Abstract
The 20S proteasome is a self-compartmentalized protease which degrades unfolded polypeptides and has been purified from eucaryotes, gram-positive actinomycetes, and archaea. Energy-dependent complexes, such as the 19S cap of the eucaryal 26S proteasome, are assumed to be responsible for the recognition and/or unfolding of substrate proteins which are then translocated into the central chamber of the 20S proteasome and hydrolyzed to polypeptide products of 3 to 30 residues. All archaeal genomes which have been sequenced are predicted to encode proteins with up to ∼50% identity to the six ATPase subunits of the 19S cap. In this study, one of these archaeal homologs which has been named PAN for proteasome-activating nucleotidase was characterized from the hyperthermophile Methanococcus jannaschii. In addition, the M. jannaschii 20S proteasome was purified as a 700-kDa complex by in vitro assembly of the α and β subunits and has an unusually high rate of peptide and unfolded-polypeptide hydrolysis at 100°C. The 550-kDa PAN complex was required for CTP- or ATP-dependent degradation of β-casein by archaeal 20S proteasomes. A 500-kDa complex of PAN(Δ1–73), which has a deletion of residues 1 to 73 of the deduced protein and disrupts the predicted N-terminal coiled-coil, also facilitated this energy-dependent proteolysis. However, this deletion increased the types of nucleotides hydrolyzed to include not only ATP and CTP but also ITP, GTP, TTP, and UTP. The temperature optimum for nucleotide (ATP) hydrolysis was reduced from 80°C for the full-length protein to 65°C for PAN(Δ1–73). Both PAN protein complexes were stable in the absence of ATP and were inhibited by N-ethylmaleimide and p-chloromercuriphenyl-sulfonic acid. Kinetic analysis reveals that the PAN protein has a relatively high Vmax for ATP and CTP hydrolysis of 3.5 and 5.8 μmol of Pi per min per mg of protein as well as a relatively low affinity for CTP and ATP with Km values of 307 and 497 μM compared to other proteins of the AAA family. Based on electron micrographs, PAN and PAN(Δ1–73) apparently associate with the ends of the 20S proteasome cylinder. These results suggest that the M. jannaschii as well as related archaeal 20S proteasomes require a nucleotidase complex such as PAN to mediate the energy-dependent hydrolysis of folded-substrate proteins and that the N-terminal 73 amino acid residues of PAN are not absolutely required for this reaction.
Energy-dependent proteolysis is not only vital to elimination of defective cellular proteins but is also central to regulation of cell division, metabolism, transcription, and other essential cellular functions (12, 22). A small group of related energy-dependent proteases have been identified from several diverse organisms; these proteases include Lon, FtsH (HflB), ClpAP, ClpXP, HslUV (ClpYQ), and the 26S proteasome (4, 39). Although this group of proteins shares limited primary sequence identity, the proteases have converged to a universal structure in which relatively nonspecific proteolytic active sites are compartmentalized from the cell by narrow openings (37, 67). The current model is that the degradation of biological substrates by this group of proteases requires additional energy-dependent proteins or protein domains for the recognition and/or unfolding of substrate proteins (21, 29, 65). Some of these energy-dependent components may not only participate in proteolysis but also have independent roles as chaperones (60). Thus, the energy-dependent component of these proteases may provide a proofreading step following the initial binding of protein substrates and enable the cell to distinguish between proteins destined for refolding, disaggregation, or destruction (23).
Some energy-dependent proteases are organized in a symmetry mismatch, including ClpAP and ClpXP which are complexes of the hexameric ClpA and ClpX ATPases interfaced with the ClpP protease of two heptameric rings (5). It is postulated that hydrolysis of nucleotides by the energy-dependent component results in rotation at the ATPase-protease interface (5) and that this rotation facilitates the processive degradation of substrate proteins (63). This mismatch, however, does not appear to be universal, since the fully functional Lon and FtsH proteases assemble into homomultimeric complexes with the ATPase and proteolytic components encoded by a single gene (10, 47).
Energy-dependent proteases, including Lon and proteasome complexes, are predicted from the genome sequences of archaea (8, 32, 34, 57). Little is known, however, about the biochemistry or biological significance of these energy-dependent proteolytic pathways in this unusual group of organisms. 20S proteasomes have been purified from diverse archaea (3, 13, 42, 74) and appear to have a self-compartmentalized structure which requires polypeptides to be at least partially unfolded prior to hydrolysis (72). This suggests that the archaeal 20S component alone, as purified, has little biological significance in native protein hydrolysis and suggests that additional components are necessary for protein degradation at the optimal growth temperature of these organisms. The related eucaryal 20S proteasome is fully functional in degrading proteins but only when associated with an energy-dependent 19S cap regulatory complex or an eight-subunit derivative of this complex, designated the base domain (21). Six of these base domain subunits are ATPases which are members of the AAA family (named AAA for ATPases associated with a variety of cellular activities) (46) and have recently been named Rpt proteins (for regulatory particle triple-A) (16). It is postulated that these energy-dependent subunits are responsible for the unfolding and/or translocation of substrate proteins into the central proteolytic chamber of the 20S proteasome.
Closely related homologs of the eucaryal ATPase proteasome subunits are predicted from the genome sequences of the archaea (8, 32, 34, 57). One homolog from Methanococcus jannaschii (MJ1176) has recently been purified from recombinant Escherichia coli and named PAN for proteasome-activating nucleotidase (84). This protein was purified as a 650-kDa complex of an N-terminal polyhistidine-tagged form of PAN in association with a derivative of PAN which had a deletion of residues 1 to 73 [PAN(Δ1–73)]. The purified PAN complex had ATPase activity and activated the energy-dependent degradation of proteins by 20S proteasomes from Methanosarcina thermophila and Thermoplasma acidophilum by four- to ninefold (84). Characteristic of AAA proteins, the deduced sequence of PAN reveals a highly conserved Walker A motif (211-GPPGTGKT-218) which is predicted to be involved in coordination of Mg2+ and formation of hydrogen bonds with nucleotide triphosphates including the β- and γ-phosphates (54, 66). A modified Walker B motif or “DEAD” box which is also predicted to be involved in Mg2+ binding and ATP hydrolysis is conserved and includes residues 270-DEID-273 of PAN (9). Additionally, a SRH or second region of homology motif [(T/S)-(N/S)-X5-D-X-A-X2-R-X2-R-X-(D/E)] which distinguishes AAA proteins from the broader family of Walker-type ATPases is also conserved and spans residues 315 to 333 of PAN. The SRH motif appears to be important in ATP hydrolysis but not ATP binding (31). The PAN sequence also has a highly charged N-terminal coiled-coil spanning residues 49 to 83 which is conserved in other AAA proteins (84). This coiled-coil may play an important role in regulating nucleotide hydrolysis, protein-protein interaction, and/or other activities as proposed for other AAA proteins including the ATPase subunits of the 26S proteasome as well as ClpA and ClpB (38, 51, 56, 68).
In this communication, the biochemical and physical properties of the 20S proteasome and PAN proteins of M. jannaschii are presented. Our results demonstrate that this 20S proteasome requires a nucleotidase complex such as PAN to mediate the energy-dependent hydrolysis of folded-substrate proteins. The N-terminal coiled-coil region of PAN is not required for this reaction and is not needed for association of the PAN protein into an ∼12-subunit complex. However, deletion of the N-terminal 73 residues does appear to influence the biochemical properties of nucleotide hydrolysis mediated by the PAN protein.
MATERIALS AND METHODS
Materials.
Biochemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Other organic and inorganic chemicals were from Fisher Scientific (Atlanta, Ga.) and were analytical grade. Restriction endonucleases and DNA-modifying enzymes were from New England BioLabs (Beverly, Mass.) or Promega (Madison, Wis.). Oligonucleotides were from Genemed Synthesis (San Francisco, Calif.). Polyvinylidene difluoride membranes were from MicroSeparations (Westborough, Mass.). The M. thermophila and T. acidophilum 20S proteasomes were purified as previously described (41, 55).
Strains and media.
Escherichia coli strains included TB-1 F− ara Δ(lac-proAB) rpsL (Strr) [φ80dlacΔ(lacZ)M15] thi hsdR(rK− mK+) (New England BioLabs) and BL21 (DE3) F− ompT [lon] hsdSB(rB− mB−) (an E. coli B strain) with DE3, a λ prophage carrying the T7 RNA polymerase gene (59). Strains were grown in Luria-Bertani (LB) medium or LB medium supplemented with 50 mg of spectinomycin per liter, 100 mg of ampicillin per liter, and/or 50 mg of kanamycin per liter as needed. M. jannaschii strain JAL-1 was grown to mid-log phase with H2 and CO2 by the method of Boone et al. (6).
Enzyme assays.
Protein concentrations were determined by the bicinchoninic acid method (58) (Pierce, Rockford, Ill.) for the PAN proteins and by the Coomassie blue dye-binding method (7) (Bio-Rad, Hercules, Calif.) for the 20S proteasome, using bovine serum albumin as the standard. Peptide-hydrolyzing activity was assayed by the release of 7-amino-4-methylcoumarin (fluorescence) or β-naphthylamine (absorbance) as previously described (41, 42, 74). Protein hydrolysis was monitored as the generation of α-amino groups using fluorescamine as previously described (74) with methionine as a standard. The release of inorganic phosphate (Pi) was measured using malachite green with modifications as previously described (35, 36). For analysis of various nucleotide-hydrolyzing activities, enzyme (2 μg per ml) was incubated with 1 mM nucleotide in 25 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] at pH 8.0 with 100 mM NaCl, 10 mM MgCl2 (TES-NaCl-MgCl2 buffer) at 80°C for PAN or 65°C for PAN(Δ1–73). For determination of the kinetic parameters, enzyme (4 μg per ml) was incubated with ATP or CTP at 0.05, 0.1, 0.25, 0.5, and 1 mM using similar assay conditions. Initial velocity was determined by measuring the release of Pi in triplicate using four time points from 0 to 6 min. Initial velocity was plotted as a function of substrate concentration by the Lineweaver-Burk method (11). Similar results were obtained using Michaelis-Menten and Eadie-Hofstee plots (11). The effects of nucleotide diphosphates were measured by preincubating the enzyme (2 μg per ml) with or without 1 mM CDP or ADP for 15 min in TES-NaCl-MgCl2 buffer at 21°C before addition of 1 mM ATP or CTP and then assayed as described above. The pH optimum of enzymes was measured using the following 25 mM buffers: 2-(N-morpholino)ethanesulfonic acid (MES) (pH 5 to 7), TES (pH 7 to 8), N-tris(hydroxymethyl)methylglycine (Tricine) (pH 8 to 8.8), and 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (CAPSO) (pH 8.9 to 10).
Protein techniques.
Molecular masses of purified protein subunits were estimated by reducing and denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% polyacrylamide gels which were then stained with Coomassie blue R-250. The molecular mass standards were phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa) (Bio-Rad). N-terminal sequencing by Edman degradation was as previously described using purified proteins separated by SDS-PAGE (74).
Native molecular masses of the complexes were determined by applying purified protein to a Superose 6 HR 10/30 column calibrated with the molecular mass standards serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa) (Sigma). Nondenaturing native gel electrophoresis was performed using a 5% resolving and 4% stacking gel of 375 mM Tris-HCl at pH 8.8 (Bio-Rad) with a running buffer containing 25 mM Tris, 192 mM glycine, 10 mM MgCl2, and 1 mM ATP at pH 8.3. ATPase activity was visualized in the gel as previously described (49) with the following modifications. After separation of the proteins by electrophoresis, the gels were incubated with or without 10 mM N-ethylmaleimide (NEM) at 55°C in 50 mM Tris buffer at pH 8.0 containing 1 mM ATP, 10 mM MgCl2, 100 mM NaCl, and 0.1 mM lead nitrate.
Cloning and site-directed mutagenesis of the M. jannaschii proteasome genes.
PCRs were performed using a Gene Cycler (Bio-Rad) to amplify DNA fragments containing M. jannaschii genes encoding the α and β subunits of the 20S proteasome and PAN protein. Oligonucleotide primers, template plasmid DNA, and cloning procedures are shown in Table 1. The DNA sequences of the fragments generated by PCR amplification were verified by sequence analysis of both strands of plasmid DNA (DNA Sequencing Facility, Department of Microbiology and Cell Science, University of Florida, Gainesville).
TABLE 1.
Plasmids used in this study
| Plasmid | Phenotype or genotype; oligonucleotides for PCR amplification or site-directed mutagenesis | Source or reference |
|---|---|---|
| pET24b | Kmr; expression vector | Novagen |
| pET15b | Apr; expression vector | Novagen |
| pSJS1240 | Spr; E. coli ileX and argU | 33 |
| pAMJL73 | Apr; 2.385 kb of M. jannaschii genomic DNA bases 524549 to 526933 of L77117 in pUC18; carries complete ORF MJ0591 (α subunit of 20S proteasome) | ATCCa |
| pAMJKH57 | Apr; 2.036 kb of M. jannaschii genomic DNA bases 1182020 to 1184055 of L77117 in pUC18; carries complete ORF MJ1237 (β subunit of 20S proteasome) | ATCC |
| pAMJHW03 | Apr; 2.157 kb of M. jannaschii genomic DNA bases 1114515 to 1116671 of L77117 in pUC18; carries complete ORF MJ1176 (proteasome-activating nucleotidase) | ATCC |
| pMjα | Kmr; 795-bp NdeI-to-EcoRI fragment generated by PCR amplification using pAMJL73; carries complete MJ0591 ligated into pET24b; produces the α subunit of the 20S proteasome; 5′-ctgcccatatgcaaatggtacctcc-3′ and 5′-gcgccgaattcttattcttcttgtt-3′ (NdeI and EcoRI sites indicated) | This study |
| pMjβ(Δpro) | Kmr; 669-bp NdeI-to-EcoRI fragment generated by PCR amplification using pAMJKH57; carries MJ1237 truncated at the 5′ end by 15 bp ligated into pET24b; produces the mature β subunit with an N terminus corresponding to Thr7 of the protein deduced from MJ1237; 5′-ctgcccatatgacaacaaccgttgg-3′ and 5′-gccgagaattcttatttgctctttc-3′ (NdeI and EcoRI sites indicated) | This study |
| pMjPAN1 | Apr; 1.6-kb NdeI-to-EcoRV fragment of M. jannaschii genomic DNA ligated into the HincII site of pUC19; carries 84 bp upstream and 191 bp downstream of the complete MJ1176 | This study |
| pMjPAN2 | Kmr; 1.6-kb EcoRI-to-HindIII fragment of pMjPAN1 ligated into pET24b; produces PAN and PAN(Δ1–73) | This study |
| pMjPAN3 | Apr; 1,312-bp fragment generated by PCR amplification using pAMJHW03; carries MJ1176 ligated into pUC19 using HincII; 5′-ccgcccatatggtttttgaagaatt-3′ and 5′-gcggcgaattcttatctgtagagaa-3′ (NdeI and EcoRI sites indicated) | This study |
| pMjPAN5 | Apr; 1,302-bp NdeI-to-EcoRI fragment of pMjPAN3 ligated into pET15b; produces full-length PAN with His6 at the N terminus and PAN(Δ1–73) | This study |
| pMjPAN6 | Apr; site-directed mutation of pMjPAN3 which disrupts the internal Shine-Dalgarno site; 5′-gagcttgaacgtgaaaatttacagttaatg-3′ (altered bases indicated) | This study |
| pMjPAN7 | Kmr; 1,302-bp NdeI-to-EcoRI fragment of pMjPAN6; carries the complete MJ1176 with the disrupted internal Shine-Dalgarno site; produces PAN of 47 kDa | This study |
| pMjPAN8 | Kmr; 1,083-bp NdeI-to-EcoRI fragment generated by PCR amplification using pAMJHW03; carries MJ1176 with a 219-bp 5′ deletion ligated into pET24b; produces PAN(Δ1–73); 5′-tccatatgaaagaaaatgagattttaa-3′ and 5′-gcggcgaaattcttatctgtagagaa-3′ (NdeI and EcoRI sites indicated) | This study |
ATCC, American Type Culture Collection.
Oligonucleotide-directed mutagenesis was performed using the Morph site-specific plasmid DNA mutagenesis kit as recommended by the supplier (5 Prime→3 Prime, Boulder, Colo.). Double-stranded plasmid pMjPAN1 was used as a template for annealing of the oligonucleotide 5′-GAGCTTGAAcGtGAaAATTTACAGTTAATG-3′, where the bold bases are the conservative codon replacements used to alter an internal ribosomal binding site of MJ1176. Mutations were confirmed by DNA sequence analysis.
Expression of M. jannaschii genes in E. coli.
The M. jannaschii genes encoding the subunits of the 20S proteasome and PAN protein were expressed separately in E. coli BL21(DE3) using the bacteriophage T7 RNA polymerase-promoter system and plasmids listed in Table 1, as previously described (41, 59) (Novagen, Madison, Wis.). An E. coli host strain containing the ileX and argU genes, encoding tRNAAUA and tRNAAGA/AGG, on plasmid pSJS1240 (33) was used to maximize the overall yield of M. jannaschii protein.
Purification of recombinant 20S proteasome and α and β(Δpro) proteins.
E. coli(pMjα or pMjβΔpro) cells were thawed in 6 volumes (wt/vol) of 10 mM sodium phosphate buffer at pH 7.2 containing 1 mM dithiothreitol (DTT) and passed through a French pressure cell at 20,000 lb/in2. This was followed by centrifugation at 16,000 × g for 30 min at 4°C. For purification of the 20S proteasome, cell lysates containing the α and β(Δpro) proteins were mixed at a 1:1 stoichiometry to a final concentration of ∼10 mg of protein per ml, heated to 85°C for 15 min, and then chilled to 0°C for 30 min. The heated sample was centrifuged at 15,000 × g for 30 min at 4°C, which removed the majority of E. coli protein contaminants. The supernatant at 2.4 mg of protein per ml was concentrated by dialysis against PEG 8000 in bovine serum albumin (BSA)-treated dialysis tubing (cutoff, 3.5 kDa) (Pierce) to ∼13 mg of protein per ml. The sample (50 mg) was added to 10 ml hydroxyapatite (Bio-Rad) equilibrated with 10 mM sodium phosphate buffer at pH 7.2 containing 5 mM DTT. Contaminating proteins were removed with 250 mM sodium phosphate, and samples which hydrolyzed N-succinyl–Leu–Leu–Val–Tyr–7-amino-4-methylcoumarin (Suc-LLVY-Amc) were obtained after elution with 500 to 750 mM sodium phosphate buffer at pH 7.2 containing 5 mM DTT. These fractions were dialyzed against 20 mM sodium phosphate buffer at pH 7.2 with 5 mM DTT for 18 h at 4°C and centrifuged at 15,000 × g for 30 min at 4°C. The protein was concentrated as described above to a final concentration of 6 to 10 mg of protein per ml. The proteasome was further purified after gel filtration in a Superose 6 HR 10/30 column (2.5 by 28.5 cm) (Pharmacia) equilibrated with 20 mM Tris buffer at pH 7.2 with 150 mM NaCl, 5 mM DTT, and 10% glycerol. The 700-kDa fractions hydrolyzing Suc-LLVY-Amc were collected and determined to be pure by reducing SDS-PAGE as well as by transmission electron microscopy to ensure that E. coli membrane vesicles were not contaminating the 20S proteasome sample. For purification of the α and β(Δpro) proteins, cell lysate containing the individual proteins was heated, centrifuged, concentrated, and applied to a Superose 6 column as described above.
Purification of recombinant PAN proteins.
For purification of N-terminal six-histidine-tagged PAN (His6-PAN), E. coli(pMjPAN5) cells were thawed in 6 volumes (wt/vol) of 20 mM Tris buffer at pH 7.9 containing 5 mM imidazole and passed through a French pressure cell at 20,000 lb/in2. This was followed by centrifugation at 16,000 × g for 30 min at 4°C. Sample was applied to a Ni2+-Sepharose column (4.8 by 0.8 cm) (Pharmacia) equilibrated with 20 mM Tris buffer at pH 7.9 containing 5 mM imidazole and then washed with 10 ml of 20 mM Tris buffer at pH 7.9 containing 60 mM imidazole. The His6-PAN protein was eluted with 10 ml of 20 mM Tris buffer at pH 7.9 containing 500 mM imidazole, pooled, and dialyzed against 25 mM Tris buffer at pH 7.5. Fractions with ATP-hydrolyzing activity at 65°C were pooled and determined to be pure by reducing SDS-PAGE.
Purification of PAN proteins without the His6 tag was monitored by ATP-hydrolyzing activity at 65°C. E. coli(pMjPAN2, pMJPAN7, or pMJPAN8) cells were thawed in 6 volumes (wt/vol) of 20 mM Tris buffer at pH 8.0 containing 1 mM DTT (hereafter referred to as Tris pH 8.0 buffer) and passed through a French pressure cell at 20,000 lb/in2. This was followed by centrifugation at 16,000 × g for 30 min at 4°C. The supernatant was heated to 85°C for 15 min, chilled to 0°C for 15 min, and centrifuged at 10,000 × g for 30 min at 4°C. Samples were applied to a HiLoad Q-Sepharose 26/10 column (2.5 by 28.5 cm) (Pharmacia) equilibrated in Tris pH 8.0 buffer and were eluted with a linear NaCl gradient (0 to 500 mM NaCl in 240 ml of Tris pH 8.0 buffer) at 375 mM NaCl. Fractions were pooled, concentrated by dialysis against PEG 8000, and applied to a Superose 6 HR 10/30 column equilibrated with Tris pH 8.0 buffer with 150 mM NaCl and 10% glycerol. Fractions were pooled and applied to a QHyperD (Bio-Rad) or MonoQ HR 5/5 column (Pharmacia) equilibrated with Tris pH 8.0 buffer with 100 mM NaCl. The proteins were eluted with a linear NaCl gradient (100 to 500 mM NaCl in 10 ml of Tris pH 8.0 buffer) at 370 mM NaCl. Full-length PAN protein was further purified by application to a hydroxyapatite column (Bio-Rad) equilibrated with 10 mM NaPO4 buffer at pH 7.5 containing 1 mM DTT and 10% glycerol (hereafter referred to as NaPO4 pH 7.5 buffer) which was then developed with a linear gradient (10 to 250 mM NaPO4 pH 7.5 buffer in 30 ml). The PAN protein, which eluted at 225 mM NaPO4 pH 7.5 buffer, was dialyzed against 50 mM Tris buffer at pH 7.5 containing 100 mM NaCl, 1 mM DTT, and 10% glycerol at 4°C for 18 h. The protein was then applied to a Sephadex G-25 (Sigma) column equilibrated with 50 mM Tris buffer at pH 7.5 containing 100 mM NaCl, 1 mM DTT, and 10% glycerol to remove all detectable Pi. The PAN and PAN(Δ1–73) proteins were determined to be pure by reducing SDS-PAGE and stored under liquid N2 without significant loss of activity.
Production of antibodies against PAN(Δ1–73) and 20S proteasome proteins.
The purified recombinant PAN(Δ1–73) protein of M. jannaschii and the α and β proteins of the M. thermophila 20S proteasome (41) were applied to a reducing SDS–12% polyacrylamide gel. Gel fragments containing the individual proteins (300 μg) were used to generate polyclonal antibodies in rabbits with Freund's adjuvant according to the supplier (Cocalico Biologicals, Reamstown, Pa.).
Purification and analysis of PAN and 20S proteasome proteins from M. jannaschii.
M. jannaschii cultures were chilled to 0°C, cells were harvested aerobically by centrifugation at 5,000 × g for 15 min at 4°C, and cell material (0.5 g per liter) was stored at −70°C. Cells were thawed in 17 volumes (wt/vol) of 50 mM Tris buffer at pH 8.0 with 150 mM NaCl, 10 mM MgCl2, 1 mM ATP, 10 mM β-mercaptoethanol, 10% glycerol, and 0.1% Triton X-100. The cell suspension was passed through a French pressure cell twice at 10,000 lb/in2 and then centrifuged at 15,000 × g for 30 min at 4°C. Cell lysate was applied to a Superose 6 HR 10/30 column equilibrated with Tris pH 8.0 buffer with 150 mM NaCl and 10% glycerol followed by ion-exchange chromatography in a MonoQ HR 5/5 column equilibrated with Tris pH 8.0 buffer with 100 mM NaCl and 10% glycerol which was then developed with a linear NaCl gradient (0.1 to 1 M NaCl in 15 ml of Tris pH 8.0 buffer). In some experiments, ion-exchange chromatography preceded gel filtration. Protein fractions were analyzed for NEM-inhibitable hydrolysis of ATP as described above and analyzed by immunoblotting after denaturing and reducing SDS–12% PAGE (27). PAN complex was also analyzed by immunoblotting of proteins separated by nondenaturing gel electrophoresis and capillary transferred to polyvinylidene difluoride membranes using 10 mM 2-(N-morpholino)ethanesulfonic acid buffer at pH 6.0 with 20% methanol. Primary antibodies as described above and goat anti-rabbit immunoglobulin G (IgG) linked to alkaline phosphatase were used for detection (Southern Biotechnology Associates, Birmingham, Ala.).
Electron microscopy of 20S proteasome and PAN proteins.
Recombinant M. jannaschii PAN or PAN(Δ1–73) complexes and the 20S proteasome were incubated separately and in equimolar ratios at 21 or 50°C for 15 min in TES-NaCl-MgCl2 buffer with 1 mM ATP. Proteins were placed on 200-mesh grids coated with Formvar or carbon film, floated on water to remove salts, fixed with 2% cacodylate-buffered glutaraldehyde, briefly stained with 1% aqueous uranyl acetate, and treated with 0.01% bacitracin to improve spreading as previously described (74). Samples were viewed and photographed on a Zeiss EM-10CA transmission electron microscope operated at 80 kV. Stereo pairs were taken at an original magnification of ×100,000 using the Zeiss goniometer stage tilted at ±10°.
RESULTS AND DISCUSSION
Translation limits heterologous synthesis of M. jannaschii 20S proteasome and proteasome-activating nucleotidase (PAN) proteins.
Three open reading frames (ORFs) which code for putative proteins involved in energy-dependent protein degradation were identified from the genome sequence of the methanoarchaeon M. jannaschii (8). These ORFs include MJ0591 and MJ1237, which are predicted to encode the α and β subunits of a 20S proteasome as well as MJ1176 which has similarity to the ATPase (Rpt) subunits of the eucaryal 26S proteasome (8, 84). To further investigate archaeal proteasome-mediated protein degradation, these ORFs were separately cloned and expressed in E. coli. When an E. coli host strain with wild-type levels of tRNA was used, the yield of recombinant M. jannaschii protein was less than 0.5% of the total cell protein as determined by SDS-PAGE and immunoblotting. Analysis of the codon composition of the three ORFs revealed that up to 12% of the codons required either tRNAAUA or tRNAAGA/AGG for translation, which are rare in E. coli. Therefore, a compatible plasmid (pSJS1240) (Table 1) with the E. coli ileX and argU genes encoding these rare tRNAs was introduced into the host (33). In the presence of plasmid pSJS1240, the overall yield of M. jannaschii protein was increased to as much as 10% of total soluble protein, confirming that the rare tRNAs limit the high-level production of these proteins in E. coli.
Molecular masses of recombinant 20S proteasome subunits are identical to those produced in vivo.
The purified α and β(Δpro) proteins of the M. jannaschii 20S proteasome, synthesized in recombinant E. coli, migrated as 34- and 29-kDa proteins as determined by reducing SDS-PAGE (Fig. 1A). The β(Δpro) protein was produced from a derivative of MJ1237 which no longer codes for the putative β propeptide which spans amino acid residues 2 to 6. Whether the N-formyl-Met of the M. jannaschii β(Δpro) is removed by the aminopeptidase of E. coli to expose a putative active-site Thr, residue 7 of the full-length β protein, remains to be determined. The molecular masses calculated from the ORFs encoding the α and β(Δpro) proteins are each 5 kDa less than those estimated by SDS-PAGE. We are unaware of any covalent modifications occurring in E. coli which would increase the molecular mass of proteins by 5 kDa. Therefore, it is more likely that the 20S proteasome proteins from this hyperthermophile are not fully unfolded after boiling for 15 min in the presence of 0.6 M β-mercaptoethanol and 2% SDS, and the partially folded structure is retarding the migration of the α and β(Δpro) proteins during separation compared to that of the proteins used to estimate their molecular mass. Immunoblot analysis of M. jannaschii cell lysate separated by SDS-PAGE revealed that both subunits of the 20S proteasome are produced in this archaeon and are the same molecular mass as those purified from recombinant E. coli. Therefore, MJ0591 and MJ1237 have been designated the psmA and psmB genes (for proteasome A and B), which is consistent with the nomenclature of the related genes from M. thermophila (42).
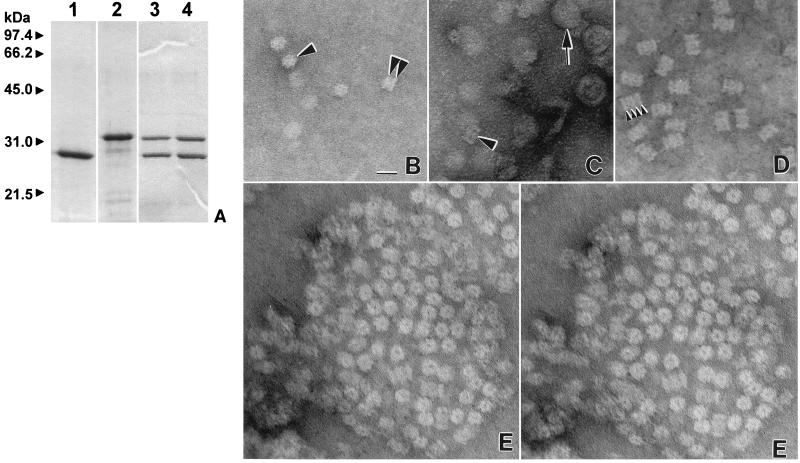
FIG. 1.
M. jannaschii 20S proteasome proteins. (A) Proteins (2 μg per lane) purified from recombinant E. coli, separated by reducing SDS–12% PAGE, and stained with Coomassie blue. Lanes: 1, β(Δpro) protein; 2, α protein; 3, 20S proteasome of 700 kDa; 4, proteasome fraction of ∼1.5 MDa. The positions of molecular mass standards are indicated to the left of the gel. (B to E) Transmission electron micrographs of the proteasome proteins. (B) End-on rings (single arrowhead) and sidewise paired rings (double arrowheads) of the α protein. (C) Proteasome fraction of ∼1.5 MDa composed of equimolar ratios of α and β(Δpro) proteins (arrowhead) which purify with membrane vesicles (arrow). (D) Side views of views of 20S proteasomes of 700 kDa with arrowheads indicating the four stacked rings of the cylindrical particle. (E) Stereo pair depicting a multilevel symmetrical array of the 20S proteasomes of 700 kDa after incubation at 65°C in Tris buffer at pH 7.2 with 150 mM NaCl, 1 mM ATP, and 10 mM MgCl2. Protein arrays were not observed under similar conditions in the absence of 150 mM NaCl or heating to 65°C. Bar, 20 nm.
Assembly of the α and β(Δpro) proteins of the 20S proteasome.
The individual α and β(Δpro) proteins of the M. jannaschii 20S proteasome did not hydrolyze the peptide substrate Suc-LLVY-Amc (Table 2). The β(Δpro) protein formed low-molecular-mass complexes of monomers to trimers, as estimated by nondenaturing electrophoresis and gel filtration, which were not visible by electron microscopy (data not shown). This is analogous to the related β subunits of the 20S proteasomes of M. thermophila, T. acidophilum, and the eubacterium Rhodococcus erythropolis which do not self-assemble into distinct complexes and remain inactive even in the absence of the propeptide (41, 55, 81, 83). The M. jannaschii α protein, in contrast, appeared to form cylinders of double stacked protein discs of 10-nm diameter which did not have visible central channels (Fig. 1B). This spontaneous self-assembly has been observed for other archaeal and eucaryal α-type proteins, including the α proteins of M. thermophila and T. acidophilum as well as the human α7 (HsC8) and Trypanosoma brucei α5 proteins which spontaneously form single, double, and/or four-stacked protein rings when produced in E. coli (19, 20, 41, 78, 83). Self-assembly of the α1 and α2 proteins of the eubacterium R. erythropolis, however, depends on the presence of β-type proteins and suggests distinct differences in 20S proteasome assembly among these organisms (81).
TABLE 2.
Chymotrypsin-like activities of the purified M. jannaschii proteasomes and in vitro assembly of the α and β(Δpro) proteins
| Protein or proteasome | Preincubation temp (°C)a | Activityb |
|---|---|---|
| α | 0, 37, 65, 85 | UDc |
| β(Δpro) | 0, 37, 65, 85 | UD |
| α-β(Δpro)d | 0 | UD |
| α-β(Δpro) | 37 | 31.3 ± 1.55 |
| α-β(Δpro) | 65 | 48.2 ± 0.74 |
| α-β(Δpro) | 85 | 47.7 ± 3.01 |
| 700-kDa proteasomes | 0 | 106 ± 2.2 |
| ∼1.5-MDa proteasomes | 0 | 137 ± 8.6 |
Proteins (0.75 mg of protein per ml of Tris buffer) were preincubated at the indicated temperatures for 15 min.
Activity measured in micromoles per minute per milligram of protein. Hydrolysis of Suc-LLVY-Amc was measured at 37°C using 20 μM substrate and 0.06 mg of purified protein in 1 ml of Tris buffer with 6% (vol/vol) dimethyl sulfoxide.
UD, undetectable.
α-β(Δpro), equimolar ratios of the separately purified α and β(Δpro) proteins mixed prior to preincubation.
After mixing equimolar ratios of the concentrated M. jannaschii α and β(Δpro) proteins and preincubating at 37 to 85°C, the proteasome proteins hydrolyzed the peptide substrate Suc-LLVY-Amc at 37°C (Table 2) (Materials and Methods). Specific activity was significantly reduced when the proteins were mixed at a 12-fold or greater dilution prior to preincubation. Furthermore, a 34% reduction in activity was observed when the temperature of preincubation was reduced from 85 to 37°C, and activity was undetectable when incubated at 0°C prior to assay (Table 2). These results suggest that the α and β(Δpro) subunits are assembling into active 20S proteasome complexes and that this in vitro assembly is dependent on protein concentration and temperature. Similar results have been observed for the M. thermophila and R. erythropolis α and β proteins which require incubation at 35 to 45°C for autocatalytic assembly into functional 20S proteasomes in vitro (41, 81). This has also been observed for in vitro assembly of the α and β proteins of T. acidophilum which require incubation at 65°C (40) or pH conditions which unfold and refold the proteins (55). Thus, among the archaea and eubacteria, it appears that the rate of autocatalytic assembly of α and β proteins into active 20S proteasome complexes is influenced by temperature and is often enhanced at the growth temperature optimum of the organism. Whether the rate of hydrolysis of the β propeptide is influenced by temperature remains to be determined. However, the rate of interaction and/or folding of α and β during in vitro assembly appears to be influenced by temperature, since assembly of even α and β(Δpro) proteins which no longer have the propeptide, as determined by N-terminal sequencing, are stimulated by heating (41). Interestingly, the presence of the β propeptide significantly reduced the efficiency of in vitro assembly of the M. jannaschii proteins compared to those of M. thermophila and R. erythropolis (data not shown) (41, 81).
Biophysical properties of the 20S proteasome complex.
An active M. jannaschii proteasome of 700 kDa was purified from the assembly and contains equimolar ratios of the α and β(Δpro) proteins based on SDS-PAGE (Fig. 1A) (Table 2). Electron microscopy of the proteasome reveals a four-stacked ring structure of 12 by 17 nm with a central channel which has structural similarities to other 20S proteasomes (Fig. 1D) (3, 14, 41, 45, 62, 74). Based on analogy to the T. acidophilum 20S proteasome (25), it is likely that the α protein of M. jannaschii forms the outer protein rings of the cylinder as well as the distinct central openings. If so, assembly of the α disc-like complexes with the β(Δpro) protein may involve conformational changes in α which generate the central portal and 2-nm increase in diameter of the 20S proteasome (Fig. 1B and E). An additional fraction of at least 1.5 MDa which was active in peptide hydrolysis was purified from the assembly mixture and contained equimolar ratios of the α and β(Δpro) proteins as well as vesicles (Fig. 1A and C). Whether two 700-kDa proteasomes associate into this larger complex and are not efficiently separated from vesicles or whether single 700-kDa proteasome particles associate directly with vesicles remains to be determined. However, the ∼1.5-MDa proteasome fraction was 1.3-fold more active in Suc-LLVY-Amc hydrolysis than the 700-kDa proteasomes (Table 2). These results suggest that changes in protein conformation may enhance peptide-hydrolyzing activities of the 20S proteasome. Incubation of the 700-kDa proteasomes in the presence of buffer with 150 mM NaCl at 65°C resulted in the formation of symmetrical arrays of the 20S proteasome in end-on views as observed on electron micrographs (Fig. 1E). Stereo pairs of these arrays (Fig. 1E) reveal that the proteasomes are not in a planar arrangement but instead are multilevel. It is likely that heating the proteins in the presence of salt promotes ionic and/or hydrophobic interactions between the outer walls of the 20S cylinder. Using similar methods, the spontaneous formation of arrays has not been observed for the 20S proteasome of M. thermophila and, thus, does not appear to be a universal feature of these proteins (40).
Multisubstrate and proteolytic activities of the 20S proteasome.
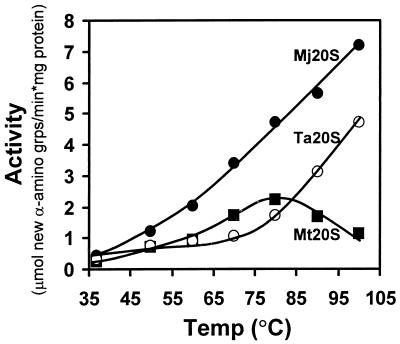
The M. jannaschii 20S proteasome degraded β-casein (Fig. 2). The rate of protein hydrolysis increased linearly to at least 100°C (Fig. 2) and was not influenced by ATP (data not shown). The highest rate of β-casein hydrolysis measured was 1.5 and 3.2 times faster than that of the 20S proteasomes of T. acidophilum and M. thermophila (Fig. 2). To our knowledge, this is the highest reported rate of protein hydrolysis catalyzed by a 20S proteasome (1, 13, 41, 74). At the extreme assay temperatures used in this experiment, it is likely that β-casein is thermally denatured, and additional proteins such as chaperones are not needed to unfold this polypeptide for entry into the central proteolytic chamber of the 20S proteasome.
FIG. 2.
Effect of temperature on β-casein degradation by archaeal 20S proteasomes. Hydrolysis of bovine β-casein was measured at the temperatures indicated using 0.2 mg of substrate protein and 0.05 mg of purified 20S proteasome in 1 ml of 20 mM Tris–1 mM DTT (pH 7.2). Abbreviations: Mj20S, Ta20S, and Mt20S, M. jannaschii, T. acidophilum, and M. thermophila 20S proteasomes, respectively; grps, groups.
The peptide-hydrolyzing activity of the M. jannaschii 20S proteasome was measured using synthetic substrates (Table 3). High rates of cleavage carboxyl to the acidic residue glutamate were observed with the substrate carbobenzoxy–Leu–Leu–Glu–β-naphthylamide (Cbz-LLE-βNa). This peptidylglutamyl-peptide hydrolyzing (PGPH) or caspase-like activity was optimal at 90°C with a narrow pH optimum of 6.5 (Table 3). The PGPH activity was not influenced by ATP, GTP, or MgCl2. However, similar to the M. thermophila 20S proteasome, the PGPH activity was reduced by at least 70% after incubation with 2% SDS. The M. jannaschii 20S proteasome also had significant chymotrypsin-like (CL) activity as measured using Suc-LLVY-Amc and Suc-Ala-Ala-Phe-Amc (Table 3). The hydrolysis of Suc-LLVY-Amc was optimal at 90°C and had a rather broad pH optimum of pH 7 to 8 (Table 3). The CL activity was not influenced by ATP, GTP, or MgCl2; however, it was reduced up to twofold by addition of 0.08 to 2% SDS and stimulated 1.7-fold by 300 mM KCl. Interestingly, the CL activities of the T. acidophilum and M. thermophila 20S proteasomes differ somewhat in that they are stimulated ca. twofold by addition of 0.02 to 2% SDS (1, 41). Low levels of trypsin-like activity were also observed for the M. jannaschii 20S proteasome (Table 3). This low rate of cleavage carboxyl to arginine residues is common to other archaeal and eubacterial 20S proteasomes (1, 41, 82) but contrasts with eucaryal 20S proteasomes which have significantly higher trypsin-like activity (52).
TABLE 3.
Multisubstrate activities of the M. jannaschii 20S proteasome
| Activitya | Substrateb | Activityc |
|---|---|---|
| PGPH | Cbz-Leu-Leu-Glu-βNa | 4,120 ± 40d |
| 483 ± 10e | ||
| 770 ± 10f | ||
| Ac-Tyr-Val-Ala-Asp-Amc | 115 ± 8.8 | |
| Ala-Glu-Amc | UDi | |
| CL | Suc-Leu-Leu-Val-Tyr-Amc | 746 ± 39 |
| 3,160 ± 120g | ||
| 830 ± 10h | ||
| Suc-Ala-Ala-Phe-Amc | 772 ± 54 | |
| Suc-Ile-Ile-Trp-Amc | 208 ± 33 | |
| Suc-Leu-Tyr-Amc | 58 ± 3 | |
| TL | Benz-Phe-Val-Arg-Amc | 31 ± 0.1 |
| Boc-Phe-Ser-Arg-Amc | 17 ± 0.8 | |
| Gly-Gly-Arg-Amc | UD | |
| SNAAP | Suc-Ile-Ala-Amc | UD |
PGPH, peptidyl-glutamyl peptide hydrolyzing; CL, chymotrypsin-like; TL, trypsin-like; SNAAP, small neutral amino acid preferring.
Cbz, carbobenzoxy; β-Na, β-naphthylamide; Ac, acetyl; Amc, 4-methyl-coumaryl-7-amide; Suc, succinyl; Boc, tert-butyloxycarbonyl.
Activity measured in micromoles of product per minute per milligram of protein. Unless otherwise indicated, peptide hydrolysis was measured at 60°C using 150 μM substrate and 0.7 μg of purified protein in 1 ml of Tris buffer (pH 7.0) with 6% (vol/vol) dimethyl sulfoxide.
Assayed at 90°C.
eAssayed in 25 mM TES buffer, pH 7.0.
Assayed in 25 mM MES buffer, pH 6.5.
Assayed with 20 μM substrate at 90°C.
Assayed with 20 μM substrate in 25 mM TES buffer, pH 7.5–0.4% DMSO.
UD, undetectable.
The multisubstrate activity of the M. jannaschii 20S proteasome is similar to that of M. thermophila which hydrolyzes Cbz-LLE-βNa at a rate that is 30 to 60% higher than the CL substrates Suc-LLVY and Suc-Ala-Ala-Phe-Amc (41). This differs, however, from 20S proteasomes isolated from other archaea and actinomycetes which exhibit very little PGPH activity (1, 3, 13, 45, 62, 74). Similar to the high rate of proteolysis catalyzed by the M. jannaschii 20S proteasome, the optimal CL and PGPH activities for this enzyme are more than 7 to 9 times higher than those of the M. thermophila 20S proteasome (41), and, to our knowledge, are the highest reported for 20S proteasomes in the absence of effector molecules. The differences in pH optima for the PGPH and CL activities for these methanoarchaeal 20S proteasomes suggest that protonation of the active site or pH-induced changes in the conformation of the 20S particle has a significant influence on the preference of peptide substrate.
Purification of the proteasome-activating nucleotidase (PAN).
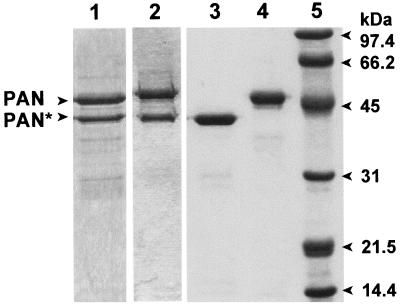
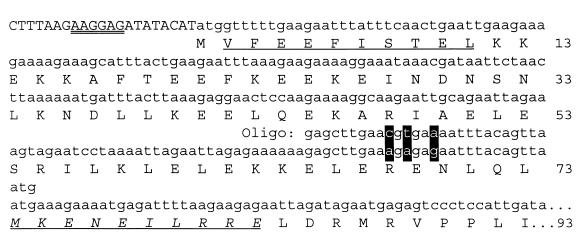
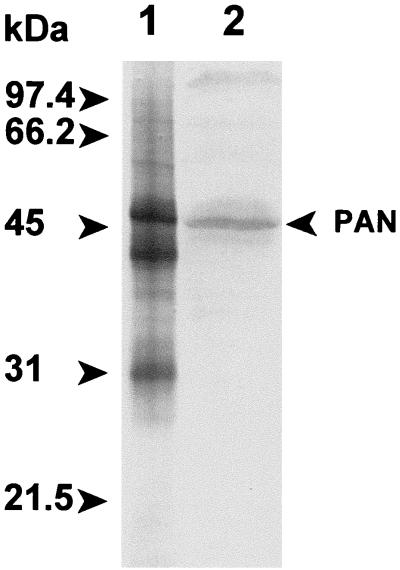
Transcription of the putative pan gene in E. coli resulted in the synthesis of three proteins of 47, 39, and 30 kDa which were not apparent in the absence of the coding sequence for this protein. This suggested that either the protein produced from the M. jannaschii gene induced the overproduction of E. coli stress response proteins or recombinant PAN proteins of variable molecular mass were generated. The 47- and 39-kDa proteins were stable after heating the cell lysate to 85°C and copurified after gel filtration as a 600-kDa fraction which hydrolyzed ATP at a rate of 1.8 μmol of Pi per min per mg of protein at 65°C (Fig. 3, lane 1). Protein sequence analysis revealed that the first 10 residues of the 47-kDa protein were identical to residues 2 to 11 of the PAN protein sequence deduced from DNA (Fig. 4). The first 10 residues of the 39-kDa subunit were found to be identical to residues 74 to 83 of the predicted PAN protein (Fig. 4). This suggests that there is an internal ribosome binding site recognized by E. coli within the full-length mRNA which accounts for the production of the shorter 39-kDa protein in E. coli. In addition, it is likely that the aminopeptidase of E. coli cleaved the N-terminal methionine residue of the full-length PAN protein; however, the N-terminal methionine of the 39-kDa PAN was not accessible for cleavage by the host peptidase. Using an alternative approach, the coding sequence for PAN was positioned in frame to produce full-length PAN with an N-terminal six-histidine tag (His6-PAN). Similar to the study by Zwickl et al. (84), the 39-kDa PAN copurified with the full-length His6-PAN after Ni2+ chromatography (Fig. 3, lane 2). However, in our study, the 39-kDa PAN appeared to copurify at an equimolar ratio with the full-length His6-PAN compared to a much lower ratio for the previously purified complex (84). The His6-PAN complex, of our studies, hydrolyzed ATP at a rate which was over 60% less than that of the untagged PAN complex described above. In addition, the His6-PAN complex was less stable than the untagged PAN complex after heating to 85°C or long-term storage at 4°C. These results suggest that the N-terminal His6 tag may influence PAN stability and activity.
FIG. 3.
M. jannaschii PAN proteins from recombinant E. coli. Protein (2 μg) was separated by reducing SDS–12% PAGE and stained with Coomassie blue. Lanes: 1, full-length PAN and PAN(Δ1–73) from recombinant E. coli/pMjPAN2; 2, His6-tagged PAN and PAN(Δ1–73) from recombinant E. coli/pMjPAN5; 3, PAN(Δ1–73) from recombinant E. coli/pMjPAN8; 4, full-length PAN from recombinant E. coli/pMjPAN7; 5, molecular mass standards with the masses indicated to the right of the gel.
FIG. 4.
Site-directed mutagenesis of the Shine-Dalgarno site located within the M. jannaschii pan gene recognized in E. coli. Capitalized nucleotides are from plasmid pET24b, and lowercase nucleotides are from the pan gene. The Shine-Dalgarno site of pET24b used for synthesis of the full-length PAN protein in recombinant E. coli is double underlined. The nucleotides targeted and the oligonucleotide (Oligo) used for site-directed mutagenesis are highlighted and indicated above the DNA coding region, respectively. Amino acid residues identical to the N-terminal protein sequence determined by Edman degradation of the purified 47-kDa PAN are underlined, whereas those of the purified 39-kDa PAN(Δ1–73) are underlined and italicized. Amino acid positions of the PAN protein deduced from the chromosomal DNA are indicated on the right.
To purify the full-length PAN protein alone, the internal ribosomal binding site of the ORF which was recognized by E. coli was altered by site-directed mutagenesis at nucleotides A202C, A204T, and G207A (Fig. 4). This destroyed the Shine-Dalgarno consensus sequence while preserving the amino acid sequence of the PAN protein. To separately produce a PAN(Δ1–73) protein, a PCR was used to generate a 219-bp deletion at the 5′ end of the gene, and the Met74 ATG codon was positioned 8 bp downstream of the ribosomal binding site in the expression plasmid. Thus, the full-length PAN and PAN(Δ1–73) protein with a 73-amino acid N-terminal deletion were produced separately and purified to homogeneity (Fig. 3).
PAN produced in M. jannaschii.
Immunoblot analysis revealed that only the PAN protein of 47 kDa is detectable in M. jannaschii cells when grown on H2 and CO2 at 65°C (Fig. 5). These results demonstrate that the pan gene is expressed in M. jannaschii and generates the full-length protein. Partial purification of PAN from M. jannaschii cell lysate reveals that the protein is associated in a distinct 550-kDa complex as determined by gel filtration, electrophoresis, and immunoblot analysis (data not shown). It remains to be determined if the 550-kDa PAN complex purified from M. jannaschii is a homooligomer or associates with additional proteins.
FIG. 5.
PAN produced in M. jannaschii. Proteins were separated by reducing SDS–12% PAGE and analyzed by Western blot using anti-PAN(Δ1–73) antibodies. Lanes: 1, total protein (1 to 2 μg) produced in recombinant E. coli/pMjPAN2 which synthesizes PAN proteins of 47, 39, and 30 kDa; 2, M. jannaschii cell lysate (20 μg) which produces only the full-length PAN of 47 kDa.
Biophysical properties of recombinant PAN.
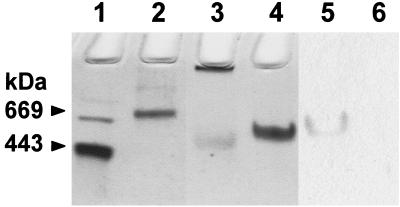
The full-length PAN and PAN(Δ1–73) proteins synthesized in recombinant E. coli each form homooligomeric complexes of 550 and 500 kDa, respectively, as determined by gel filtration. The results suggest that PAN alone is able to form a complex of ∼12 subunits and does not require the first 73 amino acids for this association. The PAN complexes were highly stable in the absence of ATP which contrasts with related proteins such as ClpA, ClpX, and NEM-sensitive fusion protein (NSF) which require nucleotide binding for complex stability (24, 26, 43). The PAN proteins were also stable after heating to 85°C in polypropylene tubes; however, they were readily inactivated when incubated in glass tubes much like ClpX (75). Nondenaturing PAGE reveals that each PAN complex migrated as a distinct band, which was in good agreement with its estimated high molecular mass (Fig. 6). The separated band of PAN protein catalyzed ATP hydrolysis and was inhibited by NEM, as determined by in-gel staining for inorganic phosphate (Fig. 6). A significant fraction of PAN(Δ1–73), however, did not enter the gel, which suggests that it is somewhat prone to aggregation under the electrophoretic conditions used (Fig. 6). When PAN(Δ1–73) is maintained in optimal buffer and salt conditions, however, the protein does not aggregate based on gel filtration chromatography (data not shown). Preincubation of either PAN protein with NEM at 65°C prior to electrophoresis did not influence their migration which contrasts with the related HslU of E. coli which dissociates into monomers in the presence of NEM (79).
FIG. 6.
Native gel separation of M. jannaschii PAN proteins. Lanes: 1, apoferritin of 443 kDa; 2, thyroglobulin of 669 kDa; 3, PAN(Δ1–73) of 500 kDa based on gel filtration; 4 to 6, full-length PAN of 550 kDa based on gel filtration. The proteins were stained with Coomassie blue (lanes 1 to 4) or stained for the generation of Pi from ATP in the absence (lane 5) or presence (lane 6) of 10 mM NEM.
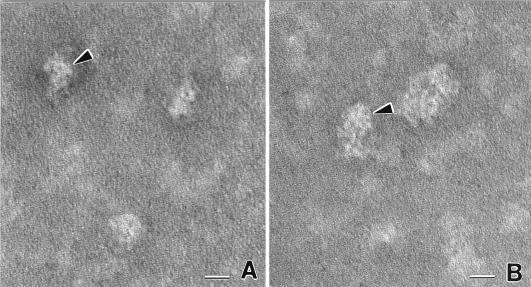
The ca. 12-subunit configuration of PAN suggested that it may associate as two stacked hexameric rings similar to other related ATPases including the mammalian NSF (26), Thermoplasma VAT (48), and Rhodococcus ARC complexes (76). Transmission electron micrographs of the recombinant PAN proteins revealed particles of 10 to 14 nm in diameter which were somewhat ring-like but not distinct hexameric or symmetrical complexes (Fig. 7). Similar results were recently described for the complex of His6-PAN and PAN(Δ1–73) (84). The reason for the observed asymmetrical PAN complexes remains to be determined.
FIG. 7.
Transmission electron micrographs of M. jannaschii PAN. The arrowhead in panel A indicates side view in which PAN resembles a comma-shaped complex. The arrowhead in panel B points to a more face-on view in which the head of the comma is at top. Distinct individual subunits are seen as white globular components of each assembly. Larger clusters probably represent random associations created when sample was dried for transmission electron microscopy observation. Bars, 10 nm.
Biochemical properties of PAN.
PAN(Δ1–73) was used as a comparison to the full-length protein to examine the influence of the predicted N-terminal coiled-coil, which spans residues 49 to 83, on the biochemical properties of PAN. Both the full-length PAN and PAN(Δ1–73) proteins have optimal ATPase activity at pH 7 to 8 and are more active at pH 8 to 10 (50 to 75% of the optimum) than at pH 5.5 (5 to 35% of the optimum). The ATPase activity of the full-length PAN protein is optimal at 80°C which is close to the optimal growth temperature of 85°C for M. jannaschii. In contrast, the ATPase activity of PAN(Δ1–73) is optimal at 65°C. The ATPase activity of the recently characterized complex of His6-PAN and PAN(Δ1–73) is optimal at 73°C (84) which is between the optimal temperature for the PAN and PAN(Δ1–73) complexes.
The PAN proteins, in our study, had reduced ATPase activity after incubation in salt-free buffer prior to assay. The ATPase activity of the full-length PAN protein was stimulated to as much as twofold by the addition of salt at concentrations of up to 3.5 M NaCl and 2.5 M KCl; above these concentrations, activity was reduced. Similar results were observed for PAN(Δ1–73), however, activity was reduced at salt concentrations above 2 M. A previous study (84) demonstrated that the ATPase activity of the complex of His6-PAN and PAN(Δ1–73) required Mg2+ and was inhibited by EDTA. In this study, the activities of the PAN complexes were also inhibited by the Mg2+-chelator EDTA as well as the sulfhydryl-blocking agents NEM and para-chloromercuriphenyl-sulfonic acid (PCMS) (Table 4). These activities were not influenced by NaN3 which is a known inhibitor of H+-transporting ATPase proteins (Table 4). These results are consistent with other AAA proteins which catalyze Mg2+-dependent ATP hydrolysis and are inhibited by NEM (2, 17, 28, 38, 50, 53, 61, 73, 76). It is possible that a cysteine residue, such as Cys384 in the C terminus of PAN which is conserved in some AAA proteins, is modified by these sulfhydryl-blocking agents. ATP did not appear to protect PAN from NEM inhibition, unlike some AAA proteins including Saccharomyces cerevisiae CDC48 (17) (data not shown).
TABLE 4.
Effects of various compounds on the ATPase activity of the M. jannaschii PAN and PAN(Δ1–73) proteins
| Effector | Concn (mM) | Relative activitya
|
|
|---|---|---|---|
| PAN | PAN(Δ1–73) | ||
| None | 100 ± 6 | 100 ± 5 | |
| EDTA | 10 | 8 ± 3 | UDd |
| SDS | 0.1 | 63 ± 5 | |
| 0.2 | 3 ± 4 | ||
| NEMb | 10 | UD | 15 ± 4 |
| 5 | 23 ± 1 | ||
| 1 | 81 ± 8 | ||
| PCMSc | 0.05 | UD | UD |
| NaN3 | 5 | 90 ± 2 | 74 ± 5 |
ATPase activity is expressed as a percentage of the control reaction with no added effector. The enzyme (2 μg per ml) was preincubated with or without the effector in 100 mM NaCl, 10 mM MgCl2, 25 mM TES (pH 8.0) at 21°C for 15 min before addition of 1 mM ATP and incubation at 80°C (PAN) or 65°C [PAN(Δ1–73)].
NEM, N-ethylmaleimide.
PCMS, p-chloromercuriphenyl-sulfonic acid.
UD, undetectable.
The PAN and PAN(Δ1–73) proteins have extremely high Vmax values for ATP hydrolysis (Table 5), like the E. coli ClpA protein (30). These values are also somewhat comparable to the S. cerevisiae CDC48 protein, which has a Vmax half that of full-length PAN (17). Many complexes containing related AAA proteins including the mammalian 26S proteasome and 19S cap (PA700) proteins, however, have Vmax values that are >100-fold lower than those of PAN (28). The affinities of the PAN proteins for ATP (Table 5) are similar to those of E. coli ClpX and yeast CDC48 which have Km values of 500 to 550 μM for ATP (17, 69) and are somewhat comparable to other AAA proteins with Km values for ATP of 650 μM for the mammalian NSF protein (44), 210 μM for E. coli ClpA (30), 280 μM for E. coli HslU (80), and 200 μM for R. erythropolis ARC (76). However, compared to PAN, the affinity for ATP of E. coli ClpB is 2.2-fold lower (77), and the affinity for ATP is from 6.3- to 100-fold higher for the yeast S4 (Rpt2) protein (38), mammalian 26S proteasome and 19S cap proteins (28), E. coli Lon protease (71), and E. coli FtsH protease (64).
TABLE 5.
Kinetic parameters of the M. jannaschii PAN and PAN(Δ1–73)
Km measured in micromolar of ATP or CTP.
Vmax measured in micromoles of Pi released per minute per milligram of protein.
The Vmax values for the CTP-hydrolyzing activity of the PAN and PAN(Δ1–73) proteins were 1.7- to 2-fold higher than the values for ATP-hydrolyzing activity (Table 5). The PAN complex purified from M. jannaschii had a similar ratio of ∼2:1 for CTP- and ATP-hydrolyzing activities, suggesting that this is not an artifact of the recombinant proteins (data not shown). The affinity of the full-length protein for CTP was ∼60% higher than for ATP, whereas deletion of the N-terminal 73 residues of PAN resulted in approximately equal affinities of the protein for CTP and ATP (Table 5). The full-length PAN appeared to only hydrolyze ATP and CTP compared to the PAN(Δ1–73) protein which catalyzed the hydrolysis of ITP, GTP, TTP, and UTP at 92, 73, 37, and 19% the rate of ATP, respectively. These results suggest that modification of the N terminus has a significant influence on the nucleotide specificity of the PAN protein. Hydrolysis of CTP and ATP by the PAN proteins was inhibited 70 to 100% in the presence of equimolar ratios of ADP. In contrast, CDP inhibited the hydrolysis of CTP 77 to 84% compared to at most a 13% inhibition of ATP hydrolysis.
Similar to these results, the recently reported complex of His6-PAN and PAN(Δ1–73) also catalyzes the hydrolysis of GTP and UTP at 58% and CTP at 205% the rate of ATP (84). In addition, the mammalian 26S proteasome and 19S cap (PA700) as well as the E. coli Lon protease hydrolyze ATP, CTP, UTP, and GTP (28, 70). High rates of CTP and ATP hydrolysis have also been demonstrated for the R. erythropolis ARC and E. coli FtsH proteins (64, 76).
The reason for the generally high rate of nucleotide hydrolysis mediated by PAN compared to the majority of other AAA proteins which have been characterized remains to be established. It is possible that the ATP-to-ADP ratios in hyperthermophiles such as M. jannaschii differ from other organisms which have been used as sources of AAA proteins. Alternatively, the high-level synthesis of PAN in recombinant E. coli enabled its rapid purification and may have minimized steps which would have otherwise reduced the ATP-hydrolyzing activity of this enzyme. It is also possible that the M. jannaschii PAN complex is not a homooligomer of 12 subunits and that nucleotide hydrolysis is stimulated by increased self-association of PAN in the recombinant protein complex. It is also possible that the ATPase activity of PAN may represent a futile reaction in vitro which results from the absence of physiological partners such as the putative 20S proteasome; however, it appears that the presence of β-casein and 20S proteasome actually stimulates PAN ATPase activity (as described below).
PAN is required for ATP-dependent hydrolysis by archaeal 20S proteasomes.
The PAN proteins were investigated for activation of protein hydrolysis mediated by archaeal 20S proteasomes. Both full-length PAN and PAN(Δ1–73) complexes stimulated the degradation of β-casein by the M. jannaschii 20S proteasome by 1.4 ± 0.04- to 2.2 ± 0.19-fold within 15 min, this stimulation was completely dependent on the presence of either ATP or CTP (data not shown). No significant energy-dependent hydrolysis of β-casein was observed in the presence of ADP, and stimulation was optimal at molar ratios ranging from 0.5:1 to 4:1 of PAN and 20S proteasome complexes (data not shown). Furthermore, the PAN (Fig. 8) and PAN(Δ1–73) complexes (data not shown) stimulated by 2- to 4-fold the degradation of β-casein by the M. thermophila 20S proteasome in the presence of ATP, but not ADP. Under these assay conditions, the rate of ATP hydrolysis by PAN was not influenced by the addition of the synthetic peptide Suc-LLVY-Amc or the 20S proteasome alone but was enhanced by the addition of β-casein by 1.22 ± 0.07-fold or β-casein with the 20S proteasome by 1.38 ± 0.08-fold (data not shown). In addition, CTP substituted for ATP in stimulation of protein hydrolysis (data not shown), which is consistent with other energy-dependent proteases including the eucaryotic 26S proteasome (18), E. coli Lon (70), E. coli FtsH (64), and the complex of M. jannaschii His6-PAN and PAN(Δ1–73) with the T. acidophilum 20S proteasome (84).
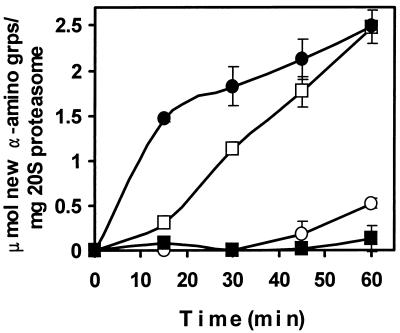
FIG. 8.
β-Casein degradation by the 20S proteasome is stimulated by PAN. PAN and M. thermophila 20S proteasome were mixed together in a 2:1 molar ratio in 25 mM TES buffer (pH 8) with 10 mM MgCl2, 1 mM nucleotide, and 2 mg of β-casein per ml and incubated at 55°C. The amounts of α-amino groups (grps) generated by protein hydrolysis were determined. Data presented are the averages of three separate experiments. Symbols: ●, PAN and 20S proteasome with ATP; ○, 20S proteasome with ATP; ■, PAN and 20S proteasome with ADP; □, PAN and 20S proteasome with AMP-PNP.
Interestingly, 5′-adenylyl β,γ-imidophosphate (AMP-PNP) which is not hydrolyzed by PAN protein (data not shown) also supported PAN activation of β-casein degradation by ca. fourfold (Fig. 8). However, the mechanism of degradation appeared to be qualitatively different. When ATP was hydrolyzed there was a rapid increase in the generation of free α-amino groups compared to the significant lag phase observed when AMP-PNP was present (Fig. 8). Although these results contrast with those of the recent study which demonstrate that the complex of His6-PAN/PAN(Δ1–73) in the presence of AMP-PNP does not activate the breakdown of proteins by the T. acidophilum 20S proteasome (84), the methods used to monitor protein hydrolysis are different. In this study, hydrolysis was measured by the production of new α-amino groups using fluorescamine, whereas the previous study measured conversion of β-[14C]casein into acid-soluble products (84). Together, these results suggest that the presence of a nonhydrolyzable ATP analogue, such as AMP-PNP, supports a single cleavage or a limited number of cleavages of β-casein, which results in the generation of large, acid-precipitable fragments with new amino groups much like the Lon protease of E. coli (15). In analogy to Lon, it is possible that ATP hydrolysis is required for the PAN-stimulated processive degradation of proteins to acid-soluble products by the 20S proteasome, whereas ATP binding to PAN supports limited hydrolysis of proteins by the 20S proteasome (15). However, further studies are needed to confirm this possibility.
PAN associates with archaeal 20S proteasomes.
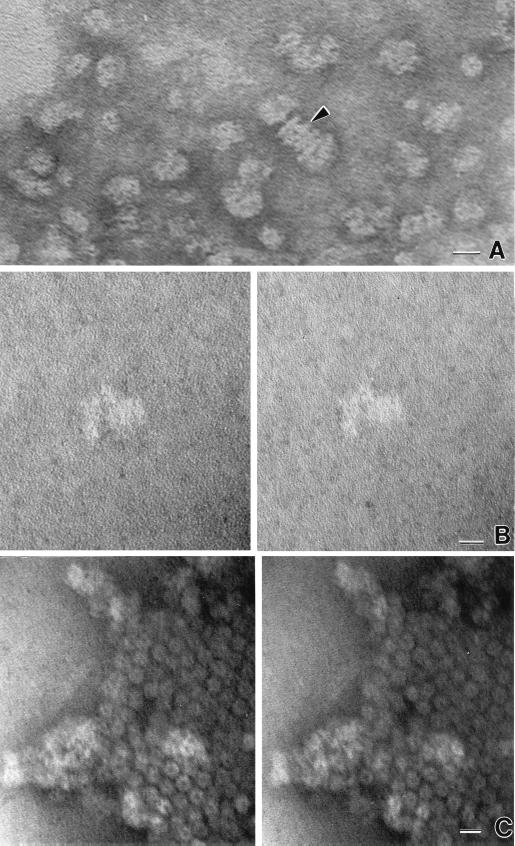
The purified M. jannaschii 20S proteasome and PAN proteins were mixed at equimolar ratios and then incubated in the presence of ATP and Mg2+ at 50°C. Electron micrographs of these mixtures reveal particles which resemble the 20S core capped either singly or doubly on each end by PAN protein complexes (Fig. 9). The singly capped proteasome-associated PAN complexes were ∼24 nm long and 12 to 14 nm wide. These micrographs are consistent with the ability of the PAN proteins to stimulate proteolysis by 20S proteasomes and suggest that archaeal 20S proteasomes may interact with PAN to form a complex which resembles the 26S proteasome subcomplex recently described for S. cerevisiae (21). The efficiency of reconstitution of these complexes, however, was less than 0.5%, suggesting that the interaction between the proteins may be transitory and not stable during electron microscopy, additional factors are necessary for stable complex formation, and/or other possibilities yet to be determined.
FIG. 9.
Transmission electron micrographs of reconstituted 20S proteasome and PAN proteins. (A) Untilted view of particle generated during reconstitution of the PAN(Δ1–73) and 20S proteasome. The arrowhead indicates cylindrical 20S proteasome assembly with apparent assemblies of PAN(Δ1–73) at both ends. (B) Stereo view of a single 20S proteasome cylinder with a putative PAN assembly at the left end. (C) Stereo view of 20S proteasome arrays with indistinct patchy material on top of arrays in center and at left, which probably represent PAN complexes. Bars, 20 nm.
ACKNOWLEDGMENTS
We are greatly indebted to J. M. Flanagan at the Brookhaven National Laboratories, D. R. Boone at Portland State University, and A. L. Goldberg at Harvard Medical School for supportive discussions. We thank D. Williams for help with transmission electron microscopy and F. Davis and J. Shelton for DNA sequencing. We also thank R. Kim and S.-H. Kim for their generosity in sending plasmid pSJS1240 and E. Seemüller and W. Baumeister for providing plasmid DNA encoding the 20S proteasome of T. acidophilum.
This work was supported in part by the Institute of Food and Agricultural Sciences Center for Biomass Programs.
Footnotes
Journal series R07342 of the Florida Agricultural Experiment Station.
REFERENCES
- 1.Akopian T N, Kisselev A F, Goldberg A L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- 2.Babst M, Sato T K, Banta L M, Emr S D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer M W, Halio S B, Kelly R M. Purification and characterization of a proteasome from the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 1997;63:1160–1164. doi: 10.1128/aem.63.3.1160-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 5.Beuron F, Maurizi M R, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 6.Boone D R, Johnson R L, Liu Y. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl Environ Microbiol. 1989;55:1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann D, Sutton G G, Blake J A, FitzGerald M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Burke M, Rajasekharan K N, Maruta S, Ikebe M. A second consensus sequence of ATP-requiring proteins resides in the 21-kDa C-terminal segment of myosin subfragment 1. FEBS Lett. 1990;262:185–188. doi: 10.1016/0014-5793(90)80185-l. [DOI] [PubMed] [Google Scholar]
- 10.Chin D T, Goff S A, Webster T, Smith T, Goldberg A L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988;263:11718–11728. [PubMed] [Google Scholar]
- 11.Cornish-Bowden A. Fundamentals of enzyme kinetics. London, United Kingdom: Portland Press; 1995. [Google Scholar]
- 12.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 13.Dahlmann B, Kuehn L, Grizwa A, Zwickl P, Baumeister W. Biochemical properties of the proteasome from Thermoplasma acidophilum. Eur J Biochem. 1992;208:789–797. doi: 10.1111/j.1432-1033.1992.tb17249.x. [DOI] [PubMed] [Google Scholar]
- 14.Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- 15.Edmunds T, Goldberg A L. Role of ATP hydrolysis in the degradation of proteins by protease La from Escherichia coli. J Cell Biochem. 1986;32:187–191. doi: 10.1002/jcb.240320304. [DOI] [PubMed] [Google Scholar]
- 16.Finley D, Tanaka K, Mann C, Feldmann H, Hochstrasser M, Vierstra R, Johnston S, Hampton R, Haber J, McCusker J, Silver P, Frontali L, Thorsness P, Varshavsky A, Byers B, Madura K, Reed S I, Wolf D, Jentsch S, Sommer T, Baumeister W, Goldberg A, Fried V, Rubin D M, Toh-e A. Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- 17.Fröhlich K U, Fries H W, Peters J M, Mecke D. The ATPase activity of purified CDC48p from Saccharomyces cerevisiae shows complex dependence on ATP-, ADP-, and NADH-concentrations and is completely inhibited by NEM. Biochim Biophys Acta. 1995;1253:25–32. doi: 10.1016/0167-4838(95)00136-i. [DOI] [PubMed] [Google Scholar]
- 18.Ganoth D, Leshinsky E, Eytan E, Hershko A. A multicomponent system that degrades proteins conjugated to ubiquitin. Resolution of factors and evidence for ATP-dependent complex formation. J Biol Chem. 1988;263:12412–12419. [PubMed] [Google Scholar]
- 19.Gerards W L, de Jong W W, Bloemendal H, Boelens W. The human proteasomal subunit HsC8 induces ring formation of other α-type subunits. J Mol Biol. 1998;275:113–121. doi: 10.1006/jmbi.1997.1429. [DOI] [PubMed] [Google Scholar]
- 20.Gerards W L, Enzlin J, Haner M, Hendriks I L, Aebi U, Bloemendal H, Boelens W. The human α-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the β-type subunits HsDelta or HsBPROS26. J Biol Chem. 1997;272:10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- 21.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman S. Regulation by proteolysis: developmental switches. Curr Opin Microbiol. 1999;2:142–147. doi: 10.1016/S1369-5274(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 24.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 25.Grizwa A, Baumeister W, Dahlmann B, Kopp F. Localization of subunits in proteasomes from Thermoplasma acidophilum by immunoelectron microscopy. FEBS Lett. 1991;290:186–190. doi: 10.1016/0014-5793(91)81256-8. [DOI] [PubMed] [Google Scholar]
- 26.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. Immunoblotting; pp. 269–309. [Google Scholar]
- 28.Hoffman L, Rechsteiner M. Nucleotidase activities of the 26S proteasome and its regulatory complex. J Biol Chem. 1996;271:32538–32545. doi: 10.1074/jbc.271.51.32538. [DOI] [PubMed] [Google Scholar]
- 29.Hoskins J R, Pak M, Maurizi M R, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang B J, Woo K M, Goldberg A L, Chung C H. Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988;263:8727–8734. [PubMed] [Google Scholar]
- 31.Karata K, Inagawa T, Wilkinson A J, Tatsuta T, Ogura T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J Biol Chem. 1999;274:26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- 32.Kawarabayasi Y, Sawada M, Horikawa H, Haidawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahasi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F T, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 33.Kim R, Sandler S J, Goldman S, Yokota H, Clark A J, Kim S-H. Overexpression of archaeal proteins in E. coli. Biotechnol Lett. 1998;20:207–210. [Google Scholar]
- 34.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 35.Kodama T, Fukui K, Kometani K. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem. 1986;99:1465–1472. doi: 10.1093/oxfordjournals.jbchem.a135616. [DOI] [PubMed] [Google Scholar]
- 36.Lanzetta P, Alvarez L, Reinach P, Candia O. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 37.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 38.Lucero H, Chojnicki E, Mandiyan S, Nelson H, Nelson N. Cloning and expression of a yeast gene encoding a protein with ATPase activity and high identity to the subunit 4 of the human 26S protease. J Biol Chem. 1995;270:9178–9184. doi: 10.1074/jbc.270.16.9178. [DOI] [PubMed] [Google Scholar]
- 39.Lupas A, Flanagan J M, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 40.Maupin-Furlow, J. A. 1999. Unpublished results.
- 41.Maupin-Furlow J A, Aldrich H C, Ferry J G. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J Bacteriol. 1998;180:1480–1487. doi: 10.1128/jb.180.6.1480-1487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maupin-Furlow J A, Ferry J G. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J Biol Chem. 1995;270:28617–28622. doi: 10.1074/jbc.270.48.28617. [DOI] [PubMed] [Google Scholar]
- 43.Maurizi M R, Singh S K, Thompson M W, Kessel M, Ginsburg A. Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and ClpP. Biochemistry. 1998;37:7778–7786. doi: 10.1021/bi973093e. [DOI] [PubMed] [Google Scholar]
- 44.Morgan A, Dimaline R, Burgoyne R D. The ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF) is regulated by soluble NSF attachment proteins. J Biol Chem. 1994;269:29347–29350. [PubMed] [Google Scholar]
- 45.Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuwald A F, Aravind L, Spouge J L, Koonin E V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 47.Ogura T, Tomoyasu T, Yuki T, Morimura S, Begg K J, Donachie W D, Mori H, Niki H, Hiraga S. Structure and function of the ftsH gene in Escherichia coli. Res Microbiol. 1991;142:279–282. doi: 10.1016/0923-2508(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 48.Pamnani V, Tamura T, Lupas A, Peters J, Cejka Z, Ashraf W, Baumeister W. Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett. 1997;404:263–268. doi: 10.1016/s0014-5793(97)00138-5. [DOI] [PubMed] [Google Scholar]
- 49.Peters J M, Harris J R, Lustig A, Müller S, Engel A, Volker S, Franke W W. Ubiquitous soluble Mg(2+)-ATPase complex. A structural study. J Mol Biol. 1992;223:557–571. doi: 10.1016/0022-2836(92)90670-f. [DOI] [PubMed] [Google Scholar]
- 50.Peters J M, Walsh M J, Franke W W. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richmond C, Gorbea C, Rechsteiner M. Specific interactions between ATPase subunits of the 26 S protease. J Biol Chem. 1997;272:13403–13411. doi: 10.1074/jbc.272.20.13403. [DOI] [PubMed] [Google Scholar]
- 52.Rivett A J. Proteasomes: multicatalytic proteinase complexes. Biochem J. 1993;291:1–10. doi: 10.1042/bj2910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roggy J L, Bangs J D. Molecular cloning and biochemical characterization of a VCP homolog in African trypanosomes. Mol Biochem Parasitol. 1999;98:1–15. doi: 10.1016/s0166-6851(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 54.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 55.Seemüller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 56.Seol J H, Yoo S J, Kang M S, Ha D B, Chung C H. The 65-kDa protein derived from the internal translational start site of the clpA gene blocks autodegradation of ClpA by the ATP-dependent protease Ti in Escherichia coli. FEBS Lett. 1995;377:41–43. doi: 10.1016/0014-5793(95)01306-7. [DOI] [PubMed] [Google Scholar]
- 57.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 59.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki C K, Rep M, Maarten van Dijl J, Suda K, Grivell L A, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 61.Tagaya M, Wilson D W, Brunner M, Arango N, Rothman J E. Domain structure of an N-ethylmaleimide-sensitive fusion protein involved in vesicular transport. J Biol Chem. 1993;268:2662–2666. [PubMed] [Google Scholar]
- 62.Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, De Mot R, Baumeister W. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 63.Thompson M W, Singh S K, Maurizi M R. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli: requirement for the multiple array of active sites in ClpB but not ATP hydrolysis. J Biol Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 64.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Melderen L, Gottesman S. Substrate sequestration by a proteolytically inactive lon mutant. Proc Natl Acad Sci USA. 1999;96:6064–6071. doi: 10.1073/pnas.96.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Hartling J A, Flanagan J M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 68.Wang W, Chevray P M, Nathans D. Mammalian Sug1 and c-Fos in the nuclear 26S proteasome. Proc Natl Acad Sci USA. 1996;93:8236–8240. doi: 10.1073/pnas.93.16.8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waxman L, Goldberg A L. Protease La from Escherichia coli hydrolyzes ATP and proteins in a linked fashion. Proc Natl Acad Sci USA. 1982;79:4883–4887. doi: 10.1073/pnas.79.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waxman L, Goldberg A L. Protease La, the lon gene product, cleaves specific fluorogenic peptides in an ATP-dependent reaction. J Biol Chem. 1985;260:12022–12028. [PubMed] [Google Scholar]
- 72.Wenzel T, Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 73.Whiteheart S W, Rossnagel K, Buhrow S A, Brunner M, Jaenicke R, Rothman J E. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson H L, Aldrich H C, Maupin-Furlow J A. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 76.Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Müller S A, Engel A, De Mot R, Baumeister W. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- 77.Woo K M, Kim K I, Goldberg A L, Ha D B, Chung C H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 78.Yao Y, Toth C R, Huang L, Wong M L, Dias P, Burlingame A L, Coffino P, Wang C C. α5 subunit in Trypanosoma brucei proteasome can self-assemble to form a cylinder of four stacked heptamer rings. Biochem J. 1999;344:349–358. doi: 10.1042/0264-6021:3440349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoo S J, Kim H H, Shin D H, Lee C S, Seong I S, Seol J H, Shimbara N, Tanaka K, Chung C H. Effects of the cys mutations on structure and function of the ATP-dependent HslVU protease in Escherichia coli. The Cys287 to Val mutation in HslU uncouples the ATP-dependent proteolysis by HslvU from ATP hydrolysis. J Biol Chem. 1998;273:22929–22935. doi: 10.1074/jbc.273.36.22929. [DOI] [PubMed] [Google Scholar]
- 80.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M-S, Tanaka K, Goldberg A L, Chung C H. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 81.Zühl F, Seemüller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418:189–194. doi: 10.1016/s0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- 82.Zühl F, Tamura T, Dolenc I, Cejka Z, Nagy I, De Mot R, Baumeister W. Subunit topology of the Rhodococcus proteasome. FEBS Lett. 1997;400:83–90. doi: 10.1016/s0014-5793(96)01403-2. [DOI] [PubMed] [Google Scholar]
- 83.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 84.Zwickl P, Ng D, Woo K M, Klenk H-P, Goldberg A L. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26S proteasome, activates protein breakdown by 20S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]