Abstract
REV-ERBα and REV-ERBβ are heme regulated nuclear receptors that are known to regulate metabolic pathways. We previously demonstrated that treatment of mice with synthetic REV-ERB agonists suppressed plasma cholesterol levels and the hepatic levels of the rate limiting enzyme in cholesterol biosynthesis (3-hydroxy-3-Mmethylglutaryl-CoA reductase). Here, we characterize the role of REV-ERB on the cholesterol biosynthetic pathway in greater detail. The REV-ERB agonist SR9009 reduced plasma cholesterol levels in both wild type C57Bl/6 and low density lipoprotein receptor (LDLR) null mice as well as reducing the expression of an array of genes within the cholesterol biosynthetic pathway. Consistent with this data, we observed increased expression of these genes in mice deficient in expression of Rev-erbα. Analysis of global run-on and deep sequencing (GRO-Seq) and chromatin immunoprecipitation deep sequencing (ChIP-Seq) data revealed that Rev-erb directly binds to the majority of genes involved in cholesterol biosynthesis and directly suppresses their expression. This study reveals insight into the complex mechanism by which Rev-erb directly and indirectly (via inhibition of Srebf2 expression) regulates cholesterol biosynthesis and provides information of how cholesterol levels are regulated in a circadian fashion. Additionally, these studies suggest that targeting Rev-erb may be an effective method for suppressing LDL cholesterol levels in the clinic.
Keywords: Nuclear receptor, Rev-erb, Cholesterol synthesis, statins, atherosclerosis
1. Introduction
Cholesterol is an essential component of the cell membrane and an important metabolic precursor in biosynthetic pathways, including steroid hormone, vitamin D and bile acid synthesis. Significant pathological conditions, including atherosclerosis, result from an imbalance in cholesterol metabolism making it crucial for tight regulation of cholesterol homeostasis in order to maintain human health [1]–[6]. This regulation is achieved via complex interplay of multiple factors that control endogenous cholesterol biosynthesis, absorption, elimination and plasma transport pathways. The liver is a central site for the regulation of these pathways and one of the principal sites for the biosynthesis of cholesterol. Cholesterol biosynthesis occurs through a multistep pathway involving numerous enzymes. Targeting the rate-limiting enzyme of this pathway (3-hydroxy-3-methylglutaryl-CoA reductase; HMGCR) with statins has been successfully utilized to therapeutically reduce low-density lipoprotein cholesterol levels and risk of atherosclerosis. However, many patients with hypercholesterolemia are unable to reach target LDL cholesterol levels on statins and others are unable to maintain statin therapy due to side effects indicating that additional cholesterol lowering drugs are needed [7], [8].
Nuclear receptors are a superfamily of ligand-activated transcription factors that regulate essential physiological processes such as growth, development and metabolic homeostasis. Rev-erbα and Rev-erbβ (Nr1d1 and Nr1d2) are nuclear receptors that serve as receptors for heme [9]–[11]. The Rev-erbs bind to specific DNA response elements and repress the transcription of target genes by either competing with retinoic acid-related orphan receptors (RORs) for binding to their DNA response elements or by directly repressing transcription via recruitment of corepressor complex in a heme-dependent manner [12], [13] [14]. Studies over the last decade have made clear the important role of the Rev-erbs in metabolic pathways. In the liver, the Rev-erbα and Rev-erbβ cistromes are enriched for genes involved in lipid metabolism [15], [16]. Studies in mice deficient in Rev-erb expression display deregulated lipid metabolism and hepatic steatosis [17]. In addition to an increase in liver triglycerides and increase in plasma cholesterol levels were also observed in Rev-erb deficient mice. Delezie et al. demonstrated that there is a significant increase in low-density lipoprotein (LDL) and total cholesterol in Rev-erbα−/− mice [18].
Consistent with these findings we recently demonstrated that SR9009, a Rev-erb agonist, reduces total plasma cholesterol and triglyceride levels in vivo [19]. Our results suggested the involvement of Rev-erbs in the cholesterologenesis process and regulation of important cholesterol biosynthesis enzymes such as HMGCR, however the effect of these ligands on the expression of other cholesterol biosynthetic enzymes was not investigated. Moreover, these effects were initially examined in HDL dominant wild type and diet induced obese mice and not a LDL cholesterol dominant model such as the LDLR null mouse model.
Here, we examine the effects of gain of function (REV-ERB agonist in wild-type and LDLR null mice) and loss of function (Rev-erbα null mice) on the expression of genes encoding cholesterol biosynthetic enzymes in the liver. Based on our results indicating a broad effect of REV-ERB on expression of enzymes encoding cholesterol biosynthetic genes we further examined the mechanism by which REV-ERB coordinately regulates this pathway using GRO-Seq and ChiP-Seq data.
2. Materials and Methods
2.1 Animals and treatment
All procedures were approved and conducted in accordance to the Scripps Florida Institutional Animal Care and Use Committee and Saint Louis University Institutional Animal Care and Use Committee. Twelve wild type C57Bl/6J mice were obtained from Jackson laboratory and maintained on chow diet. The Rev-erb agonist SR9009 was formulated in 15% cremophor. 100mg/kg (milligram drug per kilogram body weight) SR9009 or vehicle was administered twice a day intraperitoneally for ten days. Body weight and food intake was monitored daily.
Twenty homozygous LDL receptor deficient (ldlr−/−) male mice were obtained from Jackson Laboratory (Bar Harbor, ME) at the age of 12 weeks and were housed individually in 12 hour light/dark cycles. The mice were allowed to acclimate for a week and were maintained on standard chow diet. During the experiment the mice were fed Western diet containing 0.5% (w/w) cholesterol and 42% calories from fat (w/w) (Research diets, NJ). The Rev-erb agonist SR9009 was formulated in 15% cremophor (C5135, Sigma-Aldrich, St. Louis, MO). 100mg/kg SR9009 or vehicle was administered twice a day intraperitoneally for eight weeks. Body weight and food intake was monitored daily.
Rev-erbα knockout mice were bred in house and housed in 12 hour light/dark cycles. Eleven-week old Rev-erbα−/− and wildtype littermate control were sacrificed and tissues were collected at the age of 11 weeks (n = 4 per group).
2.2 Plasma lipid analysis
Mice were euthanized after 5 hour fasting and blood was collected via cardiac puncture. Concentration of plasma total cholesterol, HDL cholesterol, LDL cholesterol, triglyceride, glucose and liver enzymes were assessed using COBAS clinical chemistry analyzer (Randox Daytona).
2.3 Quantitative Real-Time PCR
Total RNA was isolated from mouse liver using Trizol-isopropanol extraction (15596026, Life Technologies, Carlsbad, CA). RNA was reverse-transcribed to make cDNA using qScript™ cDNA Synthesis Kit (101414-098, VWR, Radnor, PA) according to the manufacturer’s instructions. Real-time PCR was performed using a SYBR-green PCR master mix kit (4472913, Life Technologies, Carlsbad, CA). Primers were purchased from Integrated DNA Technologies (Coralville, IA).
2.4 Primer sequences
|
|
| Cyp51-F CCCTCAGACGGTGGCAGGGT |
|
|
| Cyp51-R GTCCAAGCGCTCTGCCCAGG |
|
|
| Dhcr7-F GGGCTGCAAGCCTGGCTCATT |
|
|
| Dhcr7-R TGCGAACGTGGACACGGCAT |
|
|
| Fdft1-F CCTTGCCCTCAGCAGCCTGG |
|
|
| Fdft1-R GCACGCTGCCAGTGGCTACA |
|
|
| Fdps-F ACAACCGGGGTTTGACCGTG |
|
|
| Fdps-R TGTCAGGGCCCGCTGAAGAC |
|
|
| Idi1-F GGCCATGACACTCAACCCAGC |
|
|
| Idi1-R TGGGCCAAACATCCAAGGGCT |
|
|
| mLss-F CGTTGCTGACTGCACAGCCG, |
|
|
| Lss-R ACAGCATCGCACAGCCGCTC |
|
|
| Mvd-F TCGAGTGTGATGGGCAGCCA |
|
|
| Mvd-R CAGAAACCAGCGGGGAACCG |
|
|
| Mvk-F AGGTCCCGCGGAGTACCAAG |
|
|
| Mvk-R CTAGCACGCGCTCACACTCC |
|
|
| Pmvk-F GGAAGGCGTGTCCCAGCCTA |
|
|
| Pmvk-R CCCGTTGCTGTCGACTCTGC |
|
|
| Sc5dl-F TAGGGGCCTGCACCACAGACT |
|
|
| Sc5dl-R GACTTGCAAACGGCGTGGGGA |
|
|
| Sqle-F CCGTTTACAGCCAGGCGAGC, |
|
|
| Sqle-R ACTGATGGACACGGGCCTCT; |
|
|
| Rev-erb alpha-F GGGCACAAGCAACATTACCA |
|
|
| Rev-erb alpha-R CACGTCCCCACACACCTTAC |
|
|
| Rev-erb beta-F TGGGACTTTTGAGGTTTTAATGG |
|
|
| Rev-erb beta-R GTGACAGTCCGTTCCTTTGC |
|
|
| Abcg5-F AGCGTCAGCAACCGTGTC |
|
|
| Abcg5-R AGCAGCGTGGTCTTCCCT |
|
|
| Srebp2-F CATCTGCCGGTGGTGGACGT, |
|
|
| Srebp2-R GCGCACAGCTGCATCGTCTC |
|
|
| Hmgcr-F ATGGCTGGGAGCATAGGCGG |
|
|
| Hmgcr-R CTGCATCCTGGCCACATGCG |
|
|
| Hmgcs1-F AGGAACGTGGTATCTGGTCA |
|
|
| Hmgcs1-R TGTGTTACTATGCACGAGCC |
|
|
| Sc4mol-F TCAGGCTCCATTTGGAATCG |
|
|
| Sc4mol-R TCCAGCAAACGTATGGTCAC |
|
|
| Gapdh-F GCCAAGGCTGTGGGCAAGGT |
|
|
| Gapdh – R TCTCCAGGCGGCACGTCAGA |
|
|
| Abca1-F GGCTCCTCCCTGTTTTTGAA |
|
|
| Abca1-R GAACTGAGGGACGATTCCAC |
|
|
| Abcg1-F CAGCTACTCTGCCGCAATGA |
|
|
| Abcg1-R TGGGCCTCTGTGAAGTTGTT |
|
|
| Abcg8-F CCAAATGTCACTCGGAGAGG |
|
|
| Abcg8-R CGCGTAGATGATGACGTAGG |
|
|
Relative mRNA levels were calculated using the 2−ΔΔCt method and normalized against Gapdh.
2.5 Western Blotting
Mouse liver was homogenized and lysed in RIPA buffer (sc-24948, Santa Cruz Biotechnology, Santa Cruz, CA) containing protease inhibitor (11836153001, Sigma-Aldrich, St. Louis, MO) to prepare protein according to the manufacturer’s instruction. After protein quantification, 25 μg of protein samples were prepared in Laemmli buffer. Protein was separated using 4–15% protein gel (4561083, Bio-Rad, Mississauga, ON, Canada) and transferred to PVDF membranes using transfer pack (170-4156, Bio-Rad, Mississauga, ON, Canada). The membranes were incubated in 5% milk in TBST for 1 hour at room temperature. Membranes were incubated overnight at 4 degrees C in 5% BSA (BP1600, Fisher Scientific, Fair Lawn, NJ) containing primary antibody directed against HMGCR (1:1000; ab174830, Abcam), HMGCS1 (1:1000; ab155787, Abcam, Cambridge, MA), SREBP2 (1:1000; 10007663, Cayman Chemical Company, Ann Arbour, MI) or β-actin (1:1000; sc-130656, Santa Cruz Biotechnology, Santa Cruz, CA). After incubation, the membranes were washed in TBST and appropriate HRP-coupled secondary antibody (1:5000; 7074S, Cell Signaling Technology, Danvers, MA) diluted in 5% milk in TBST was used.
2.6 Cell culture and treatment
HepG2 cells (HB-8065) were obtained from ATCC and cultured in Advanced minimum essential medium (Advanced MEM, Life Technologies Inc., MD) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a humidified incubator under 5% CO2. Cells were treated with 10 μM SR9011 or DMSO control for 24 hours prior to analysis of gene expression.
2.7 Cholesterol Biosynthesis Assay
HepG2 cells were treated with SR9009 for 24 hours followed by 3-hour incubation in 1 μCi/ml of 14C sodium acetate (Perkin Elmer, Waltham, MA) for in vitro analysis of cholesterol synthesis. AR-2000 radio-TLC Imaging Scanner was used to analyze radioisotope incorporation in cholesterol to determine cholesterol biosynthesis.
2.7 Statistical Analysis
Data are expressed as mean +/− SEM. Student’s t-test was used to calculate statistical significance. P<0.05 was considered significant.
2.8 GRO-Seq/ChIP-Seq datasets and analysis
Raw GRO-Seq and ChIP-Seq datasets [17], [20], [21] were downloaded from NIH GEO and SRA public servers (http://www.ncbi.nlm.nih.gov/sra). The accession numbers of these datasets are SRP057271/GSE67962, SRP008783/GSE26345 and SRP044381/GSE59486. FASTQ files were converted from the downloaded files using fastq-dump (version 2.3.5). Reads were mapped to mouse mm10 genome using STAR (version 2.4.2a). The generated BAM files were analyzed using HOMER software (version 4.7.2) [22] and R statistical packages (version 3.2.0) to identify binding peaks, quantify expression/binding signals and produce histogram plots. Tag directories were first created from the mapped reads using makeTagDirectory. Next, for GRO-Seq analysis, the reads were quantified using analyzeRNA.pl, followed by TMM normalization and differential expression analysis by edgeR. Both normalization and differential expression analysis were carried out using getDiffExpression.pl in the HOMER package. For ChIP-Seq analysis, Rev-erb binding sites were determined by peak calling using findPeaks.pl (FDR <= 0.001, fold over input > = 4, Poisson p <= 0.0001). Binding signals were quantified by annotatePeaks.pl and plotted using R. De novo motif discovery and motif enrichment analyses were carried out using findMotifsGenome.pl. All analyses were performed with default parameter settings. To visualize ChIP-Seq signals, bedgraph files were generated using makeUCSCfile and loaded into IGV (Integrative Genomics Viewer) (https://www.broadinstitute.org/igv/). P-values of gene-list enrichment were calculated using binomial tests with R packages.
2.9 Gene Ontology (GO)
Gene Ontology and gene-list enrichment analysis of Rev-erb target genes were performed with ToppGene server (http://toppgene.cchmc.org) as previously reported [23].
3. Results
3.1 Rev-erb regulates cholesterol biosynthesis
To understand the role of the Rev-erbs in regulation of the expression genes encoding enzymes required for cholesterol biosynthesis in vivo we used C57Bl/6J male mice and treated them with the synthetic Rev-erb agonist SR9009 (100mg/kg, I.P. b.i.d.) for ten days. Body weight was unaffected by the treatment (Figure 1a). Since the liver is the major site for cholesterol biosynthesis, we examined the expression of cholesterol biosynthesis pathway genes in this tissue. Expression of key genes such as Hmgcr and other intermediate genes in the pathway such as mevalonate diphosphate decarboxylase (Mvd), methylsterol monooxygenase 1 (Sc4mol) and mevalonate kinase (Mvk) were significantly reduced in SR9009 treated animal compared to vehicle (Figure 1b). Based on our previous studies we expected a reduction in total plasma cholesterol levels in these mice [19]. We observed a reduction in total cholesterol (43%), HDL-C (42%) and triglycerides (39%) with SR9009 treatment (Figure 1c). LDL cholesterol levels were undetectable (data not shown).
Figure 1.
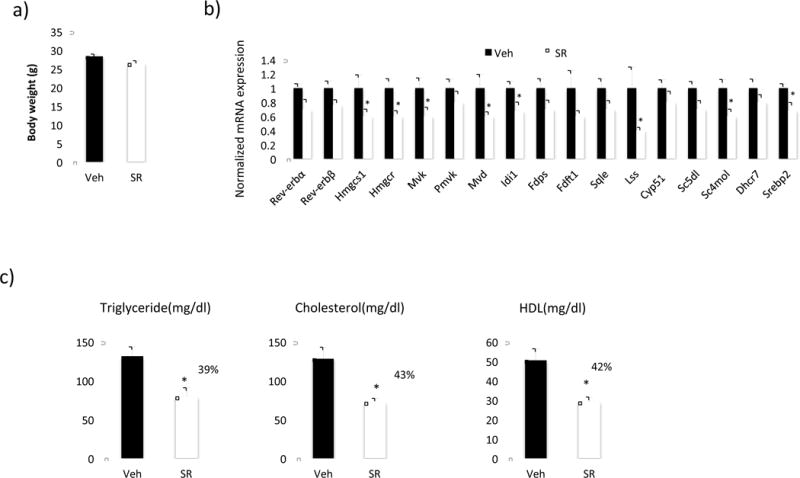
Rev-erb agonist SR9009 reduces the expression of cholesterol biosynthesis pathway genes and plasma cholesterol levels in C57Bl/6 mice. Male mice (n=6) were treated with SR9009 (100mg/kg, I.P. b.i.d.) for ten days. a) Body weight of mice at the end of the treatment. B) mRNA expression of cholesterol biosynthesis pathway genes in mouse liver. mRNA was normalized to Gapdh. c) Plasma levels of triglyceride, total cholesterol, LDL-cholesterol and HDL-cholesterol. Data are presented as the mean ± SE. Student’s t-test was used to calculate statistical significance. * indicates a p-value < 0.05.
While wild-type C57Bl/6J (WT) mice can provide some insight into cholesterol metabolism, there are major differences in HDL and LDL cholesterol levels in mice vs. humans. Mice that are genetically modified and under specific diet conditions can acquire plasma lipid profile similar to that of human and these mice are commonly used to study metabolic disorders including atherosclerosis [24]. Next, to investigate the ability of the Rev-erbs to regulate cholesterol biosynthesis in mouse model of elevated serum cholesterol similar to humans, we used LDL receptor deficient mice. These mice were fed with a high cholesterol diet and were treated with SR9009 (100mg/kg, twice a day) for a period of eight weeks. Similar to our observation in WT mice, gene expression analysis in liver of these mice showed reduction of cholesterol biosynthesis genes (Figure 2b). Plasma cholesterol analysis showed significant reduction in total cholesterol (36%), LDL-C (41%) and triglycerides (40%) in response to SR9009 treatment (Figure 2c). HDL-C levels were not affected by SR9009 treatment. The body weight was not affected by the treatment (Figure 2a). These data suggest that the Rev-erbs regulate cholesterol biosynthesis genes in vivo and pharmacological activation of these receptors improves the plasma lipid profile significantly. We also examined the hepatic expression of cholesterol transporters (abca1, abcg1, abcg5, and abcg8) in response to SR9009 treatment in WT and LDLR null mice. There were no changes in response to SR9009 in the LDLR null mice and only abca1 expression was altered in WT mice (a reduction of ~25%) (data not shown).
Figure 2.
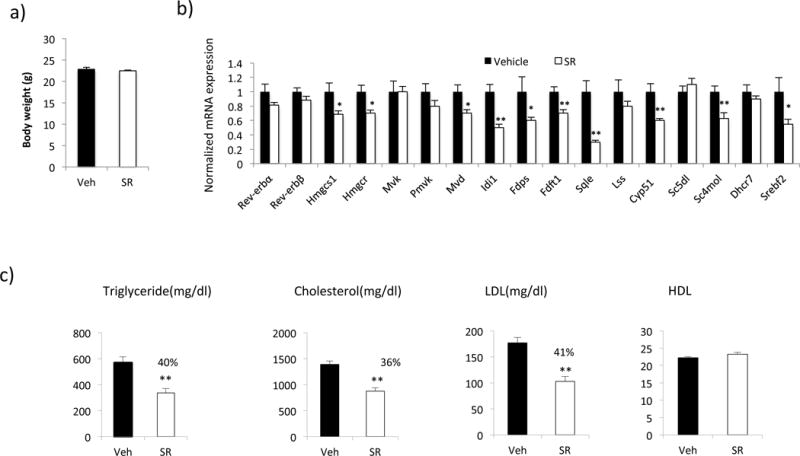
Rev-erb agonist SR9009 reduces the expression of cholesterol biosynthesis pathway genes and plasma cholesterol levels LDLR−/− mice. Male mice (n=11) were placed in high cholesterol diet (0.5%) and treated with SR9009 (100mg/kg, I.P. b.i.d.) for two months. a) Body weight of mice at the end of the treatment. B) mRNA expression of cholesterol biosynthesis pathway genes in mouse liver. mRNA was normalized to Gapdh. c) Plasma levels of triglyceride, total cholesterol, LDL-cholesterol and HDL-cholesterol. Data are presented as the mean ± SE. Student’s t-test was used to calculate statistical significance. * indicates a p-value < 0.05.
Previous data demonstrated that Rev-erb deficiency leads to disrupted lipid metabolism [15]. To test whether cholesterol biosynthetic enzyme gene expression is altered in Rev-erbα−/− mice, we examined liver samples and found significant upregulation of an array of cholesterol biosynthesis genes (Table 1). This increase in expression in the absence of Rev-erbα demonstrates an important role of the Rev-erbs in suppression of expression of these genes. These data, taken together, indicate that Rev-erb plays a role in suppression of several cholesterologenic enzyme genes and that pharmacological activation of Rev-erb leads to further suppression of these genes, which correlates with reduced plasma cholesterol levels.
Table 1.
Hepatic Expression of Cholesterologic Enzyme Genes In WT vs. Rev-erba KO mice
| Genes | Wildtype Control | Rev-erb α KO |
|---|---|---|
| Cyp51 | 1 ± 0.09 | 0.86 ± 0.10 |
| Dhcr7 | 1 ± 0.07 | 1.47 ± 0.22* |
| Fdft1 | 1 ± 0.12 | 0.92 ± 0.14 |
| Fdps | 1 ± 0.15 | 1.03 ± 0.24 |
| Hmgcr | 1 ± 0.09 | 1.71 ± 0.40* |
| Hmgcs1 | 1 ± 0.17 | 1.56 ± 0.23* |
| Msmol1 | 1 ± 0.14 | 1.20 ± 0.16 |
| Pmvk | 1 ± 0.11 | 1.63 ± 0.16* |
| Lss | 1 ± 0.12 | 1.92 ± 0.29* |
| Mvd | 1 ± 0.10 | 1.65 ± 0.47 |
| Mvk | 1 ± 0.13 | 0.96 ± 0.16 |
| Sc5dl | 1 ± 0.04 | 1.02 ± 0.15 |
| Sqle | 1 ± 0.16 | 1.07 ± 0.24 |
| Idi1 | 1 ± 0.24 | 1.57 ± 0.29* |
Gene expression of cholesterol biosynthesis pathway genes in mouse liver from 11 week old Rev-erb α- deficient (KO) mice and wild-type (WT) mice (n = 4). mRNA was normalized to Gapdh. Data are presented as the mean ± SE. Student’s t-test was used to calculate statistical significance.
indicates a p-value < 0.05.
We examined the levels of protein of several of the enzymes to confirm the effects of SR9009 treatment in the ldlr−/− mice. Levels of HMGCS1 were significantly suppressed by treatment with SR9009 (Figure 3a). There was a trend in reduction of the accumulation of HMGCR by treatment with SR9009 (Figure 3b). Additionally, levels of cleaved form of Sterol regulatory element-binding protein 2 (SREBP2) were also inhibited in SR9009 treated mice (Fig. 3c). Previously, we demonstrated that hepatic Srebp2 gene expression was decreased in mice treated with SR9009 [19]. SREBP2 is a master transcription factor that regulates the array of genes encoding cholesterol biosynthetic genes [25], [26] thus it is possible that REV-ERB may be regulating the expression of genes encoding cholesterol biosynthetic enzymes indirectly (via regulation of SREBP2) or a combination of indirect and direct mechanisms (via regulation of SREBP2 and direct regulation of the cholesterol biosynthetic enzyme genes). We attempted to quantitate the uncleaved form of SREBP2 as well but were unsuccessful using three distinct antibodies (Abcam ab30682, Cayman 10007663 and Santa Cruz sc-271616). However, the reduction in the cleaved, nuclear form of SREBP2 is consistent with the reduction in expression of its key target genes we observed.
Figure 3.
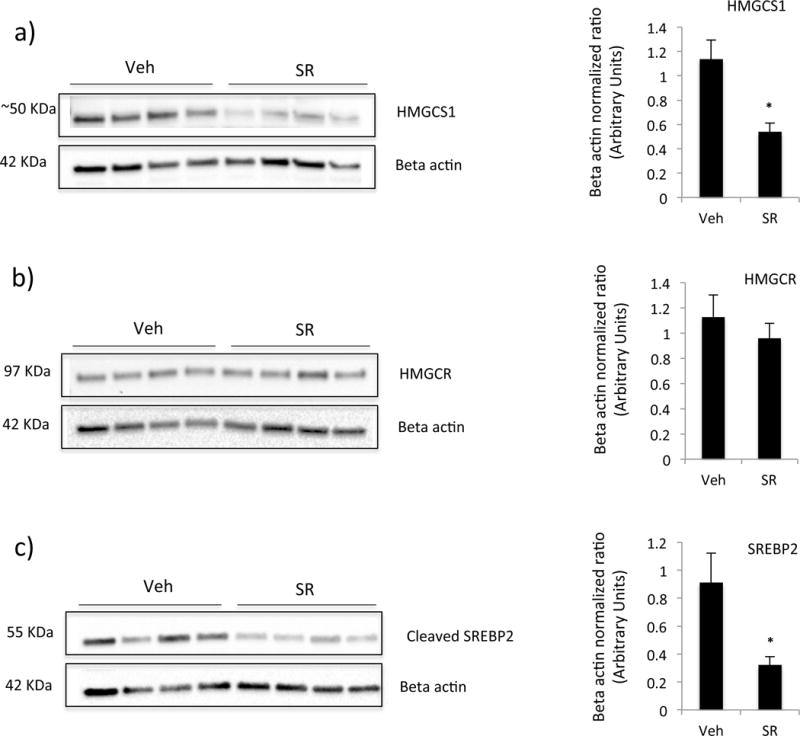
Western blot analysis of liver extract from LDLR−/− mice treated with SR9009 (100mg/kg, I.P. b.i.d.) or vehicle for two months. Data are presented as the mean ± SE. Student’s t-test was used to calculate statistical significance. n=4, * indicates a p-value < 0.05.
We also assessed the ability of pharmacological activation of Rev-erb to suppress cholesterologenesis in the HepG2 cell line. As shown in Fig. 4a, treatment of HepG2 cells with SR9009 led to a reduction in de novo cholesterol biosynthesis as detected by incorporation of radiolabeled acetate into cholesterol. Similar decreases were observed in the expression of cholesterologenic enzyme gene expression in the HepG2 cells in response to drug relative to the in vivo studies in both WT and LDLR null mice (Fig. 4b).
Figure 4.
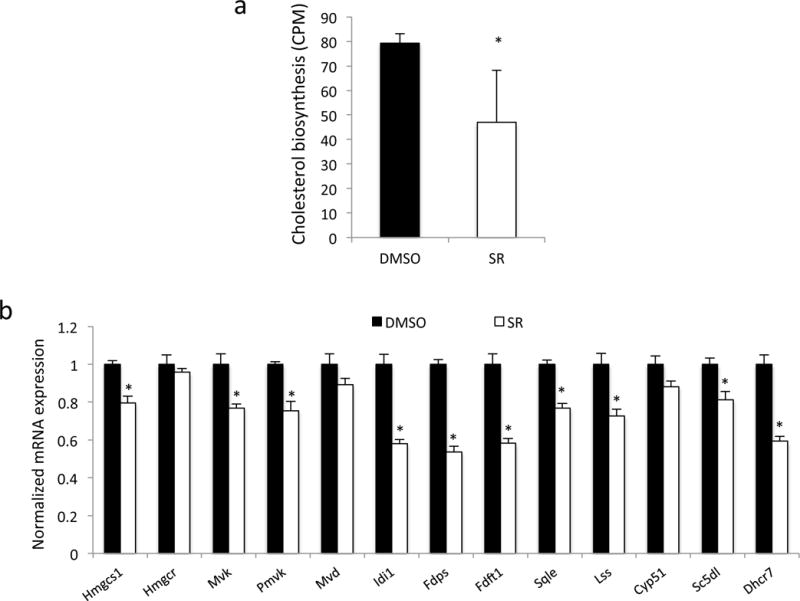
Analysis of cholesterol biosynthesis in HepG2 cells in response to pharmacological activation of Rev-erb. a) Results of a de novo cholesterol biogenesis assay in HepG2 cells treated with Rev-erb agonist or vehicle (DMSO) for 24 h prior to assessment of radiolabeled acetate into cholesterol by TLC. b) Level of expression of cholesterologenic enzyme genes in response to treatment of Rev-erb agonist or vehicle (DMSO) for 24h. n=3, * indicates a p-value < 0.05.
3.2 Rev-erb represses transcription of cholesterol biosynthesis genes
To test whether the reduced expression of cholesterol biosynthetic enzyme genes could be a consequence of direct regulation by the Rev-erbs, we analyzed previously reported global run-on sequencing (GRO-seq) datasets derived from Rev-erbα-deficient mice (KO) and wild-type (WT) mice at Zeitgeber time 10 (ZT10) [21]. Raw GRO-seq reads were normalized using TMM (trimmed mean of M-values) [27]. Fold changes in transcription of genes between Rev-erbα KO and WT mice were calculated by EdgeR [28]. Clock, a direct target gene of the Rev-erbs [29], was used as a positive control and displayed significant upregulation upon loss of Rev-erbα (Figure 5a; top right panel). GRO-seq analysis indicated that nearly all cholesterol biosynthetic enzyme genes (16 out of 17) were significantly up-regulated in Rev-erbα KO mice (p = 3.5e-07) (Figure 5a; left top panel). Using an independent analysis, we confirmed that the total up-regulated genes upon Rev-erbα loss were highly enriched with the cholesterol biosynthetic genes (Figure 5a; bottom panel). Rev-erb expression displays a circadian rhythm of expression (high at ZT10 and low at ZT22) and this oscillation influences the behavior of its target genes. A similar analysis of GRO-seq data was carried out to compare signals at ZT10 and ZT22 in WT mice. Recapitulating the results observed with the Rev-erbα KO mice, nascent transcription rates of cholesterol biosynthetic enzyme genes were increased at ZT22 (low Rev-erb and thus limited suppression) compared to ZT10 (high Rev-erb and significant suppression), respectively (p = 1.2e-05) (Figure 5b; top panel). Using an independent analysis, we confirmed that the total up-regulated genes when Rev-erb levels are low were highly enriched with the cholesterol biosynthesis genes (Figure 5b; bottom panel). Comparison of these data sets clearly demonstrates that when Rev-erb expression is low 16 of 17 of the genes in the cholesterologenic pathway are upregulated suggesting an important role for Rev-erb in suppressing this pathway. Interestingly, Cyp51 was the cholesterol biosynthetic gene that was the outlier in both the KO and circadian analysis, which suggests that a distinct mechanism of regulation than the other 16 cholesterologenic genes. Srebf2 expression is upregulated when Rev-erbα expression is low in both data sets, thus it is still unclear if Rev-erb may be also having direct effects on the expression of the other genes. This is examined by assessing ChIP seq data described below.
Figure 5.
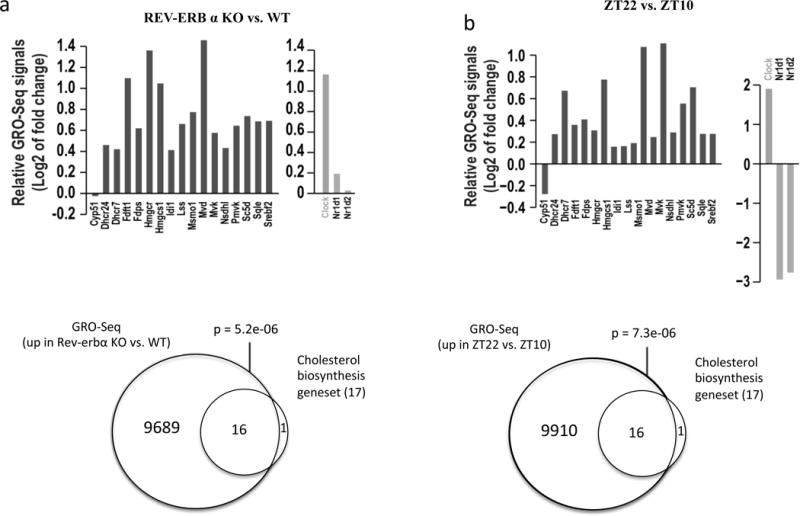
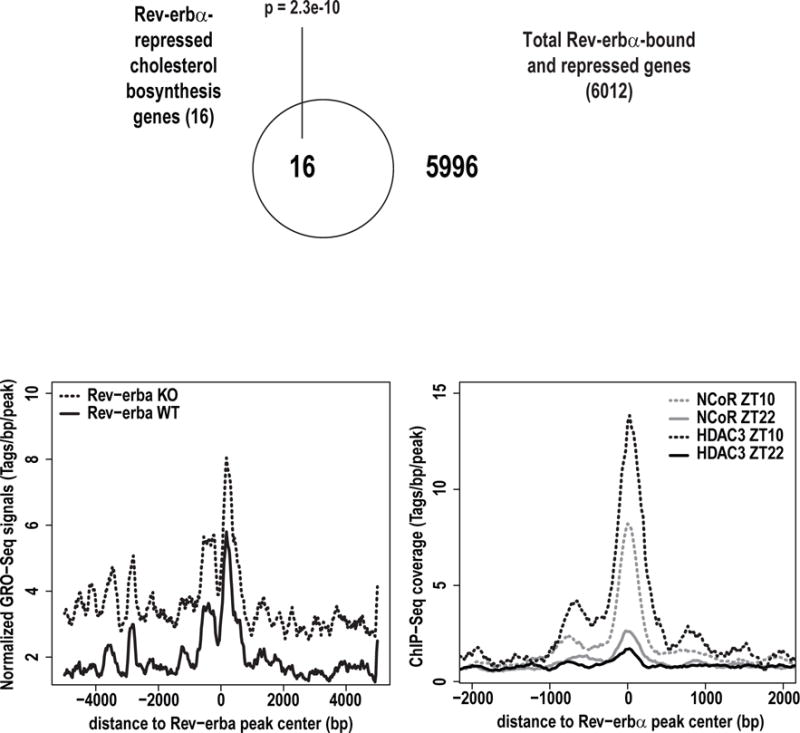
Rev-erbα represses transcription of cholesterol biosynthesis pathway genes. a) Comparison of global run-on sequencing (GRO-seq) signals from mouse liver between Rev-erbα deficient (KO) mice and wild-type (WT) mice. b) Comparison of GRO-seq signals from mouse liver between ZT10 and ZT22 time points in wild-type C57Bl/6J mice. c) Comparison of genes bound and repressed by Rev-erb based on ChIP-seq and GRO-seq results. d) Normalized GRO-seq signals of cholesterol biosynthesis pathway genes in Rev-erbα KO vs. WT mice at ZT22. e) Analysis of available HDAC3 and NCoR ChIP-Seq datasets derived from mouse liver at ZT10 and ZT22 time points.
3.3 Rev-erb represses transcription by recruiting HDAC3 and NCoR
To understand the mechanism by which Rev-erb downregulated cholesterol biosynthetic enzyme genes, we asked whether the Rev-erbs directly bound to these genes. Comparison of genes bound and repressed by Rev-erbα based on ChIP-seq [20] and GRO-seq results revealed 6012 functional and direct target genes of Rev-erb. These genes included the 16 above-characterized Rev-erb-downregulated cholesterol biosynthetic genes, revealing a highly significant enrichment (p=2.3e-10) (Figure 5c). Binding of Rev-erb to these genes inhibited nascent transcription at the binding sites as shown by increased GRO-Seq signals in the Rev-erb KO vs. WT mice (Figure5).
In order for Rev-erb to suppress the expression of its target genes it must recruit the NCoR/HDAC3 corepressor complex [14]. Thus, we analyzed HDAC3 and NCoR ChIP-Seq datasets [17] derived from mouse liver at ZT10 and ZT22 time points in order to assess the ability of Rev-erb to recruit this complex at the two time points (ZT10 high Rev-erb vs. ZT22 low Rev-erb). Fig. 5e illustrates the binding signals of HDAC3 and NCoR flanking the Rev-erb binding sites at the 16 cholesterol biosynthetic enzyme genes at the two times. HDAC3 and NCoR occupancy is highest at ZT10 when Rev-erb expression is peaking while HDAC3 and NCoR occupancy is lowest at ZT22 when Rev-erb expression is at its lowest (Figure 5e). These results are consistent with a model that Rev-erb represses the expression of these 16 cholesterologenic genes directly by facilitating recruitment of NCoR and HDAC3.
3.4 The Rev-erb DNA response element plays a major role in recruiting Rev-erb to cholesterol biosynthetic enzyme genes
A recent study suggests two distinct mechanisms by which Rev-erb is recruited to its target genes: a Rev-erb DNA binding domain (DBD) dependent mechanism where Rev-erb directly binds to its cognate DNA response element (RORE or RevDR2) and DBD independent mechanism by tethering to tissue-specific transcription factors [20]. The relative contribution of these mechanisms to Rev-erb recruitment to cholesterol biosynthetic enzyme genes, however, is not known. Our de novo motif discovery of Rev-erb-bound loci showed a significant enrichment of the AGGTCA consensus nuclear receptor motif whose sequence matches the cognate binding site of RORα, which is also recognized by the Rev-erbs (Figure 6). We also observed an enrichment of the consensus-binding motif of the liver-specific transcription factor HNF6, however the significance of the ROR binding motif (p~10−8) was much greater than the HNF6 motif (p=0.001) (Figure 6). A detailed analysis showed that the majority of the genes (12 out of 16) contained a nuclear receptor binding response element in the Rev-erb binding sites. Among these genes, Srebp2 and Hmgcr contained Rev-erb binding sites carrying both ROR and HNF6 motifs (Figure 6). These enzymes regulate key steps of the pathway and co-localization of RORE and liver-specific factor binding elements suggests a potential cooperative function of these distinct transcription factors in regulating important metabolic pathways. In fact, our observation of a significant overlap in Rev-erb and HNF6 binding sites in cholesterologenic enzyme genes is consistent with a recent study indicating that these two factors coordinately regulate lipid metabolism in the liver [30].
Figure 6.
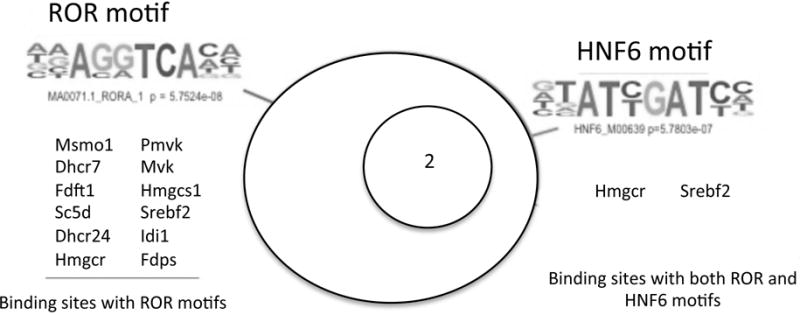
De novo motif discovery of Rev-erb-bound loci. a) Cholesterol biosynthesis genes were bound and repressed by Rev-erbα. The binding sites of Rev-erbα of these genes were enriched with consensus nuclear receptor binding motifs, such as the ROR binding site, and to a lesser extent, binding sites of pioneering tissue-specific factors, such as HNF6.
Finally, we performed global analysis of functional and direct target genes of Rev-erbα. Gene Ontology analysis indicated that these genes were enriched with genes involved in the regulation of cholesterol biosynthesis, and that perturbation of these genes was associated with cholesterol related abnormal mouse phenotype and human disease (Figure 7), consistent with important biological function of the Rev-erbs in regulating cholesterol genes in liver.
Figure 7.
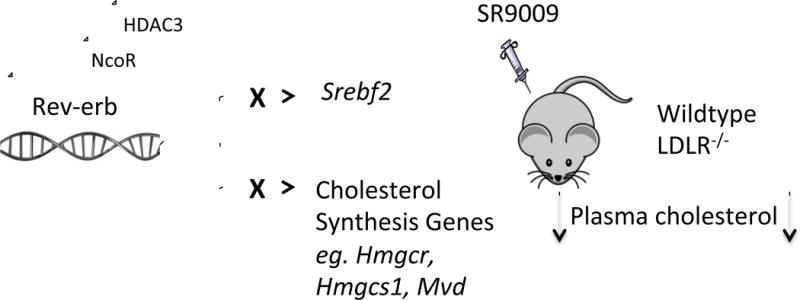
Global analysis of functional and direct target genes of Rev-erbα. Comparison of genes bound and repressed by Rev-erbα, based on ChIP-Seq and GRO-Seq results, revealed 6012 functional and direct target genes of Rev-erbα. Gene-Ontology analysis indicated that these genes were enriched with genes involved in the regulation of cholesterol biosynthesis, and that perturbation of these genes is associated with cholesterol related abnormal mouse phenotype and human disease.
4. Discussion
The Rev-erb nuclear receptors have been demonstrated to play a significant role in regulation of lipid metabolism [16]–[18]. Previously, we demonstrated that pharmacological activation of Rev-erb led to reduction in plasma cholesterol levels in mice [19] and others have demonstrated that mice with genetic deletion of Rev-erb display elevated plasma cholesterol levels [18]. The purpose of this study was to determine the role of Rev-erb in regulation of the entire cholesterol biosynthesis pathway and to identify the mechanism of Rev-erb action. Here, we demonstrate that Rev-erb coordinately regulates the vast majority of genes that encode important enzymes in cholesterol biosynthesis pathway. Pharmacological modulation of the receptor activity using a synthetic agonist resulted in significant reduction of expression of the majority of hepatic cholesterol biosynthetic enzyme genes confirming suppressive role of the Rev-erbs in cholesterol biosynthesis pathway. These findings suggest important role of the Rev-erbs in regulating the cholesterol synthesis pathway and potentially in maintaining cholesterol levels in the body. This was indeed observed in vivo where we saw large reduction in total cholesterol as well as LDL and triglycerides levels with synthetic Rev-erb agonist. This is particularly interesting because cholesterol-lowering effects were observed not only in wild type mice but also in a mouse that mimics human lipid profile (LDLR null mice) and is a model for dyslipidemia and metabolic disease such as atherosclerosis. These mice have highly elevated plasma lipids and the reduction of 36% and 41% in total cholesterol and LDL cholesterol were observed upon activation of the Rev-erb. A previous study reported an increase in plasma LDL and cholesterol in Rev-erb KO mice which further supports our finding that Rev-erbs negatively regulate cholesterol levels [18].
By analyzing data sets from previously reported GRO-seq and ChIP-seq studies we illustrate that binding of Rev-erbα to genes that encode cholesterol biosynthetic genes inhibits nascent transcription. This was particularly apparent in data from Rev-erbα null mice where loss of the receptor resulted in an increase in transcription of these genes. Although the vast majority of Rev-erb regulated genes within this pathway behaved in this manner, transcription of Cyp51 did not increase in the absence of Rev-erbα. This was unexpected given our results from gene expression studies showing a suppressive effect of Rev-erb activation. However, comparing our results from WT and LDLR−/− mice treated with SR9009 we observed expression of Cyp51 gene was not significantly reduced in wild type mice. It is possible that this discrepancy could be due to differences in mouse model and their metabolic states since LDLR−/− mice have altered cholesterol homeostasis and these mice were also under high cholesterol diet during the experiment period.
To carry out their transcriptional repressive function, Rev-erbs recruit the NCoR/HDAC3 co-repressor complex to Rev-erb response elements in promoters and enhancers of target genes [14]. Consistent with this existing model, we found strong reduction in occupancies of HDAC3 and NCoR in cholesterol biosynthesis genes in the absence of Rev-erbα suggesting recruitment of NCoR/HDAC3 co-repressor complexes by Rev-erb to repress cholesterol gene transcription. A previous study has shown the importance of circadian genomic recruitment of HDAC3 by Rev-erbα in liver lipid metabolism [16]. Co-localization of HDAC3 and Rev-erbα was found at 130 lipid related genes at ZT10 and deletion of HDAC3 or Rev-erbα in liver was shown to cause fatty liver and hepatic steatosis emphasizing the fact that HDAC3 recruitment by Rev-erb and histone deacetylation is important for normal hepatic lipid homeostasis [16], [17]. Consistent with their findings we show an effect of Rev-erb on liver lipid metabolism, but more specifically on genes important in de novo cholesterol biosynthesis.
Using de novo motif analysis we found that the Rev-erb DNA response element plays a major role in recruiting the receptor to cholesterol biosynthesis genes. While binding to response elements and recruitment of co-repressors is well-established mechanism for transcriptional repression of target genes by Rev-erb, a recent study suggests that Rev-erb regulates clock and metabolic genes by two distinct mechanisms [20]. This study revealed that the Rev-erbs can either bind to DNA directly or interact indirectly through other transcription factors allowing regulation of genes in a tissue-specific manner. Interestingly, our analysis revealed that two key genes Hmgcr and Srebf2 were enriched in both Rev-erb and liver-specific HNF6 binding sites. Co-localization of both Rev-erb and HNF6 response elements at these regulatory regions suggests potential cooperative function of these distinct transcription factors in modulating important steps of this metabolic pathway and provide a potential mechanism for distinctions in hepatic vs. non-hepatic cholesterol biosynthesis.
It is interesting to note that in addition to regulation of the expression of the genes encoding the majority of cholesterol synthetic enzymes, Rev-erb also regulates the expression of SREBP2, which itself is a master transcriptional regulator of these genes [31]. Our results show that Rev-erb leads to reduction in accumulation of the nuclear form of the SREBP2 protein, reduction in Srebf2 gene expression and downregulation of SREBP2 target genes as well as increase in nascent Srebf2 mRNA transcription in the absence of Rev-erbα. Under SR9009-treated conditions one would expect hepatic cholesterol levels to be reduced and that the cleaved form of SREBP2 may be elevated to compensate for the decreased cholesterol levels. However, the levels of the cleaved form of SREBP2 remained lower in SR9009-treated mice relative to vehicle-treated mice. The decrease in cleaved SREBP2 levels is consistent with our data demonstrating a broad reduction in the levels of expression of cholesterologenic enzyme gene expression under these conditions. Consistent with previous findings with SREBP2 inhibitors, downregulation of multiple enzymes in cholesterol biosynthesis pathway could be due to direct inhibition of SREBP-2 by Rev-erb [32]. This suggests that Rev-erb regulation of the genes encoding the biosynthetic enzymes is two fold: direct regulation and indirect regulation via modulation of SREBP2 expression. Since SREBP2 enhances its own transcription through SREs in its promoter in a feed-forward loop, transcriptional repression by Rev-erb could inhibit this process as well [33]. This is particularly interesting because many studies have demonstrated that SREBP2 inhibitors have beneficial effects in treating metabolic syndrome [32], [34], [35]. One study demonstrated, by direct comparison to a HMGCR inhibitor, that SREBP2 inhibitor compound was more effective in reducing lipid level, fatty acid synthesis, triglyceride synthesis and in improving insulin sensitivity in mice [32]. Given the additional unmet need to optimize and improve the existing treatment because of high residual risk, side effects and tolerance issues, these studies have shown that more effective ways of targeting lipids compared to statins are possible and need to be explored.
One class of lipid lowering agents that acts through cholesterol biosynthesis pathway is the HMGCR inhibitors, statins. By inhibiting HMGCR and lowering the cholesterol pool in the liver this class of drug upregulates LDL receptors through feedback mechanism resulting in clearance of LDL from the circulation. Our studies show that similar to statins, Rev-erb targets the cholesterol biosynthetic pathway. However our observed cholesterol-lowering effects using Rev-erb agonist were carried out in mice lacking LDL-receptors. This indicates that unlike statins, the cholesterol-lowering effects of Rev-erb agonists observed in our experiments are independent of LDL-receptor function. These findings suggest that other important cholesterol homeostasis pathways may also be affected by Rev-erb activation. Experiments where hematopoietic deficiency of Rev-erb was shown to accelerate atherogenesis was also carried out in LDL receptor deficient model [36]. The use of other models such as ApoE−/− mice may help distinguish whether additional cholesterol lowering effects could be obtained when LDL-receptor is present.
The regulation of rhythmic expression of cholesterol biosynthesis genes by Rev-erbs indicates that these nuclear receptors are critical links between circadian regulation and metabolism. We and others have previously shown Rev-erbs play an anti-inflammatory role and alter macrophage phenotype [36]–[38]. Our new results suggest that these anti-inflammatory effects combined with the ability to reduce cholesterol levels and biosynthesis could have vast implications in metabolic syndrome and cardiovascular disorders such as atherosclerosis. In conclusion, our findings reveal mechanistic insights into the complex interplay between circadian factors and peripheral cholesterol biosynthesis in maintaining cholesterol levels and provide evidence towards the possibility of pharmacological targeting of Rev-erbs in the treatment of metabolic disorders.
Acknowledgments
The authors would like to thank Chun Guo for technical assistance, Dr. Melissa Kazantzis for plasma cholesterol analysis and Dr. Cyrielle Billon for helpful discussions. This publication was supported by American Heart Association Predoctoral Fellowship (SS) and the National Institutes of Health (MH093429).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflicts of interest.
References
- 1.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 2.Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, et al. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med. 1994;330(2):107–13. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 3.Kritchevsky D. Cholesterol and atherosclerosis. Am J Clin Nutr. 1962;10(19):269–276. doi: 10.1093/ajcn/10.4.269. [DOI] [PubMed] [Google Scholar]
- 4.Gylling H, Hallikainen M, Kolehmainen M, Toppinen L, Pihlajamäki J, Mykkänen H, et al. Cholesterol synthesis prevails over absorption in metabolic syndrome. Transl Res. 2007;149(6):310–316. doi: 10.1016/j.trsl.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Di Paolo G, Kim T-W. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011 March;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med. 2004;10(12):1352–8. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
- 7.Murphy SA, Cannon CP, Wiviott SD, McCabe CH, Braunwald E. Reduction in Recurrent Cardiovascular Events With Intensive Lipid-Lowering Statin Therapy Compared With Moderate Lipid-Lowering Statin Therapy After Acute Coronary Syndromes: From the PROVE IT–TIMI 22 (Pravastatin or Atorvastatin Evaluation and Infection Therapy–Thrombolysis In Myocardial Infarction 22) Trial. J Am Coll Cardiol. Dec 15;54(25):2358–2362. doi: 10.1016/j.jacc.2009.10.005. AD. [DOI] [PubMed] [Google Scholar]
- 8.Ahn CH, Choi SH. New Drugs for Treating Dyslipidemia: Beyond Statins. Diabetes Metab J. 2015 Apr;39(2):87–94. doi: 10.4093/dmj.2015.39.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989;9(3):1128–36. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 12.Forman BM, Chen J, Blumberg B, Kliewer S, Henshaw R, Ong ES, et al. Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8(9):1253–61. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 13.Adelmant G, Bègue A, Stéhelin D, Laudet V. A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. Proc Natl Acad Sci U S A. 1996;93(8):3553–8. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin L, Lazar M. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 15.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(2011):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delezie J, Dumont S, Dardente H, Oudart H, Grechez-Cassiau A, Klosen P, et al. The nuclear receptor REV-ERB is required for the daily balance of carbohydrate and lipid metabolism. The FASEB Journal. 2012;26:3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 19.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, et al. Regulation of Circadian Behavior and Metabolism by Synthetic REV-ERB Agonists. Nature. 2012 Mar;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, et al. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348(6242):1488–92. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potteaux S, Ait-Oufella H, Mallat Z. Mouse models of atherosclerosis. Drug Discov Today Dis Model. 2007;4(4):165–170. [Google Scholar]
- 25.Ericsson J, Jackson SM, Lee BC, Edwards PA. Sterol regulatory element binding protein binds to a cis element in the promoter of the farnesyl diphosphate synthase gene. Proc Natl Acad Sci USA. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan GM, Jiang GJ, Koch RL, Shechter I. Molecular-Cloning and Functional-Analysis of the Promoter of the Human Squalene Synthase Gene. J Biol Chem. 1995;270:21958–21965. doi: 10.1074/jbc.270.37.21958. [DOI] [PubMed] [Google Scholar]
- 27.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crumbley C, Burris TP. Direct regulation of CLOCK expression by REV-ERB. PLoS One. 2011;6(3):e17290. doi: 10.1371/journal.pone.0017290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Fang B, Damle M, Guan D, Li Z, Kim Y, et al. HNF6 and Rev-erb α integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 2016;30:1636–1644. doi: 10.1101/gad.281972.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton JD. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans. 2002;30:1091–1095. doi: 10.1042/bst0301091. [DOI] [PubMed] [Google Scholar]
- 32.Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, et al. Inhibition of SREBP by a Small Molecule, Betulin, Improves Hyperlipidemia and Insulin Resistance and Reduces Atherosclerotic Plaques. Cell Metab. 2011 Jan;13(1):44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271(43):26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 34.Kamisuki S, Mao Q, Abu-Elheiga L, Gu Z, Kugimiya A, Kwon Y, et al. A Small Molecule That Blocks Fat Synthesis By Inhibiting the Activation of SREBP. Chem Biol. 2009 Aug;16(8):882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, Xiaoli, Zong H, Abdulla A, Yang EST, Wang Q, et al. Inhibition of SREBP Transcriptional Activity by a Boron-Containing Compound Improves Lipid Homeostasis in Diet-Induced Obesity. Diabetes. 2014 Jul;63(7):2464–2473. doi: 10.2337/db13-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma H, Zhong W, Jiang Y, Fontaine C, Li S, Fu J, et al. Increased atherosclerotic lesions in LDL receptor deficient mice with hematopoietic nuclear receptor Rev-erbα knock- down. J Am Heart Assoc. 2013;2:e000235. doi: 10.1161/JAHA.113.000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitaula S, Billon C, Kamenecka TM, Solt LA, Burris TP. Suppression of atherosclerosis by synthetic REV-ERB agonist. Biochem Biophys Res Commun. (0) doi: 10.1016/j.bbrc.2015.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]


