Abstract
Diffusion-weighted magnetic resonance imaging (dMRI) is a non-invasive imaging technique that provides information about the barriers to the diffusion of water molecules in tissue. In the brain, this information can be used in several important ways, including to examine tissue abnormalities associated with brain disorders and to infer anatomical connectivity and the organization of white matter bundles through the use of tractography algorithms. However, dMRI also presents certain challenges. For example, historically, the biological validation of tractography models has shown only moderate correlations with anatomical connectivity as determined through invasive tract-tracing studies. Some of the factors contributing to such issues are low spatial resolution, low signal-to-noise ratios, and long scan times required for high-quality data, along with modeling challenges like complex fiber crossing patterns. Leveraging the capabilities provided by an ultra-high field scanner combined with denoising, we have acquired whole-brain, 0.58 mm isotropic resolution dMRI with a 2D-single shot echo planar imaging sequence on a 10.5 Tesla scanner in anesthetized macaques. These data produced high-quality tractograms and maps of scalar diffusion metrics in white matter. This work demonstrates the feasibility and motivation for in-vivo dMRI studies seeking to benefit from ultra-high fields.
Keywords: Diffusion MRI, Structural connectivity, Nonhuman primate, Tractography
1. Introduction
Diffusion-weighted magnetic resonance imaging (dMRI) is a non-invasive neuroimaging modality that is sensitive to the restriction of water molecules within tissue (Le Bihan et al., 1986). In the brain, dMRI has been used to study pathology in multiple disease states, including Alzheimer’s Disease, Parkinson’s Disease and schizophrenia (Granberg et al., 2017; Kamagata et al., 2020; Kubicki et al., 2007). More specifically, diffusion tensor imaging (DTI) uses this information to characterize the microstructure and primary fiber orientation present in each voxel (Le Bihan et al., 2001; Mori and Zhang, 2006). Other, more recent models, including constrained spherical deconvolution (CSD) (Tournier et al., 2008) and neurite orientation dispersion and density imaging (NODDI) (Zhang et al., 2012), also estimate multiple fiber orientations as well as axonal density in brain tissue. These estimates can then be utilized by tractography algorithms to visualize white matter pathways (Behrens et al., 2003; Tournier et al., 2019) and estimate structural brain connectivity.
The majority of dMRI brain studies (in both humans and nonhuman primates) have been conducted at 3 Tesla, whether in individual laboratories or through large-scale efforts like the human connectome project (HCP) (Autio et al., 2020; Sotiropoulos et al., 2013; Tounekti et al., 2018). Such studies can be more than adequate for some applications of dMRI, such as the assessment of between-population differences in white matter tissue properties. However, structural connectivity estimates derived from such dMRI data through tractography are often inaccurate, producing not only false connections, but also failing to reproduce known ones (Azadbakht et al., 2015; Bullock et al., 2022; Donahue et al., 2016; Grier et al., 2020; Maier-Hein et al., 2019; Reveley et al., 2015b; Schilling et al., 2019; Shen et al., 2019; Thomas et al., 2014). One contributing factor to these inaccuracies is the large dMRI voxel size relative to an axon or axonal fasciculi. An average axon is < 1 μm in diameter (Liewald et al., 2014), whereas a typical isotropic dMRI voxel is ≳1.5 mm. This means a single voxel can contain millions of axons with various orientations or exhibit partial volume effects that can confound tractography estimates (Alexander et al., 2001). One potential solution to this dilemma is improved spatial resolution, which requires higher signal-to-noise ratio (SNR). Substantial SNR gains can be achieved by using higher magnetic fields (Pohmann et al., 2016; Tavaf et al., 2021; Ugurbil, 2014, 2014; Vaughan et al., 2001), despite the accompanying decrease in T 2, which is disadvantageous for diffusion encoding with respect to SNR. Contemporary techniques, such as using single echoes (rather than two) enabled by removal of eddy current effects through image processing (Andersson and Sotiropoulos, 2016; Sotiropoulos et al., 2013), acquiring highly undersampled data along the phase encoding direction using parallel imaging and partial Fourier (Vu et al., 2015), and using high performance gradients (Setsompop et al., 2013; Sotiropoulos et al., 2013) make it possible to achieve sufficiently short echo-times (TEs) for diffusion encoding to exploit the intrinsic SNR gains of high magnetic fields (Uǧurbil et al., 2013). SNR can be further improved by adding a denoising step when preprocessing dMRI data (Moeller et al., 2021; Veraart et al., 2016b; Veraart et al., 2016c). Additional challenges of increased B0 and B1 inhomogeneity and increased tissue energy absorption at the higher magnetic fields are also becoming tractable using new techniques, in particular parallel transmit coils and parallel transmit RF pulse design (Uğurbil, 2018) and optimized RF pulses (Wu et al., 2018) As a result, 7 Tesla is increasingly prevalent in dMRI applications in humans (Gulban et al., 2018; Kida et al., 2016; Lenglet et al., 2012; Moeller et al., 2021; Sotiropoulos et al., 2016; Vu et al., 2015; Wang et al., 2019; Zitella et al., 2015).
Higher field scanners have also successfully been used to conduct dMRI studies, with improved spatial resolution, SNR, and/or contrast-to-noise ratio (CNR) in smaller mammals and ex vivo tissue (Sakai et al., 2020; Schaeffer et al., 2017). Ex vivo dMRI scans of the human brain can be collected on small-bore, very high field scanners typically reserved for animal studies. On such systems, voxel dimensions of a few hundred microns can be achieved, although they suffer from reduced CNR because of the properties of fixed tissue, often requiring scan times of hours to days (Bastiani et al., 2016; McNab et al., 2009; Roebroeck et al., 2019; Seehaus et al., 2015). In vivo dMRI scans of small animals, such as mice, have achieved similarly impressive resolutions (Alomair et al., 2015; Yon et al., 2020). Indeed, higher spatial resolution (0.5 mm vs 1 mm isotropic) achieved by limiting the diffusion encoding to low b values improves estimates of smaller fibers and reduces partial volume effects, which has been shown to improve tractography detection of U-fibers (Tounekti et al., 2018). Increased resolution could also improve gray matter applications of dMRI, such as distinguishing cortical regions and defining microstructure (Ganepola et al., 2018; Truong et al., 2014). However, tractography based on extremely high resolution dMRI data can still generate erroneous pathways, especially if the loss of SNR that accompanies higher resolutions pushes the data to a noise dominated, low SNR regime; for example, ex vivo diffusion MRI data acquired at 0.13 mm isotropic resolution produced more erroneous streamlines in multiple fiber pathways than data acquired at lower resolutions (Calabrese et al., 2014). Clearly, the optimal protocol for collecting high resolution data will require a balance between total scan time, angular resolution and b-values. Data obtained with both high and low resolutions can be combined to exploit advantages inherent in the two different data sets (Sotiropoulos et al., 2016).
The challenges and benefits of higher field strengths change according to the species studied, but the macaque is a frequently used neurobiological model for the human brain because of its size and the human-macaque cortical homologies not present in rodents (Hutchison and Everling, 2012; Laubach et al., 2022; Mars et al., 2016; Petrides and Pandya, 2002; Preuss, 1995; Watson and Platt, 2012). Furthermore, because of these similarities, the macaque brain is also used to study a wide array of mechanisms that model brain disorders and treatments (Heilbronner and Chafee, 2019; Johnson et al., 2021; Johnson and Slopsema, 2017; Nelson and Winslow, 2009; Vitek and Johnson, 2019; Worlein, 2014; Xiao et al., 2016). Unlike humans, macaques can undergo invasive procedures, such as tract-tracing, optogenetics, chemogenetics, and single-unit recordings (Cushnie et al., 2020; De et al., 2020; Galvan et al., 2017; Raper et al., 2019; Upright et al., 2018; Watanabe et al., 2020). There is a wealth of anatomical tract-tracing data available for the macaque, including CoCoMac and Core-Nets (Bakker et al., 2012; Markov et al., 2014; Stephan et al., 2001), which have been used to validate and refine diffusion tractography in macaques (Azadbakht et al., 2015; Donahue et al., 2016; van den Heuvel et al., 2015). The combination of strong homology to the human brain and the ability to employ invasive procedures to modulate neural activity and determine anatomical connectivity means that high-resolution in vivo dMRI of the macaque brain would be invaluable.
While very high resolution dMRI has been acquired with ex vivo scans in both humans and macaques (Azadbakht et al., 2015; Calabrese et al., 2015; Fritz et al., 2019; Sébille et al., 2019), there are significant benefits to acquiring high resolution scans in vivo. For example, individualized surgical planning for deep brain stimulation (DBS) in humans and macaques can benefit from in vivo dMRI for targeting (Davidson et al., 2020; Hunsche et al., 2013; Patriat et al., 2018; Plantinga et al., 2018; Johnson et al., 2016). Similarly, preoperative dMRI scans can guide intracranial electrode implantation in macaques (Saalmann et al., 2012). Other intraoperative surgeries in humans and/or macaques could benefit from accurate dMRI data on white matter organization and connectivity, including the current push for adeno-associated virus-mediated gene therapies (Hudry and Vandenberghe, 2019; Weiss et al., 2022). The use of in vivo dMRI also provides the opportunity to perform longitudinal studies that monitor normal development, the progress of various disease states or as a biomarker for neurodegeneration (Andica et al., 2020; Baldaranov et al., 2017; de Groot et al., 2016; Fields, 2008; Huber et al., 2018; Krogsrud et al., 2016; Lövdén et al., 2010; Mackey et al., 2012). These approaches are only possible with the use of in vivo dMRI acquisitions, which could be improved with higher resolution scans.
Here, we detail our efforts to acquire high-quality dMRI at 10.5 Tesla in anesthetized rhesus and long-tailed macaques. Macaques are too large for ultrahigh field, small-bore preclinical scanners often used for in vivo studies of smaller model organisms. The 10.5T/88 cm bore magnet utilized in the present study currently represents the highest field, large bore scanner in the world capable of scanning macaques (Yacoub et al., 2022) and humans (He et al., 2020). Our approach produces, to our knowledge, the highest spatial and angular resolution in vivo macaque dMRI data (0.58 mm isotropic voxels) to date and is presented as proof-of-concept for this type of data collection.
2. Materials and methods
2.1. Subjects
We obtained data from 5 female macaque monkeys (4 Macaca fascicularis and 1 Macaca mulatta). Weights ranged from 3.2 kg to 7.8 kgs. Experimental procedures were carried out in accordance with the University of Minnesota Institutional Animal Care and Use Committee and the National Institute of Health standards for the care and use of nonhuman primates. All subjects were pair-housed in a light and temperature-controlled colony room. Animals were fed and watered ad libitum. None of the animals had any prior implants or cranial surgery performed.
2.2. Data acquisition
All data were acquired on a passively shielded, 10.5 Tesla, 88 cm horizontal bore magnet with 60 cm accessible bore within the gradients and bore liner. This magnet was interfaced with a Siemens (Erlangen, Germany) console and electronics (Magnetom 10.5T Plus) and Siemens SC72 gradient sub-system operating with a maximum slew rate of 200 mT/m/s, and maximal gradient strength of 70 mT/m. The 10.5T system operates on the E-line (E12U) platform, comparable to clinical platforms (3T Prisma/Skyra, 7T Terra). As such, the user interface and pulse sequences were identical to those running on clinical platforms. A custom designed RF coil (Supplementary Figure 1a) with an 8-channel transmit/receive dipole array consisting of 18 cm long end-loaded dipoles combined with a close-fitting 16-channel loop receive only array head cap, and an 8-channel loop receive only array of 50 × 100 mm size located under the chin (Lagore et al., 2021; Yacoub et al., 2022) was used in all experiments; thus, the total number of receive channels in this coil is equal to 32. The 16 close fitting head cap receive only loops are divided into 14 individual receive loops of 37 mm combined with 2 larger ear loops of 80 mm. All receiver loops were arranged in an overlapping configuration for nearest neighbor de-coupling. The combined 32 receive channels support 3 fold acceleration in the phase encode direction. The coil holder was designed to be semi-stereotaxic, capable of holding the head of the animal in a stable, centered position via customized ear bars. The receive elements were modelled to adhere as close to the surface of the animals’ skulls as possible. Transmit phases were fine-tuned for excitation uniformity for one representative animal and the calculated phases were then used for all subsequent acquisitions. Magnetic field homogenization (B0 shimming) was performed using a customized field of view with the Siemens internal 3D mapping routines. Multiple iterations of the shims (using the cardiac adjusted FOV shim parameters) were performed and manual fine adjustments performed on each animal. High-order shim elements (3rd order) were disregarded for these procedures.
On scanning days, anesthesia was induced by intramuscular injection of atropine (0.5 mg/kg), dexmedetomidine (13 μg/kg) and ketamine hydrochloride (7.5 mg/kg). Animals were transported to the anteroom of the scanner and intubated with an endotracheal tube. Anesthesia was maintained using 1.5–2% isoflurane mixed with oxygen (2 L/h flow rate at 100%).
Animals were placed in sphinx position onto a custom-built coil bed with integrated ear bars for pseudo-stereotactic head fixation as described previously (Lagore et al., 2021; Yacoub et al., 2022). Rectal temperature, respiration rate, end-tidal carbon dioxide, electrocardiogram and oxygen saturation were monitored using an MRI compatible monitor (IRADIMED 3880 MRI Monitor, USA). Temperature was maintained with a circulating water bath, chemical heating pads and blankets for thermal insulation.
2.3. Anatomical scan
T1w images were acquired on all subjects with a 3D-MPRAGE sequence and T2w images were acquired on 3 subjects with a 3D T2 SPACE sequence. T1w: FOV: 131 × 150 mm2; matrix size of 280 × 320 (0.5 mm isotropic resolution); TR/TE of 3300/3.56 ms. T2w: FOV: 128 × 150 mm2; matrix size of 260 × 320 (0.5 mm isotropic resolution); TR/TE of 3000/100 ms; flip angle of 120 deg. In order to perform structural image processing for subjects without a T2w structural image, the bias field was corrected with the N4biascorrection tool as implemented in ANTS (Tustison et al., 2010). For subjects with a T2w structural image (not always acquired due to early termination of the scan because of animal monitoring issues), a T1w/T2w ratio image was generated by skull stripping both the T1w and T2w images, registering the T2w to the T1w space and then dividing the T1w by the T2w image.
2.4. dMRI acquisition and preprocessing
Pilot studies (not presented here) were performed in order to optimize the custom coil for the 10.5T magnet (Lagore et al., 2021). Testing was performed to achieve the most stable readout direction which resulted in pure z direction. This meant that the smallest field of view for whole brain coverage could be obtained in a sphinx position. A stable and reliable positioning of the animal was of utmost importance for reproducible and reliable data quality. Sequence optimization was performed within the limits set by our gradient and SAR monitoring. In short, we aimed at achieving the highest possible resolution resolvable using a single shot that allowed for coverage of the whole macaque brain and remained manageable with post-hoc distortion correction. Our coil geometry allows roughly 3x in-plane acceleration (GRAPPA in our case) for the small volume needed, limiting the overall matrix size we can achieve. Segmented encoding approaches will likely yield higher resolutions, at the cost of q-space sampling efficiency, but require extreme stability of the animal.”
The dMRI sequence utilized was a 2D single shot EPI acquired with reverse phase encoding head/foot (H/F), foot/head (F/H) consistent with the requirements for FSL Topup/Eddy offline EPI distortion correction (Andersson et al., 2003). B1 + field optimization was performed using a vendor provided flip angle mapping sequence and the power was calibrated for each subject. Precalibrated fixed phases were used in all subjects. Each scan included 3 or 4 pairs (H/F and F/H) of 114 diffusion-weighted volumes (TR/TE of 8270/78.4 ms; matrix size, 260 × 144 × 75; 6/8 phase partial Fourier, 0.58 mm isotropic resolution), contingent upon maintenance of acceptable vital signs. An iPAT acceleration of 3 was used without any slice acceleration. Each multi-shell acquisition contained 7 b = 0 s/mm2 vol (one every 16 vol), 54 b = 1000s/mm2 vol and 53 b = 2000s/mm2 images sampled on the whole sphere. Total scan time for the diffusion acquisition was approximately 2 h and 16 min.
Diffusion image preprocessing was performed using a custom pipeline incorporating components from FSL (Jenkinson et al., 2012), MRtrix3 (Tournier et al., 2019), and Freesurfer (Fischl, 2012). Dicoms were converted directly into the MRtrix image format and then back-converted to NIFTI files with the corresponding bvecs/bvals files. The sphinx orientation headers were corrected with Freesurfer’s mri_convert with–sphinx option for further processing with FSL. All acquisitions were visually inspected for gross movement and other artifacts that would prevent further analysis. Data was denoised with MRtrix dwidenoise and Gibbs ringing artifacts were corrected using MRtrix3 mrdegibbs. To increase the SNR for the estimation of the susceptibility induced off-resonance field, an average b0 image for each PE direction was generated and then passed to the TOPUP tool (Andersson et al., 2003) and implemented in FSL (Smith et al., 2004). Eddy current correction was performed using the FSL tool Eddy_Cuda9.1 (Andersson and Sotiropoulos, 2016) using the default settings with the following modifications (nvoxhp = 10,000 and –repol (Andersson et al., 2016)). After distortion and eddy current corrections, data was converted back to the MRtrix image format to estimate diffusion tensors and generate fractional anisotropy (FA), apparent diffusion coefficient (ADC) maps and RGB maps of the principal direction of diffusion using the MRtrix dwi2tensor and tensor2metric tools (Tournier et al., 2019). A visual summary of this pipeline is provided in Supplemental Figure 1B
SNR was quantified by drawing 5 ROIs on the FA map for each animal–Supplemental Figure 2. One gray matter ROI was drawn in the cortex. Two white matter ROIs were drawn (one in the corpus callosum and one in the anterior limb of the internal capsule). One mixed ROI was drawn on the descending posterior corona radiata. A final ROI was drawn outside of the brain to quantify the standard deviation of the noise. The mean signal intensity was measured in each ROI in data that had not been denoised or had Gibbs ringing corrected. SNR was calculated by dividing the mean signal within each ROI by the standard deviation of the noise in each volume. The calculated SNR values from each volume were averaged into a single value for each shell for each subject. These values are reported in Tables 1–2. The same ROIs were also used to calculate fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD) and radial diffusivity (RD). The mean and standard deviation of these metrics are reported in Table 3 – 4.
Table 1.
Signal-to-Noise Ratios 0.58 mm isotropic resolution diffusion weighted images in select regions prior to denoising and Gibb’s ringing removal.
| b = 0 s/mm2 | b = 1000s/mm2 | b = 2000s/mm2 | |
|---|---|---|---|
|
| |||
| Corpus Callosum | 17.01 ± 6.95 | 7.36 ± 2.25 | 5.58 ± 1.57 |
| ALIC | 19.66 ± 4.33 | 9.82 ± 2.13 | 5.68 ± 1.1 |
| dpcr | 13.52 ± 1.76 | 7.23 ± 2.13 | 5.3 ± 1.43 |
| gray matter | 14.7 ± 2.23 | 8.28 ± 1.10 | 5.56 ±.63 |
Table 2.
Signal-to-Noise Ratios 0.58 mm isotropic resolution diffusion weighted images in select regions after denoising and Gibb’s ringing removal.
| b = 0 s/mm 2 | b = 1000s/mm 2 | b = 2000s/mm 2 | |
|---|---|---|---|
|
| |||
| Corpus Callosum | 29.18 ± 16.42 | 12.67 ± 7.2 | 12.41 ± 7.53 |
| ALIC | 34.4 ± 20.96 | 32.91 ± 22.90 | 9.73 ± 5.21 |
| dpcr | 22.30 ± 15.42 | 12.96 ± 8.74 | 5.3 ± 1.43 |
| gray matter | 26.33 ± 16.93 | 15.03 ± 10.07 | 9.97 ± 6.63 |
Table 3.
Scalar Diffusion Metrics prior to denoising and Gibb’s ringing removal. Data reported as mean ± standard deviation.
| FA | MD (x10ˆ − 3)mm 2 /s | RD (x10ˆ − 3)mm 2 /s | AD (x10ˆ − 3)mm 2 /s | |
|---|---|---|---|---|
|
| ||||
| Corpus Callosum | 0.691 ± 0.041 | 0.743 ± 0.095 | 0.420 ± 0.058 | 1.379 ± 0.194 |
| ALIC | 0.637 ± 0.065 | 0.537 ± 0.087 | 0.288 ± 0.025 | 1.034 ± 0.225 |
| dpcr | 0.548 ± 0.041 | 0.600 ± 0.024 | 0.405 ± 0.040 | 0.992 ± 0.061 |
| gray matter | 0.128 ± 0.007 | 0.701 ± 0.038 | 0.655 ± 0.065 | 0.791 ± 0.045 |
Table 4.
Scalar Diffusion Metrics after denoising and Gibb’s ringing removal. Data reported as mean ± standard deviation.
| FA | MD (x10ˆ − 3)mm 2 /s | RD (x10ˆ − 3)mm 2 /s | AD (x10ˆ − 3)mm 2 /s | |
|---|---|---|---|---|
|
| ||||
| Corpus Callosum | 0.691 ± 0.041 | 0.721 ± 0.095 | 0.420 ± 0.062 | 1.325 ± 0.181 |
| ALIC | 0.630 ± 0.070 | 0.508 ± 0.078 | 0.277 ± 0.018 | 0.997 ± 0.205 |
| dpcr | 0.521 ± 0.041 | 0.583 ± 0.023 | 0.410 ± 0.023 | 0.928 ± 0.043 |
| gray matter | 0.118 ± 0.009 | 0.685 ± 0.033 | 0.644 ± 0.031 | 0.766 ± 0.038 |
2.5. Tractography
Tractography was performed with the MRtrix3 software package (Tournier et al., 2019). First, the response functions were estimated from the preprocessed diffusion images with dwi2response using the “dhollander” algorithm for multishell data. The response functions were then used to estimate fiber orientation directions (FODs) with a constrained spherical deconvolution (CSD) using dwi2fod with the msmt_csd algorithm that generated three separate FODs maps (white matter, gray matter and CSF) (Jeurissen et al., 2014; Tournier et al., 2004). FODs were normalized for intensity using mtnormalize (Raffelt et al., 2017). For fiber tracking, tckgen was used with the iFOD2 model (Tournier et al., 2010). Whole brain tractograms were generated from a white matter mask. Individual tracts were assessed with seeds manually drawn on each subject’s FA map. Seeds were defined as the injection sites for previously described pathways as determined by anatomical tract-tracing to assess the quality of the tractography. No waypoints or exclusion masks were used for these tractograms.
3. Results
3.1. Anatomical images
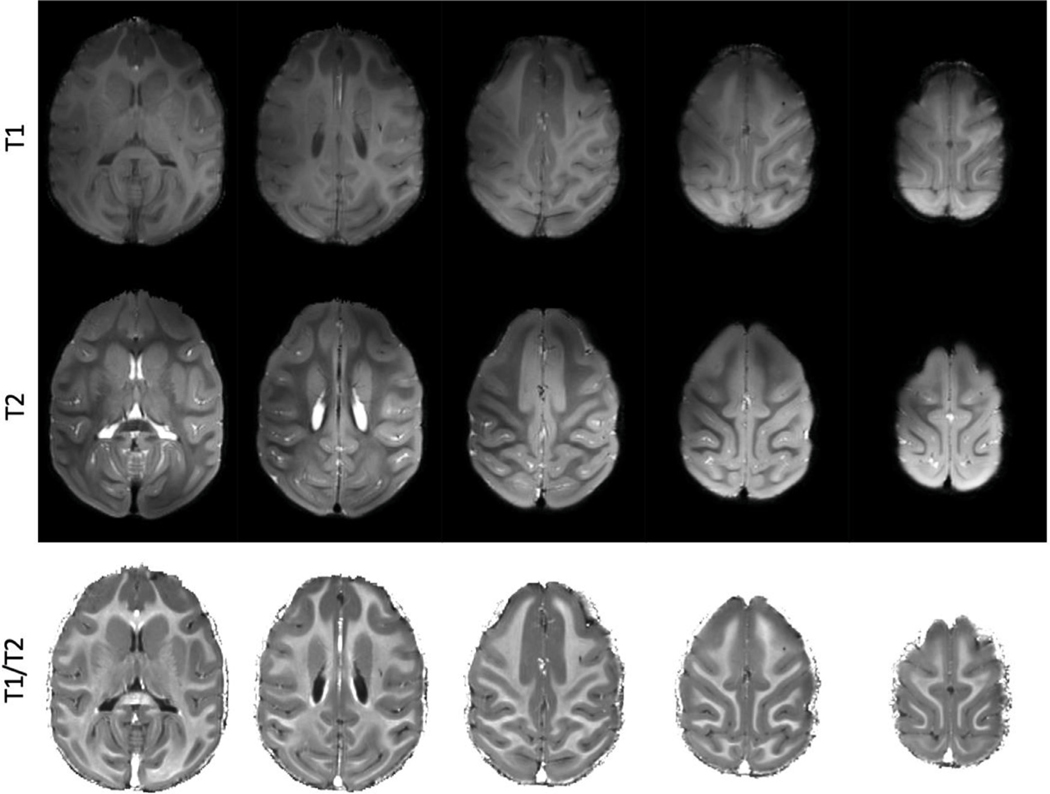
Anatomical imaging of the whole macaque brain was performed using our custom 32 channel receive (32Rx) coil. Selected images are shown in Fig. 1. Cortical, subcortical, and white matter structures are all clearly visible. A more extensive discussion of the anatomical acquisition can be found here (Lagore et al., 2021).
Fig. 1. Structural images of the macaque brain acquired at 10.5T.
Top panel shows T1w images. Middle panel shows T2w images. The bottom panel shows a T1w/T2w ratio image.
3.2. dMRI
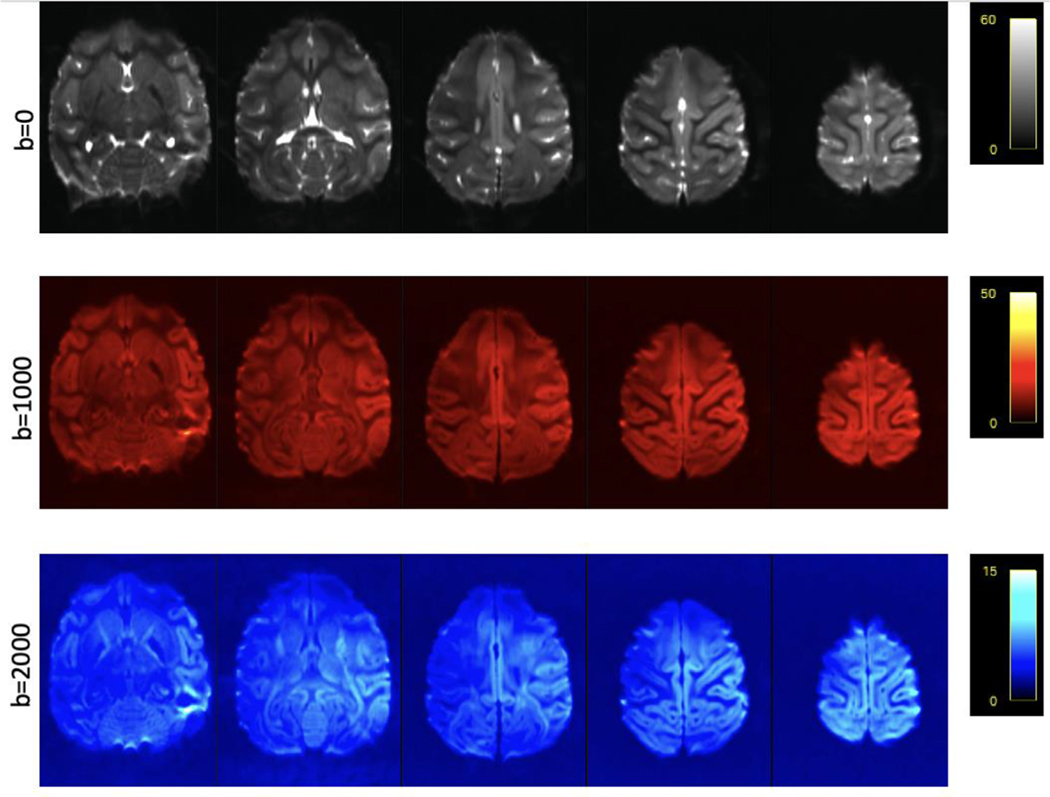
SNR was relatively homogeneous throughout the brain. Average SNR maps for each shell (prior to denoising and Gibbs ringing removal) are shown in Fig. 2 and Supplemental Figures 3–6. SNR values for each region of interest before and after denoising/Gibbs ringing removal are reported in Table 1 and Table 2 respectively. On average, SNR was increased by approximately 75% after denoising and removal of Gibbs ringing.
Fig. 2. Signal-to-noise ratio maps for 0.58 mm isotropic resolution dMRI data acquired at 10.5T from a single subject, I.
The average SNR for each shell is shown. Selected axial slices show a relatively homogenous SNR through the entire brain.
3.3. Denoising
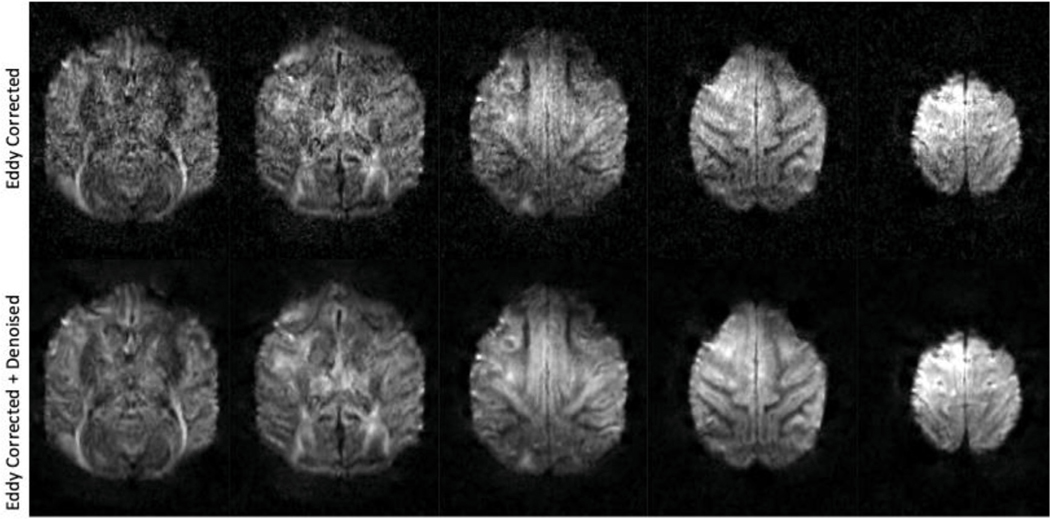
dMRI has an intrinsically low SNR (relative to other MRI modalities). The SNR is proportional to the voxel volume at a fixed field strength. When increasing the resolution of the diffusion scan, there is a corresponding reduction in SNR. To maintain SNR at high resolution (especially at higher b-values which provide higher diffusion weighting, but at the penalty of increased noise), a denoising algorithm was applied to these data. Prior to any interpolation, the dwidenoise tool in MRtrix based on the MPPCA algorithm (Veraart et al., 2016b, 2016c) was applied to the data. Fig. 3 shows the dMRI data after eddy-current distortions correction (top half) and eddy-current distortion corrected plus dwidenoise denoising (bottom half). Note that the more superficial slices (thus closer to the coil with higher SNR) show less improvement with denoising, though the boundaries of cortical sulci become more defined. Deeper structures show much cleaner boundaries. This is readily apparent in the optic radiation. Some sulci that are not well-defined in the eddy-current corrected only data can be visualized more readily in the denoised dataset.
Fig. 3. Effect of MPPCA denoising on diffusion weighted images.
The top panel shows 5 axial slices of data processed through Eddy and Topup. The bottom panel shows Eddy corrected with additional denoising.
3.4. Gibbs ringing artifact correction
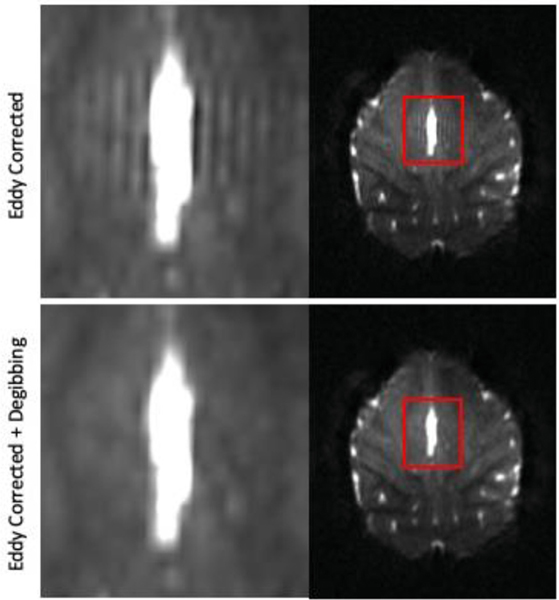
Gibbs ringing is an imaging artifact observed at boundaries between tissue types. This artifact manifests as parallel lines adjacent to high contrast tissue boundaries in the brain, often observed near the ventricle and cortex/CSF interface. It is important to correct these artifacts as they can significantly alter modeling of the diffusion tensor and kurtosis parameters (Kellner et al., 2016; Perrone et al., 2015; Veraart et al., 2016a). Gibbs ringing is a truncation artifact caused by partial k-space reconstructions and it can be corrected with the MRtrix tool mrdegibbs (Kellner et al., 2016). One way to minimize the appearance of Gibbs ringing is to increase the matrix size of the acquisition. However, while this would minimize the ringing artifact, it is not ideal given the already long readout time, which would increase further along with geometric distortions, signal dropouts, and echo time. Fig. 4 shows the removal of Gibbs ringing. This artifact is clearly visible in the b0 images and where the enlarged area demonstrating Gibbs ringing is highlighted with red boxes. The ringing artifact is clearly visible in the upper panel near the ventricles and cortical surface. The lower panel shows the data with the ringing removed by the MRtrix tool, mrdegibbs.
Fig. 4. Removal of Gibbs ringing artifact.
The top panel shows parallel lines in the b = 0 image immediately adjacent to a ventricle/gray matter interface. The bottom panel shows the data after being processed with mrdegibbs. The ringing artifact is removed, while retaining the integrity of the original data.
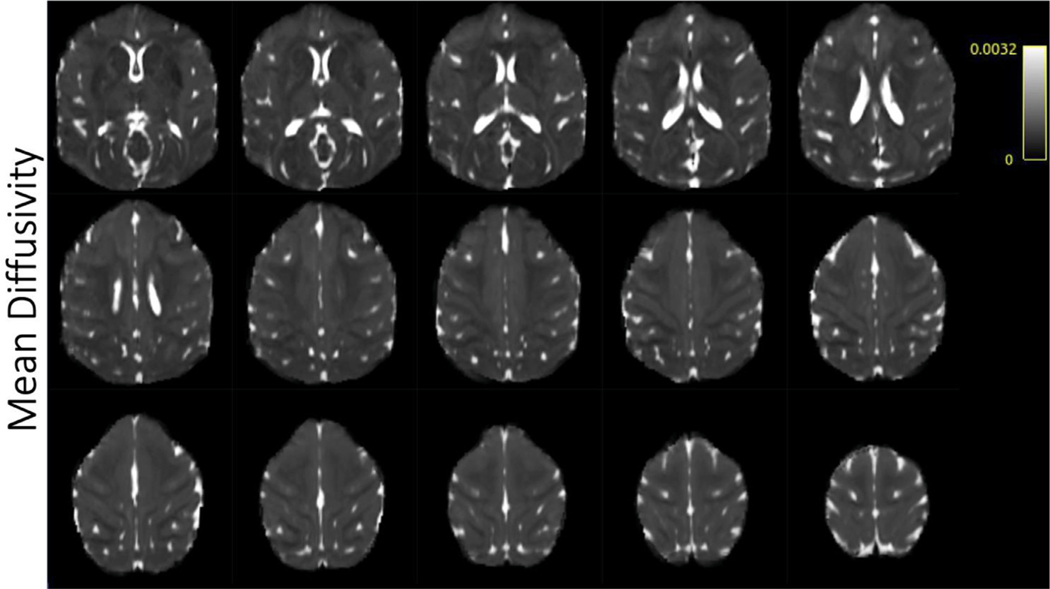
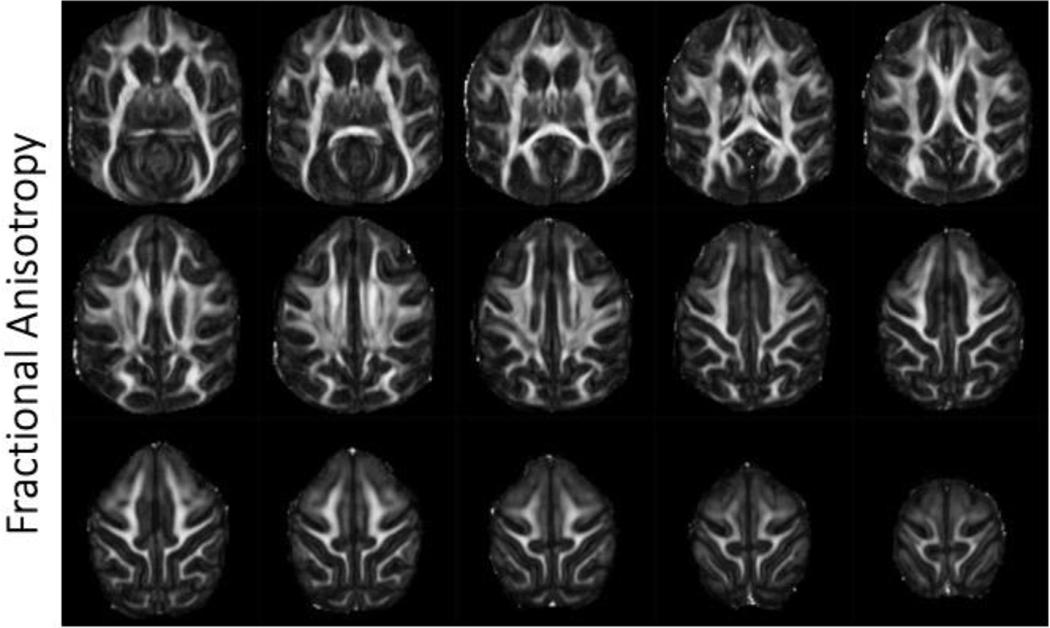
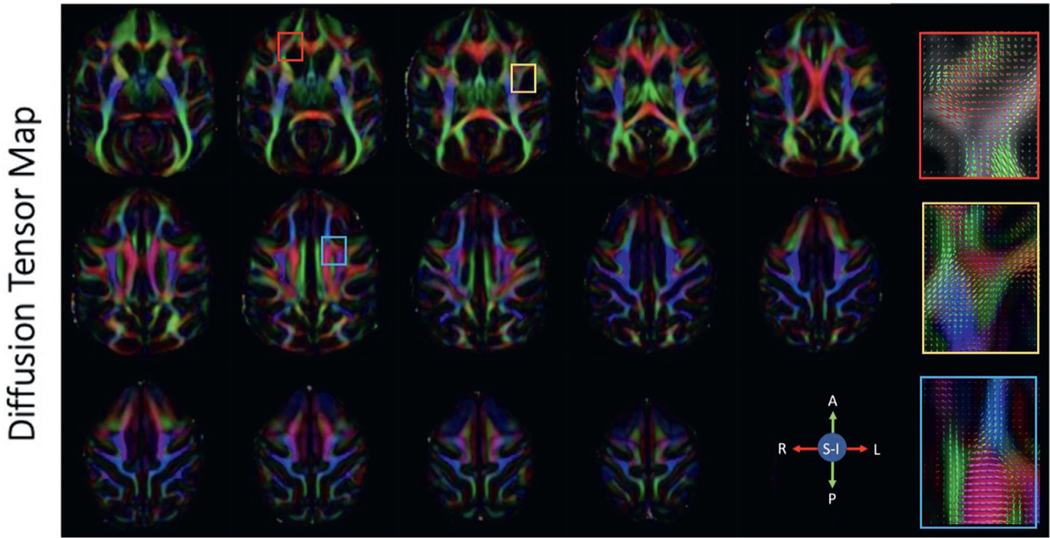
3.5. Fractional anisotropy/MD/DTI
Fractional anisotropy (Fig. 5), mean diffusivity (Fig. 6), and color-coded fractional anisotropy (Fig. 7) maps were generated after denoising and Gibb’s ringing removal. Scalar metrics calculated before and after denoising/Gibbs ringing removal are summarized in Tables 3 and 4, respectively. Exemplar fiber orientation distribution functions are displayed in the colored boxes in Fig. 7. The high resolution scans allow for differentiation of smaller fiber bundles and structures that are often affected by partial volume effects. These include U-fibers and gray matter/white matter boundaries in the cortex.
Fig. 5.
Selected axial slices showing mean diffusivity in a single subject acquired at 0.58 mm isotropic resolution.
Fig. 6. Fractional anisotropy.
Selected axial slices showing fractional anisotropy in a single subject acquired at 0.58 mm isotropic resolution.
Fig. 7. Diffusion tensor maps.
Selected axial slices showing the principal diffusion direction in a single subject acquired at 0.58 mm isotropic resolution. Red, green and blue represent fibers traveling right-left, anterior-posterior and superior-inferior respectively. The colored boxes correspond to the fiber orientation distribution functions displayed on the right.
3.6. Tractography
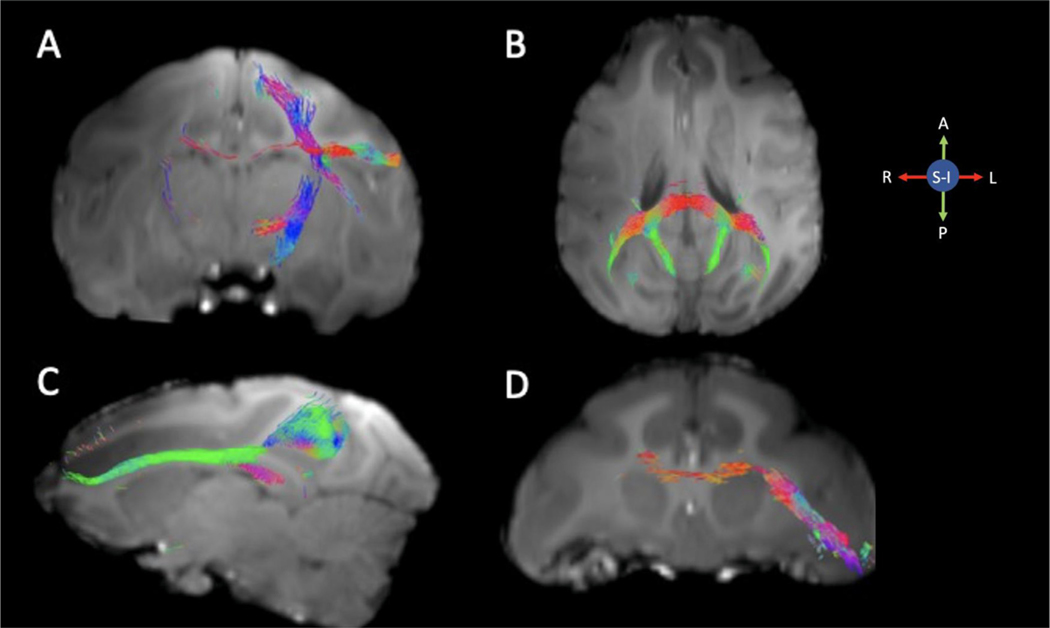
Fig. 8 shows whole brain streamlines generated from the white matter FODs. To demonstrate the feasibility of tractography at 10.5T with 0.58 mm isotropic resolution, Fig. 9 shows tractography derived from well-described pathways with tract-tracing validation from the literature. The tractography seeds are shown in supplemental figure 7. In Fig. 9 A, a seed was drawn in the motor cortex (Case 24 from (Schmahmann and Pandya, 2006)). Fibers can be seen clearly descending in the internal capsule, as well as crossing the corpus callosum, travelling in the superior longitudinal fasciculus, and traversing toward the external capsule. Although all of these belong to the correct bundles, we also see some streamlines descending in the contralateral internal capsule, which represents a false positive. In Fig. 9 B, a seed was placed in the white matter of the caudal corpus callosum (connectivity previously defined in (Kennedy et al., 1986; Kennedy and Dehay, 1988; Sunderland, 1940)), resulting in streamlines reaching large portions of the parietal and occipital lobes, while avoiding more frontal and ventral cerebral cortical regions. In Fig. 9 C, a seed was placed in the precuneus (Case 17 from (Schmahmann and Pandya, 2006)). Here, streamlines can be seen traversing the dorsal cingulum bundle (but not temporal or subgenual components, (Heilbronner and Haber, 2014) and the superior longitudinal fasciculus I. Finally, in Fig. 9 D, a seed was placed in the ventrolateral prefrontal cortex (Case 33 from (Schmahmann and Pandya, 2006)). Streamlines can be seed traveling dorsally and medially, around the striatum, to reach the corpus callosum and cross to the contralateral hemisphere.
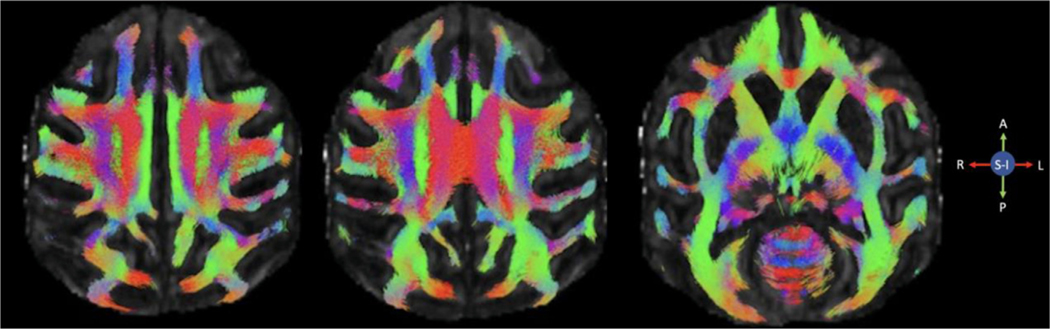
Fig. 8. Representative whole brain tractograms from a single subject.
Selected axial slices showing whole brain streamlines.
Fig. 9. Tractography in dMRI data collected at 10.5T.
Tractography from a single subject with seeds in the motor cortex (A), caudal corpus callosum (B), precuneus (C), and ventrolateral prefrontal cortex (D). Streamlines are overlaid on a T1 image registered to the diffusion space.
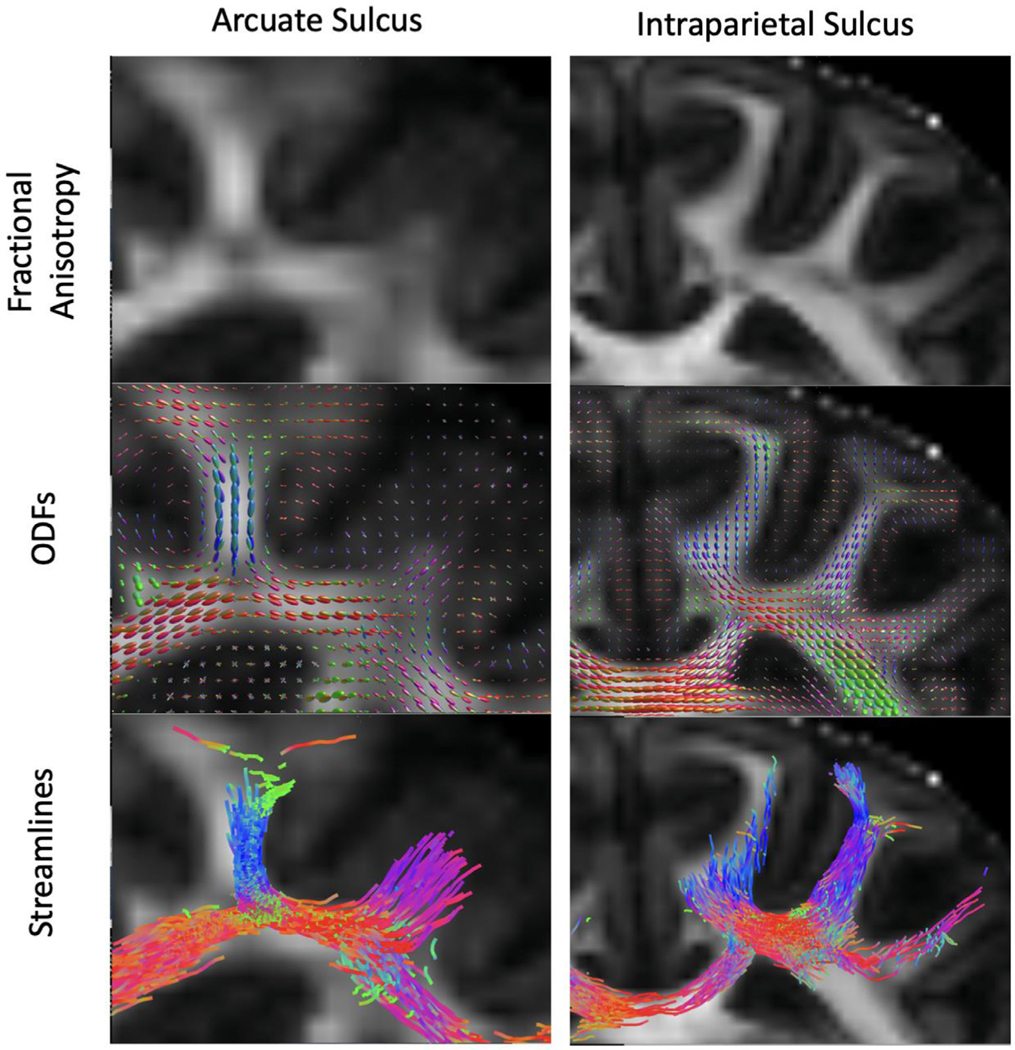
3.7. U fibers
Sulci present specific issues for tractography; namely, at the fundus, it is difficult to track both U-fibers (local association fibers coursing around the sulcus) and the deep, penetrating fibers that represent longer-range connections (Reveley et al., 2015a). Here, we seeded the fundus of the arcuate sulcus and the intraparietal sulcus. Fig. 10 shows both U-fibers and deep penetrating fibers. Thus, with this acquisition protocol, we are able to visualize the U-fibers described in ex vivo scans (Oishi et al., 2011) and with in vivo 3D-msEPI acquisitions (Tounekti et al., 2018). Supplemental figures 8 and 9 demonstrate consistency across multiple subjects for the same U-fibers in the arcuate and intraparietal sulci, respectively.
Fig. 10.
U-fibers. Fractional anisotropy maps for the arcuate and intraparietal sulci that contain short association fibers, also known as U-fibers. The middle panel shows the ODFs in the corresponding region. The lower panel displays streamlines generated from a seed that straddles the gray matter/white matter boundary as described by Reveley (Reveley et al., 2015a).
4. Discussion
Diffusion neuroimaging is a noninvasive imaging modality that provides information about the diffusion of water molecules in tissue in both clinical and nonclinical populations. Of particular relevance to neuroscience is the ability to model white matter in the brain from these properties and to generate estimates of anatomical connectivity from tractography. Such connectivity measures are expected to improve with higher spatial resolutions. However, going beyond the typically supramillimeter resolutions commonly employed today to submillimeter resolution dMRI is challenging due to the reduction in SNR caused by the correspondingly smaller voxel volume. Utilizing an ultra-high field magnet is one method to improve the SNR when compared to the same voxel size acquired at lower fields, assuming the echo time for diffusion encoding, TE, is not undesirably long. Macaques, like humans, are too large for other high field, small-bore MRIs that have been used extensively with smaller mammals (Motovylyak et al., 2019; Schaeffer et al., 2017; Schilling et al., 2017; Wu and Zhang, 2016). Here, we present, for the first time, in vivo dMRI of macaque brains imaged on a large bore 10.5 Tesla scanner capable of human imaging. We have obtained multishell, high angular resolution, 0.58 mm isotropic resolution data. Utilizing a combination of freely available software packages, we have generated tractography maps that recapitulate known white matter pathways when compared to established anatomical data.
Using a single shot 2D EPI protocol in the anesthetized macaque, we were able to collect up to 4 repeated reverse phase encoded acquisitions (34 min each) of high angular resolution, multi-shell data to increase the SNR. We did not note any motion artifacts or corruption of data resulting from physiological motion of the brain, which can be a problem with 3D-msEPI acquisitions (Liu et al., 2016). Single shot EPI acquisitions are prone to have artifacts (image distortion and signal loss) due to magnetic field inhomogeneities (Andersson et al., 2003); these become more pronounced with increasing static magnetic field used for the study and long echo train lengths required for high resolution scans. Nevertheless, we found the distortions to be well corrected by the FSL Topup/Eddy tools (Supplemental Figure 10).
Macaque imaging comes with unique challenges and oftentimes requires specific tools and significant adaptations for optimal results. One major hurdle is that the macaque head is significantly larger than those of rodents, marmosets, or squirrel monkeys (preventing the use of high field, small bore MRIs with significantly stronger gradients), but are approximately 10 times smaller than the human brain (Donahue et al., 2018). This means that higher spatial resolution is required to resolve similar structures in the macaque brain compared to humans. Along with the aforementioned SNR limitation, another problem is the lack of a commercially available coils specific to macaque imaging, particularly at ultra-high fields. A fully custom 32 channel head coil was developed for the 10.5T scanner, which required extensive optimization (Lagore et al., 2021). This coil has also been used to study resting-state functional MRI in macaques, which provides the opportunity to perform multimodal imaging studies within-subject at 10.5T (Yacoub et al., 2022).
The majority of whole brain submillimeter resolution macaque dMRI studies have been performed ex vivo. The long scan times and lack of breathing-related motion artifacts means that ex vivo scans can achieve stunning results. Ex vivo scans are capable of attaining resolutions as high as 0.13 mm (Calabrese et al., 2014). (Calabrese et al., 2015) used a 7T scanner with a single shell (b = 1500s/mm 2), 12 diffusion directions, and a total scan time of 46 h per subject. Ex vivo whole brain coverage at 0.2 mm isotropic resolution has also been achieved with an 11.4T magnet and a diffusion weighted 3D segmented EPI acquisition with 3 shells over 73 h (Sébille et al., 2019). Another ex vivo study conducted at 4.7T collected dMRI data on 2 brains, one imaged at 0.8 mm isotropic resolution with 4 averages of 61 directions each over 64 h and a second imaged at 0.43 mm isotropic resolution with 3D-EPI with 120 directions over 27 h (Azadbakht et al., 2015). Resolutions as high as 0.4 mm isotropic have been reported for ex vivo human whole brain coverage with a 53 hour acquisition (Fritz et al., 2019). Calabrese et al. also conducted an ex vivo study holding total scan time constant at 60 h, while varying the number of b-values, spatial and angular resolution (Calabrese et al., 2014). This study generated datasets acquired between 0.13 and 0.6 mm isotropic resolution. While these studies have provided valuable information regarding the limits of dMRI, complementary protocols that take advantage of the non-invasive nature of the technique are needed. Additionally, fixation alters the fundamental diffusion properties of brain tissues (Shatil et al., 2018), further limiting the translation of these techniques.
In vivo macaque dMRI with 1–1.5 mm isotropic resolution has been widely reported and easily attainable with most 3T scanners and commercially available coils (Feng et al., 2017; Liu et al., 2016; Meng et al., 2018; Rane and Duong, 2011; Shen et al., 2019; Shimony et al., 2018; Weiss et al., 2022). 1.1 mm isotropic dMRI data has been acquired with a knee coil (Shimony et al., 2018). Utilizing a standard head coil, 1.5 mm isotropic resolution diffusion MRI was collected on ten macaques in vivo; this dataset was further refined by registering the high resolution ex vivo dMRI to the in vivo diffusion space to generate a population average of white matter tracks (Feng et al., 2017). Liu et al. also acquired 1 mm isotropic dMRI with a custom built 3 channel coil (Liu et al., 2016). Another study performed with a pediatric head coil acquired 1 mm isotropic dMRI data as well as 0.5 mm isotropic T1 and T2 images to generate a population average white matter and gray matter maps (Weiss et al., 2022). However, there has been only a small number of in vivo dMRI studies with submillimeter isotropic resolution in anesthetized macaques, mostly at 3T. Autio et al. (2020), presented 0.9 mm dMRI data acquired on a 3T scanner with 2D-single shot EPI with simultaneous multi-slice accelerations (Autio et al., 2020). This study used the same b-value scheme (b = 1000, 2000 and 3000 s/mm 2) as in the HCP but utilized a much higher number of diffusion directions (500 vs 270 in the HCP) to improve the data quality in a 30 min acquisition. Another study acquired dMRI at 0.7 mm isotropic resolution on a 3T scanner, with a custom made, subject specific implanted 8-channel coil array using a single shell (b = 1000s/mm2) 256 direction acquisition (Janssens et al., 2012). The coil array was attached to the animal’s skull with ceramic screws and encased in dental acrylic during a standard headpost surgery. While the implanted coil produces very high-quality data with 3 h of acquisition, the invasive nature of the coil is a significant limitation for a non-invasive imaging modality. Finally, another study reported 0.5 mm isotropic resolution (Tounekti et al., 2018) utilizing 3D multi-shot echo planar imaging on a 3T clinical scanner with 30 diffusion directions applied with a b = 1000s/mm2 and a total acquisition time of 130 min. While 3D-msEPI is capable of increasing resolution, it is also highly susceptible to physiological motion artifacts. In Tounekti et al. (2018), approximately 8% of diffusion weighted images were discarded due to artifacts. At 7T, Wang et al. present the optimization of readout segmented echo planar imaging (rsEPI) for whole macaque brain coverage. A single 30 direction, b = 1000s/mm2 diffusion scan was collected at 0.8 mm isotropic resolution in a single phase encode direction over 45 min (Wang et al., 2019). As noted above, with the exception of Autio et al. (2020), all of these studies used single b value of 1000s/mm2 for diffusion weighting. Higher b values lead to SNR loss not only because of the higher diffusion weighting but also the longer TE that is required to acquire the data. Since the TE for the entire experiment is set by the TE for the highest b value employed, in a multi-shell experiment, the lower b value shell also suffers SNR losses. Avoiding higher b values in a multi-shell experiment, on the other hand, negatively impacts the quality of tractography estimates significantly (Sotiropoulos et al., 2013)
The brief summary of submillimeter resolution macaque dMRI given above highlights the challenges of high resolution in vivo dMRI protocols that often require tradeoffs among acquisition time, acceleration factors, angular resolution, maximum b values employed, and potentially invasive procedures. The work presented in our study is non-invasive, acquired with both high spatial and angular resolution using two shells (b = 1000 and 2000s/mm2) and a 2D spin echo sequence in approximately two hours. The preprocessed data produces high quality scalar metric maps and tractography streamlines.
Tractography allows for the visualization of white matter pathways and estimation of connectivity strength between brain regions in vivo and non-invasively (Lenglet et al., 2012; Soares et al., 2013). dMRI is currently the only technique available that can estimate structural connectivity in human brains in vivo (dissection and polarized imaging techniques allow for ex vivo human structural connectivity calculations, but this limits our ability to assess population differences and brain-behavior relationships). Thus, optimization and validation of the technique is of the utmost importance. Future studies will compare the results of our high resolution tractography to anatomical databases, to within-subject anatomical data generated from these animals, and to lower field dMRI results. While tractography has improved dramatically with the advent of newer methodology, e.g. CSD (Tournier et al., 2007), NODDI (Zhang et al., 2012), etc., it has historically had the significant limitation that it is only moderately correlated with true anatomical connectivity (as estimated with tract-tracing) (Donahue et al., 2016; Grier et al., 2020; Shen et al., 2019; Thomas et al., 2014; van den Heuvel et al., 2015). As discussed, in the Introduction section, spatial resolution is one of the primary factors in resolving crossing fibers (Donahue et al., 2016). The inherently low SNR of diffusion MRI images, especially when high b values are employed, makes increasing the resolution challenging because as spatial resolution improves, the signal reduces even further (Jones, 2010). In this study, we utilized both SNR gains available at 10.5T magnetic field and denoising using the MPPCA approach as implemented in MRtrix (Veraart et al., 2016b, 2016c). Future diffusion data acquired on the 10.5T scanner will be acquired consistent with the requirements of the recently published Noise reduction with Distribution Corrected (NORDIC) PCA method, and we expect to see further improvements in data quality (Moeller et al., 2021). The collection of high-quality dMRI data, as with all MRI modalities, involves a complex tradeoff of many imaging parameters. Especially important are the field strength of the magnet and the gradient slew rate and amplitude. Within the possible parameter space available with current instrumentation, we opted for targets that minimized distortions as much as possible and provided sufficient SNR for reliable tract reconstruction over the entire brain. Additional significant gains will be achieved using a gradient set that can operate with substantially higher maximal gradient strength and high slew rates at 10.5T, enabling more extensive q-space sampling and additional optimization of scan parameters such as resolution, scan time, and q-space coverage. Such an optimization is expected to lead to better data quality. Ultimately, however, the validation of our data will depend on comparison with ground truth anatomical data.
Much can be learned from diffusion scans on anesthetized macaques that will improve or inform the methods when applied to the study of human brains, especially when combined with neural tract tracing experiments within the same subjects that will allow direct comparison of diffusion tractography data to known anatomical connectivity. This highlights one important advantage of using nonhuman animal models: the ability to compare to ground-truth neural tract-tracing data to assess the accuracy of dMRI (Azadbakht et al., 2015; Girard et al., 2020; Shen et al., 2019; van den Heuvel et al., 2015).
5. Conclusions
Using a 10.5T large bore MRI, a custom 32 channel coil, and a denoising approach, this study presents high spatial resolution (0.58 mm isotropic voxels), high angular resolution (115 directions), multi-shell diffusion MRI data in anesthetized macaques achieved with 2D-single shot EPI protocols in approximately 2 h. Our study demonstrates the feasibility of collecting these data at ultra-high field. Despite the challenges of the intrinsically low SNR of diffusion MRI, this acquisition protocol at 10.5T combined with denoising produces tractography results in agreement with known pathways and is capable of differentiating short association fibers in the cortex that are often lost to partial volume effects. Further optimization of acquisition protocols with the macaque model as well as significantly improved hardware, such as higher channel count receivers (48 +) and a high performance head gradient insert employed at 10.5T, and improved denoising techniques will further push the dMRI methods to truly explore the ultimate limits of in vivo 10.5T diffusion MRI.
Supplementary Material
Acknowledgements
We thank Steve Jungst for continuing support with our coils and hardware setup. We thank Hannah Lee, Jen Holmberg, Adriana Cushnie, Tanya Casta, and Megan Monko for support with animal care and data acquisition. We thank Research Animal Resources at UMN, especially Whitney McGee and Anne Merley, for helping us implement new and improved anesthesia protocols.
Funding statement
This work was supported by NIH grants R01MH118257 (SRH), P50MH119569 (SRH), P41EB027061 (CL, KU), U01EB025144 (KU), T32DA007234 to P. Mermelstein (MDG), P30DA048742 (JZ, SRH), R56EB031765 (JZ), P50NS098753 (NH), a UMN AIRP award (JZ, SRH), MnDrive Brain Conditions (SRH).
Footnotes
Ethics statement
Animal procedures described were approved by the University of Minnesota Institutional Animal Care and Use Committee and adhered to the National Institute of Health standards for the care and use of nonhuman primates. Code will be deposited on github upon manuscript acceptance. Data will be made available through the PRIMatE Data Exchange (PRIME-DE).
Credit authorship contribution statement
Mark D. Grier: Conceptualization, Methodology, Writing – original draft, Data curation, Software, Investigation. Essa Yacoub: Conceptualization, Methodology, Writing – review & editing, Investigation. Gregor Adriany: Methodology, Writing – review & editing, Investigation. Russell L. Lagore: Methodology, Writing – review & editing, Investigation. Noam Harel: Conceptualization, Methodology, Writing – review & editing, Investigation. Ru-Yuan Zhang: Methodology, Writing – review & editing, Software. Christophe Lenglet: Conceptualization, Methodology, Writing – review & editing. Kâmil Uğurbil: Conceptualization, Methodology, Writing – review & editing, Funding acquisition. Jan Zimmermann: Conceptualization, Methodology, Writing – original draft, Funding acquisition, Data curation, Software, Investigation. Sarah R. Heilbronner: Conceptualization, Methodology, Writing – original draft, Funding acquisition, Investigation.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.neuroimage.2022.119200.
References
- Alexander AL, Hasan KM, Lazar M, Tsuruda JS, Parker DL, 2001. Analysis of partial volume effects in diffusion-tensor MRI. Magn. Reson. Med doi: 10.1002/mrm.1105. [DOI] [PubMed] [Google Scholar]
- Alomair OI, Brereton IM, Smith MT, Galloway GJ, Kurniawan ND, 2015. In vivo high angular resolution diffusion-weighted imaging of mouse brain at 16.4 Tesla. PLOS ONE. doi: 10.1371/journal.pone.0130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JLR, Graham MS, Zsoldos E, Sotiropoulos SN, 2016. Incorporating outlier detection and replacement into a non-parametric framework for movement and distortion correction of diffusion MR images. Neuroimage 141, 556–572. [DOI] [PubMed] [Google Scholar]
- Andersson JLR, Skare S, Ashburner J, 2003. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20, 870–888. [DOI] [PubMed] [Google Scholar]
- Andersson JLR, Sotiropoulos SN, 2016. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andica C, Kamagata K, Hatano T, Saito Y, Ogaki K, Hattori N, Aoki S, 2020. MR biomarkers of degenerative brain disorders derived from diffusion imaging. J. Magn. Reson. Imaging 52, 1620–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autio JA, Glasser MF, Ose T, Donahue CJ, Bastiani M, Ohno M, Kawabata Y, Urushibata Y, Murata K, Nishigori K, Yamaguchi M, Hori Y, Yoshida A, Go Y, Coalson TS, Jbabdi S, Sotiropoulos SN, Kennedy H, Smith S, Van Essen DC, Hayashi T, 2020. Towards HCP-Style macaque connectomes: 24-Channel 3T multiarray coil, MRI sequences and preprocessing. Neuroimage 215, 116800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadbakht H, Parkes LM, Haroon HA, Augath M, Logothetis NK, de Crespigny A, D’Arceuil HE, Parker GJM, 2015. Validation of High-Resolution Tractography Against In Vivo Tracing in the Macaque Visual Cortex. Cereb. Cortex 25, 4299–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker R, Wachtler T, Diesmann M, 2012. CoCoMac 2.0 and the future of tract-tracing databases. Front. Neuroinform 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldaranov D, Khomenko A, Kobor I, Bogdahn U, Gorges M, Kassubek J, Müller H−P, 2017. Longitudinal diffusion tensor imaging-based assessment of tract alterations: an application to amyotrophic lateral sclerosis. front. Hum. Neurosci 11, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani M, Oros-Peusquens A−M, Seehaus A, Brenner D, Möllenhoff K, Celik A, Felder J, Bratzke H, Shah NJ, Galuske R, Goebel R, Roebroeck A, 2016. Automatic segmentation of human cortical layer-complexes and architectural areas using Ex vivo diffusion MRI and its validation. Frontiers Neurosci. doi: 10.3389/fnins.2016.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TEJ, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM, 2003. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med 50, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Bihan DL, Le Bihan D, Mangin J−F, Poupon C, Clark CA, Pappata S, Molko N, Chabriat H, 2001. Diffusion tensor imaging: concepts and applications. J. Magn. Reson. Imaging doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- Bullock DN, Hayden EA, Grier MD, Tang W, Pestilli F, Heilbronner SR, 2022. A taxonomy of the brain’s white matter: Twenty-one major tracts for the 21st century. Cerebral Cortex doi: 10.1093/cercor/bhab500, bhab500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Styner MA, Johnson GA, 2014. Investigating the tradeoffs between spatial resolution and diffusion sampling for brain mapping with diffusion tractography: Time well spent? Hum. Brain Mapp 35. doi: 10.1002/hbm.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA, 2015. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage 117, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushnie AK, El-Nahal HG, Bohlen MO, May PJ, Basso MA, Grimaldi P, Wang MZ, de Velasco Ezequiel MF, Sommer MA, Heilbronner SR, 2020. Using rAAV2-retro in rhesus macaques: promise and caveats for circuit manipulation. J. Neurosci. Methods 345, 108859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Lipsman N, Meng Y, Rabin JS, Giacobbe P, Hamani C, 2020. The use of tractography-based targeting in deep brain stimulation for psychiatric indications. Front. Hum. Neurosci 14, 588423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, El-Shamayleh Y, Horwitz GD, 2020. Fast and reversible neural inactivation in macaque cortex by optogenetic stimulation of GABAergic neurons. Elife 9. doi: 10.7554/eLife.52658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot M, Cremers LGM, Ikram MA, Hofman A, Krestin GP, van der Lugt A, Niessen WJ, Vernooij MW, 2016. White matter degeneration with aging: longitudinal diffusion MR imaging analysis. Radiology 279, 532–541. [DOI] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, Van Essen DC, 2018. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc. Natl. Acad. Sci. U. S. A 115, E5183–E5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Behrens TE, Dyrby TB, Coalson T, Kennedy H, Knoblauch K, Van Essen DC, Glasser MF, 2016. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J. Neurosci 36, 6758–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD, 2008. White matter in learning, cognition and psychiatric disorders. Trends Neurosci 31, 361–370. doi: 10.1016/J.TINS.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, 2012. Freesurfer. Neuroimage doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz FJ, Sengupta S, Harms RL, Tse DH, Poser BA, Roebroeck A, 2019. Ultrahigh resolution and multi-shell diffusion MRI of intact ex vivo human brains using k-dSTEAM at 9.4T. Neuroimage 202, 116087. [DOI] [PubMed] [Google Scholar]
- Galvan A, Stauffer WR, Acker L, El-Shamayleh Y, Inoue K−I, Ohayon S, Schmid MC, 2017. Nonhuman primate optogenetics: recent advances and future directions. J. Neurosci 37, 10894–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganepola Tharindu, Nagy Zoltan, Ghosh Aurobrata, Papadopoulo Theodore, Alexander Daniel C., Sereno Martin I., 2018. Using diffusion MRI to discriminate areas of cortical grey matter. Neuroimage 182 (November), 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G, Caminiti R, Battaglia-Mayer A, St-Onge E, Ambrosen KS, Eskildsen SF, Krug K, Dyrby TB, Descoteaux M, Thiran J−P, Innocenti GM, 2020. On the cortical connectivity in the macaque brain: a comparison of diffusion tractography and histological tracing data. Neuroimage doi: 10.1016/j.neuroimage.2020.117201. [DOI] [PubMed] [Google Scholar]
- Granberg T, Fan Q, Treaba CA, Ouellette R, Herranz E, Mangeat G, Louapre C, Cohen-Adad J, Klawiter EC, Sloane JA, Mainero C, 2017. In vivo characterization of cortical and white matter neuroaxonal pathology in early multiple sclerosis. Brain 140, 2912–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier MD, Zimmermann J, Heilbronner SR, 2020. Estimating brain connectivity with diffusion-weighted magnetic resonance imaging: promise and peril. Biol Psychiatry Cogn. Neurosci. Neuroimaging 5, 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulban OF, De Martino F, Vu AT, Yacoub E, Ugurbil K, Lenglet C, 2018. Cortical fibers orientation mapping using in-vivo whole brain 7T diffusion MRI. Neuroimage 178, 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronner SR, Haber SN, 2014. Frontal cortical and subcortical projections provide a basis for segmenting the cingulum bundle: Implications for neuroimaging and psychiatric disorders. J. Neurosci 34. doi: 10.1523/JNEUROSCI.5459-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ertürk MA, Grant A, et al. , 2020. First in-vivo human imaging at 10.5T: imaging the body at 447 MHz. Magn. Reson. Med 84 (1), 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronner SR, Chafee MV, 2019. Learning how neurons fail inside of networks: non-human primates provide critical data for psychiatry. Neuron 102, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber E, Donnelly PM, Rokem A, Yeatman JD, 2018. Rapid and widespread white matter plasticity during an intensive reading intervention. Nat. Commun 9, 2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry E, Vandenberghe LH, 2019. Therapeutic AAV gene transfer to the nervous system: a clinical reality. Neuron 102, 263. [DOI] [PubMed] [Google Scholar]
- Hunsche S, Sauner D, Runge MJR, Lenartz D, El Majdoub F, Treuer H, Sturm V, Maarouf M, 2013. Tractography-guided stimulation of somatosensory fibers for thalamic pain relief. Stereotact. Funct. Neurosurg doi: 10.1159/000350024. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Everling S, 2012. Monkey in the middle: why non-human primates are needed to bridge the gap in resting-state investigations. Front. Neuroanat 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens T, Keil B, Farivar R, McNab JA, Polimeni JR, Gerits A, Arsenault JT, Wald LL, Vanduffel W, 2012. An implanted 8-channel array coil for high-resolution macaque MRI at 3T. Neuroimage doi: 10.1016/j.neuroimage.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. Neuroimage doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jeurissen B, Tournier J−D, Dhollander T, Connelly A, Sijbers J, 2014. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411–426. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Slopsema JP, 2017. Deep brain stimulation, in: Neuroprosthetics: Theory and Practice. pp. 762–789. [Google Scholar]
- Johnson LA, Aman JE, Yu Y, Escobar Sanabria D, Wang J, Hill M, Dharnipragada R, Patriat R, Fiecas M, Li L, Schrock LE, Cooper SE, Johnson MD, Park MC, Harel N, Vitek JL, 2021. High-frequency oscillations in the pallidum: a pathophysiological biomarker in parkinson’s disease? Mov. Disord 36, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Nebeck SD, Muralidharan A, Johnson MD, Baker KB, Vitek JL, 2016. Closed-loop deep brain stimulation effects on parkinsonian motor symptoms in a non-human primate – is beta enough? Brain Stimulat. doi: 10.1016/j.brs.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, 2010. Diffusion MRI. Oxford University Press. [Google Scholar]
- Kamagata K, Andica C, Hatano T, Ogawa T, Takeshige-Amano H, Ogaki K, Akashi T, Hagiwara A, Fujita S, Aoki S, 2020. Advanced diffusion magnetic resonance imaging in patients with Alzheimer’s and Parkinson’s diseases. Neural Regener. Res 15, 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner E, Dhital B, Kiselev VG, Reisert M, 2016. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn. Reson. Med 76, 1574–1581. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Dehay C, 1988. Functional implications of the anatomical organization of the callosal projections of visual areas V1 and V2 in the macaque monkey. Behav. Brain Res 29, 225–236. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Dehay C, Bullier J, 1986. Organization of the callosal connections of visual areas v1 and v2 in the macaque monkey. J. Comp. Neurol doi: 10.1002/cne.902470309. [DOI] [PubMed] [Google Scholar]
- Kida I, Ueguchi T, Matsuoka Y, Zhou K, Stemmer A, Porter D, 2016. Comparison of diffusion-weighted imaging in the human brain using readout-segmented EPI and PROPELLER turbo spin echo with single-shot EPI at 7 T MRI. Investig. Radiol doi: 10.1097/rli.0000000000000248. [DOI] [PubMed] [Google Scholar]
- Krogsrud SK, Fjell AM, Tamnes CK, Grydeland H, Mork L, Due-Tønnessen P, Bjørnerud A, Sampaio-Baptista C, Andersson J, Johansen-Berg H, Walhovd KB, 2016. Changes in white matter microstructure in the developing brain–a longitudinal diffusion tensor imaging study of children from 4 to 11years of age. Neuroimage 124, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki M, McCarley R, Westin C−F, Park H−J, Maier S, Kikinis R, Jolesz FA, Shenton ME, 2007. A review of diffusion tensor imaging studies in schizophrenia. J. Psychiatr. Res 41, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagore RL, Moeller S, Zimmermann J, DelaBarre L, Radder J, Grant A, Ugurbil K, Yacoub E, Harel N, Adriany G, 2021. An 8-dipole transceive and 24-loop receive array for non-human primate head imaging at 10.5 T. NMR Biomed. doi: 10.1002/nbm.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubach M, Amarante L, Kyra Swanson T, White SR, 2022. n.d. What, if anything, is rodent prefrontal cortex? 10.31234/osf.io/c2a79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M, 1986. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 161, 401–407. [DOI] [PubMed] [Google Scholar]
- Lenglet C, Abosch A, Yacoub E, de Martino F, Sapiro G, Harel N, 2012. Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PLoS One 7. doi: 10.1371/journal.pone.0029153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewald D, Miller R, Logothetis N, Wagner H−J, Schüz A, 2014. Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque. Biol. Cybern 108, 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhao X, Ma Y, Tang X, Gao J−H, 2016. DWI using navigated interleaved multi-shot EPI with realigned GRAPPA reconstruction. Magn. Reson. Med 75, 280–286. [DOI] [PubMed] [Google Scholar]
- Lövdén M, Bodammer NC, Kühn S, Kaufmann J, Schütze H, Tempelmann C, Heinze H−J, Düzel E, Schmiedek F, Lindenberger U, 2010. Experience-dependent plasticity of white-matter microstructure extends into old age. Neuropsychologia 48, 3878–3883. [DOI] [PubMed] [Google Scholar]
- Mackey AP, Whitaker KJ, Bunge SA, 2012. Experience-dependent plasticity in white matter microstructure: reasoning training alters structural connectivity. Front. Neuroanat 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde J−C, Côté M−A, Garyfallidis E, Zhong J, Chamberland M, Yeh F−C, Lin Y−C, Ji Q, Reddick WE, Glass JO, Chen DQ, Feng Y, Gao C, Wu Y, Ma J, He R, Li Q, Westin C−F, Deslauriers-Gauthier S, González JOO, Paquette M, St-Jean S, Girard G, Rheault F, Sidhu J, Tax CMW, Guo F, Mesri HY, Dávid S, Froeling M, Heemskerk AM, Leemans A, Boré A, Pinsard B, Bedetti C, Desrosiers M, Brambati S, Doyon J, Sarica A, Vasta R, Cerasa A, Quattrone A, Yeatman J, Khan AR, Hodges W, Alexander S, Romascano D, Barakovic M, Auría A, Esteban O, Lemkaddem A, Thiran J−P, Cetingul HE, Odry BL, Mailhe B, Nadar MS, Pizzagalli F, Prasad G, Villalon-Reina JE, Galvis J, Thompson PM, De Santiago Requejo F, Laguna PL, Lacerda LM, Barrett R, Dell’Acqua F, Catani M, Petit L, Caruyer E, Daducci A, Dyrby TB, Holland-Letz T, Hilgetag CC, Stieltjes B, Descoteaux M, 2019. Author correction: the challenge of mapping the human connectome based on diffusion tractography. Nat. Commun 10, 5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey-Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, Van Essen DC, Kennedy H, 2014. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb. Cortex 24, 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Verhagen L, Gladwin TE, Neubert F-X, Sallet J, Rushworth MFS, 2016. Comparing brains by matching connectivity profiles. Neurosci. Biobehav. Rev 60, 90. doi: 10.1016/J.NEUBIOREV.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab Jennifer A., Jbabdi Saâd, Deoni Sean C.L., Douaud Gwenaëlle, Behrens Timothy E.J., Miller Karla L., 2009. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage 46 (3), 775–785. [DOI] [PubMed] [Google Scholar]
- Meng Y, Hu X, Zhang X, Bachevalier J, 2018. Diffusion tensor imaging reveals microstructural alterations in corpus callosum and associated transcallosal fiber tracts in adult macaques with neonatal hippocampal lesions. Hippocampus 28, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller Steen, Pisharady Pramod Kumar, Ramanna Sudhir, Lenglet Christophe, Wu Xiaoping, Dowdle Logan, Yacoub Essa, Uğurbil Kamil, Akçakaya Mehmet, 2021. NOise reduction with distribution corrected (NORDIC) PCA in dMRI with complex–valued parameter-free locally low-rank processing. N NeuroImage 226 (February), 117539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Zhang J, 2006. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51, 527–539. [DOI] [PubMed] [Google Scholar]
- Motovylyak A, Skinner NP, Schmit BD, Wilkins N, Kurpad SN, Budde MD, 2019. Longitudinal in vivo diffusion magnetic resonance imaging remote from the lesion site in rat spinal cord injury. J. Neurotrauma doi: 10.1089/neu.2018.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EE, Winslow JT, 2009. Non-human primates: model animals for developmental psychopathology. Neuropsychopharmacology 34, 90–105. [DOI] [PubMed] [Google Scholar]
- Oishi K, Huang H, Yoshioka T, Ying SH, Zee DS, Zilles K, Amunts K, Woods R, Toga AW, Pike GB, Rosa-Neto P, Evans AC, van Zijl PCM, Mazziotta JC, Mori S, 2011. Superficially located white matter structures commonly seen in the human and the macaque brain with diffusion tensor imaging. Brain Connect. 1, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriat R, Cooper SE, Duchin Y, Niederer J, Lenglet C, Aman J, Park MC, Vitek JL, Harel N, 2018. Individualized tractography-based parcellation of the globus pallidus pars interna using 7T MRI in movement disorder patients prior to DBS surgery. Neuroimage 178, 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone D, Aelterman J, Pi ž urica A, Jeurissen B, Philips W, Leemans A, 2015. The effect of Gibbs ringing artifacts on measures derived from diffusion MRI. Neuroimage 120, 441–455. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN, 2002. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and corticocortical connection patterns in the monkey. Eur. J. Neurosci 16, 291–310. [DOI] [PubMed] [Google Scholar]
- Pohmann R, Speck O, Scheffler K, 2016. Signal-to-noise ratio and MR tissue parameters in human brain imaging at 3, 7, and 9.4 tesla using current receive coil arrays. Magn. Reson. Med 75. doi: 10.1002/mrm.25677. [DOI] [PubMed] [Google Scholar]
- Plantinga BR, Temel Y, Duchin Y, Uluda ğ K, Patriat R, Roebroeck A, Kuijf M, Jahanshahi A, ter Haar Romenij B, Vitek J, Harel N, 2018. Individualized parcellation of the subthalamic nucleus in patients with Parkinson’s disease with 7T MRI. Neuroimage 168. doi: 10.1016/j.neuroimage.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM, 1995. Do rats have prefrontal cortex? the rose-woolsey-akert program reconsidered. J. Cogn. Neurosci doi: 10.1162/jocn.1995.7.1.1. [DOI] [PubMed] [Google Scholar]
- Raffelt D, Dhollander T, Tournier J−D, Tabbara R, Smith RE, Pierre E, Connelly A, 2017. Bias field correction and intensity normalisation for quantitative analysis of apparent fibre density. In Proc. ISMRM 26, 3541. [Google Scholar]
- Rane S, Duong TQ, 2011. Comparison of in vivo and Ex vivo diffusion tensor imaging in rhesus macaques at short and long diffusion times. Open Neuroimag J. doi: 10.2174/1874440001105010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper J, Murphy L, Richardson R, Romm Z, Kovacs-Balint Z, Payne C, Galvan A, 2019. Chemogenetic inhibition of the amygdala modulates emotional behavior expression in infant rhesus monkeys. eNeuro 6. doi: 10.1523/ENEURO.0360-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley C, Seth AK, Pierpaoli C, Silva AC, Yu D, Saunders RC, Leopold DA, Ye FQ, 2015. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc. Natl. Acad. Sci. U. S. A 112, E2820–E2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroeck A, Miller KL, Aggarwal M, 2019. Ex vivo diffusion MRI of the human brain: technical challenges and recent advances. NMR Biomed. 32, e3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalmann YB, Pinsk MA, Wang L, Li X, Kastner S, 2012. The pulvinar regulates information transmission between cortical areas based on attention demands. Science 337, 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Hata J, Shintaku Y, Ohta H, Sogabe K, Mori S, Okano HJ, Hamada Y, Hirabayashi T, Minamimoto T, Sadato N, Okano H, Oishi K. The japan monkey centre primates brain imaging repository of high-resolution postmortem magnetic resonance imaging: the second phase of the archive of digital records. bioRxiv 2020.08.23.263517; doi: 10.1101/2020.08.23.263517. [DOI] [PubMed] [Google Scholar]
- Schaeffer DJ, Adam R, Gilbert KM, Gati JS, Li AX, Menon RS, Everling S, 2017. Diffusion-weighted tractography in the common marmoset monkey at 9.4T. J. Neurophysiol 118, 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K, Gao Y, Stepniewska I, Choe AS, Landman BA, Anderson AW, 2017. Reproducibility and variation of diffusion measures in the squirrel monkey brain, in vivo and ex vivo. Magn. Reson. Imaging 35, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Nath V, Hansen C, Parvathaneni P, Blaber J, Gao Y, Neher P, Aydogan DB, Shi Y, Ocampo-Pineda M, Schiavi S, Daducci A, Girard G, Barakovic M, Rafael-Patino J, Romascano D, Rensonnet G, Pizzolato M, Bates A, Fischi E, Thiran J−P, Canales-Rodríguez EJ, Huang C, Zhu H, Zhong L, Cabeen R, Toga AW, Rheault F, Theaud G, Houde J−C, Sidhu J, Chamberland M, Westin C−F, Dyrby TB, Verma R, Rathi Y, Irfanoglu MO, Thomas C, Pierpaoli C, Descoteaux M, Anderson AW, Landman BA, 2019. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 185, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN, 2006. Fiber Pathways of the Brain doi: 10.1093/acprof:oso/9780195104233.001.0001. [DOI] [Google Scholar]
- Sébille SB, Rolland A−S, Welter M−L, Bardinet E, Santin MD, 2019. Post mortem high resolution diffusion MRI for large specimen imaging at 11.7 T with 3D segmented echo-planar imaging. J. Neurosci. Methods doi: 10.1016/j.jneumeth.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Seehaus A, Roebroeck A, Bastiani M, Fonseca L, Bratzke H, Lori N, Vilanova A, Goebel R, Galuske R, 2015. Histological validation of high-resolution DTI in human post mortem tissue. Front. Neuroanat 9, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatil AS, Uddin MN, Matsuda KM, Figley CR, 2018. Quantitative MRI changes due to progressive formalin fixation in whole human brain specimens: longitudinal characterization of diffusion, relaxometry, and myelin water fraction measurements at 3T. Front. Med 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Goulas A, Grayson DS, Eusebio J, Gati JS, Menon RS, McIntosh AR, Everling S, 2019. Exploring the limits of network topology estimation using diffusion-based tractography and tracer studies in the macaque cortex. Neuroimage 191, 81–92. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM, 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 (Suppl 1), S208–S219. [DOI] [PubMed] [Google Scholar]
- Soares JM, Marques P, Alves V, Sousa N, 2013. A hitchhiker’s guide to diffusion tensor imaging. Front Neurosci doi: 10.3389/fnins.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, Glasser MF, Hernandez M, Sapiro G, Jenkinson M, Feinberg DA, Yacoub E, Lenglet C, Essen DC, Van Ugurbil K, Behrens TEJ, 2013. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 80, 125–143. doi: 10.1016/J.NEUROIMAGE.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Hernández-Fernández M, Vu AT, Andersson JL, Moeller S, Yacoub E, Lenglet C, Ugurbil K, Behrens TEJ, Jbabdi S, 2016. Fusion in diffusion MRI for improved fibre orientation estimation: an application to the 3T and 7T data of the human connectome project. Neuroimage 134, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kötter R, 2001. Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Philos. Trans. R. Soc. Lond. B Biol. Sci 356, 1159–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland S, 1940. The distribution of commissural fibres in the corpus callosum in the macaque monkey. J. Neurol. Psychiatry 3, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaf N, Lagore RL, Jungst S, Gunamony S, Radder J, Grant A, Moeller S, Auer-bach E, Ugurbil K, Adriany G, Van de Moortele PF, 2021. A self-decoupled 32-channel receive array for human-brain MRI at 10.5 T. Magn. Reson. Med 86. doi: 10.1002/mrm.28788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C, 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc. Natl. Acad. Sci. U. S. A 111, 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tounekti S, Troalen T, Bihan-Poudec Y, Froesel M, Lamberton F, Ozenne V, Cléry J, Richard N, Descoteaux M, Ben Hamed S, Hiba B, 2018. High-resolution 3D diffusion tensor MRI of anesthetized rhesus macaque brain at 3T. Neuroimage 181, 149–161. [DOI] [PubMed] [Google Scholar]
- Tournier J−D, Calamante F, Connelly A, 2010. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. In: Proceedings of the International Society for Magnetic Resonance in Medicine, p. 1670. [Google Scholar]
- Tournier J−D, Calamante F, Connelly A, 2007. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459–1472. [DOI] [PubMed] [Google Scholar]
- Tournier J−D, Calamante F, Gadian DG, Connelly A, 2004. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. Neuroimage 23, 1176–1185. [DOI] [PubMed] [Google Scholar]
- Tournier J−D, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh C−H, Connelly A, 2019. MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroim-age 202, 116137. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Yeh CH, Calamante F, Cho KH, Connelly A, Lin CP, 2008. Resolving crossing fibres using constrained spherical deconvolution: validation using diffusion-weighted imaging phantom data. Neuroimage 42, 617–625. doi: 10.1016/J.NEUROIMAGE.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Truong Trong-Kha, Guidon Arnaud, Song Allen W., 2014. Cortical Depth Dependence of the Diffusion Anisotropy in the Human Cortical Gray Matter in Vivo. PLoS ONE 9 (3), e91424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U ğurbil K, 2018. Imaging at ultrahigh magnetic fields: history, challenges, and solutions. Neuroimage 168, 7–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurbil K, 2014. Magnetic resonance imaging at ultrahigh fields. IEEE Transactions on Biomedical Engineering doi: 10.1109/tbme.2014.2313619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U ǧurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JLR, Behrens TEJ, Glasser MF, Van Essen DC, Yacoub E, 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage 80. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upright NA, Brookshire SW, Schnebelen W, Damatac CG, Hof PR, Browning PGF, Croxson PL, Rudebeck PH, Baxter MG, 2018. Behavioral effect of chemogenetic inhibition is directly related to receptor transduction levels in rhesus monkeys. J. Neurosci 38, 7969–7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, de Reus MA, Feldman Barrett L, Scholtens LH, Coopmans FMT, Schmidt R, Preuss TM, Rilling JK, Li L, 2015. Comparison of diffusion tractography and tract-tracing measures of connectivity strength in rhesus macaque connectome. Hum. Brain Mapp 36, 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan JT, Garwood M, Collins CM, Liu W, DelaBarre L, Adriany G, Andersen P, Merkle H, Goebel R, Smith MB, Ugurbil K, 2001. 7T vs. 4T: RF power, homogeneity, and signal-to-noise comparison in head images. Magn Reson Med 46, 24–30. [DOI] [PubMed] [Google Scholar]
- Veraart J, Fieremans E, Jelescu IO, Knoll F, Novikov DS, 2016a. Gibbs ringing in diffusion MRI. Magn. Reson. Med 76, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraart J, Fieremans E, Novikov DS, 2016b. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med doi: 10.1002/mrm.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraart J, Novikov DS, Christiaens D, Ades-Aron B, Sijbers J, Fieremans E, 2016c. Denoising of diffusion MRI using random matrix theory. Neuroimage 142, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek JL, Johnson LA, 2019. Understanding Parkinson’s disease and deep brain stimulation: role of monkey models. Proceedings Nat. Acad. Sci doi: 10.1073/pnas.1902300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu AT, Auerbach E, Lenglet C, Moeller S, Sotiropoulos SN, Jbabdi S, Andersson J, Yacoub E, Ugurbil K, 2015. High resolution whole brain diffusion imaging at 7T for the human connectome project. Neuroimage 122, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wang D, Zhang J, Bai R, Qian M, Sun Y, Lu Y, Zhang X, 2019. Evaluation of submillimeter diffusion imaging of the macaque brain using readout-segmented EPI at 7 T. IEEE Transac. Biomed. Engin doi: 10.1109/tbme.2019.2899132. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Sano H, Chiken S, Kobayashi K, Fukata Y, Fukata M, Mushiake H, Nambu A, 2020. Forelimb movements evoked by optogenetic stimulation of the macaque motor cortex. Nat. Commun 11, 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KK, Platt ML, 2012. Of mice and monkeys: using non-human primate models to bridge mouse-and human-based investigations of autism spectrum disorders. J. Neurodev. Disord 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AR, Liguore WA, Domire JS, Button D, McBride JL, 2022. n.d. Intrastriatal AAV2.retro administration leads to extensive retrograde transport in the rhesus macaque brain: implications for disease modeling and therapeutic development. doi: 10.1101/2020.01.17.910893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlein JM, 2014. Nonhuman primate models of depression: effects of early experience and stress. ILAR J. 55, 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Zhang J, 2016. In vivo mapping of macroscopic neuronal projections in the mouse hippocampus using high-resolution diffusion MRI. NeuroImage. doi: 10.1016/j.neuroimage.2015.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Auerbach EJ, Vu AT, Moeller S, Lenglet C, Schmitter S, Van de Moortele P−F, Yacoub E, U ğurbil K, 2018. High-resolution whole-brain diffusion MRI at 7T using radiofrequency parallel transmission. Magn. Reson. Med 80, 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zitella LM, Duchin Y, Teplitzky BA, Kastl D, Adriany G, Yacoub E, Harel N, Johnson MD, 2016. Multimodal 7T imaging of thalamic nuclei for preclinical deep brain stimulation applications. Front. Neurosci 10, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub E, Grier MD, Auerbach EJ, Lagore RL, Harel N, Ugurbil K, Adriany G, Zilverstand A, Hayden BY, Heilbronner SR, Zimmermann J, 2022. n.d. Ultra-high field (10.5 T) resting state fMRI in the macaque. doi: 10.1101/2020.05.21.109595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon M, Bao Q, Chitrit OJ, Henriques RN, Shemesh N, Frydman L, 2020. High-resolution 3D in vivo brain diffusion tensor imaging at ultrahigh fields: following maturation on juvenile and adult mice. Front Neurosci doi: 10.3389/fnins.2020.590900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC, 2012. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016. [DOI] [PubMed] [Google Scholar]
- Zitella LM, Xiao Y, Teplitzky BA, Kastl DJ, Duchin Y, Baker KB, Vitek JL, Adriany G, Yacoub E, Harel N, Johnson MD, 2015. In vivo 7T MRI of the non-human primate brainstem. PLoS ONE 10, e0127049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.