum, which is increasingly recognised as a pathogen in neonates and immunocompromised patients.18 Bocavirus, which has been identified as a cause of severe pneumonia only in the past 5 years,14 was identified in 3% of children.
Previous work has demonstrated that mNGS can enable pathogen identification in patients who have received antibiotics before sample collection,6,19,20 because microbial nucleic acid persists even if organisms are no longer viable. Reflecting this, we found that mNGS identified microbes missed by culture-based methods and identified putative bacterial LRTI pathogens in 46% of patients with suspected LRTI but negative culture-based clinical testing.
Both viral and bacterial LRTI were more common in autumn and winter months, consistent with previous reports,5 and probably reflecting greater transmission due to increased indoor exposures and the commonality of viral-bacterial co-infections. Together, our findings emphasise that detection of a viral pathogen alone does not rule out bacterial pneumonia in this population, even in the context of negative bacterial cultures. The high prevalence of co-infections also suggests the potential clinical use of mNGS for both microbiological diagnosis and antimicrobial stewardship, given the capacity of this technology to simultaneously detect bacterial and viral pathogens without culture.
Clinical deployment of mNGS for infectious disease diagnosis is increasingly common21 and has affected patient care by identifying novel and diagnostically challenging pathogens missed by standard testing modalities.21 Although mNGS has been clinically validated for pathogen detection from sterile site fluids such as cerebrospinal fluid, clinical use for microbiologically complex respiratory samples is still in the early stages.
We found that certain microbes were typically associated with LRTI, such as respiratory syncytial virus and human metapneumovirus, in line with previous studies demonstrating that these pathogens are more common in community-acquired pneumonia and symptomatic patients with upper respiratory tract infections than in asymptomatic controls.14,22,23 Bocavirus, which isn’t commonly the cause of severe pneumonia in children, was also associated with LRTI.24
Rhinovirus was the most frequently detected pathogen in patients with no evidence of LRTI (30 [25%]), consistent with previous studies;22,25 conversely respiratory syncytial virus was rarely detected. Children without LRTI who had respiratory syncytial virus detected had lower viral loads than those with respiratory syncytial virus-related LRTI, a difference that was not observed for rhinovirus, potentially reflecting greater overall virulence of respiratory syncytial virus versus rhinovirus. Consistent with this finding, patients with rhinovirus had shorter durations of overall hospital and paediatric ICU stays.
S aureus was detected in 12% of children without LRTI, suggesting that incidental detection of this pathogen in the lower airway might be underappreciated. Frequent detection of incidental pathogen carriage and multiple putative pathogens detected among patients with LRTI in our cohort emphasises the need for better methods, such as host transcriptional profiling,6 to assess the significance of pathogens identified by highly sensitive molecular assays. Understanding pathogen colonisation in asymptomatic patients is relevant to clinical decision making and antimicrobial stewardship.
Although the multicentre design of this study provided a more representative sample, it resulted in variability of viral PCR testing platforms, and clinical microbiological culturing and reporting techniques differed between sites.26 The use of a single mNGS assay might partially offset this variability, although variability in clinical testing probably affected results. Although mNGS identified most clinically detected pathogens, differences between clinical testing and mNGS results were also observed, particularly because the rules-based model identifies dominant pathogens, while other pathogens detected by mNGS are eliminated. Differences might also be attributable to variability in clinical microbiological practices between sites, lack of quantitative cultures, and differences in specimen types assayed by viral PCR (upper airway) versus mNGS (lower airway). Reflecting this, the sensitivity of mNGS increased from 63% to 87% if clinical culture data from a single study site and lower respiratory viral PCR were used instead as a reference. Additionally, although previous work has shown comparable performance between DNA sequencing and RNA sequencing for pathogen detection, we cannot rule out the possibility that our findings might have slightly differed if DNA sequencing data were used.
Distinguishing between pathogens and commensals is a challenge for LRTI diagnostics and, although the rules-based computational model provides a framework for doing so, the model has limitations. High taxonomic abundance or airway microbiome dominance does not necessarily equate to pathogenicity, and some bacteria (eg, S pneumoniae and H influenzae) can contextually be found as either commensals or pathogens. Further, some pathogens might lead to disease without being dominant in the airway.
Given that tracheal aspirate represents a mixture of microbes from throughout the lung, differences exist between tracheal aspirate specimens and those obtained by bronchoalveolar lavage. Previous work, however, has demonstrated compositional similarity of the lung microbiome between these specimen types in subjects with LRTI, whose microbial communities are typically characterised by dominant pathogens.27 In line with this finding, we observed that patients with definite LRTI had lower alpha diversity of their bacterial airway microbiome than those without LRTI. Although we observed compositional differences between groups, we did not observe a significant difference in beta diversity, which might relate to our observation of frequent incidental carriage of potentially pathogenic bacteria in the no evidence group.
Our findings might have been influenced by selection bias during patient enrolment and observer bias during LRTI adjudication. Enrolment occurred before the COVID-19 pandemic, and thus we were unable to assess prevalence of SARS-CoV-2 or the effect of infection control measures on community transmission of other pathogens. Finally, because this study was conducted on samples from the lower respiratory tracts of intubated patients, it was not possible to include a control group of healthy individuals.
In summary, we assessed the lower respiratory microbial landscape of severe LRTI. Overall, respiratory syncytial virus accounted for a disproportionate burden of disease, H influenzae and M catarrhalis were more common than previously recognised, and atypical microbes were identified. Incidental carriage of potentially pathogenic bacteria was frequent in uninfected controls, and viral or bacterial co-infections were identified in over half the patients. Together, these results advance our understanding of LRTI microbial epidemiology. Incorporating host transcriptional profiling with broadrange mNGS pathogen detection in future studies might improve the ability to distinguish true infection from colonisation, which has implications for diagnosis and antimicrobial stewardship. Furthermore, mNGS holds promise for enhancing emerging pathogen surveillance and for characterising the cause of LRTI in other patients at high-risk and understudied patient populations.
Supplementary Material
Research in context
Evidence before this study
We searched PubMed on Feb 11, 2021, for surveillance studies of lower respiratory tract infection (LRTI) in critically ill paediatric patients. Search terms were “lower respiratory tract infection,” “pediatric,” “epidemiology,” and “critical” with no date or language restrictions. Although 29 studies were identified, only two had clear applicability to the clinical question of LRTI surveillance in critically ill children. These studies are the EPIC study of community-acquired pneumonia requiring hospitalisation in US children and the PERCH study of severe pneumonia requiring hospitalisation in children younger than 5 years in Africa and Asia. Few studies have employed metagenomic next generation sequencing (mNGS) for microbial detection, and none have focused exclusively on critically ill children with LRTI requiring mechanical ventilation.
Added value of this study
This study advances our understanding of LRTI microbial epidemiology in a previously understudied population who face a high risk of adverse outcomes. We report the largest LRTI surveillance study of critically ill, mechanically ventilated children, and the only one thus far employing mNGS in addition to standard of care clinical infectious disease diagnostics. The combination of clinician-ordered diagnostics and lower respiratory mNGS enabled a microbiological diagnosis in 90% of cases, providing a more comprehensive and unbiased surveillance of paediatric LRTI than previous studies. Our findings demonstrate that respiratory syncytial virus might be disproportionately responsible for severe LRTI in children; Haemophilus influenzae and Moraxella catarrhalis might be more common than previously recognised; and viral-bacterial co-infection might occur at high rates in this population. Additionally, our findings suggest that detection of potentially pathogenic bacteria in mechanically ventilated children without LRTI is frequent.
Implications of all the available evidence
These results have implications for both preventive and therapeutic efforts. The potentially high prevalence of viral-bacterial co-infections in our cohort emphasises that detection of a respiratory virus alone does not rule out concomitant bacterial pneumonia. This study confirms the disproportionate burden of respiratory syncytial virus in critically ill children, emphasising an unmet need for vaccination and therapeutics targeting this viral pathogen. This work highlights the widespread detection of potentially pathogenic microbes in children admitted to an intensive care unit, even in the absence of LRTI. The frequent detection of incidental pathogen carriage, as well as the potential for co-infections with multiple pathogens, emphasises the need for better diagnostic methods, such as combined metagenomics microbe detection and host transcriptional profiling, to assess the significance of pathogens identified by highly sensitive molecular assays.
Acknowledgments
This work was supported in part by the following cooperative agreements from NICHD, NHLBI, and NIH: UG1HD083171 (PMM), 1R01HL124103 (PMM), UG1HD049983 (JC), UG01HD049934 (RWR, CL, and JMD), UG1HD083170 (MWH), UG1HD050096 (KLM), UG1HD63108 (AFZ), UG1HD083116 (AS), UG1HD083166 (PSM), UG1HD049981 (MMP), and K23HL138461-01A1 (CRL). The investigators thank all patients and their families for participating in this project. We also would like to acknowledge the contributions from investigators at the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, USA. Additionally, we would like to acknowledge the contributions of principal investigators, co-investigators, research coordinators, and allied research personnel at the following sites: Children’s Hospital of Colorado, Aurora, CO; Children’s Hospital of Michigan, Detroit, MI; Children’s Hospital of Philadelphia, Philadelphia, PA; Children’s National Medical Center, Washington, DC; Nationwide Children’s Hospital, Columbus, OH; Mattel Children’s Hospital, University of California Los Angeles, Los Angeles, CA; Children’s Hospital of Pittsburgh, University of Pittsburgh Medical Center, Pittsburgh, PA; Benioff Children’s Hospitals, San Francisco, San Francisco, CA; University of Utah; and Data Coordinating Center, Salt Lake City, Utah.
Footnotes
Declaration of interests
JK reports support from Genentech, outside the submitted work. LA reports funding from Pfizer, outside the submitted work. ABM reports grants from the Francis Family Foundation and the US National Institutes of Health (NIH) Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), outside the submitted work. EAFS reports grants from the US NIH during the conduct of the study. EAFS reports grants, personal fees, and non-financial support from AstraZeneca, Merck, Regeneron, Pfizer, and Roche; personal fees, non-financial support, and other support from AbbVie; personal fees from Alere; other support from GSK; grants from Johnson and Johnson; and grants and non-financial support from Novavax, outside the submitted work. MWH reports grants from NIH NICHD, during the conduct of the study; and personal fees from La Jolla Pharmaceuticals, outside the submitted work. AFZ received NICHD funding through the Collaborative Pediatric Critical Care Research Network during the conduct of the study. KLM reports grants from NIH, during the conduct of the study. AS reports grants from NIH, outside the submitted work. MMP reports grants from NIH, during the conduct of the study. PSM reports grants from NIH NICHD, during the conduct of the study. JMD reports grants from NIH, during the conduct of the study. MSZ reports grants from National Heart, Lung, and Blood Institute (NHLBI; K23HL146936), outside the submitted work. PMM reports grants from NIH, during the conduct of the study. All other authors declare no competing interests.
Contributor Information
Alexandra Tsitsiklis, Department of Medicine, Division of Infectious Diseases, University of California San Francisco, San Francisco, CA, USA.
Christina M Osborne, Section of Critical Care Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA; Section of Infectious Diseases, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Jack Kamm, Chan Zuckerberg Biohub, San Francisco, CA, USA.
Kayla Williamson, Department of Biostatistics and Informatics, University of Colorado, Colorado School of Public Health, Aurora, CO, USA.
Katrina Kalantar, Chan Zuckerberg Biohub, San Francisco, CA, USA.
Gytis Dudas, Gothenburg Global Biodiversity Centre, Gothenburg, Sweden.
Saharai Caldera, Department of Medicine, Division of Infectious Diseases, University of California San Francisco, San Francisco, CA, USA; Chan Zuckerberg Biohub, San Francisco, CA, USA.
Amy Lyden, Chan Zuckerberg Biohub, San Francisco, CA, USA.
Michelle Tan, Chan Zuckerberg Biohub, San Francisco, CA, USA.
Norma Neff, Chan Zuckerberg Biohub, San Francisco, CA, USA.
Victoria Soesanto, Department of Biostatistics and Informatics, University of Colorado, Colorado School of Public Health, Aurora, CO, USA.
J Kirk Harris, Section of Pulmonary Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Lilliam Ambroggio, Section of Emergency Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA; Section of Hospital Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Aline B Maddux, Section of Critical Care Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Todd C Carpenter, Section of Critical Care Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Ron W Reeder, Department of Pediatrics, University of Utah, Salt Lake City, UT, USA.
Chris Locandro, Department of Pediatrics, University of Utah, Salt Lake City, UT, USA.
Eric A F Simões, Section of Infectious Diseases, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Matthew K Leroue, Section of Critical Care Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA.
Mark W Hall, Department of Pediatrics, Division of Critical Care Medicine, Nationwide Children’s Hospital, Columbus, OH, USA.
Athena F Zuppa, Anesthesiology and Critical Care Medicine, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Joseph Carcillo, Department of Pediatrics, University of Pittsburgh, Pittsburgh, PA, USA.
Kathleen L Meert, Department of Pediatrics, Children’s Hospital of Michigan, Central Michigan University, Detroit, MI, USA.
Anil Sapru, Department of Pediatrics, University of California Los Angeles, Los Angeles, CA, USA.
Murray M Pollack, Department of Pediatrics, Children’s National Hospital and George Washington School of Medicine and Health Services, Washington, DC, USA.
Patrick S McQuillen, Department of Pediatrics, University of California San Francisco, San Francisco, CA, USA.
Daniel A Notterman, Department of Molecular Biology, Princeton University, Princeton, NJ, USA.
J Michael Dean, Department of Pediatrics, University of Utah, Salt Lake City, UT, USA.
Matt S Zinter, Department of Pediatrics, University of California San Francisco, San Francisco, CA, USA.
Brandie D Wagner, Department of Biostatistics and Informatics, University of Colorado, Colorado School of Public Health, Aurora, CO, USA.
Joseph L DeRisi, Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, CA, USA; Chan Zuckerberg Biohub, San Francisco, CA, USA.
Peter M Mourani, Section of Critical Care Medicine, Department of Pediatrics, University of Colorado School of Medicine and Children’s Hospital Colorado, Aurora, CO, USA; Department of Pediatrics, Section of Critical Care Medicine, University of Arkansas for Medical Sciences and Arkansas Children’s Hospital, Little Rock, AR, USA.
Charles R Langelier, Department of Medicine, Division of Infectious Diseases, University of California San Francisco, San Francisco, CA, USA; Chan Zuckerberg Biohub, San Francisco, CA, USA.
Data sharing
Deidentified individual patient data and raw fastq microbial sequencing alignments are available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA748764. Viral phylogenetic data are publicly accessible on the Global Initiative on Sharing All Influenza Data website at https://www.gisaid.org/.28 Code for the background correction algorithm is available at https://github.com/czbiohub/idseqr/. Code for the rules-based model is available at https://github.com/alexandra-t/rules_based_model.
References
- 1.WHO. The top 10 causes of death. 2018. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed Aug 1, 2021).
- 2.Khemani RG, Smith L, Lopez-Fernandez YM, et al. Paediatric acute respiratory distress syndrome incidence and epidemiology (PARDIE): an international, observational study. Lancet Respir Med 2019; 7: 115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien KL, Baggett HC, Brooks WA, et al. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 2019; 394: 757–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson RS, Asaro LA, Hutchins L, et al. Risk factors for functional decline and impaired quality of life after pediatric respiratory failure. Am J Respir Crit Care Med 2019; 200: 900–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015; 372: 835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA 2018; 115: e12353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mick E, Kamm J, Pisco AO, et al. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat Commun 2020; 11: 5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mourani PM, Sontag MK, Williamson KM, et al. Temporal airway microbiome changes related to ventilator associated pneumonia in children. Eur Respir J 2020; 57: 2001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsalik EL, Henao R, Nichols M, et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Science Translational Medicine 2016; 8: 322rall. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalantar KL, Carvalho T, de Bourcy CFA, et al. IDseq-an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 2020; 9: giaalll. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung NC, Miasojedow B, Startelc M, Gambin A. Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data. BMC Bioinformatics 2019; 20 (suppl 15): 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack MM, Holubkov R, Glass P, et al. Functional Status Scale: new pediatric outcome measure. Pediatrics 2009; 124: el8–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon TD, Haaland W, Hawley K, Lamblca K, Mangione-Smith R. Development and validation of the pediatric medical complexity algorithm (PMCA) version 3.0. Acad Pediatr 2018; 18: 577–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlaberg R, Queen K, Simmon K, et al. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J Infect Dis 2017; 215: 1407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghazaly M, Nadel S. Characteristics of children admitted to intensive care with acute bronchiolitis. Eur J Pediatr 2018; 177: 913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorburn K, Harigopal S, Reddy V, Taylor N, van Saene HKF. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006; 61: 611–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winstead JM, McKinsey DS, Tasker S, De Groote MA, Baddour LM. Meningococcal pneumonia: characterization and review of cases seen over the past 25 years. Clin Infect Dis 2000; 30: 87–94. [DOI] [PubMed] [Google Scholar]
- 18.Waites KB, Katz B, Schelonka RL. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin Microbiol Rev 2005; 18: 757–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langelier C, Zinter MS, Kalantar K, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med 2017; 197: 524–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinter MS, Dvorak CC, Mayday MY, et al. Pulmonary metagenomic sequencing suggests missed infections in immunocompromised children. Clin Infect Dis 2018; 64: 1847–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 2019; 380: 2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byington CL, Ampofo K, Stockmann C, et al. Community surveillance of respiratory viruses among families in the Utah better identification of germs-longitudinal viral epidemiology (BIG-LoVE) study. Clin Infect Dis 2015; 61: 1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Self WH, Williams DJ, Zhu Y, et al. Respiratory viral detection in children and adults: comparing asymptomatic controls and patients with community-acquired pneumonia. J Infect Dis 2016; 213: 584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicente D, Cilla G, Montes M, Pérez-Yarza EG, Pérez-Trallero E. Human bocavirus, a respiratory and enteric virus. Emerg Infect Dis 2007; 13: 636–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin ET, Kuypers J, Chu HY, et al. Heterotypic infection and spread of rhinovirus A, B, and C among childcare attendees. J Infect Dis 2018; 218: 848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prinzi AM, Parker SK, Curtis DJ, Ziniel SI. The pediatric endotracheal aspirate culture survey (PETACS): examining practice variation across pediatric microbiology laboratories in the United States. J Clin Microbiol 2020; 59: e02232–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalantar KL, Moazed F, Christenson SC, et al. Metagenomic comparison of tracheal aspirate and mini-bronchial alveolar lavage for assessment of respiratory microbiota. Am J Physiol Lung Cell Mol Physiol 2019; 316: L578–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 2017; 1: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified individual patient data and raw fastq microbial sequencing alignments are available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA748764. Viral phylogenetic data are publicly accessible on the Global Initiative on Sharing All Influenza Data website at https://www.gisaid.org/.28 Code for the background correction algorithm is available at https://github.com/czbiohub/idseqr/. Code for the rules-based model is available at https://github.com/alexandra-t/rules_based_model.


