Abstract
An early event in the induction of the SOS system of Escherichia coli is RecA-mediated cleavage of the LexA repressor. RecA acts indirectly as a coprotease to stimulate repressor self-cleavage, presumably by forming a complex with LexA. How complex formation leads to cleavage is not known. As an approach to this question, it would be desirable to identify the protein-protein interaction sites on each protein. It was previously proposed that LexA and other cleavable substrates, such as phage λ CI repressor and E. coli UmuD, bind to a cleft located between two RecA monomers in the crystal structure. To test this model, and to map the interface between RecA and its substrates, we carried out alanine-scanning mutagenesis of RecA. Twenty double mutations were made, and cells carrying them were characterized for RecA-dependent repair functions and for coprotease activity towards LexA, λ CI, and UmuD. One mutation in the cleft region had partial defects in cleavage of CI and (as expected from previous data) of UmuD. Two mutations in the cleft region conferred constitutive cleavage towards CI but not towards LexA or UmuD. By contrast, no mutations in the cleft region or elsewhere in RecA were found to specifically impair the cleavage of LexA. Our data are consistent with binding of CI and UmuD to the cleft between two RecA monomers but do not provide support for the model in which LexA binds in this cleft.
The SOS regulatory system controls the response of Escherichia coli to treatments that damage DNA or inhibit DNA replication (12, 30). During normal cell growth, LexA protein represses a set of about 20 genes. Inducing treatments generate an inducing signal that activates another regulatory protein, RecA. Activated RecA in turn mediates the cleavage of LexA, inactivating it and leading to derepression of the SOS regulon. In vitro, RecA can be activated by forming a ternary complex with single-stranded DNA and a nucleoside triphosphate such as ATP, dATP, or ATP(S). In this complex, RecA forms a helical filament along the single-stranded DNA. It is likely that this complex also represents the activated in vivo form of RecA.
Although interaction of LexA with activated RecA triggers the cleavage reaction, many lines of evidence indicate that RecA does not act as a true protease but instead causes LexA to cleave itself (28). LexA can undergo self-cleavage in vitro in a reaction termed autodigestion (28). This reaction cuts the same bond as in RecA-mediated cleavage; moreover, mutations that inhibit RecA-mediated cleavage also prevent autodigestion. Hence, we believe that the actual chemistry of catalysis is carried out by groups in LexA, not in RecA, and we term activated RecA a “coprotease” to emphasize its indirect role in promoting cleavage.
Activated RecA can also mediate the cleavage of two other groups of proteins. The first is a group of temperate phage repressors, exemplified by λ CI repressor, which are cleaved in lysogens upon SOS-inducing treatments (43). Cleavage of CI is far slower than that of LexA. If DNA damage is severe, CI cleavage leads to prophage induction. The second set of substrates is a set of mutagenesis proteins, exemplified by the host UmuD protein, that are activated by specific cleavage to perform specific roles in SOS mutagenesis. Again, UmuD cleavage is slower than that of LexA, so that mutagenesis only takes place in severely damaged cells. The cleavage reactions of λ CI and UmuD appear to be entirely parallel mechanistically to that of LexA. Both proteins undergo self-cleavage, and the residues involved in cleavage are conserved in CI and UmuD. Hence, it is believed that RecA acts indirectly to stimulate these reactions as well.
It is not yet clear how RecA stimulates cleavage. Our evidence with LexA favors a conformational model in which RecA stabilizes a reactive conformation of LexA (44). However, it remains possible that RecA makes a more direct contribution to the chemistry of bond breakage. One analogy can be made with GTPase-activating proteins (GAPs), which greatly stimulate the GTPase activity of Ras and other small G proteins by contributing groups to the active site of this reaction (47). One approach to distinguishing these models is to identify the binding sites for LexA and other cleavable proteins on the RecA protein, and the work described here was carried out with this goal.
Two previous lines of evidence have suggested that LexA, CI, and UmuD interact at different sites in RecA. First, several recA mutant proteins appear to exhibit specific defects for cleaving some but not all substrates (see below), suggesting that these alleles affect residues that contact some substrates but not others. Second, many λ CI mutations that block RecA-mediated cleavage in vivo were isolated (13, 14); biochemical analysis showed that 9 of 15 mutant proteins were not impaired for autodigestion. These findings are consistent with the model in which these nine mutations affect residues that interact with RecA, although this has not been shown directly. Strikingly, these mutations do not affect residues that are conserved in other cleavable proteins, suggesting that the RecA-binding site on λ CI is not conserved in these other proteins.
In this work, we have sought to identify the LexA binding site in RecA by site-directed mutagenesis. Although several recA alleles show specific defects for cleavage of certain substrates, none of these is specifically defective for LexA cleavage; that is, no recA mutant protein yet characterized is defective for promoting LexA cleavage and proficient for the other functions of RecA. Our approach was to use the crystal structure of RecA as a guide to identify potential residues and to test a particular model for the location of the binding site for cleavable substrates.
Story et al. (53) determined the structure of a helical filament of RecA and proposed that the cleavable proteins bind to a cleft located between two adjacent RecA monomers in this filament (Fig. 1). This model was based on the fact that two particular recA alleles change residues located in this cleft and affect cleavage reactions. These alleles are recA1734, which changes Arg243 to Leu (8), and recA91, which changes Gly229 to Ser (39). recA1734 and two other recA alleles at Arg243 (recA433 and recA435 [9]) confer defects for cleavage of UmuD and phage φ80 repressor but allow cleavage of LexA and λ CI. The recA91 allele is also reportedly defective for cleavage of φ80 repressor and normal for λ CI (39), although no data have been published for this mutant allele. The findings that these mutant proteins are specifically defective for cleavage of some but not all substrates suggests, first, that the mutant RecA protein can be activated; second, that the mutations are likely to affect a protein-protein interaction directly; and third, that not all cleavable proteins bind at the same site, as discussed above. In any case, the location of recA1734 and recA91 in the cleft between two monomers suggests that UmuD and φ80 repressor, at least, bind in this cleft.
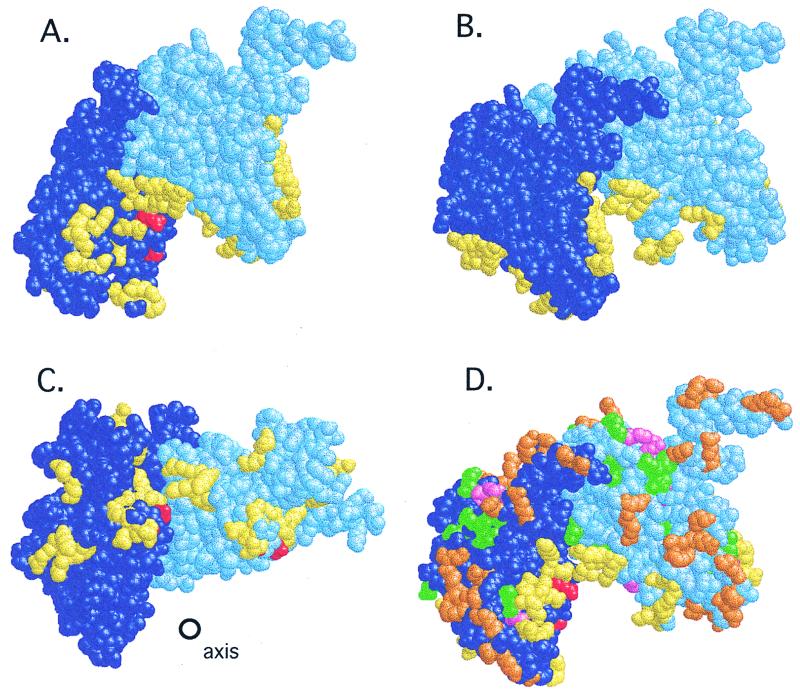
FIG. 1.
Location of site-directed changes in the RecA structure. Shown are two adjacent monomers from the RecA filament (see the text), colored dark and light blue. In the view in panel D, the helix axis lies directly behind the interface between the two monomers and is oriented vertically (not shown). The views in panels A and B are that of panel D rotated by 30° or −30° along the y axis; that in panel C is the view of panel D rotated −90° along the y axis so that the cleft is viewed from beneath; the helix axis is shown. (A to C) Mutational changes in the cleft. Residues Arg243 and Gly229 are shown in red; residues changed in the cleft (identified in Table 3) are shown in yellow. (D) Locations of other mutations and changes on the outer surface of the filament. Mutations destroying RecA function (listed in Table 2) are shown in pink; those that leave LexA cleavage intact (Table 2) are shown in green; changes made in the present work (Table 3) are shown in orange; yellow and red are as in panels A to C. RasMol scripts generating these views are available at http://www.biochem.arizona.edu/Little/Littlelab.htm.
Another supportive line of evidence for this model came from image reconstructions of electron micrographs of LexA bound to a RecA filament (62). In these images, extra electron density, attributed to LexA, was found in an area that corresponds roughly to the locations of Arg243 and Gly229. In addition, LexA appeared to contact two adjacent RecA monomers and possibly made contact across the groove with a RecA subunit in the next turn of the helix.
In this work, we have tested the model in which LexA, UmuD, and λ CI bind in the cleft by using site-directed mutagenesis to change most of the polar residues in the cleft to alanine. In addition, we changed many other residues located on the surface of the crystal structure. Mutant proteins were then tested in vivo for the ability to support specific cleavage and to carry out other functions of RecA.
MATERIALS AND METHODS
Materials.
Chemicals were from Sigma, Fisher Chemicals, and U.S. Biochemicals. The restriction enzymes, T4 DNA polymerase, and T4 DNA ligase were from New England Biolabs, Promega, and Boehringer Mannheim. Protran nitrocellulose was from Schleicher & Schuell. Anti-rabbit immunoglobulin G-horseradish peroxidase (IgG-POD) was from Boehringer Mannheim, and the ECL Western blotting detection system was from Amersham. Rabbit antibodies against LexA were described previously (26); rabbit antibodies against RecA and λ CI were made by similar methods. Affinity-purified rabbit antibody against UmuD was a generous gift from Roger Woodgate. Oligonucleotides for site-directed mutagenesis and DNA sequencing were from the Midland Certified Reagent Company or the Division of Biotechnology at the University of Arizona. The Altered Sites site-directed mutagenesis kit was from Promega. [35S]methionine was from ICN.
Computer modeling of the RecA crystal structure.
Coordinates for the RecA monomer were obtained from the Brookhaven Protein Data Base (PDB file 2REB). A model of the RecA filament was produced by using the sixfold helical symmetry of the monomers in the filament; we are grateful to Sue Roberts for doing this transformation. This RecA filament model was then examined by using the Insight II and RasMol version 2.5 programs (46).
Bacterial and phage growth.
Luria broth and tryptone growth media were prepared as described previously (34) with antibiotic concentrations as described elsewhere (33). Phage λ growth was as described previously (48). M9 minimal medium for pulse labeling was also described previously (26).
Bacterial strains.
The derivatives of E. coli K-12 used are listed in Table 1.
TABLE 1.
Bacterial strains, phage, and plasmids
| Strain, phage, or plasmid | Relevant genotype or description | Vector | Source or reference |
|---|---|---|---|
| Strain | |||
| ES1301 | lacZ53 mutS201::Tn5 thyA36 rha5 metB1 deoC | Promega | |
| GY7066 | metB thi pyrE lacMS286 (φ80dIIlacBKI) Δ(srl-recA)306::Tn10 sfiB114 | 8 | |
| JAM267 | JL2460/pJAM13 | This work | |
| JAM444 | JL2460/pJAM92 | This work | |
| JAM485 | GY7066/pJAM93 | This work | |
| JL783 | AB1157 lexA+ Δ(srl-recA)306::Tn10/F′ lacIq | 31 | |
| JL1434 | lexA71::Tn5 sulA211 supE Δ(lacIPOZYA)169 pro+/F′ lacIq lacZΔM15::Tn9 | 25 | |
| JL1447 | lexA71::Tn5 sulA211 supE Δ(lacIPOZYA)169 pro+ Δ(srl-recA)306::Tn10 (λsulA::lacZ cIind−)/F′ lacIq lacZΔM15::Tn9 | 52 | |
| JL2460 | lexA71::Tn5 sulA211 umuDC+ supE Δ(lacIPOZYA)169 pro+ Δ(srl-recA)306:: Tn10/F′ lacIq lacZΔM15::Tn9 | This worka | |
| Phage λ MMS885 | b1453 Δ(att-gam) Spi− χDcI857 | K. Knight | |
| Plasmids | |||
| pAlter | Plasmid for mutagenesis | pBR322 | Promega |
| pATT49 | lacp/o::lexAEK45 | pBR322 | A. Thliverisb |
| pBR322 | Cloning vector; bla tet | 3 | |
| pET5a UmuD | T7p::umuD | pET5a (pBR322 derivative) | G. Walker |
| pET5a UmuD′ | T7p::umuD′ | pET5a | G. Walker |
| pFG600 | lacp/o::λ cI | pBR322 | F. Gimble |
| pGB2 | Low-copy-number vector; Spcr | 4 | |
| pJAM13 | lacp/o::λ cI | pGB2 | This work |
| pJAM20 | Middle portion of recA | pAlter | This work |
| pJAM30 | C-terminal portion of recA | pAlter | This work |
| pJAM50 | N-terminal portion of recA | pAlter | This work |
| pJAM92 | lacp/o::lexAEK45 | pGB2 | This work |
| pJAM93 | lacI+ | pGB2 | This work |
| pJWL319 | lacI+ | pBR322 | This work |
| pTRecA220 | tacp/o::recA+ bla | pBR322 | 49 |
JL1434 was transduced to tetracycline resistance by P1 grown on JL1447.
pATT49 is an EK45 derivative of pJWL184 (24) made by A. Thliveris (personal communication) by replacing the MluI-BamHI interval of pJWL184 with a fragment carrying the EK45 mutation.
Construction of plasmids.
pFG600 was made by Fred Gimble (personal communication); it contains a lacp/o::λ cI fusion from pKB280 (2), a silent SphI site at the codons for residues 92 and 93 of cI, and 264 bp of λ DNA downstream of cI, including a PstI site and ending at a ClaI site, cloned into the EcoRI and ClaI sites of pBR322. pJAM13 carried a lacP/O::λ cI operon fusion on pGB2, a low-copy-number vector compatible with pBR322, and was made by cloning a 1,033-bp EcoRI-PstI fragment from pFG600 into pGB2. pJAM92 carried a lacP/O::lexAEK45 fusion and was made by cloning a 1,152-bp EcoRI-HindIII fragment of pATT49 into pGB2. pJWL319 was made by R. Ramage by cloning EcoRI-HincII and HincII-MspI fragments from pMC9 (35) into pBR322 cut with EcoRI and ClaI; the resulting insert contained the wild-type lacI promoter and lacI gene, starting at a HincII site at −50 relative to the lacI transcript and ending at an MspI site within the lacZYA promoter. pJAM93 carried lacI+ on pGB2 and was constructed by subcloning the EcoRI-HindIII region containing lacI+ from pJWL319. Plasmids for site-directed mutagenesis were made by cloning regions of recA from pTrecA220 into pAlter1. pJAM20 carried a 546-bp PstI-EcoRI fragment (amino acids 79 to 260). pJAM30 carried a 960-bp PstI-NruI fragment (amino acids 79 to 352) cloned into pAlter1 digested with SmaI and PstI. pJAM50 contained a 273-bp HindIII-PstI fragment (amino acids 1 to 76).
Site-directed mutagenesis.
Site-directed mutagenesis was done as described in the Promega Altered Sites II kit technical manual (revised August 1994). Regions of the recA gene from pTRecA220 were subcloned into pAlter1 for mutagenesis (see above). Mutagenic oligonucleotides were designed to create or destroy restriction sites so that the presence of the mutations could be detected using restriction digests (details available upon request). Plasmids with the desired change were sequenced to confirm the presence of the mutation. The region carrying the desired mutation was then subcloned into pTrecA220. The entire subcloned region was sequenced to ensure that no other changes were present. Mutations are referred to by the one-letter amino acid code of the mutated residue followed by its residue number. Since each mutant has more than one mutation, the changes are combined into one “word.” Thus, the mutation R105K106 has Arg105 and Lys106 changed to Ala.
Expression of RecA.
Wild-type and mutant RecA proteins were expressed from the pTrecA220 plasmid and derivatives. The level of RecA expressed from pTrecA220 is about equal to that of RecA expressed from the wild-type gene on the chromosome after 15 min of growth in the presence of the SOS-inducing drug nalidixic acid (data not shown). Levels of RecA for each mutant strain were determined by Western analysis in derivatives of JL2460.
Analysis of cleavage using Western blots.
To analyze LexA cleavage, we used a mutant LexA protein, LexAEK45, in order to uncouple LexA cleavage from potential effects due to differing levels of other SOS genes. This mutant does not bind the normal LexA DNA binding site (55); hence, differences in levels of cleavage do not affect expression of the SOS genes. A low-copy-number plasmid carrying a lacP::lexAEK45 fusion, pJAM92, was transformed into a lexA-defective or lexA(Def), strain, JL2460, and this strain, JAM444, was used for the LexA cleavage assays. Strains derived from JL2460 and JAM267 were used for analysis of UmuD and CI cleavage, respectively. The Western blotting protocol was as described previously with minor changes (52). Anti-rabbit immunoglobulin G-POD from Boehringer Mannheim was used as a secondary antibody. Chemiluminescence detection was done with the Amersham ECL Western blotting detection system according to the instructions provided.
UV sensitivities of cells containing recA mutations.
Two different assays were done to examine the UV sensitivities of the different recA mutations in JL2460-derived strains. For a semiquantitative assay, cells were grown to mid-exponential phase (≈2 × 108 cells/ml) in L broth. Five microliters of culture was spread on tryptone plates in a streak running the length of the plate. Zones of the streak were subjected to varying doses of UV radiation by shadowing them with an opaque card. After incubation overnight at 37°C, the relative sensitivities of the recA mutants were gauged by the amount of growth in areas that received different doses of UV radiation. To measure quantitative survival curves (40), cultures were grown to mid-exponential phase (≈2 × 108 cells/ml) in tryptone broth. The cultures were diluted 1:100 in cold 10 mM MgSO4. All steps after UV exposure were done in dim light. Five milliliters of diluted culture was placed in a glass petri dish and exposed to UV light. After various doses, aliquots were removed from the culture. The aliquots were then diluted in 10 mM MgSO4, and 100 μl of the appropriate dilutions was spread on λ plates. After incubation at 37°C for ≈48 h, colonies were counted, and the surviving fraction was calculated.
Recombination assays.
Two different assays were used to assess recombination defects. In the first assay, λ red− gam− mutants are dependent on functional RecA for packaging their DNA, and consequently for the formation of plaques (51). Derivatives of JL2460 containing plasmids with wild-type recA, no recA, or the recA mutations were grown to mid-exponential phase (≈2 × 108 cells/ml) in tryptone broth. The cultures were spun down and resuspended in an equal volume of 10 mM MgSO4. Cells (300 μl) were mixed with 3 ml of BBL top agar and plated on tryptone plates. Several different dilutions of the red− gam− phage, λMMS885, were spotted onto the lawns. The plates were incubated overnight at 37°C, and phage growth was assessed. In the second assay, intrachromosomal recombination was examined as described previously (17), except that the plates were incubated at 37°C for approximately 4 days. Plasmids containing recA+, the mutant recA alleles, or no recA gene, were transformed into JAM485, a derivative of GY7066, which contains two nonoverlapping lac deletions on the chromosome. The number of lac+ papillae per colony for ≈10 colonies about 0.5 cm in diameter was determined. The average number of lac+ papillae per colony is reported below.
Analysis of LexA cleavage by RecA mutant proteins using 35S pulse labeling.
Analysis of LexA cleavage by 35S pulse labeling was done as described previously (26) except that nalidixic acid (50 μg/ml) was used as an inducing treatment instead of UV light and the distribution of radioactivity was determined using a PhosphorImager. In addition, the plasmid carrying the LexAEK45 gene used for Western analysis did not produce enough protein to be seen in this assay (data not shown). Instead, derivatives of strain JL783 were used. This strain carries the wild-type lexA gene under the lexA promoter on the chromosome. Since LexA represses its own promoter in this situation, cells with RecA mutants that cannot cleave LexA well will produce less LexA during labeling than recA+ cells.
RESULTS
Choice of residues in recA for mutagenesis.
At the outset of this work, we made several assumptions in designing our approach to the analysis of RecA interaction with its substrates. First, we assumed that the residues directly involved in the interaction could be changed without affecting other functions of RecA. Second, although the crystal structure is of a filament that is probably not the active form (61), we assumed that most of the residues on the surface of this structure would also be on the surface of the active RecA filament. Third, we assumed that the binding site is located on a part of the structure that is visible rather than in a disordered region that is not seen in the structure (subsequent work by others [37] suggests that this assumption may be flawed). Finally, we assumed that weaker binding would result in slower cleavage and that this would result in differences in LexA levels that could be detected by our assays.
As discussed above, Story et al. (53) proposed that LexA, and the other cleavable proteins, bind in the cleft formed between two adjacent monomers in the RecA filament. To test this hypothesis, residues in the cleft were targeted for site-directed mutagenesis. Amino acids located around Arg243 were of particular interest, since this residue has been implicated in interactions with UmuD (8, 9) (Fig. 1).
In addition, other surface residues of RecA were examined as possible sites for interaction. These were chosen based on two criteria. First, the locations of previously characterized mutations let us rule out portions of the surface. These included mutations that destroy or reduce all RecA functions. Such changes may interfere, for instance, with the ability of RecA to form activated filaments. Other mutations appear not to affect LexA cleavage, although they may be defective for other functions, suggesting that these regions of RecA do not interact with LexA (Table 2).
TABLE 2.
Previously characterized recA mutations
| Residue | Change in amino acid | Allele no. | Reference |
|---|---|---|---|
| Mutations abolishing RecA functiona | |||
| 51 | Leu to Phe | 13 | 21 |
| 60 | Arg to Cys | 56 | 21 |
| 97 | His to Ala | None | 38 |
| 182 | Leu to Gln | 1623 | 32 |
| 183 | Lys to Met | 2183 | 1 |
| 211 | Gly to Ala | 611 | 22 |
| 216 | Lys to Ala | None | 49 |
| 217 | Phe to Ala | None | 49 |
| 222 | Arg to Ala | None | 49 |
| 225 | Ile to Val | 142 | 8 |
| 248 | Lys to Ala | None | 38 |
| 264 | Tyr to Gly | None | 11 |
| 284 | Ile to Asp | 2284 | 1 |
| Mutations allowing LexA cleavageb | |||
| 28 | Glu to Lys | 718 | 54 |
| 37 | Val to Met | 803 | 21 |
| 38 | Glu to Lys | 730, 1211 | 21, 58 |
| 39 | Thr to Ile | 1235 | 58 |
| 44 | Ser to Leu | None | R. Devoret, personal communication |
| 112 | Asp to Gly | None | R. Devoret, personal communication |
| 117 | Ser to Phe | 1730 | 7 |
| 119 | Pro to Ser | 432 | 10 |
| 139 | Asp to Gly | 1626 | 32 |
| 145 | Ser to Thr | 1647 | 32 |
| 169 | Arg to Cys | 1203 | 58 |
| 179 | Ala to Val | 1212 | 58 |
| 184 | Gln to Lys | 1202 | 58 |
| 187 | Thr to Ala | 1625 | 32 |
| 213 | Asn to Lys | 1634 | 32 |
| 247 | Val to Met | None | R. Devoret, personal communication |
| 257 | Glu to Pro | 801 | 56 |
| 275 | Val to Asp | 1620 | 32 |
| 283 | Leu to Glu | 2283E | 1 |
| 301 | Gly to Ser | 1601 | 58 |
| C terminus | 25-amino-acid deletion | 5327 | 16 |
Alleles containing changes that make nonfunctional RecA proteins.
Alleles supporting LexA cleavage; some alleles are defective for other RecA functions. In this category, only alleles with nonconservative changes were considered, as slight changes to the residue may not produce observable effects. Only changes in residues visible in the crystal structure are listed.
Second, residues close to the axis of the filament were not considered. Since this region binds to the DNA and ATP necessary for activation, mutations in this region would have a higher probability for disrupting the functions necessary for forming active filaments. Also, since filaments are believed to form before RecA and LexA interact, these areas of activated RecA may be less accessible to LexA. Again, recent work (19) suggests that this assumption may be faulty (see Discussion).
Mutagenesis of recA.
For each mutation, two, or in one case three, residues that are grouped spatially in the crystal structure were changed to alanine. We reasoned that if the RecA interaction with its coprotease substrates is dependent on several relatively weak interactions, multiple changes would more likely yield a cleavage-defective phenotype. In general, polar residues were chosen for mutagenesis. In one case, two hydrophobic residues in the cleft were changed. Residues targeted for mutagenesis are shown in Fig. 1 and listed in Table 3. Targeted residues were changed (see Materials and Methods) to alanine, a residue whose small side chain should reduce contacts without perturbing the overall folding of the protein. Similar approaches have been used to examine other protein-protein interactions (60).
TABLE 3.
Summary of mutant protein functions
| Mutation | RecA protein levelc | UV resistanced | Recombinatione
|
Basal LexA levelf | Cleavageg
|
|||
|---|---|---|---|---|---|---|---|---|
| λred− gam− | Intrachromosomal | LexA | λ CI | UmuD | ||||
| ΔrecA | None | S | NG | <0.07 | ++++ | − | − | − |
| recA+ | ++++ | R | G | 27 | ++ | WT | WT | WT |
| Clefta | ||||||||
| R105K106 | ++++ | R | G | 25 | +++ | S. def | S. def | WT |
| T242R243 | +++ | S+ | G | 0.2 | ++ | WT | Def | − |
| K232E235 | +++ | R | G | 27 | +++ | S. def | Def | Hyper |
| E233N236 | +++ | R | G | 27 | ++ | S. def | WT | WT |
| I102Y103 | ++++ | S | G− | <0.07 | +++ | S. def | Def | Def |
| K280E281K282 | ++++ | R | G | 33 | +++ | S. def | Def | S. def |
| K256Q257 | ++++ | R | G | 2.4 | ++ | WT | Constit | WT |
| K310D311 | ++++ | R | G | 24 | ++ | S. def | Def | WT |
| E285K286 | < | R− | G | 28 | +++ | S. def | S. def | S. def |
| E259Q261 | +++ | R | G | 25 | ++ | WT | Constit | WT |
| Other surfacesb | ||||||||
| E4K8 | ++++ | R | G | 27 | ++ | WT | Def | WT |
| Q16K19 | ++++++ | R | G | 18 | +++ | Def | WT | WT |
| R33D36 | +++ | R | G | 26 | ++ | WT | WT | WT |
| R85E86 | ++++ | R | G | 20 | ++ | WT | WT | WT |
| R176K177 | ++++ | S | NG | <0.07 | ++++ | − | − | − |
| E296K297 | ++ | R | G | 27 | +++ | S. def | WT | WT |
| Q300N304 | ++++ | R− | G | 6.1 | + | Constit | Constit | Constit |
| E314K317 | + | R | G | 26 | ++ | WT | S. def | WT |
| K294E318 | < | R | G | 35 | ++ | WT | S. def | WT |
| K321E325 | ++++ | R | G | 25 | ++ | WT | WT | WT |
Mutations affecting residues in the cleft.
Mutations affecting residues lying on the surface of the filament.
RecA protein levels: <, less than 10% RecA protein relative to wild type (WT); +, 10 to 25% WT; ++, 25 to 50%; +++, 50 to 75%; ++++, 75 to 100%.
UV resistance. Results of the UV sensitivity streak test are shown. R, WT cell growth; R−, slightly sensitive; S, like the ΔrecA control; S+, slightly more resistant than the ΔrecA strain.
Recombination. Growth of λred− gam− is indicated by G for WT growth, G− for growth of fewer and smaller plaques, and NG for no plaque growth. The numbers for intrachromosomal recombination indicate the average number of lac+ papillae per colony.
Basal LexA level. The relative levels of LexAEK45 are given. ++++, level with no RecA; +++, somewhat more than in the recA+ host; ++, level in a host with recA+; +, less than in recA+ host.
Cleavage by Western blotting. WT, rate of cleavage similar to that seen with WT RecA; S. def, somewhat defective; Def, some cleavage was observed but considerably less than WT; −, no cleavage detected; Constit, constitutive cleavage (that is, cleavage in the absence of nalidixic acid); Hyper, hypercleavable (that is, an increased rate relative to WT in the presence of nalidixic acid).
Cellular levels of RecA mutant proteins.
To uncouple levels of RecA from SOS induction, the RecA mutant proteins were expressed from a multicopy plasmid under the tac promoter. We compared the levels of mutant RecA proteins with that of the wild type using Western analysis (data not shown; summarized in Table 3). The levels of most mutant proteins were similar to those of the wild type. Several mutants had lower levels of RecA. K294E318 and E285K286 had very low levels; however, K294E318 was like the wild type for almost all the tested functions of RecA described below, so even this low protein level did not appear to impair RecA function in these assays. Q16K19 had elevated levels of protein.
UV sensitivity and recombination of mutants.
To test whether the mutations affected other functions of RecA, mutants were examined for defects in DNA repair and homologous recombination. Because some of these mutants might be defective for cleavage of LexA, and therefore unable to derepress the DNA repair genes of the SOS system, UV light sensitivity was tested in strains without functional LexA to avoid complications due to SOS regulation, using a semiquantitative test (see Materials and Methods) for UV sensitivity (summarized in Table 3). Most of the mutants were similar to the wild type. Two, I102Y103 and R176K177, were completely defective, and T242R243 was also very sensitive.
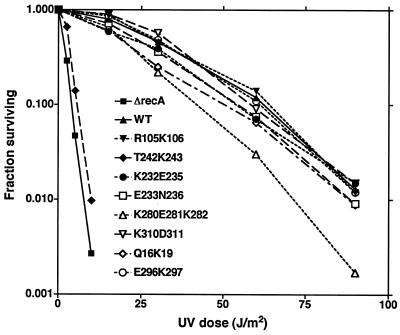
Tests of cleavage described in the following sections identified several mutations that conferred changes in cleavage but that appeared normal by the above-mentioned assay for UV sensitivity. The UV sensitivities of these mutants were examined by using a more quantitative UV survival assay (Fig. 2). T242R243 was included for comparison to previously characterized recA alleles changed at R243, and, as was noted above, it was very sensitive to UV. K280E281K282 also appeared to be slightly sensitive; however, the other mutants appeared to be similar to wild-type RecA, suggesting they had no major defects in DNA repair functions.
FIG. 2.
UV survival curves of wild type and selected mutants. Derivatives of JL2460 carrying plasmids with the indicated recA alleles are shown; the ΔrecA host carried pBR322. Cells were grown and irradiated with various doses of UV light as described in Materials and Methods; the fraction surviving was determined as described. The legend inset in the graph identifies the recA allele carried by each strain.
Two tests were used to examine recombination. The first test asked whether a λ red− gam− mutant could form plaques on lawns of hosts making mutant RecA proteins. This phage requires RecA-promoted recombination to make packageable DNA (51); plaque formation denotes recombination (Table 3). By this test, almost all the mutants supported recombination. Indeed, several mutants, such as T242R243 and I102Y103, that were very sensitive to UV nonetheless supported plaque formation. The difference in the two assays may be due to the fact that the red− gam− phage test is specific for recombination while survival of DNA damage requires other RecA functions. It is known that other recA mutations considered recombination defective, as judged by Hfr crosses, can support the growth of red− gam− phage (D. G. Ennis, personal communication). This suggests that phage growth requires only low levels of recombination.
In the second assay, we used a strain containing two nonoverlapping lac deletions and scored lac+ papillae that result from intrachromosomal recombination (Table 3). Several of the mutants that were similar to the wild type in the λ red− gam− assay showed reduced levels of recombination in this assay, presumably because this assay is more sensitive to small differences in recombination.
λ CI cleavage.
Mutants were assessed for cleavage of λ CI protein. Host cells lacked LexA function to avoid complications due to variations in induction of SOS function and were treated with nalidixic acid to induce the SOS system. The ability of the mutant RecA proteins to cleave CI was based on the amount of intact protein after 40 min of growth in nalidixic acid. Cells with or without treatment were harvested and analyzed by Western analysis. Typical Western blots are shown in Fig. 3, and the data are summarized in Table 3.
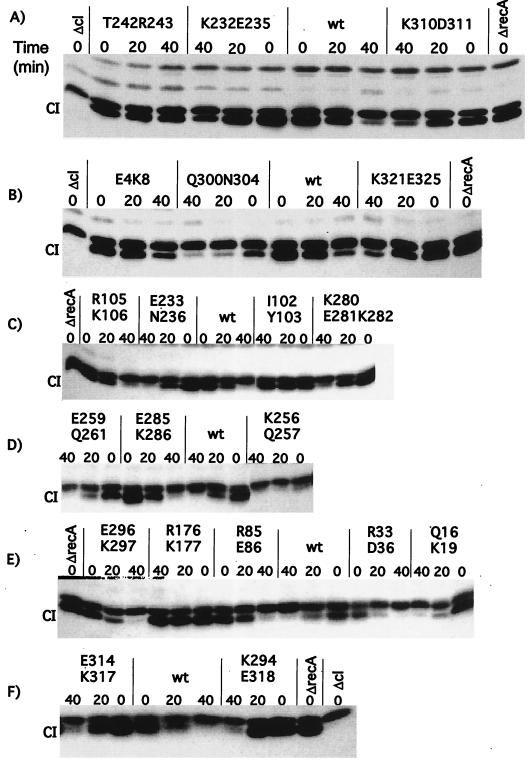
FIG. 3.
In vivo cleavage of λ CI repressor by mutant RecA proteins. Derivatives of JAM267 carrying pTrecA220 or mutant derivatives were treated as described in Materials and Methods. The strain marked ΔrecA carried pBR322. Each panel represents the results of a representative experiment. For each mutant strain, a time course is shown. The lanes marked “0” contain samples taken without treatment; those marked “20” and “40” were taken 20 and 40 min after the addition of nalidixic acid (50 μg/ml). The samples were analyzed by Western blotting with antibody against λ CI (see Materials and Methods). The name of the mutation is shown above each set of samples. The location of intact CI is shown; the band just above CI is a cross-reacting protein. wt, wild type.
The mutants had a range of phenotypes. Over half of the mutant proteins were able to cleave CI at least as well as wild-type RecA. Three mutants, K256Q257, E259Q261, and Q300N304 were constitutive for cleavage (Fig. 3B and D); E259Q261 was weaker than the others. All of the other mutant proteins, except R176K177, retained some ability to cleave CI, although six were noticeably defective. Somewhat unexpected was the defect in cleavage of T242R243 (Fig. 3A). Other alleles of RecA with changes at residue 243 are able to support λ prophage induction, a process that requires the cleavage of ≈90% of the CI (2a). We surmise (but have not tested directly) that the additional presence of the T242 change makes this mutant protein more defective for CI cleavage than those changed only at R243.
UmuD cleavage.
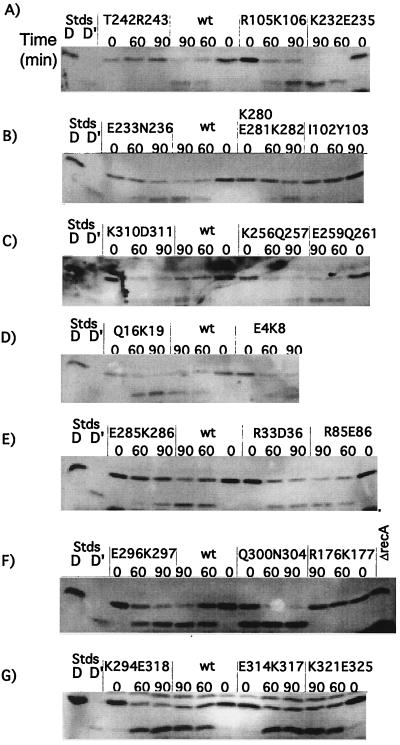
The ability of the RecA mutants to mediate cleavage of UmuD to UmuD′ was examined. UmuD was expressed from the chromosomal umuD gene, which was derepressed due to the presence of a lexA(Def) mutation. The cleavage product, UmuD′, was visible in the Western blots, and the ratio of UmuD to UmuD′ was useful for assessing cleavage. Since UmuD cleavage is slower than that of CI, cleavage was assessed at later time points. Typical results are shown in Fig. 4 and summarized in Table 3.
FIG. 4.
In vivo cleavage of UmuD by mutant RecA proteins. Derivatives of JL2460 carrying pTrecA220 or mutant derivatives were grown and treated as described in Materials and Methods; the strain marked ΔrecA carried pBR322. Each panel represents the results of a representative experiment. For each mutant strain, a time course is shown. Lanes marked “D” and “D′” are standards (Stds) from cells overproducing UmuD and UmuD′, respectively, and serve as markers. The lanes marked “0” contain samples taken without treatment; those marked “60” and “90” were taken 60 and 90 min after addition of nalidixic acid (50 μg/ml). The samples were analyzed by Western blotting with antibody to UmuD, as described in Materials and Methods. The name of the mutation is shown above each set of samples. wt, wild type.
Almost all of the mutant proteins cleaved UmuD like wild-type RecA. Only five showed any defects for UmuD cleavage. As expected from other studies of mutant proteins changed at residue 243, T242R243 was completely defective for cleavage (Fig. 4A). K232E235 displayed a “hypercleavable” phenotype (Fig. 4A). Cells with this mutant protein appeared to have all of the UmuD processed to UmuD′ by the 60-min time point, whereas wild-type RecA had about equal amounts of UmuD and UmuD′.
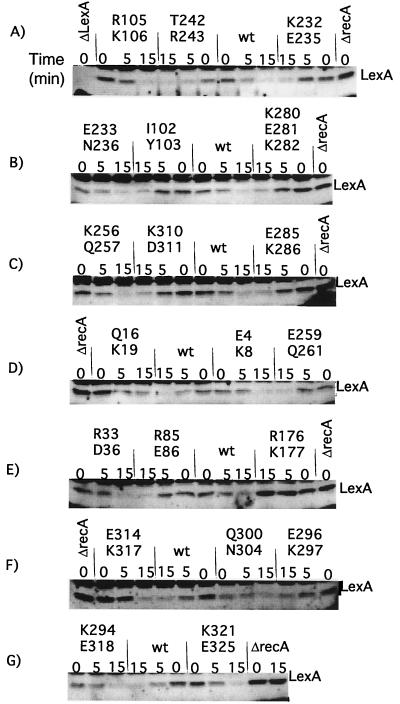
LexA cleavage.
The ability of the mutant RecA proteins to support cleavage of LexA was examined by Western analysis, using a mutant LexA protein, LexAEK45. Because LexA cleavage is rapid, samples were taken 5 and 15 min after the addition of nalidixic acid. Unlike the cases of λ CI and UmuD, analysis of LexA cleavage involves a complication. In cells with the recA+ plasmid, some of the LexAEK45 was cleaved even in the absence of inducing treatment, as can be seen by comparing the amount of intact LexA in the ΔrecA strain with the amount in the wild-type recA strain at time zero in Fig. 5. This is referred to as “basal cleavage” and has been seen in many other studies (19, 26, 29, 45, 50, 52). For wild-type recA, after the addition of the inducing agent, the rate of LexAEK45 cleavage increased substantially (Fig. 5); we refer to this increased rate of cleavage as “induced cleavage.” We do not know what signals activate RecA for basal cleavage or if they are the same as those in induced cells.
FIG. 5.
In vivo cleavage of LexA by mutant RecA proteins. Derivatives of strain JAM444 carrying pTrecA220 or mutant derivatives were grown and treated as described in Materials and Methods; the strain marked ΔrecA carried pBR322. Each panel represents the results of a representative experiment. For each mutant strain, a time course is shown. The lanes marked “0” contain samples taken without treatment; those marked “5” and “15” were taken 5 and 15 min after the addition of nalidixic acid (50 μg/ml). Samples were analyzed by Western blotting with antibody to LexA, as described in Materials and Methods. The name of the mutation is shown above each set of samples. The location of intact LexA is shown; the other bands are proteins that cross-reacted with the LexA antibody. Cleavage fragments are not shown; their levels are uninformative, since LexA cleavage fragments are unstable (27) (for an example, cf. Fig. 6). As described in the text, several mutants had less basal cleavage than the wild type. The amount of basal LexA cleavage did not appear to depend on the amount of RecA mutant protein. Q16K19 had very low levels of basal cleavage yet had higher levels of mutant RecA. On the other hand, K294E318 protein was present at very low levels and yet had the same amount of basal cleavage of LexA as the wild type (wt). The fact that the amount of basal cleavage did not correlate with the amount of RecA suggests that the extent of basal cleavage was limited by the amount of signal in the cell and/or by the ability of RecA protein to become activated by it.
Basal cleavage complicates the interpretation of mutant phenotypes. As described below, several mutant proteins were defective for basal cleavage but supported at least some induced cleavage. Cells with proteins defective for basal cleavage contained higher levels of intact LexA than the wild type; hence, after the cells are treated with inducing agent, it would take longer to cleave the bulk of the LexA even if the mutant proteins are cleaving LexA at the same rate as the wild type. Consequently, a direct comparison of intact LexA levels at the same time points could not always be made.
The amount of basal LexA cleavage varied among the different mutants, as can be seen by comparing the 0 time point lanes for the wild type and the mutants (Fig. 5; summarized in Table 3). For example, before the addition of nalidixic acid, Q16K19, E285K286, and K280E281K282 (Fig. 5D, C, and B, respectively) appeared to have an amount of intact LexA similar to that of the ΔrecA strain.
After the addition of nalidixic acid, the rate of wild-type cleavage increased markedly. About half the mutant proteins appeared to cleave LexA as well as the wild type. This included T242R243 (Fig. 5A), even though this mutant was very UV sensitive and defective for cleavage of CI and UmuD. This result confirms studies with other RecA alleles that are mutated at residue 243 (8, 9). Only R176K177 was completely defective for cleavage (Fig. 5E), but this protein appeared to be defective for all other RecA functions tested as well. Q16K19 (Fig. 5D) appeared to be partially defective for LexA cleavage. Eight other mutant proteins appeared to be slightly defective for cleavage; seven of these affected residues that lie in the cleft, but five also had small defects in basal cleavage.
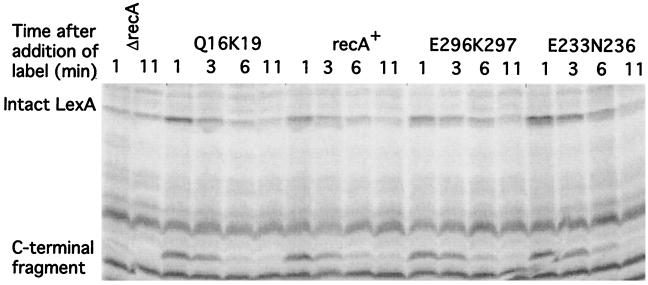
Western analysis was useful as an initial screen, but it does not examine cleavage directly. To follow the fate of newly synthesized LexA, we used a pulse-chase method to examine selected mutants. In this assay, cells were treated with nalidixic acid as before; after 30 min, they were given a pulse of [35S]methionine label, and aliquots were taken at various times after the addition of label. The cells were lysed, and LexA was precipitated with antibody and analyzed by gel electrophoresis and visualization with a PhosphorImager. We compared the rate of cleavage of wild-type LexA with those of three mutants. These were Q16K19, which appeared to give the slowest cleavage of LexA, and two representative mutants, E233N236 and E296K297, which appeared to have slight defects in cleavage. All three mutants appeared normal for cleavage of CI and UmuD. In the pulse-labeling assay, all three mutants appeared to have LexA cleavage rates similar to that of wild-type RecA (Fig. 6). We conclude that none of these mutant proteins is markedly defective for LexA cleavage, as judged by this assay.
FIG. 6.
Pulse-chase analysis of LexA cleavage for selected mutants. Derivatives of JL783 carrying pTrecA220 or mutant derivatives were grown as described in Materials and Methods; the strain marked ΔrecA carried pBR322. The cultures were treated with nalidixic acid for 30 min, followed by the addition of [35S]methionine (50 μCi/ml) and sampling at the times indicated. The identity of each set of samples is indicated above the gel. The samples were precipitated with antibody to LexA and analyzed as described in the text. The locations of intact LexA and the C-terminal cleavage product are indicated. The other bands are cross-reacting species also precipitated by the antibody. In the ΔrecA strain, less LexA is made, because LexA negatively autoregulates its own expression.
It is likely that the apparent defects of these proteins in the Western analysis resulted from differences in the amount of basal cleavage for the mutant proteins, as noted above. In addition, it is possible that some of the mutant proteins might be activated slowly in vivo. A slowly activating protein might appear defective for cleavage as judged by the Western assay but normal by the pulse-chase assay, which examined the fate of newly made LexA at a later time. It might also appear normal for cleavage of CI or UmuD, since these functions were assayed after longer treatments with nalidixic acid.
DISCUSSION
We first discuss the effects of the recA mutations on cleavage of CI and UmuD and the implications of our findings for the model in which cleavable proteins bind in the cleft region. Next we consider mechanisms by which RecA might be activated constitutively for cleavage. Then we discuss cleavage of LexA, ask why we were unable to recover LexA-specific mutant RecA proteins, and consider the possibility that the actual rate of RecA-mediated LexA cleavage might not be the rate-determining step in the LexA cleavage pathway.
Evidence for binding of λ CI and UmuD to the cleft region.
Several of the recA mutations had effects upon cleavage of λ CI and/or UmuD. Several mutations affecting residues in the cleft had modest defects in cleavage of λ CI, consistent with the model in which CI interacts with RecA in the cleft. If this is the case, the data are most consistent with the suggestion that the CI-RecA interaction involves many weak interactions and that interruption of any one or two of these has only modest effects on the strength of binding, since none of the mutations completely abolished CI cleavage.
At the same time, several of the recA mutants affecting other regions of the RecA filament also showed partial defects for CI cleavage. All three of the mutants (E4K8, E314K317, and K294E318) that were specifically defective for CI cleavage had changes in residues outside the cleft. One possibility is that CI makes contacts both within the cleft and in other regions. Another possible interpretation is that some of the changes are in residues that directly contact CI while others act indirectly, for example, by destabilizing a conformation of RecA that binds CI.
UmuD cleavage was completely defective in the T242R243 mutant, as expected from previous studies with mutations affecting Arg243 (8, 9). Less expected was the effect of the K232E235 change, which appeared to accelerate somewhat the rate of UmuD cleavage while reducing that of LexA and λ CI. Lys232 lies about 18 Å from Arg243. It is unlikely that changing residues 232 and 235 to Ala would create a new contact; more likely, these changes result in an adjustment of this part of the cleft which permits tighter binding to UmuD. It is somewhat surprising that no changes other than that in T242R243 reduced the level of UmuD cleavage, since we would expect multiple contacts at the UmuD-RecA interface. We surmise that, as discussed above for CI, UmuD interacts strongly with Arg243 but weakly with all its other contacts in the cleft.
Coprotease-constitutive mutations.
Several of our mutations, and many previously described mutations, confer constitutive activation upon RecA. For those constitutive mutations tested biochemically, this property has been found to result from a relaxed specificity for the polynucleotide and/or nucleotide cofactors that activate RecA (20, 57, 59). For a mutant protein made constitutive by this mechanism, one would expect constitutive activation for all substrates, given comparable levels of RecA (cf. reference 10). Indeed, we observed that the Q300N304 mutation had this property. By contrast, two mutations in the cleft, K256Q257 and E259Q261, were specifically constitutive for cleavage of λ CI. One plausible mechanism for specific constitutivity is that the mutations create a new contact and thereby confer extremely tight binding of the mutant protein to a particular substrate. In this mechanism, since cells contain a low level of activated RecA (as judged by the basal cleavage of LexA), a much tighter interaction would result in cleavage of that substrate. This mechanism seems unlikely in our mutations, however, given that they changed polar residues to Ala. Hence, we suggest that specific constitutivity can arise from some mechanism other than creation of a new contact.
Similarly, several changes at Pro67 or in the L1 loop (19, 37) appear to confer constitutive activation towards one substrate. While creating a new contact seems more plausible in these cases, given the chemical nature of the changes, our findings open the possibility that specific constitutivity can arise from a mechanism that does not affect the protein-protein interface directly. Detailed biochemical analysis of mutant proteins, and direct assays of interaction between RecA and its substrates, will likely be necessary to uncover the mechanistic basis for constitutive activation.
Cleavage of LexA.
RecA-dependent cleavage of LexA occurs rapidly after induction of the SOS system but can also be observed at a low rate in untreated cells (26, 45), a reaction we term basal cleavage. The molecular basis of basal cleavage is poorly understood—for instance, it is not known if the signal that activates RecA for basal cleavage differs from that which is present after SOS induction. It is also not known whether basal cleavage operates all the time in a given cell or if instead it acts sporadically. DNA replication forks in untreated cells frequently encounter sporadic damage (5, 6), some of which is repaired by RecA-dependent pathways. It is likely that RecA can be activated at such sites, resulting in basal cleavage, which might well operate only a portion of the time in any given cell.
We found that several of our mutants appeared to carry out less basal cleavage than the wild type (Table 3). Such mutant RecA proteins could interact poorly with LexA or might be unable to make effective use of the basal signals in the uninduced cell. Two of these mutants were tested for LexA cleavage by the pulse-chase assay and were found to support a normal rate of cleavage (Fig. 6). This finding favors the idea that the mutant proteins, particularly Q16K19, cannot utilize basal signals as well as the wild type.
With regard to induced LexA cleavage, however, we were unable to identify recA mutations with specific effects upon cleavage of LexA, in contrast with the specific effects of certain mutants on cleavage of CI and UmuD. In particular, we found no mutations in the cleft that had substantial effects on LexA cleavage, again in contrast to the result with CI and UmuD. Hence our work does not provide support for the proposal that LexA binds in this cleft.
The opposite conclusion was reached on the basis of electron microscopy of LexA-RecA complexes (62), which showed extra electron density in an area roughly corresponding to the cleft. We are unable to account for this disparity, but we note that the conclusions from electron micrograph image reconstruction are somewhat tentative for three reasons. First, the RecA filament was made on double-stranded DNA (dsDNA) to create a more regular structure for analysis. However, RecA complexes on dsDNA support lower rates of LexA cleavage and have a somewhat different structure than filaments made with single-stranded DNA. Second, the excess electron density corresponded to a mass that was considerably less than that of LexA, a finding attributed to incomplete occupancy of the filament. Finally, a tryptic fragment of LexA, a fragment which should bind to activated RecA, failed to do so, leaving open the possibility that the LexA-RecA interaction analyzed in this way was different from that analyzed by the in vivo and in vitro methods used here and in most previous studies of specific cleavage.
In any case, we did not identify LexA-specific mutations in the cleft or elsewhere on RecA. Why might this be the case? First, although we targeted most of the polar residues in the cleft, and 20 residues on the outer surface of the filament, we might have picked the wrong residues. Second, although we made double mutations, it is plausible for any given double that only one of the two residues is involved in the RecA-LexA interaction and that changing that one did not weaken the interaction enough to give a detectable effect. Third, as suggested for CI, if the interaction between LexA and RecA involves many weak interactions, loss of any one contact might confer a small defect. Finally, as we now discuss, one or more of our starting assumptions in targeting particular residues may have been faulty.
We assumed that the desired mutation would affect only LexA cleavage and not other functions of RecA. Such mutations might not exist. For example, recombination involves binding of a second dsDNA molecule to a RecA filament. Binding of a noncleavable LexA or the UmuD′C complex to RecA prevents RecA from binding a second strand of DNA (15, 41, 42). If the same region of RecA is involved in binding LexA and in interactions with the second strand of DNA, then certain mutations might affect both of these functions. It does seem plausible, however, that certain mutations would allow them to be separated.
We also assumed that the interaction between LexA and RecA did not occur close to the axis of the filament. However, recent work (19) has shown that one particular mutation, Pro67 to Arg, appears to have specific defects in cleaving LexA. This residue lies on the interior face of the filament (that is, on the “back side” of the molecule as seen in Fig. 1D). Whether this residue makes specific contacts with LexA is uncertain, however, since many other changes in it allow LexA cleavage, leaving open the possibility that the Pro67-to-Arg mutation works in an indirect manner. In any case, this assumption may prove to be faulty.
We further assumed that some or all of the interaction takes place in the portion of the structure that is visible. However, two internal portions of RecA, termed L1 and L2, are not seen in the structure, presumably because they form disordered loops. Recent work with residues in or near the L1 loop suggests the possibility that this loop is involved in RecA-LexA interaction (37).
Finally, we assumed that a weaker interaction between LexA and RecA would lead to detectable decreases in the rate of cleavage. That is, the rate-determining step for cleavage is conversion of free LexA into cleavage products, and decreases in the rate of this step will result in slower cleavage. In support of this assumption, previous studies have identified recA alleles with specific defects in cleavage of other substrates, and the recent studies with the Pro67-to-Arg mutant protein suggest that it is possible to identify changes in the rate of LexA cleavage. We note that mutants giving this behavior might be of the desired type but could in principle be ones that give a lower rate of cleavage by the RecA-LexA complex.
It is curious, however, that mutations in the other partner, LexA, that impair its interaction with RecA have also not been identified by assays that embodied this assumption. A large set of noninducible, or lexA(Ind−), mutations was identified by defects in RecA-mediated in vivo cleavage (24). A mutant LexA with defects in RecA-LexA interaction (a “RecA-specific” mutant) would appear to be normal, or nearly so, for autodigestion but defective for RecA-mediated cleavage. Of some 20 mutant proteins analyzed, however, none had this property (25). This finding is in striking contrast to a parallel analysis (14) of mutations in the λ CI repressor. Of the 15 mutant CI proteins studied, 9 autodigested normally or, in one case, faster than the wild type. This pattern suggests that these nine were RecA-specific mutants, although this hypothesis has not been tested directly. The large fraction of such mutations suggests that there is a qualitative difference between the pathways of cleavage of LexA and λ repressor. Although many reasons can account for the absence of such mutations in LexA, one could be, again, that a weakened interaction did not result in decreases in LexA cleavage substantial enough to give a phenotype in the screen employed in isolating those mutations.
This could occur if the RecA-LexA interaction were well above the Km. The measured Km in vitro is about 0.5 μM (23). The in vivo LexA concentration is about 1 μM (36, 45), and only about 20% of the LexA is free in the cell (45), so that in a wild-type cell the interaction should be below or near the Km. Consistent with this inference, LexA breakdown by activated RecA in the presence of chloramphenicol (45), as judged by Western blotting, showed first-order kinetics. In both our screens for mutants, however, LexA was expressed from a plasmid. In the screen for lexA(Ind−) mutations, the in vivo concentration of LexA in the absence of cleavage was 5 to 10 μM (24), so that mutant proteins with a weakened interaction could plausibly be missed. In the present work, by contrast, the LexA concentration in the absence of cleavage was roughly one-third the level in a wild-type cell (not shown), so that it should be at or below the Km.
We offer a speculation that could account for the paucity of mutations affecting the RecA-LexA interaction. Perhaps the bulk of the LexA is sequestered in the cell in some form from which it is only slowly released. This form is unlikely to be molecules nonspecifically bound to DNA, which one would expect to be released rapidly. In any case, if the rate of release from this sequestered form is far lower than the rate at which RecA mediates cleavage of the newly available molecules, small decreases in the rate at which RecA acts might not have a detectable effect on the overall rate of LexA disappearance; eventually, of course, the rate of cleavage could become so low as to be rate limiting. This model predicts that mutant forms of LexA should exist that could not be sequestered and that would exhibit faster in vivo cleavage. Although hypercleavable or Inds LexA proteins do exist, these also are cleaved faster in vitro (cf. references 18 and 44), where compartmentation should not be an issue. It remains unclear why mutations affecting this interaction are so elusive.
ACKNOWLEDGMENTS
We are grateful to Sue Roberts for help with the computer modeling of RecA; Roger Woodgate for his kind gift of UmuD antibody; Ken Knight for communication of unpublished results; Raymond Devoret, Fred Gimble, Ken Knight, and Andy Thliveris for strains; Michael Cox, Ken Knight, and Steve Kowalczykowski for helpful discussions; and Carol Dieckmann for comments on the manuscript.
This work was supported in part by NIH grant GM24178 and NSF grant MCB-9305092.
REFERENCES
- 1.Alexseyev A A, Bakhlanova I V, Zaitsev E N, Lanzov V A. Genetic characteristics of new recA mutants of Escherichia coli K-12. J Bacteriol. 1996;178:2018–2024. doi: 10.1128/jb.178.7.2018-2024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backman K, Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978;13:65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- 2a.Bailone A, Levine A, Devoret R. Inactivation of prophage λ repressor in vivo. J Mol Biol. 1979;131:553–572. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 5.Cox M M. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 6.Cox M M. Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol. 1999;63:310–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 7.Dutreix M, Burnett B, Bailone A, Radding C M, Devoret R. A partially deficient mutant, recA1730, that fails to form normal nucleoprotein filaments. Mol Gen Genet. 1992;232:489–497. doi: 10.1007/BF00266254. [DOI] [PubMed] [Google Scholar]
- 8.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ennis D G, Levine A S, Koch W H, Woodgate R. Analysis of recA mutants with altered SOS functions. Mutat Res DNA Repair. 1995;336:39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- 10.Ennis D G, Little J W, Mount D W. Novel mechanism for UV sensitivity and apparent UV nonmutability of recA432 mutants: persistent LexA cleavage following SOS induction. J Bacteriol. 1993;175:7373–7382. doi: 10.1128/jb.175.22.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag N E, McEntee K. Site-directed mutagenesis of the RecA protein of Escherichia coli. Tyrosine 264 is required for efficient ATP hydrolysis and strand exchange but not for LexA repressor inactivation. J Biol Chem. 1991;266:7058–7066. [PubMed] [Google Scholar]
- 12.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 13.Gimble F S, Sauer R T. Mutations in bacteriophage λ repressor that prevent RecA-mediated cleavage. J Bacteriol. 1985;162:147–154. doi: 10.1128/jb.162.1.147-154.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimble F S, Sauer R T. λ repressor inactivation: properties of purified Ind− proteins in the autodigestion and RecA-mediated cleavage reactions. J Mol Biol. 1986;192:39–47. doi: 10.1016/0022-2836(86)90462-6. [DOI] [PubMed] [Google Scholar]
- 15.Harmon F G, Rehrauer W M, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. 2. Inhibition of DNA strand exchange by the uncleavable LexA S119A repressor argues that recombination and SOS induction are competitive processes. J Biol Chem. 1996;271:23874–23883. [PubMed] [Google Scholar]
- 16.Horii T, Ozawa N, Ogawa T, Ogawa H. Inhibitory effects of N- and C-terminal truncated Escherichia coli recA gene products on functions of the wild-type recA gene. J Mol Biol. 1992;223:105–114. doi: 10.1016/0022-2836(92)90719-z. [DOI] [PubMed] [Google Scholar]
- 17.Ishimori K, Sommer S, Bailone A, Takahashi M, Cox M M, Devoret R. Characterization of a mutant RecA protein that facilitates homologous genetic recombination but not recombinational DNA repair: RecA423. J Mol Biol. 1996;264:696–712. doi: 10.1006/jmbi.1996.0670. [DOI] [PubMed] [Google Scholar]
- 18.Kim B, Little J W. LexA and λ CI repressors as enzymes: specific cleavage in an intermolecular reaction. Cell. 1993;73:1165–1173. doi: 10.1016/0092-8674(93)90645-7. [DOI] [PubMed] [Google Scholar]
- 19.Konola J T, Guzzo A, Gow J B, Walker G C, Knight K L. Differential cleavage of LexA and UmuD mediated by recA pro67 mutants: implications for common LexA and UmuD binding sites on RecA. J Mol Biol. 1998;276:405–415. doi: 10.1006/jmbi.1997.1531. [DOI] [PubMed] [Google Scholar]
- 20.Konola J T, Nastri H G, Logan K M, Knight K L. Mutations at Pro67 in the RecA protein P-loop motif differentially modify coprotease function and separate coprotease from recombination activities. J Biol Chem. 1995;270:8411–8419. doi: 10.1074/jbc.270.15.8411. [DOI] [PubMed] [Google Scholar]
- 21.Kowalczykowski S C. Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie. 1991;73:289–304. doi: 10.1016/0300-9084(91)90216-n. [DOI] [PubMed] [Google Scholar]
- 22.Larminat F, Cazaux C, Germanier M, Defais M. New mutations in and around the L2 disordered loop of the RecA protein modulate recombination and/or coprotease activity. J Bacteriol. 1992;174:6264–6269. doi: 10.1128/jb.174.19.6264-6269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L L. A genetic and biochemical analysis of LexA repressor cleavage in Escherichia coli K-12. Ph.D. dissertation. Tucson: University of Arizona; 1988. [Google Scholar]
- 24.Lin L L, Little J W. Isolation and characterization of noncleavable (Ind−) mutants of the LexA repressor of Escherichia coli K-12. J Bacteriol. 1988;170:2163–2173. doi: 10.1128/jb.170.5.2163-2173.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L L, Little J W. Autodigestion and RecA-dependent cleavage of Ind− mutant LexA proteins. J Mol Biol. 1989;210:439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- 26.Little J W. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983;167:791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- 27.Little J W. Variations in the stability of lexA repressor during the SOS regulatory cycle. In: Friedberg E C, Bridges B A, editors. Cellular responses to DNA damage. New York, N.Y: Alan R. Liss, Inc.; 1983. pp. 369–378. [Google Scholar]
- 28.Little J W. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little J W, Hill S A. Deletions within a hinge region of a specific DNA-binding protein. Proc Natl Acad Sci USA. 1985;82:2301–2305. doi: 10.1073/pnas.82.8.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little J W, Mount D W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 31.Little J W, Shepley D P, Wert D W. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S-K, Eisen J A, Hanawalt P C, Tessman I. recA mutations that reduce the constitutive coprotease activity of the RecA1202(Prtc) protein: possible involvement of interfilament association in proteolytic and recombination activities. J Bacteriol. 1993;175:6518–6529. doi: 10.1128/jb.175.20.6518-6529.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis R, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Miller J H, Lebkowski J S, Greisen K S, Calos M P. Specificity of mutations induced in transfected DNA by mammalian cells. EMBO J. 1984;3:3117–3121. doi: 10.1002/j.1460-2075.1984.tb02267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreau P L. Effects of overproduction of single-stranded DNA-binding protein on RecA-dependent processes in Escherichia coli. J Mol Biol. 1987;194:621–634. doi: 10.1016/0022-2836(87)90239-7. [DOI] [PubMed] [Google Scholar]
- 37.Nastri H G, Guzzo A, Lange C S, Walker G C, Knight K L. Mutational analysis of the RecA protein L1 region identifies this area as a probable part of the co-protease substrate binding site. Mol Microbiol. 1997;25:967–978. doi: 10.1111/j.1365-2958.1997.mmi533.x. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen T T, Muench K A, Bryant F R. Inactivation of the recA protein by mutation of histidine 97 or lysine 248 at the subunit interface. J Biol Chem. 1993;268:3107–3113. [PubMed] [Google Scholar]
- 39.Ogawa H, Ogawa T. General recombination: functions and structure of RecA protein. Adv Biophys. 1986;21:135–148. doi: 10.1016/0065-227x(86)90019-5. [DOI] [PubMed] [Google Scholar]
- 40.Peterson K R, Mount D W. Differential repression of SOS genes by unstable LexA41 (Tsl-1) protein causes a “split-phenotype” in Escherichia coli K-12. J Mol Biol. 1987;193:27–40. doi: 10.1016/0022-2836(87)90623-1. [DOI] [PubMed] [Google Scholar]
- 41.Rehrauer W M, Bruck I, Woodgate R, Goodmann M F, Kowalczykowski S C. Modulation of RecA nucleoprotein function by the mutagenic UmuD′C protein complex. J Biol Chem. 1998;273:32384–32387. doi: 10.1074/jbc.273.49.32384. [DOI] [PubMed] [Google Scholar]
- 42.Rehrauer W M, Lavery P E, Palmer E L, Singh R N, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. 1. LexA repressor cleavage is competitive with binding of a secondary DNA molecule. J Biol Chem. 1996;271:23865–23873. [PubMed] [Google Scholar]
- 43.Roberts J W, Devoret R. Lysogenic induction. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 123–144. [Google Scholar]
- 44.Roland K L, Smith M H, Rupley J A, Little J W. In vitro analysis of mutant LexA proteins with an increased rate of specific cleavage. J Mol Biol. 1992;228:395–408. doi: 10.1016/0022-2836(92)90829-9. [DOI] [PubMed] [Google Scholar]
- 45.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 46.Sayle R, Milner-White E J. RasMol: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 47.Scheffzek K, Ahmadian M R, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 48.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 49.Skiba M C, Knight K L. Functionally important residues at a subunit interface site in the RecA protein from Escherichia coli. J Biol Chem. 1994;269:3823–3828. [PubMed] [Google Scholar]
- 50.Slilaty S N, Little J W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith G. General recombination. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 175–209. [Google Scholar]
- 52.Smith M H, Cavenagh M M, Little J W. Mutant LexA proteins with an increased rate of in vivo cleavage. Proc Natl Acad Sci USA. 1991;88:7356–7360. doi: 10.1073/pnas.88.16.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Story R M, Weber I T, Steitz T A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 54.Sweasy J B, Witkin E M, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thliveris A T, Mount D W. Genetic identification of the DNA binding domain of Escherichia coli LexA protein. Proc Natl Acad Sci USA. 1992;89:4500–4504. doi: 10.1073/pnas.89.10.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thoms B, Wackernagel W. Suppression of the UV-sensitive phenotype of Escherichia coli recF mutants by recA(Srf) and recA(Tif) mutations requires recJ+ J Bacteriol. 1988;170:3675–3681. doi: 10.1128/jb.170.8.3675-3681.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W-B, Sassanfar M, Tessman I, Roberts J W, Tessman E S. Activation of protease-constitutive RecA proteins of Escherichia coli by all of the common nucleoside triphosphates. J Bacteriol. 1988;170:4816–4822. doi: 10.1128/jb.170.10.4816-4822.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W-B, Tessman E S. Location of functional regions of the Escherichia coli RecA protein by DNA sequence analysis of RecA protease-constitutive mutants. J Bacteriol. 1986;168:901–910. doi: 10.1128/jb.168.2.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W-B, Tessman E S, Tessman I. Activation of protease-constitutive RecA proteins of Escherichia coli by rRNA and tRNA. J Bacteriol. 1988;170:4823–4827. doi: 10.1128/jb.170.10.4823-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X, Egelman E H. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, Egelman E H. The LexA repressor binds within the deep helical groove of the activated RecA filament. J Mol Biol. 1993;231:29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]