Abstract
The current diagnosis and treatment of sarcoma continue to show limited timeliness and efficacy. In order to enable the early detection and management of sarcoma, increasing attentions have been given to the tumor microenvironment (TME). TME is a dynamic network composed of multiple cells, extracellular matrix, vasculature, and exosomes. Exosomes are nano-sized extracellular vesicles derived from various cells in the TME. The major function of exosomes is to promote cancer progress and metastasis through mediating bidirectional cellular communications between sarcoma cells and TME cells. Due to the content specificity, cell tropism, and bioavailability, exosomes have been regarded as promising diagnostic and prognostic biomarkers, and therapeutic vehicles for sarcoma. This review summarizes recent studies on the roles of exosomes in TME of sarcoma, and explores the emerging clinical applications.
Keywords: Exosomes, Tumor microenvironment, Sarcoma, Communication, Clinical application
Introduction
Sarcoma is a heterogeneous group of rare mesenchymal-derived cancers constituting only 1% of all malignancies [1]. However, some types, such as osteosarcoma and Ewing’s sarcoma, noticeable affect children and adolescents [2]. No specific diagnostic and prognostic indicators are available for most sarcomas, which is detrimental to the early detection and treatment evaluation [3, 4]. New diagnostic and therapeutic approaches are urgently needed to improve the overall survival of sarcoma patients.
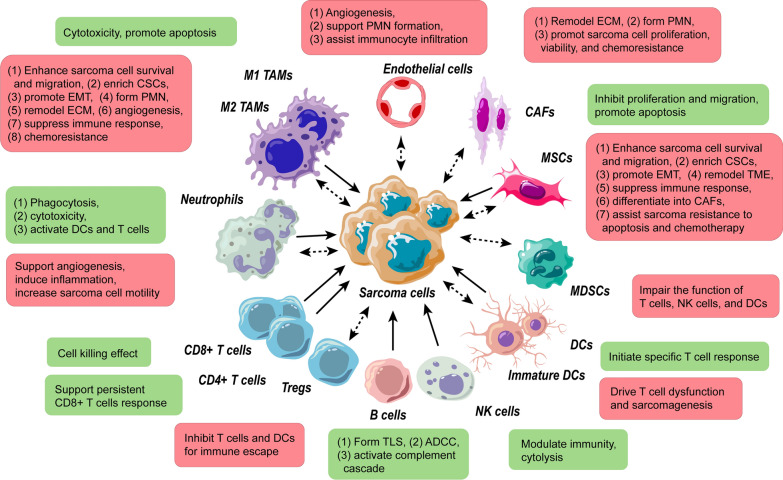
The tumor microenvironment (TME) is widely recognized as an essential modulator of cancer development, and a source for identifying potential biomarkers and therapies for a wide range of neoplasms, including hepatocellular carcinoma, colorectal cancer, pancreatic cancer, breast cancer, lung cancer, and sarcoma [5–10]. The TME of sarcoma encompasses sarcoma cells, immune cells, and stromal cells, whose fundamental role is to dynamically interact with sarcoma cells (Fig. 1). The interactions are mediated by contact- and noncontact-dependent mechanisms [11, 12]. The latter are connected by soluble molecules, including cytokines and subcellular structures, such as exosomes [13–17].
Fig. 1.
Effects of cells in TME on the progression and metastasis of sarcoma. The boxes represent the final effect on sarcoma after cellular interactions. Each red box and dotted arrow denote pro-tumor effects of microenvironment cells, and the double-head arrows indicate bidirectional actions. Green box and solid arrow represent anti-tumor effects of microenvironment cells
Exosomes, as a subtype of extracellular vesicles, are spherical particles with a diameter of 30–150 nm [18]. Exosomes are encapsulated by lipid bilayer membranes with surface proteins and enclose a cargo of biomolecules, including nucleic acids, proteins, lipids, and other bioactive substances [18–20]. The composition of exosomes varies depending on the type of parental cells. Biogenesis and secretion of exosomes are based on the endosomal system. Early endosomes are formed by the inward fusion of endocytic vesicles [21]. Early endosomes return the contents to the plasma membrane as recycling endosomes or converse into multivesicular bodies by inward invagination of endosomal membrane and cargo package into intraluminal vesicles [21]. The protein sorting of intraluminal vesicles is highly regulated by the endosomal-sorting complex required for transport-dependent or -independent pathways [22, 23]. Next, the multivesicular bodies get into the final intracellular destination, either by fusing with lysosomes to be degraded or by fusing with the plasma membrane [24]. Rab27A and Rab27B are the important mediators to induce multivesicular bodies to transfer to the cell periphery [25]. Ultimately, the soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex drives multivesicular bodies to fuse with the plasma membrane and release intraluminal vesicles as exosomes into the extracellular microenvironment [18]. Almost all types of TME cells and malignant cells can release exosomes, which are widely distributed in biofluids [19]. After release into extracellular space, exosomes can be internalized by recipient cells through several mechanisms, including phagocytosis, macropinocytosis, plasma membrane fusion, and endocytosis [26]. Uptake mechanism and amount of exosomes by recipient cells rely on recipient cells’ surface receptors and exosomal surface proteins. Hypoxia and hypoxia-related conditions, such as low pH and oxidative stress, increase exosome production and fusion efficiency via hypoxia-induced pathways and lipid content alteration of exosome membrane [27, 28]. Exosomes have now been considered as biological vehicles transporting regulatory molecules to bridge tumor-cell interactions in TME, holding key roles in immune response, tumorigenesis, dissemination, angiogenesis, and chemotherapy resistance [29–35]. Exosomes have the properties of content specificity, cell tropism, and bioavailability in different malignancies [36, 37]. Considerable studies have demonstrated that exosomes may serve as promising non-invasive diagnostic and prognostic biomarkers, as well as possible vectors for therapeutic modalities in cancer treatment [36–39].
In this review, we summarize the most recent studies on the roles of exosomes in TME of sarcoma, and discuss the potential implications in clinic.
The roles of exosomes in sarcoma TME
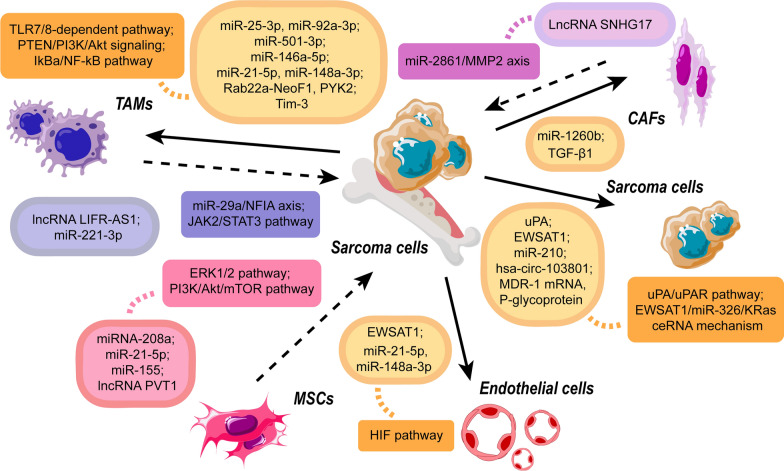
Exosomes are highly specialized entities for intercellular communications in TME. The main function of exosomes is to transfer bioactive substances (DNA, RNA, lipids, proteins, and other signaling molecules) from donor cells to recipient cells (Fig. 2). Exosomes provide stimulatory signals and attenuate inhibitory signals for sarcoma cells, resulting in the exchange of genetic information and reprogramming of target cells.
Fig. 2.
Exosomal molecules and corresponding pathways in the exosome-mediated interactions between sarcoma cells and TME cells. The dotted arrow indicates the transfer of exosomes from TME cells to sarcoma cells, while the solid arrow represents the transfer from sarcoma cells to TME cells. The oval with frame line contains the effective molecules in exosome transport. The color of each oval corresponds to the origin of the exosomes, where orange denotes sarcoma cell-derived exosomes, blue denotes TAM-derived exosomes, red denotes MSC-derived exosomes, and purple denotes CAF-derived exosomes. The box covers the corresponding pathways activated by exosomes
Exosomes mediate the macrophage and sarcoma cell interactions for immune suppression and sarcoma progress
The development of sarcoma is linked with the immune microenvironment. Although exosomal antigens can induce tumor-specific immune reaction, accumulating studies have indicated that sarcoma creates a favorable surrounding by exosome-based dynamic signaling. Noticeably, Tumor-associated macrophages (TAMs) play key roles in shaping local immune response in TME via both promoting and suppressing immunity to sarcoma.
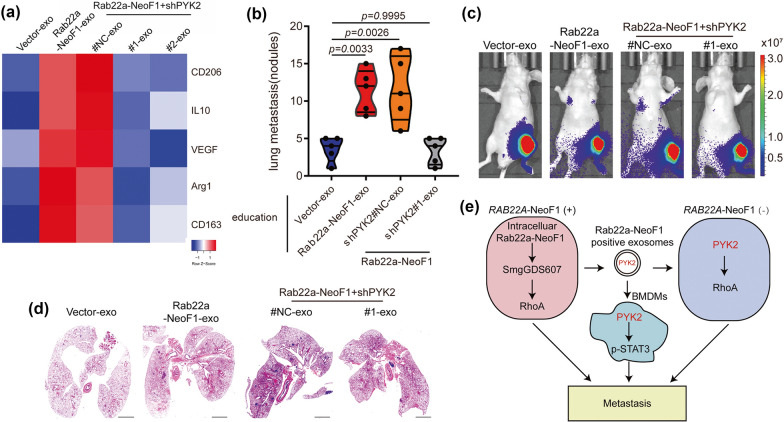
Exosomes derived from sarcoma cells could alter the differentiation and function of macrophages, leading to local and distant spread, and immune suppression (Table 1). Sarcoma-derived exosomes promote sarcoma dissemination via targeting the mononuclear macrophage system. Exosomal microRNAs and proteins from sarcoma cells promoted the expression of cytokines and matrix metalloproteinases in macrophages [40–42]. These products endowed sarcoma cells with malignant capacity and remodeled extracellular matrix for local invasion [40–42]. Particularly, exosomes from osteosarcoma cells might affect the differentiation and maturity of osteoclasts, the specialized cells originated from monocyte/macrophage lineage [43–45]. The exosomal miR-501-3p and pro-osteoclastic contents influence osteoclastogenesis, leading to sarcoma infiltration via aggravating osteolysis [43–45]. In addition to local spread, exosomes from sarcoma are strongly associated with its distant metastasis. Exosomal miR-146a-5p suppressed osteoclastogenesis to disrupt barriers in primary lesions, which was favorable to the early progression of osteosarcoma metastasis [46]. Meanwhile, in metastatic lesions, exosomal proteins Rab22a-NeoF1 and PYK2 recruited macrophages and induced M2 polarization, establishing a functional pre-metastatic niche for pulmonary metastasis of osteosarcoma (Fig. 3) [14]. Sarcoma-derived exosomes play an essential role in these metastatic processes. Sarcoma with impaired exosome release had remarkably fewer metastatic foci and smaller areas of metastatic lesions than that with normal release ability [46]. With tumor growth, hypoxia induces sarcoma release of exosomes which can alter the immune and metabolic profile of infiltrating macrophages to better evade the immune response and promote tumor progression. Hypoxic conditions substantially enhanced the levels of immunomodulatory proteins and chemokines in exosomes produced by hypoxic tumor cells, including CSF-1, CCL2, TGF-β, FTH, and FTL [47]. The hypoxia-induced exosomes mediated monocyte/macrophage recruitment to the lesions, and the M2 phenotype was more pronounced than M1 [48]. Meanwhile, these exosomes enhanced macrophage polarization towards the M2 phenotype. The polarization may be manipulated by molecular determinants, including STAT, NF-κB, PPAR, KLF, IRF, and HIF families, and miRNAs, in which the clear role of exosomes remains to be explored. Exosomes transported let-7a miRNA and suppressed insulin-Akt–mTOR signaling pathway to enhance oxidative phosphorylation and M2 polarization of infiltrating macrophages [47]. The M2 macrophages inhibited activated CD4+ T cells and had increased the expression of mitogenic, growth, angiogenic, and pro-metastatic cytokines and enzymes [49, 50]. M2 macrophages also promoted the proliferation of tumor cells and elevated the angiogenic ability of endothelial cells by favoring oxidative phosphorylation [47].
Table 1.
Effect of sarcoma-derived exosomes on the TME cells
| Sarcoma type | TME cells | Exosomal cargos | Mechanisms | Clinical significance | References |
|---|---|---|---|---|---|
| OS | Macrophages | Tim-3 | Induce conversion to M2 macrophages | Pre-metastatic lung formation↑ | [40] |
| LPS | Macrophages | miR-25-3p, miR-92a-3p | Increase IL-6 secretion via TLR7/8-dependent pathway | Progression↑ | [41] |
| OS | Osteoclasts; endothelial cells | miR-21-5p, miR-148a-3p | Promote bone remodeling; enhance angiogenesis | Metastasis↑ | [42] |
| OS | Osteoblasts; osteoclasts; endothelial cells | \ | Promote bone remodeling; facilitate angiogenesis via VEGF/ANGPT2/FGF2-mediated mechanism | Progression and metastasis↑ | [43] |
| OS | Monocytes | miR-501-3p | Promote bone remodeling via PTEN/PI3K/Akt pathway | Metastasis↑ | [45] |
| OS | Osteoclasts | miR-146a-5p | Promote bone remodeling via IkBa/NF-kB pathway | Metastasis↑ | [46] |
| OS, FS | Monocytes | \ | Transform the monocyte phenotype to inhibit CD4+ T cells | Immune suppression↑ | [49] |
| OS | Macrophages | \ | Induce conversion to M2 macrophages | Immune suppression↑ | [50] |
| OS | Macrophages | Rab22a-NeoF1, PYK2 | Activate RhoA and STAT-3 | Pre-metastatic lung formation↑ | [14] |
| EWS | CD33+ and CD14+ cells | \ | Impede dendritic cell differentiation and maturation | Immune suppression↑ | [55] |
| OS | CD11b+ Gr-1+ cells | \ | Induce CD11b+ cells infiltration to lung | Pre-metastatic lung formation↑ | [56] |
| OS | MSCs | \ | Induce MSCs phenotypic conversion | Progression and metastasis↑ | [58] |
| OS | MSCs | \ | Induce LINE-1 hypomethylations for MSCs transformation | Progression and metastasis↑ | [59] |
| MFS | Fibroblasts | miR-1260b | Decrease PCDH9 level | Progression and Infiltration↑ | [67] |
| OS | Fibroblasts | \ | Induce fibroblast phenotypic conversion | Progression↑ | [68] |
| OS | Fibroblasts | TGF-β1 | Induce myofibroblast/CAFs differentiation | Pre-metastatic lung formation↑ | [70] |
| OS | Endothelial cells; OS cells | EWSAT1 | Promote angiogenesis; regulate Akt and Erk signaling via EWSAT1/miR-326/KRas ceRNA mechanism | Progression↑ | [73] |
OS, osteosarcoma; miR, microRNA; LPS, liposarcoma; TLR7/8, toll-like receptor 7 and 8; VEGF, vascular endothelial growth factor; ANGPT2, angiopoietin 2; FGF2, fibroblast growth factor 2; PTEN, phosphatase and tensin homologue deleted on chromosome 10; PI3K, phosphoinositide 3 kinase; Akt, AKT8 virus oncogene cellular homolog; IkBa, inhibitor of NF-κB alpha; NF-κB, nuclear factor kappa B; FS, fibrosarcoma; RhoA, Ras homolog gene family, member A; STAT-3, signal transducer and activator of transcription 3; EWS, Ewing’s sarcoma; MSCs, mesenchymal stem cells; LINE-1, long interspersed element; MFS, myxofibrosarcoma; PCDH9, protocadherin 9; CAFs, cancer-associated fibroblasts; EWSAT1, Ewing sarcoma associated transcript 1; Erk, extracellular signal-regulated kinase; KRas, v-Ki-ras2Kirsten rat sarcoma viral oncogene homolog; ceRNA, competing endogenous RNA
Fig. 3.
The Rab22a-NeoF1 fusion protein in exosomes facilitates M2 polarization and induces lung metastasis. a Macrophages were incubated with exosomes, and the heatmap showed the differential expression profiles of the M2-like macrophage markers. b–d Mice were pre-treated with exosomes for 3 weeks and subsequently injected with sarcoma cells. The lung metastases were evaluated 3 weeks later. b n = 5 biologically independent mice. Data are mean ± s.d with P values. c Representative IVIS imaging. d H&E-stained lung sections. e The hypothesis of the function and mechanism of Rab22a-NeoF1 fusion protein in osteosarcoma. Rab22a-NeoF1 fusion protein activates the cellular RhoA by binding to SmgGDS607. The exosomal Rab22a-NeoF1 promotes RhoA activation by the exosomal PYK2 and facilitates the pulmonary pre-metastatic niche formation by promoting macrophage recruitment and by increasing M2 macrophages
(Reprinted with permission from Ref [14] Copyright © 2021, Li Zhong et al.)
TAMs, with M1-like and M2-like phenotypes, appear to be a double-edged sword in sarcoma progress via releasing exosomes (Table 2). In the early stage of sarcomagenesis, TME is predominantly infiltrated with M1-like macrophages. M1 TAMs-derived exosomes are immune-promoting and tumor-suppressive. Pro-inflammatory cytokines and cytotoxic factors were up-regulated in M1 TAMs-derived exosomes [51]. These exosomes could impede the activation of immune-suppressive Tregs, promote macrophage-mediated defense, and expedite sarcoma cell apoptosis [51]. However, as mentioned above, the polarization of M1 into M2 occurs during tumor progression. Thus, the immunosuppressive M2-like subtype becomes the dominant phenotype in the late stage. M2 TAMs transfer exosomal molecules, including microRNA and lncRNA to promote the survival, proliferation, and migration of sarcoma cells [52, 53]. In osteosarcoma, exosomal lncRNA LIFR-AS1 from M2 TAMs acted as a miR-29a sponge to restrain apoptosis and promote invasion of recipient tumor cells [53].
Table 2.
Effect of TME-derived exosomes on the sarcoma cells
| TME cells | Sarcoma type | Exosomal cargos | Mechanisms | Clinical significance | References |
|---|---|---|---|---|---|
| TAMs | OS | \ | Promote anti-tumor response; activate caspase-3 and -7 | Progression↓ | [51] |
| TAMs | OS | miR-221-3p | Activate JAK2/STAT3 pathway | Progression and metastasis↑ | [52] |
| TAMs | OS | lncRNA LIFR-AS1 | Activate miR-29a/NFIA axis | Progression and metastasis↑ | [53] |
| MSCs | OS | \ | Decrease hsa-miR-195 and hsa-miR-124, increase hsa-miR-148a | Progression and metastasis↑ | [60] |
| MSCs | OS | miR-21-5p | Activate PI3K/Akt/mTOR pathway | Progression↑ | [61] |
| ADSC | OS | \ | Up-regulate COLGALT2, vimentin, and MMP2/9 | Progression and metastasis↑ | [62] |
| MSCs | OS | \ | Activate Hedgehog pathway | Progression↑ | [63] |
| MSCs | OS | miRNA-208a | Activate ERK1/2 pathway | Progression and metastasis↑ | [64] |
| MSCs | ATRT | miR-155 | Down-regulate SMARCA4 | Metastasis↑ | [65] |
| MSCs | OS | lncRNA PVT1 | Increase ERG level | Progression and metastasis↑ | [66] |
| CAFs | OS | lncRNA SNHG17 | Activate miR-2861/MMP2 axis | Progression↑ | [71] |
| OS cells | OS | uPA | Activate uPA/uPAR axis | Metastasis↑ | [69] |
| EWS cells | EWS | miR-210 | Down-regulate CASP8AP2 | Progression↑ | [28] |
| OS cells | OS | hsa-circ-103801 | Reduce cisplatin sensitivity, inhibit apoptosis, and increase MRP-1 and P-glycoprotein expression | Chemoresistance↑ | [91] |
| OS cells | OS | MDR-1 mRNA, P-glycoprotein | Increase MDR-1 mRNA, P-glycoprotein | Chemoresistance↑ | [96] |
TAMs, tumor-associated macrophages; OS, osteosarcoma; miR, microRNA; JAK2, Janus Kinase 2; STAT-3, signal transducer and activator of transcription 3; lncRNA, long non-coding RNA; LIFR-AS1, Leukemia inhibitory factor receptor antisense RNA 1; NFIA, nuclear factor I A; MSCs, Mesenchymal stem/stromal cells; hsa-miR, homo sapiens microRNA; PI3K, phosphoinositide 3-kinase; Akt, AKT8 virus oncogene cellular homolog; mTOR, mammalian target of rapamycin; PIK3R1, phosphoinositide-3-kinase regulatory subunit 1; ADSC, adipose-derived mesenchymal stem cells; COLGALT2, collagen beta (1-O) galactosyltransferase 2; MMP2/9, matrix metalloproteinases-2 and-9; ERK1/2, extracellular signal-regulated kinase 1/2; ATRT, atypical teratoid/rhabdoid tumor; SMARCA4, SWI/SNF Related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4; PVT1, plasmacytoma variant translocation 1; ERG, ETS-related gene; CAFs, cancer-associated fibroblasts; SNHG17, Small nucleolar RNA host gene 17; MMP2, matrix metalloproteinase 2; uPA, urokinase-type plasminogen activator; uPAR, uPA receptor; CASP8AP2, caspase 8 associated protein 2; MRP-1, multidrug resistance-associated protein-1; MRP-1, multidrug resistance-1
Exosomes facilitate the immune cell and sarcoma cell interactions for immunoediting and sarcoma metastasis
Cancer immunoediting is the process in which the immune system can both inhibit and promote cancer progression. Cancer immunoediting includes three phases: elimination (immune activation), equilibrium, and escape phase (immunosuppression). Exosomes can mediate immunoediting and sarcoma development via linking communications between sarcoma cells and immune cells (Table 1). The involvement of macrophages has been highlighted above.
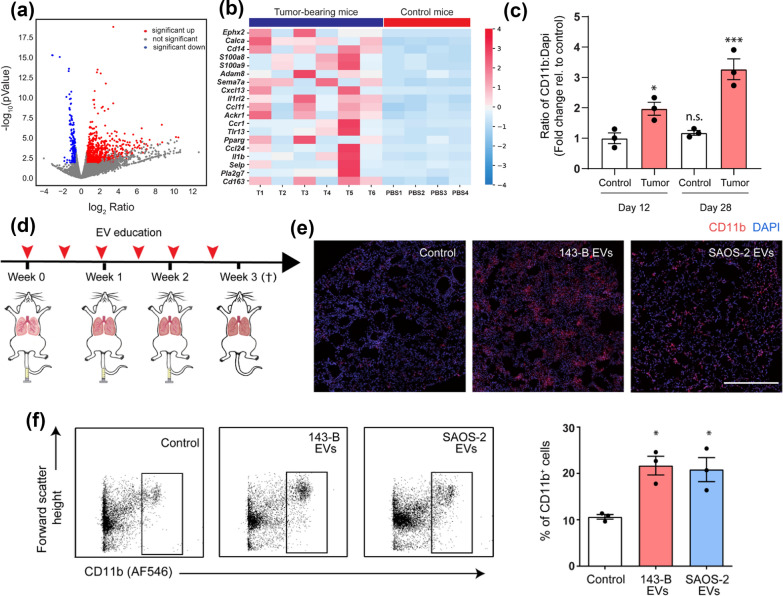
In the elimination and equilibrium phases, sarcoma-secreted exosomes containing lncRNA FOXP4-AS1 could recruit innate immune cells to TME, including Tregs, activated NK cells, and M1 macrophages [54]. The infiltrating immune cells had either tumor-promoting or tumor-suppressive effects. However, in the escape phase of immunoediting, sarcoma-derived exosomes have been shown to directly impair immune response. These exosomes induced the release of pro-inflammatory cytokines in CD33+ myeloid cells and CD14+ monocytes, and inhibited the differentiation of myeloid cells into dendritic cells [55]. The immature dendritic cells differentiated under this circumstance impeded immune reaction [55]. The immunosuppressive activities included inhibition of T cell proliferation, reduction of IFN-γ release, and induction of IL-10 and IL-6 secretion [55]. Immune cells also participate in exosome-dominated metastatic activities. Osteosarcoma-derived exosomes could recapitulate the infiltration of inflammatory CD11b+ Gr-1+ cells into the pre-metastatic lungs (Fig. 4) [56]. The formation and function of the pre-metastatic niche required a combined involvement of immunocytes and sarcoma-derived exosomes [56].
Fig. 4.
Transcriptomic profiling of pre-metastatic lungs showed an inflammatory response and myeloid cell infiltration, which was promoted by sarcoma-derived extracellular vesicles (EVs). a Volcano plot of differentially expressed genes in pre-metastatic lungs. b Heatmap representation of up-regulated genes under “inflammatory response” in tumor-bearing mice and controls. Cut-off values of fold change are > 1.5 and FDR < 0.2. c Quantification of CD11b+ cells in lungs of controls and tumor-bearing mice (day 12 and day 28). d Mice were treated with either PBS, 143-B EVs, or SAOS-2 EVs (10 µg) 2 times a week for 3 weeks. e Representative immunofluorescence images of CD11b+ cells (red) and nuclei (blue) in lungs of treated mice. f Representative flow cytometric profile (left) and quantification (right) of CD11b+ cells isolated from lungs of treated mice
(Reprinted with permission from Ref [56] Copyright © 2020, Cancers, Alekhya Mazumdar et al.)
Exosomes support the mesenchymal stem cell and sarcoma cell for sarcoma development
Mesenchymal stem cells (MSCs) are pluripotent stem cells with a remarkable capacity for self-renewal and multi-directional differentiation. As non-hematopoietic precursors, MSCs can differentiate into osteoblasts, chondroblasts, adipocytes, etc. [57]. Thus, MSCs contribute to the maintenance and regeneration of connective tissue, especially bone tissue. Exosomes take part in the interactions between sarcoma cells and MSCs.
Sarcoma-originated exosomes educate the recruited MSCs to undergo heterogeneous differentiation into a tumor-promoting phenotype (Table 1). Osteosarcoma-derived exosomes induced LINE-1 hypomethylation, leading to the epigenetic transformation of MSCs into pro-tumorigenic and pro-metastatic cancer-associated fibroblasts (CAFs) phenotype [58, 59]. The transformed MSCs had altered cytokine expression, and considerably over-expression of matrix metalloproteinases, vascular endothelial growth factors, and adhesion molecules [59]. These products contributed to extracellular matrix degrading, angiogenesis, and sarcoma invasion.
Correspondingly, MSCs can secrete abundant exosomes and act as paracrine mediators for sarcoma progression in TME (Table 2). MSC-derived exosomes assist the survival and proliferation of sarcoma cells via eluding inhibitory signals from hypoxia and chemotherapeutic agents. Serum-deprived MSCs prevented sarcoma cells from nutrient deprivation-induced death, via exosomes attenuating oxidative phosphorylation and increasing lactate uptake [60]. MSCs also facilitated the resistance of sarcoma to drug-induced apoptosis. Stressed MSCs decreased doxorubicin sensitivity of sarcoma cells through exosomes increasing transporter expression [60]. Meanwhile, MSCs-derived exosomes could also provide stimulatory signals for cancer initiation and development [61–65]. Exosomes carrying oncogenic microRNAs dysregulated proto-oncogenes and tumor suppressor genes in sarcoma cells, leading to the improvement of propagation, invasion, and migration (Fig. 5) [61, 63–65]. For instance, MSCs-derived exosomal miR-208a negatively targeted programmed cell death 4 to activate the ERK1/2 signaling pathway, thereby increasing the viability, clonogenicity, and migration of osteosarcoma cells [64]. The resultant increase in aggressiveness could be partly explained by the exosome-induced epithelial–mesenchymal transition and growth factor secretion [60, 62]. Additionally, MSC-derived exosomes also exerted pro-tumor effects through transporting oncogenic lncRNA PVT1 [66]. The exosomal PVT1 increased the level of transcription factor ERG in osteosarcoma cells [66]. ERG was previously found to be linked with bone matrix formation and sarcoma progression via activating TNSALP transcription [66].
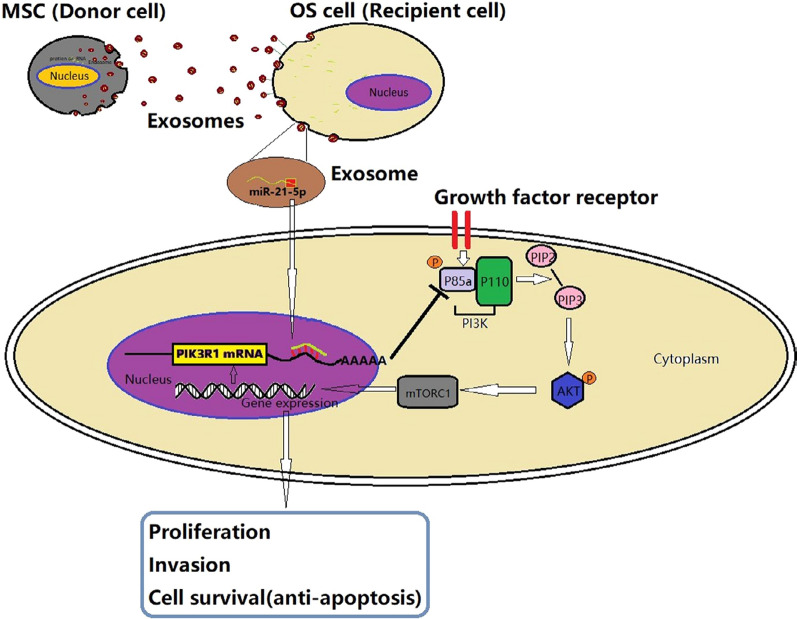
Fig. 5.
Schematic diagram of the role of exosomal miR-21-5p derived from MSCs in regulating the progression of OS cells. MSCs transported miR-21-5p to sarcoma cells via exosomes. Exosomal miR-21-5p targeted PIK3R1 gene and activated PI3K/Akt/mTOR pathway, thus resulting in sarcoma proliferation, invasion, and survival
(Reprinted with permission from Ref [61] Copyright © 2020, Journal of Cellular and Molecular Medicine, Jin Qi et al.)
Exosomes assist the fibroblast and sarcoma cell interactions for sarcoma invasive growth
CAFs are actively involved in cancerous lesions through remodeling matrix, promoting sarcoma progression, and assisting metastatic niche formation. Exosomes mediate the interactions between CAFs and sarcoma cells to support sarcoma expansion.
Sarcoma cells release exosomes to alter fibroblast phenotype, thereby promoting tumor infiltrative and metastatic activities (Table 1). The malignant transformation of neighbouring normal fibroblasts contributed to sarcoma local invasion [67, 68]. The surrounding fibroblasts absorbed exosomes from aggressive sarcoma cells and converted into a tumor-like phenotype [67, 68]. The resulting fibroblasts had enhanced proliferation and survival ability, and acquired capability to grow in an anchorage-independent manner [68]. More importantly, these adjacent transformed fibroblasts facilitated tumor growth into surrounding soft tissues, and led to frequent recurrence of infiltrative myxofibrosarcoma [67]. In addition, the activation of distant fibroblasts is one of the early events in sarcoma metastasis. Sarcoma-derived exosomes drive metastatic phenotype transformation of neighbouring sarcoma cells [69]. Meanwhile, the fibroblasts were recruited to the pre-metastatic lung by sarcoma-derived exosomes [70]. The lung fibroblasts internalized exosomes and differentiated into pro-metastatic myofibroblasts [70]. The acquisition of metastatic property of sarcoma cells and accumulation of lung fibroblasts, together with their conversion into myofibroblasts supported pulmonary metastasis of osteosarcoma [69, 70].
CAFs-derived exosomes also in turn boost the malignant behaviors of sarcoma (Table 2). The exosomes from CAFs carried lncRNA SNHG17, which up-regulated the expression of neoplastic matrix metalloproteinases in sarcoma cells [71]. Thus, the physical barrier was disrupted, and the invasive and metastatic growth of sarcoma was enhanced [71].
Exosomes expediate the endothelial cell and sarcoma cell interactions for angiogenesis
Angiogenesis refers to the ability of organisms to form neovasculature based on the original vasculature. As growing beyond its vascular supply, sarcoma mass forms hypoxic gradients and even a severely hypoxic core. Hypoxia induces an intricate intracellular signaling network in sarcoma cells which includes HIF, MAPK, PI3K, and NFĸB pathways [28, 72]. These signaling pathways are involved in the regulation of cell proliferation, metabolism, apoptosis, migration, stemness, and inflammatory response [28, 72]. For example, the elevation of miR-210 level in exosomes was dependent on HIF-1α stabilized expression in parental hypoxic cells [28]. Exosomal miR-210 was delivered to normoxic cells to promote sphere formation and enhance stemness via down-regulating proapoptotic CASP8AP2, which is pivotal to the survival of stem-like cells [28]. Although sarcoma cells in the hypoxic niche can directly deliver exosomes to nearby normoxic sarcoma cells for the acclimatization to hypoxia, the formation of neovasculature is a requirement for sarcoma growth [28]. These vessels funnel nutrients and waste metabolites from the sarcoma core, enable immune cell extravasation, and provide opportunities for tumor hematogenous metastasis.
Sarcoma cells encourage the initiation and development of angiogenesis by targeting endothelial cells in an exosome-mediated manner (Table 1). Exosomes increased the levels of angiogenic factors in TME. Sarcoma delivered microRNAs into endothelial cells through exosomes to increase angiogenic factors expression, including VEGF-A, IL-6, and IL-8 [42]. These angiogenic factors could induce neovascularization, regulate endothelial integrity, and modulate vascular permeability [42]. Besides, exosomes from sarcoma cells directly enhanced the reactivity of endothelial cells to these pro-angiogenic molecules. LncRNA and microRNA in sarcoma-derived exosomes up-regulated sensitivity and tube formation capacity of endothelial cells, which were prerequisites for the function of angiogenic factors [42, 73]. The combination of endothelial responsiveness and biosynthesis of pro-angiogenic substances leads to active angiogenesis. The hypoxia-induced dysfunctional vascularization and acquisition of epithelial-to-mesenchymal transition phenotype contribute to the promotion of tumor cell mobility and metastasis.
Clinical applications of exosomes in sarcoma
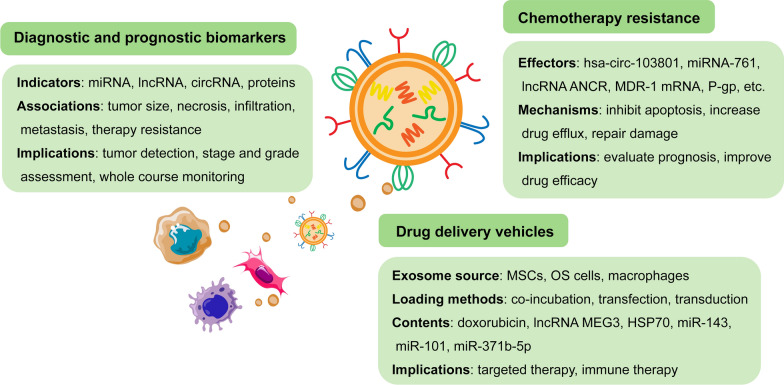
TME plays an integral part in sarcoma biology, participating in sarcoma initiation, development, and response to treatment. With the in-depth research in this emerging field, the potential clinical value of exosomes in TME has been gradually excavated. The molecular features of sarcoma-derived exosomes mirror many of the molecular features of the sarcoma they originate from, reflecting the physiological status of the sarcoma cells. Meanwhile, exosomes in TME are critical mediators in sarcomagenesis, affecting tumor progression and metastasis. Therefore, exosomes have potential roles in early diagnosis, prognosis prediction, chemosensitivity evaluation, and targeted therapy in sarcoma, which might represent an advancement in precision medicine (Fig. 6).
Fig. 6.
Clinical applications of exosomes in sarcoma management
Exosomes as diagnostic and prognostic biomarkers
Except for tumor biopsy and radiographic imaging, no clinically relevant indicator is available for the timely detection of sarcoma. As discussed above, sarcoma-derived exosomes are enriched with microRNAs, lncRNAs, circRNAs, and proteins, which are more abundant in sarcoma cells than in normal cells. The similarities between exosomes and the parental cells exhibit the potential of exosomes as biomarkers. The detection of exosomes will assist in tumor burden assessment, therapeutic responsiveness evaluation, and recurrence monitoring.
Each sarcoma is characterized by its specific nucleic acid or protein profile, which is the basis of the molecular diagnosis of sarcoma. Liquid biopsy profiling of plasma exosomes in sarcoma has been found to show great potential for tumor detection, and stage and grade assessment (Table 3). High serum levels of exosomes containing specific microRNAs are related to sarcoma status [67, 74–81]. High-throughput sequencing identified that 57 micorRNAs in serum exosomes were differentially expressed in osteosarcoma patients and controls with 20 expression being up-regulated and 37 being down-regulated [77]. The increased exosomal miR-195-3p could promote the proliferation and invasion of sarcoma cells, which further supported its significance in sarcoma detection [77]. Serum levels of exosomal miR-1260b were high in ten patients with infiltrative myxofibrosarcoma, which were more dominant in preoperative patients’ samples than those in postoperative [67]. The circulating exosomal miR-1260b levels were associated with tumor burden and the infiltrative ability of myxofibrosarcoma [67]. Serum-based circulating microRNAs are expected to serve as non-invasive and repeatable liquid biopsy indicators for tumor monitoring in sarcoma patients. Proteomic investigations of circulating exosomes revealed that some proteins were helpful in indicating tumor progression and metastasis [82–85]. For example, the level of serum exosomal PD-L1 and N-cadherin was higher in osteosarcoma patients, especially in the ones with pulmonary metastasis, than that in healthy controls [86]. The determination of cut-off values of these proteins proved to be useful in the identification and clinical staging of osteosarcoma patients (Fig. 7) [86]. Currently, based on mass spectrometry and proteome profiles, thousands of plasma exosomal proteins can be captured from a trace sample for the evaluation of osteosarcoma lung metastasis [87, 88]. Seven exosomal proteins were selected to differentiate patients with osteosarcoma from healthy controls and further distinguish between those with lung metastasis and non-lung metastasis [87]. The exosome-derived fusion transcripts and lncRNAs are also candidate diagnostic biomarkers for sarcoma [89, 90]. A deep analysis of these exosomal components and their levels will support an accurate diagnosis of sarcoma.
Table 3.
Studies on the diagnostic and prognostic values of exosomes
| Sarcoma type | Exosome source | Exosomal cargos | Sample source | Clinical application | References |
|---|---|---|---|---|---|
| MFS | Serum | miR-1260b | 15 patients, 5 non-sarcoma patients, 9 healthy controls | Diagnostic biomarker | [67] |
| DSRCT | Serum | miR-34a-5p, miR-22-3p, miR-324-5p | 3 patients, 4 healthy controls | Diagnostic biomarker | [74] |
| OS | Serum | miR-195-3p | 25 patients, 10 healthy controls | Diagnostic biomarker | [77] |
| OS | Serum | miRNAs | 5 patients | Diagnostic biomarker | [78] |
| DDLPS | Serum, tissue | miR-1246, -4532, -4454, -619-5p, and -6126 | 22 OS, 17 DDLPS, 3 EWS patients | Diagnostic biomarker | [79] |
| SS | Serum | miR-92b-3p | 12 SS patients, 12 benign tumor patients, 12 healthy controls | Diagnostic biomarker | [80] |
| RTK | Serum | miR-214-3p | 10 patients, 10 healthy controls | Diagnostic biomarker | [81] |
| OS | Serum | Proteins | 8 patients, 5 controls with fracture, 5 healthy controls | Diagnostic biomarker | [83] |
| EWS | Serum | CD99/MIC2, NGFR | 10 patients, 6 healthy controls | Diagnostic biomarker | [84] |
| GIST | Serum | KIT, SPRY4 | 9 patients, 9 healthy controls | Diagnostic and prognostic biomarker | [85] |
| OS | Serum | PD-L1, N-cadherin | 70 OS patients, 9 benign tumor patients, 22 healthy controls | Diagnostic biomarker | [86] |
| OS | Serum | Proteins | 20 lung metastasis patients, 20 non-metastasis patients, 12 healthy controls | Diagnostic biomarker | [87] |
| ARMS, SS | Serum | Fusion transcripts | 65 ARMS patients, 15 SS patients | Diagnostic biomarker | [89] |
| OS | Serum, tissue | lncRNA CASC15 | Sera from 5 patients and healthy controls; 30 OS specimens, 30 normal bone tissues | Diagnostic biomarker | [90] |
| OS | Serum | hsa-circ-103801 | 43 patients, 5 healthy controls | Prognostic biomarker, chemoresistance↑ | [91] |
| OS | Tissue | lncRNA SNHG17 | 5 pairs of tumor and tumor-free tissues | Prognostic biomarker | [75] |
| OS | Serum, tissue | miR-25-3p | Sera from 10 patients and 10 healthy controls; 45 biopsy specimens | Prognostic and diagnostic biomarker | [76, 93] |
| OS | Serum, tissue | SENP1 | Sera from 146 patients; 60 pairs of tumor and tumor-free tissues | Prognostic biomarker | [92] |
| OS | Serum | Alpha-2-macroglobulin, protein S, complement C2 | \ | Prognostic biomarker, chemoresistance↑ | [94] |
| OS | Serum | lncRNA ANCR | 10 patients, 10 chemosensitive controls | Prognostic biomarker, chemoresistance↑ | [95] |
MFS, myxofibrosarcoma; miR, microPNA; DSRCT, Desmoplastic small round cell tumor; OS, osteosarcoma; DDLPS, dedifferentiated liposarcoma; EWS, Ewing’s sarcoma; SS, synovial sarcoma; RTK, rhabdoid tumor of the kidney; NGFR, nerve growth factor receptor; GIST, gastrointestinal stromal tumor; KIT, tyrosine kinase receptor; SPRY4, Sprouty homolog 4; PD-L1, programmed death-ligand 1; ARMS, alveolar rhabdomyosarcoma; lncRNA, long noncoding RNAs; CASC15, cancer susceptibility 15; hsa, homo sapiens; SNHG17, Small nucleolar RNA host gene 17; SENP1, sentrin SUMO-specific protease 1; ANCR, Angelman syndrome chromosome region
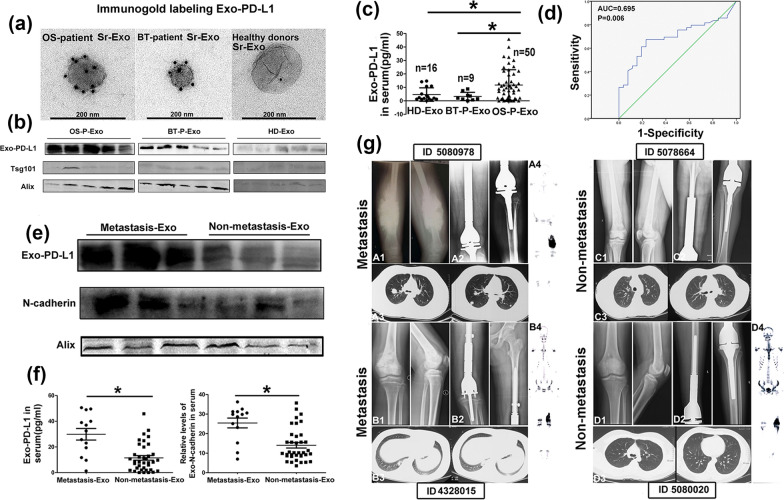
Fig. 7.
Higher level of exosomal PD-L1 in OS patients, especially those with pulmonary metastasis, compared to healthy donors and patients with benign tumor (BT). a A TEM image of exosomes immunogold-labelled with PD-L1 antibody. b Western blot analysis of PD-L1 expression in exosomes. c The levels of PD-L1 in serum exosomes from healthy donors and patients with OS and BT by ELISA. d ROC analysis showed that the AUC of serum-derived exosomal PD-L1 was 0.695, which could distinguish OS patients from healthy controls. e, f Higher expression of exosomal PD-L1 and N-cadherin in OS patients with metastasis than those without metastasis via western blot and ELISA analysis. g Radiological images of 4 OS patients with and without pulmonary metastasis
(Reprinted with permission from Ref [86] Copyright © 2020, Journal of Nanobiotechnology, Jun Wang et al.)
Accumulating studies have discovered that the molecular types and expression levels inside exosomes vary with disease progression, making them well-suited to assess prognosis and therapeutic efficacy. The levels of exosomal cargos, including lncRNAs, circRNAs, microRNAs, and proteins, had clinicopathological relevance in tumor size, necrosis rate, pulmonary metastasis, and therapy resistance (Table 3) [71, 76, 86, 91–95]. Sarcoma patients with higher expression of these exosomal contents in tissues and sera showed higher metastasis and recurrence rates, and shorter survival time [71, 86, 92, 93]. SNHG17 transcript level in the osteosarcoma tumor tissues was approximately 3 times higher than that in the normal tissues, and SNHG17 was mainly encapsulated in exosomes [71]. Patients with high expression of SNHG17 were significantly associated with poor prognosis [71]. The detection of circulating exosomes may be more valuable than that of tissue exosomes in clinical applications. A proteomic signature to discriminate between poor and good prognosis with high accuracy was revealed via using mass spectrometry to compare serum exosomes with different disease-free interval [94]. The expression levels of exosome-derived sentrin SUMO-specific protease 1 (SENP1) in patients’ plasma were significantly related to tumor stage, surgical stage, and overall survival of patients [92]. Patients with higher expression of plasma exosome-derived SENP1 had worse disease-free survival and overall survival [92]. The area under the receiver operating characteristic curve of plasma exosomal SENP1, as 1- and 3-year disease-free survival biomarkers, was 0.90 and 0.96, respectively [92]. The plasma exosome-derived SENP1 was superior to plasma SENP1 as a prognostic biomarker [92]. Up-regulated level of specific molecules in the serum exosomes from patients could also imply that sarcoma cells were resistant to chemotherapy and those patients might have a shorter survival time [91].
Exosomes as mediators of chemotherapy resistance
Drug resistance development is one of the crucial reasons for chemotherapy failure and poor prognosis in sarcoma. Exosomes typically function in the export of waste metabolites and signaling molecules from parental cells. Drug-resistant sarcoma cells are capable of shuttling chemotherapeutic agents out of cells, and delivery drug resistance molecules to tumor cells in TME via exosomes. Therefore, exosomes hold important roles in chemotherapy resistance of sarcoma.
Exosomes have been shown to transfer circRNAs, microRNAs, lncRNAs, mRNAs, and proteins from drug-resistant cells to drug-sensitive cells, thereby inducing primary drug resistance and multiple drug resistance [91, 94–97]. For instance, the multidrug-resistant osteosarcoma cells released exosomes containing multidrug resistance-1 mRNA and P-glycoprotein [96]. The exosomes could be taken up into secondary cells, facilitating the convey of doxorubicin-resistant capacity [96]. Exosomes highly enriched with hsa_circ_103801 derived from cisplatin-resistant cells also reduced drug sensitivity via inhibiting apoptosis and increasing the expression of multidrug resistance-associated protein 1 and P-glycoprotein [91]. Extracellular miR-761 from drug-resistant cells targeted three proteins, including thyroid hormone receptor interactor 6, lamin A/C, and NAD-dependent protein deacetylase sirtuin-3 [97]. Knockdown of any of these proteins in recipient cells could confer increased resistance to chemotherapeutic agents [97]. Briefly, the establishment of chemoresistance in sarcoma cells involves multiple mechanisms, including down-regulating apoptosis, increasing drug efflux, and modulating the expression of multidrug resistance substances [91, 96, 97]. Expression of the exosomal lncRNA ANCR in plasma was associated with the resistance to doxorubicin, and the lncRNA ANCR level was negatively correlated with survival time in osteosarcoma patients [95]. Therefore, detection of exosomes in serum during disease management contributed to the monitor of sarcoma chemosensitivity for better guidance of personalized chemotherapy treatment (Fig. 8) [91, 95].
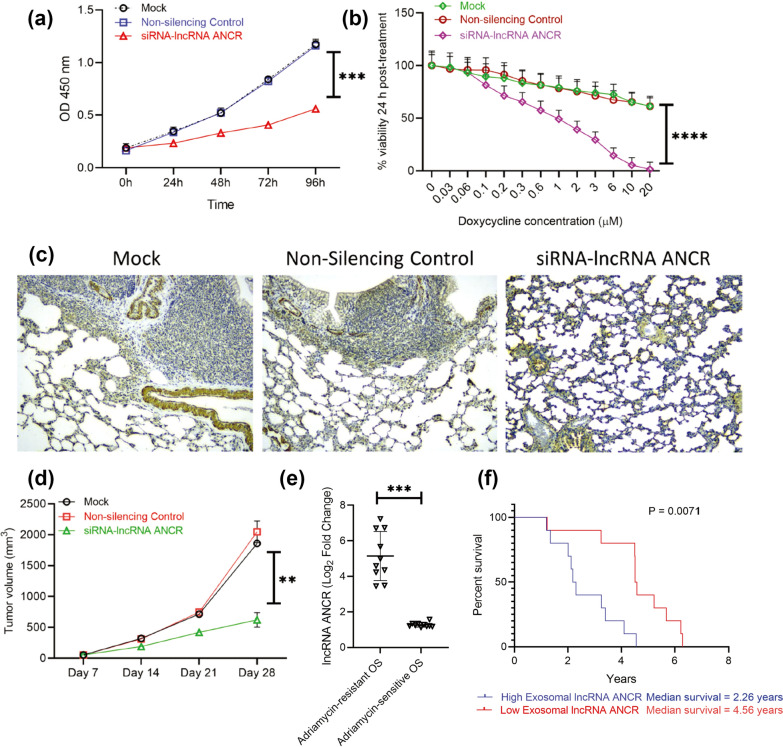
Fig. 8.
Exosomal lncRNA ANCR dictates in vitro sensitivity of sarcoma to doxycycline, expression of which is critical for drug resistance, tumor progression, and overall survival of patients. A Relative growth rate of the drug resistant cells mock-transfected, or transfected with non-silencing control siRNA, or siRNA targeting DANCR were quantified using CCK8 assay. b Cell viability of the differently transfected drug resistant cells after treated with doxycycline. c Representative IHC staining of osteocalcin. Nude mice were injected with sarcoma cells pre-treated with exosomes and added to sarcoma cells. All mice were treated with doxycycline orally once every 3 days for 4 weeks. Knockdown of DANCR made sarcoma cells sensitive to doxycycline and inhibited lung metastasis. d The tumor growth rate. e Expression of DANCR in OS patients sensitive or resistant to Adriamycin (n = 10 each) via using RT-qPCR. f Kaplan–Meyer survival curve showed significantly higher overall survival in patients with lower ANCR expression (median survival 4.56 years) than those with higher ANCR expression (median survival 2.26 years; P = 0.0071)
(Reprinted with permission from Ref [95] Copyright © 2022, Frontiers in oncology, Xin Hu et al.)
Exosomes as potential delivery vehicles of therapeutic agents
The critical functions of exosomes in sarcoma development demonstrate their potential as therapeutic vectors. Exosomes, with phospholipid bilayers and specific ligands, communicate with targeted cells in TME and directly fuse with their cytomembrane, thus increasing the internalization of the encapsulated drugs. An exosome-based delivery system is expected to improve efficacy, reduce side effects, and prolong the half-life of cancer drugs (Table 4).
Table 4.
Studies on exosomes as potential delivery vectors in sarcoma treatment
| Sarcoma type | Target cells | Exosome source | Exosomal cargos | Loading methods | Mechanisms | Loading efficiency | References |
|---|---|---|---|---|---|---|---|
| OS | Sarcoma cells | MSCs | Doxorubicin | Co-incubation | Nucleic acid synthesis ↓ | 12% | [98] |
| OS | Sarcoma cells | MSCs | miR-143 | Lipofection | Cell migration ↓ | Not mentioned | [99] |
| OS | Sarcoma cells | OS cells | lncRNA MEG3 | Lipofection | Sponging miR-185-5p | Not mentioned | [100] |
| FS | Immune cells | Macrophages | HSP70 | Incubation at 42℃ | Immune response ↑ | Not mentioned | [101] |
| OS | Sarcoma cells | MSCs | miR-101 | Lentiviral transduction | Cell migration ↓ | About 48% | [108] |
| OS | Sarcoma cells | OS cells | miR-371b-5p | Co-incubation | Cell proliferation and migration ↓ | Not mentioned | [109] |
OS, osteosarcoma; MSCs, mesenchymal stem cells; miR, microRNA; lncRNA, long non-coding RNA; FS, fibrosarcoma; HSP70, heat shock protein 70
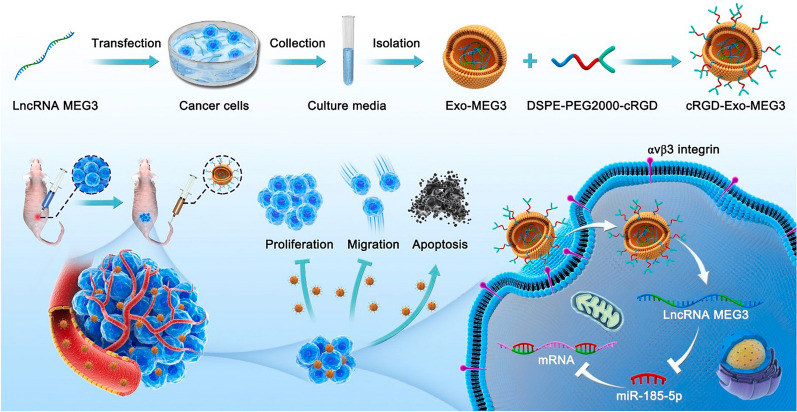
Bioactive molecules can be loaded into exosomes by directly modifying isolated exosomes or indirectly manipulating producer cells. The exosomes extracted from MSCs were loaded with doxorubicin by co-incubation with doxorubicin-HCl [98]. Doxorubicin enveloped in the exosomes showed enhanced cellular uptake efficiency and tumor-killing efficacy with lower cytotoxicity to normal cells than free drug [98]. The producer MSCs could also be engineered via transfection of miR-143 intracellularly to release miR-143-loaded exosomes [99]. The miR-143-loaded exosomes significantly reduced the migratory potential of sarcoma cells [99]. Similarly, the modified tumor cells produced lncRNA MEG3-loaded exosomes [100]. The isolated exosomes were further co-incubated with micelles containing cRGDyK to prepare cRGD-conjugated exosomes for enhanced tumor-targeting [100]. The cRGD peptide can specifically bind to αv β3 integrin, which is up-regulated on cancer cells and activated endothelial cells of growing vessels (Fig. 9). In addition to acting as drug vehicles, exosomes can also assist immunization therapy. The TAMs-derived exosomes enriched in heat shock protein 70 were a viable immunoadjuvant in sarcoma immunotherapy [101]. The number of sarcoma cells notably decreased after vaccinating HSP70-loaded exosomes [101]. Exosomal membrane-bound Staphylococcal enterotoxin B had a synergistic effect with HSP70, leading to the activation of T cells and secretion of cytokines [101].
Fig. 9.
Schematic diagram of the synthesis of cRGD-Exo-MEG3 and its application as a targeted lncRNA MEG3 delivery vectors for osteosarcoma treatment
(Reprinted with permission from Ref. [100] Copyright © 2022, Journal of Controlled Release, Xin Huang et al.)
To apply exosomes as delivery vectors, it is necessary to understand the distribution mode and location of exogenously administered exosomes in vivo. The administered exosomes mainly accumulated in the liver, spleen, lung, gastrointestinal tract, and bone marrow [102]. The surface of exosomes can be modified with specific molecules that can selectively bind to sarcoma cells for better accumulation at the tumor site. B16-BL6 exosomes were promptly cleared from the circulation after systemic administration with a circulating half-life of 2 min, and little could be detected in the serum at 4 h after injection [103]. From the pharmacokinetic profile of exosomes, intravenously administered exosomes had a half-life of 2–30 min [104]. Macrophage-mediated phagocytosis was responsible for the rapid clearance, which strongly reduced the number of available particles and limited exosome accumulation in the lesions. Engineering exosomes with antiphagocytic molecules could evade immune recognition and thus increase the circulating half-life and bioavailability of exosomes. The candidate molecules comprised CD47, CD24, CD31, β2M, and PD-L1 [105]. For instance, CD47 interacts with the immune inhibitory receptor SIRPα and can neutralize detection by phagocytic cells. The CD47-SIRPα axis inhibited macrophage activation and activated the ‘don’t eat me’ signal [105]. The exosomes with high expression of CD47 could be highly detected for a longer time. CD47 was generally up-regulated in exosomes derived from MSCs, making them suitable production cells [105].
Conclusion and future direction
Significant evidence supports that TME plays critical roles in the initiation and progression of sarcoma. Exosomes are key components of the TME and act as messengers of intercellular communications. This review brings together information on the roles of exosomes in the bidirectional crosstalk between sarcoma cells and TME cells, as well as the clinical value of exosomes. Although existing studies cannot fully elaborate the functions of exosomes, these studies highlight the increasing significance and potential applications of exosomes in sarcoma microenvironment.
However, some critical problems remain unsolved. The term, exosomes, has been widely applied to various kinds of extracellular vesicles, muddying the field and causing the research to be sometimes treated with skepticism. Exosomes can be defined as a subtype of extracellular vesicles, which are released from cells upon fusion of the multivesicular body with the plasma membrane. But this definition is not helpful for the purity and uniformity assurance of exosomes. The heterogeneity of exosomes impedes the identification of exosomes and induces intricate biological reactions, which hinders a comprehensive understanding of their biogenesis, contents, biodistribution, and roles in the TME. The size and amount of biophysically similar extracellular vesicles also make exosomes difficult to be obtained as relatively pure preparations and to characterize properly [106]. Ascribing the exquisite and specific functions of exosomes requires concrete information reporting beyond the mere description of activities in a potentially contaminated and heterogeneous preparation [106]. Many techniques have been developed to extract exosomes, such as ultracentrifugation, tangential flow filtration, ultrafiltration devices, and size exclusion chromatography. Some isolation methods may degrade the structural integrity and functionality of exosomes. There is still no established exosome isolation method as a gold standard. These factors are the obstacles in the exploration of exosome roles in tumor development and the search for specific clinical indicators. Fundamental research should be conducted to reveal the unique characteristics of exosomes to distinguish them from other extracellular vesicles and identify specific exosome populations.
In terms of exosome therapeutic applications in sarcoma, the primary consideration is the cell source that will be used as producer cells, including dendritic cells, MSCs, and patient-derived tumor cells. MSCs may be a major candidate, because MSC-derived exosomes with high CD47 expression have a long half-life. Therapeutic exosomes might also be extracted from different plants, such as ginger, grapes, and lemons [107]. However, the immunotoxicity of exosomes needs to be taken as important. Exosomes extracted from allogenic or heterologous cell sources may elicit immune responses. Although patients injected with trillions of exosomes via blood transfusion do not show immune-related toxicity, the immunogenicity of exosomes developed for sarcoma therapy needs to be thoroughly evaluated. A third limitation in exosome-based treatments is the heavy workload while unsatisfactory yield during exosome production, as well as low loading and delivery efficiency. The clinical breakthrough may depend on the advances in exosome-mimetics and genetic engineering.
These understandings of exosomes in TME of sarcoma may be enhanced through the following aspects: (1) intensive exploration of biological properties of exosomes; (2) searching for more effective methods in exosome detection and analysis; (3) comprehension of the dynamic cellular interplay in TME; (4) discovery of available biomarkers and vehicles with targetability; (5) exploitation of valid approaches to load exosomes with markers and agents; (6) optimization of the target delivery mechanisms of exosomes; (7) conducting clinical trials to confirm existing hypotheses.
Acknowledgements
Not applicable.
Author contributions
HY wrote the text, screened potentially eligible studies, extracted and analyzing data, updated reference lists. XH updated reference lists, extracted and analyzing data, drawn and modified figures, and revised the final manuscript. YW extracted and analyzed data, drawn figures, interpreted results and creating ‘Summary of findings’ tables. CT and FH revised the manuscript, and designed the review. ZD and LM conceived and designed the review, arbitrated potentially eligible studies, and revised the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to publication.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huali Ye and Xin Hu contributed equally as co-first authors
Contributor Information
Zhenfeng Duan, Email: zxd221@med.miami.edu.
Li Min, Email: minli1204@scu.edu.cn.
References
- 1.Zhu MMT, Shenasa E, Nielsen TO. Sarcomas: immune biomarker expression and checkpoint inhibitor trials. Cancer Treat Rev. 2020;91:102115. doi: 10.1016/j.ctrv.2020.102115. [DOI] [PubMed] [Google Scholar]
- 2.Molina ER, Chim LK, Barrios S, Ludwig JA, Mikos AG. Modeling the tumor microenvironment and pathogenic signaling in bone sarcoma. Tissue Eng Part B Rev. 2020;26:249–271. doi: 10.1089/ten.teb.2019.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gronchi A, Miah AB, Dei Tos AP, Abecassis N, Bajpai J, Bauer S, et al. Soft tissue and visceral sarcomas: ESMO-EURACAN-GENTURIS clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2021;32:1348–1365. doi: 10.1016/j.annonc.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Strauss SJ, Frezza AM, Abecassis N, Bajpai J, Bauer S, Biagini R, et al. Bone sarcomas: ESMO-EURACAN-GENTURIS-ERN PaedCan Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2021;32:1520–1536. doi: 10.1016/j.annonc.2021.08.1995. [DOI] [PubMed] [Google Scholar]
- 5.Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18:130. doi: 10.1186/s12943-019-1047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasprzak A. The role of tumor microenvironment cells in colorectal cancer (CRC) Cachexia. Int J Mol Sci. 2021;22:1565. doi: 10.3390/ijms22041565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farajzadeh VS, Keshavarz-Fathi M, Silvestris N, Argentiero A, Rezaei N. The role of inflammatory cytokines and tumor associated macrophages (TAMs) in microenvironment of pancreatic cancer. Cytokine Growth Factor Rev. 2018;39:46–61. doi: 10.1016/j.cytogfr.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Nascimento C, Ferreira F. Tumor microenvironment of human breast cancer, and feline mammary carcinoma as a potential study model. Biochim Biophys Acta Rev Cancer. 2021;1876:188587. doi: 10.1016/j.bbcan.2021.188587. [DOI] [PubMed] [Google Scholar]
- 9.Li M, Liu L, Dong M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer. 2021;20:22. doi: 10.1186/s12943-021-01312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye H, He X, Feng X. Developing neobavaisoflavone nanoemulsion suppresses lung cancer progression by regulating tumor microenvironment. Biomed Pharmacother. 2020;129:110369. doi: 10.1016/j.biopha.2020.110369. [DOI] [PubMed] [Google Scholar]
- 11.Berger-Achituv S, Brinkmann V, Abed UA, Kuhn LI, Ben-Ezra J, Elhasid R, et al. A proposed role for neutrophil extracellular traps in cancer immunoediting. Front Immunol. 2013;4:48. doi: 10.3389/fimmu.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mcewen A, Emmanuel C, Medbury H, Leick A, Walker DM, Zoellner H. Induction of contact-dependent endothelial apoptosis by osteosarcoma cells suggests a role for endothelial cell apoptosis in blood-borne metastasis. J Pathol. 2003;201:395–403. doi: 10.1002/path.1457. [DOI] [PubMed] [Google Scholar]
- 13.Curino A, Mitola DJ, Aaronson H, Mcmahon GA, Raja K, Keegan AD, et al. Plasminogen promotes sarcoma growth and suppresses the accumulation of tumor-infiltrating macrophages. Oncogene. 2002;21:8830–8842. doi: 10.1038/sj.onc.1205951. [DOI] [PubMed] [Google Scholar]
- 14.Zhong L, Liao D, Li J, Liu W, Wang J, Zeng C, et al. Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct Target Ther. 2021;6:59. doi: 10.1038/s41392-020-00414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jablonska J, Wu CF, Andzinski L, Leschner S, Weiss S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-β. Int J Cancer. 2014;134:1346–1358. doi: 10.1002/ijc.28551. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhao C, Li Y, Wen J, Wang S, Wang D, et al. Osteosarcoma exocytosis of soluble LGALS3BP mediates macrophages toward a tumoricidal phenotype. Cancer Lett. 2022;528:1–15. doi: 10.1016/j.canlet.2021.12.023. [DOI] [PubMed] [Google Scholar]
- 17.Liang X, Guo W, Ren T, Huang Y, Sun K, Zhang H, et al. Macrophages reduce the sensitivity of osteosarcoma to neoadjuvant chemotherapy drugs by secreting Interleukin-1 beta. Cancer Lett. 2020;480:4–14. doi: 10.1016/j.canlet.2020.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomed. 2020;15:6917–6934. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min L, Shen J, Tu C, Hornicek F, Duan Z. The roles and implications of exosomes in sarcoma. Cancer Metastasis Rev. 2016;35:377–390. doi: 10.1007/s10555-016-9630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juan T, Furthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. 2018;74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 23.Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei X, Li Z, Yang D, Kong X, Lu X, Shen Y, et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J Extracell Vesicles. 2021;10:e12153. doi: 10.1002/jev2.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(19–30):1–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 26.Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. 2014;289:22258–22267. doi: 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kling MJ, Chaturvedi NK, Kesherwani V, Coulter DW, Mcguire TR, Sharp JG, et al. Exosomes secreted under hypoxia enhance stemness in Ewing's sarcoma through miR-210 delivery. Oncotarget. 2020;11:3633–3645. doi: 10.18632/oncotarget.27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang M, Chen X, Wang L, Qin L, Wang H, Sun Z, et al. Cancer-derived exosomal TRIM59 regulates macrophage NLRP3 inflammasome activation to promote lung cancer progression. J Exp Clin Cancer Res. 2020;39:176. doi: 10.1186/s13046-020-01688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Zhang K, Zhi Y, Wu Y, Chen B, Bai J, et al. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin Transl Med. 2021;11:e478. doi: 10.1002/ctm2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, et al. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020;27:3258–3272. doi: 10.1038/s41418-020-0575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong L, Pu Y, Zhang L, Qi Q, Xu L, Li W, et al. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles promote lung adenocarcinoma growth by transferring miR-410. Cell Death Dis. 2018;9:218. doi: 10.1038/s41419-018-0323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia Y, Ding X, Zhou L, Zhang L, Yang X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene. 2021;40:246–261. doi: 10.1038/s41388-020-01486-7. [DOI] [PubMed] [Google Scholar]
- 34.Mao Y, Wang Y, Dong L, Zhang Y, Zhang Y, Wang C, et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res. 2019;38:389. doi: 10.1186/s13046-019-1384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shan G, Gu J, Zhou D, Li L, Cheng W, Wang Y, et al. Cancer-associated fibroblast-secreted exosomal miR-423-5p promotes chemotherapy resistance in prostate cancer by targeting GREM2 through the TGF-β signaling pathway. Exp Mol Med. 2020;52:1809–1822. doi: 10.1038/s12276-020-0431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Li J, Gu P, Fan X. The application of nanoparticles in cancer immunotherapy: targeting tumor microenvironment. Bioact Mater. 2021;6:1973–1987. doi: 10.1016/j.bioactmat.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou L, Wang W, Wang F, Yang S, Hu J, Lu B, et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol Cancer. 2021;20:57. doi: 10.1186/s12943-021-01352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nie H, Xie X, Zhang D, Zhou Y, Li B, Li F, et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale. 2020;12:877–887. doi: 10.1039/C9NR09011H. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Z, Wang L, Wu C, Huang L, Ruan Y, Xue W. Tumor-derived exosomes induced M2 macrophage polarization and promoted the metastasis of osteosarcoma cells through Tim-3. Arch Med Res. 2021;52:200–210. doi: 10.1016/j.arcmed.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 41.Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 2017;77:3846–3856. doi: 10.1158/0008-5472.CAN-16-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raimondi L, De Luca A, Gallo A, Costa V, Russelli G, Cuscino N, et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis. 2020;41:666–677. doi: 10.1093/carcin/bgz130. [DOI] [PubMed] [Google Scholar]
- 43.Ucci A, Cappariello A, Ponzetti M, Tennant F, Loftus AEP, Shefferd K, et al. Anti-osteoblastogenic, pro-inflammatory and pro-angiogenic effect of extracellular vesicles isolated from the human osteosarcoma cell line MNNG/HOS. Bone. 2021;153:116130. doi: 10.1016/j.bone.2021.116130. [DOI] [PubMed] [Google Scholar]
- 44.Garimella R, Washington L, Isaacson J, Vallejo J, Spence M, Tawfik O, et al. Extracellular membrane vesicles derived from 143B osteosarcoma cells contain pro-osteoclastogenic cargo: a novel communication mechanism in osteosarcoma bone microenvironment. Transl Oncol. 2014;7:331–340. doi: 10.1016/j.tranon.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin L, Wang H, Guo W, He E, Huang K, Zhao Q. Osteosarcoma-derived exosomal miR-501-3p promotes osteoclastogenesis and aggravates bone loss. Cell Signal. 2021;82:109935. doi: 10.1016/j.cellsig.2021.109935. [DOI] [PubMed] [Google Scholar]
- 46.Araki Y, Aiba H, Yoshida T, Yamamoto N, Hayashi K, Takeuchi A, et al. Osteosarcoma-derived small extracellular vesicles enhance tumor metastasis and suppress osteoclastogenesis by miR-146a-5p. Front Oncol. 2021;11:667109. doi: 10.3389/fonc.2021.667109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park JE, Dutta B, Tse SW, Gupta N, Tan CF, Low JK, et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene. 2019;38:5158–5173. doi: 10.1038/s41388-019-0782-x. [DOI] [PubMed] [Google Scholar]
- 48.Yakupova EI, Maleev GV, Krivtsov AV, Plotnikov EY. Macrophage polarization in hypoxia and ischemia/reperfusion: insights into the role of energetic metabolism. Exp Biol Med. 2022;247:958–971. doi: 10.1177/15353702221080130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luong N, Lenz JA, Modiano JF, Olson JK. Extracellular vesicles secreted by tumor cells promote the generation of suppressive monocytes. ImmunoHorizons. 2021;5:647–658. doi: 10.4049/immunohorizons.2000017. [DOI] [PubMed] [Google Scholar]
- 50.Wolf-Dennen K, Gordon N, Kleinerman ES. Exosomal communication by metastatic osteosarcoma cells modulates alveolar macrophages to an M2 tumor-promoting phenotype and inhibits tumoricidal functions. Oncoimmunology. 2020;9:1747677. doi: 10.1080/2162402X.2020.1747677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K, An J, Yang S, Park S, Lee J, Chae H, et al. Influence of canine macrophage-derived extracellular vesicles on apoptosis in canine melanoma and osteosarcoma cell lines. Anticancer Res. 2021;41:719–730. doi: 10.21873/anticanres.14823. [DOI] [PubMed] [Google Scholar]
- 52.Liu W, Long Q, Zhang W, Zeng D, Hu B, Liu S, et al. miRNA-221-3p derived from M2-polarized tumor-associated macrophage exosomes aggravates the growth and metastasis of osteosarcoma through SOCS3/JAK2/STAT3 axis. Aging. 2021;13:19760–19775. doi: 10.18632/aging.203388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Yu Y, Wang J, Han Y, Ren T, Huang Y, et al. Macrophages-derived exosomal lncRNA LIFR-AS1 promotes osteosarcoma cell progression via miR-29a/NFIA axis. Cancer Cell Int. 2021;21:192. doi: 10.1186/s12935-021-01893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong J, Wu L, Huang L, Wu C, Liu Z, Deng W, et al. LncRNA FOXP4-AS1 promotes progression of ewing sarcoma and is associated with immune infiltrates. Front Oncol. 2021;11:718876. doi: 10.3389/fonc.2021.718876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gassmann H, Schneider K, Evdokimova V, Ruzanov P, Schober SJ, Xue B, et al. Ewing sarcoma-derived extracellular vesicles impair dendritic cell maturation and function. Cells-Basel. 2021;10:2081. doi: 10.3390/cells10082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazumdar A, Urdinez J, Boro A, Arlt MJE, Egli FE, Niederöst B, et al. Exploring the role of osteosarcoma-derived extracellular vesicles in pre-metastatic niche formation and metastasis in the 143-B xenograft mouse osteosarcoma model. Cancers. 2020;12:3457. doi: 10.3390/cancers12113457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortini M, Avnet S, Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. doi: 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 58.Lagerweij T, Pérez-Lanzón M, Baglio SR. A preclinical mouse model of osteosarcoma to define the extracellular vesicle-mediated communication between tumor and mesenchymal stem cells. J Vis Exp. 2018;135:e56932. doi: 10.3791/56932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mannerström B, Kornilov R, Abu-Shahba AG, Chowdhury IM, Sinha S, Seppänen-Kaijansinkko R, et al. Epigenetic alterations in mesenchymal stem cells by osteosarcoma-derived extracellular vesicles. Epigenetics-Us. 2019;14:352–364. doi: 10.1080/15592294.2019.1585177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallabhaneni KC, Hassler M, Abraham A, Whitt J, Mo Y, Atfi A, et al. Mesenchymal stem/stromal cells under stress increase osteosarcoma migration and apoptosis resistance via extracellular vesicle mediated communication. PLoS ONE. 2016;11:e166027. doi: 10.1371/journal.pone.0166027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi J, Zhang R, Wang Y. Exosomal miR-21-5p derived from bone marrow mesenchymal stem cells promote osteosarcoma cell proliferation and invasion by targeting PIK3R1. J Cell Mol Med. 2021;25:11016–11030. doi: 10.1111/jcmm.17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Chu Y, Li K, Zhang G, Guo Z, Wu X, et al. Exosomes secreted by adipose-derived mesenchymal stem cells foster metastasis and osteosarcoma proliferation by increasing COLGALT2 expression. Front Cell Dev Biol. 2020;8:253. doi: 10.3389/fcell.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi J, Zhou Y, Jiao Z, Wang X, Zhao Y, Li Y, et al. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth through hedgehog signaling pathway. Cell Physiol Biochem. 2017;42:2242–2254. doi: 10.1159/000479998. [DOI] [PubMed] [Google Scholar]
- 64.Qin F, Tang H, Zhang Y, Zhang Z, Huang P, Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J Cell Physiol. 2020;235:4734–4745. doi: 10.1002/jcp.29351. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y, Nguyen PNN, Ma H, Ho W, Chen Y, Chien Y, et al. Tumor mesenchymal stromal cells regulate cell migration of atypical teratoid rhabdoid tumor through exosome-mediated miR155/SMARCA4 pathway. Cancers. 2019;11:720. doi: 10.3390/cancers11050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao W, Qin P, Zhang D, Cui X, Gao J, Yu Z, et al. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183–5p. Aging (Albany, NY) 2019;11:9581–9596. doi: 10.18632/aging.102406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morita T, Fujiwara T, Yoshida A, Uotani K, Kiyono M, Yokoo S, et al. Clinical relevance and functional significance of cell-free microRNA-1260b expression profiles in infiltrative myxofibrosarcoma. Sci Rep-Uk. 2020;10:9414. doi: 10.1038/s41598-020-66120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urciuoli E, Giorda E, Scarsella M, Petrini S, Peruzzi B. Osteosarcoma-derived extracellular vesicles induce a tumor-like phenotype in normal recipient cells. J Cell Physiol. 2018;233:6158–6172. doi: 10.1002/jcp.26464. [DOI] [PubMed] [Google Scholar]
- 69.Endo-Munoz L, Cai N, Cumming A, Macklin R, Merida De Long L, Topkas E, et al. Progression of osteosarcoma from a non-metastatic to a metastatic phenotype is causally associated with activation of an autocrine and paracrine uPA axis. PLoS ONE. 2015;10:e133592. doi: 10.1371/journal.pone.0133592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazumdar A, Urdinez J, Boro A, Migliavacca J, Arlt MJE, Muff R, et al. Osteosarcoma-derived extracellular vesicles induce lung fibroblast reprogramming. Int J Mol Sci. 2020;21:5451. doi: 10.3390/ijms21155451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao A, Zhao Z, Liu W, Cui X, Wang N, Wang Y, et al. Carcinoma-associated fibroblasts promote the proliferation and metastasis of osteosarcoma by transferring exosomal LncRNA SNHG17. Am J Transl Res. 2021;13:10094–10111. [PMC free article] [PubMed] [Google Scholar]
- 72.Meng W, Hao Y, He C, Li L, Zhu G. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer. 2019;18:57. doi: 10.1186/s12943-019-0982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao SC, Huang JY, Wei ZY, Li ZX, Guo SC. EWSAT1 acts in concert with exosomes in osteosarcoma progression and tumor-induced angiogenesis: the "double stacking effect". Adv Biosyst. 2020;4:2000152. doi: 10.1002/adbi.202000152. [DOI] [PubMed] [Google Scholar]
- 74.Colletti M, Paolini A, Galardi A, Paolo VD, Pascucci L, Russo I, et al. Expression profiles of exosomal miRNAs isolated from plasma of patients with desmoplastic small round cell tumor. Epigenomics-Uk. 2019;11:489–500. doi: 10.2217/epi-2018-0179. [DOI] [PubMed] [Google Scholar]
- 75.Avnet S, Lemma S, Cortini M, Di Pompo G, Perut F, Lipreri MV, et al. The release of inflammatory mediators from acid-stimulated mesenchymal stromal cells favours tumour invasiveness and metastasis in osteosarcoma. Cancers. 2021;13:5855. doi: 10.3390/cancers13225855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8:33375–33392. doi: 10.18632/oncotarget.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye Z, Zheng Z, Peng L. MicroRNA profiling of serum exosomes in patients with osteosarcoma by high-throughput sequencing. J Invest Med. 2020;68:893–901. doi: 10.1136/jim-2019-001196. [DOI] [PubMed] [Google Scholar]
- 78.Cuscino N, Raimondi L, De Luca A, Carcione C, Russelli G, Conti L, et al. Gathering novel circulating exosomal microRNA in osteosarcoma cell lines and possible implications for the disease. Cancers (Basel) 2019;11:1924. doi: 10.3390/cancers11121924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohama I, Asano N, Matsuzaki J, Yamamoto Y, Yamamoto T, Takahashi RU, et al. Comprehensive serum and tissue microRNA profiling in dedifferentiated liposarcoma. Oncol Lett. 2021;22:623. doi: 10.3892/ol.2021.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M, et al. Circulating microRNA-92b-3p as a novel biomarker for monitoring of synovial sarcoma. Sci Rep. 2017;7:14634. doi: 10.1038/s41598-017-12660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaoka B, Nagasaki-Maeoka E, Uekusa S, Hidaka A, Hijikata K, Kato R, et al. Exosomal miR-214-3p as a potential novel biomarker for rhabdoid tumor of the kidney. Pediatr Surg Int. 2021;37:1783–1790. doi: 10.1007/s00383-021-04989-w. [DOI] [PubMed] [Google Scholar]
- 82.Jerez S, Araya H, Thaler R, Charlesworth MC, López-Solís R, Kalergis AM, et al. Proteomic analysis of exosomes and exosome-free conditioned media from human osteosarcoma cell lines reveals secretion of proteins related to tumor progression. J Cell Biochem. 2017;118:351–360. doi: 10.1002/jcb.25642. [DOI] [PubMed] [Google Scholar]
- 83.Brady JV, Troyer RM, Ramsey SA, Leeper H, Yang L, Maier CS, et al. A preliminary proteomic investigation of circulating exosomes and discovery of biomarkers associated with the progression of osteosarcoma in a clinical model of spontaneous disease. Transl Oncol. 2018;11:1137–1146. doi: 10.1016/j.tranon.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samuel G, Crow J, Klein JB, Merchant ML, Nissen E, Koestler DC, et al. Ewing sarcoma family of tumors-derived small extracellular vesicle proteomics identify potential clinical biomarkers. Oncotarget. 2020;11:2995–3012. doi: 10.18632/oncotarget.27678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Atay S, Wilkey DW, Milhem M, Merchant M, Godwin AK. Insights into the proteome of gastrointestinal stromal tumors-derived exosomes reveals new potential diagnostic biomarkers. Mol Cell Proteomics. 2018;17:495–515. doi: 10.1074/mcp.RA117.000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang J, Zhang H, Sun X, Wang X, Ren T, Huang Y, et al. Exosomal PD-L1 and N-cadherin predict pulmonary metastasis progression for osteosarcoma patients. J Nanobiotechnol. 2020;18:151. doi: 10.1186/s12951-020-00710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han Z, Peng C, Yi J, Wang Y, Liu Q, Yang Y, et al. Matrix-assisted laser desorption ionization mass spectrometry profiling of plasma exosomes evaluates osteosarcoma metastasis. iScience. 2021;24:102906. doi: 10.1016/j.isci.2021.102906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han Z, Yi J, Yang Y, Li D, Peng C, Long S, et al. SERS and MALDI-TOF MS based plasma exosome profiling for rapid detection of osteosarcoma. Analyst. 2021;146:6496–6505. doi: 10.1039/D1AN01163D. [DOI] [PubMed] [Google Scholar]
- 89.Stegmaier S, Sparber-Sauer M, Aakcha-Rudel E, Munch P, Reeh T, Feuchtgruber S, et al. Fusion transcripts as liquid biopsy markers in alveolar rhabdomyosarcoma and synovial sarcoma: a report of the Cooperative Weichteilsarkom Studiengruppe (CWS) Pediatr Blood Cancer. 2022;69:e29652. doi: 10.1002/pbc.29652. [DOI] [PubMed] [Google Scholar]
- 90.Zhang H, Wang J, Ren T, Huang Y, Yu Y, Chen C, et al. LncRNA CASC15 is upregulated in osteosarcoma plasma exosomes and CASC15 knockdown inhibits osteosarcoma progression by regulating miR-338-3p/RAB14 axis. Onco Targets Ther. 2020;13:12055–12066. doi: 10.2147/OTT.S282053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan Y, Lin Y, Mi C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol Int. 2021;45:858–868. doi: 10.1002/cbin.11532. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, Wu J, Song S, Chen H, Hu Y, Xu B, et al. Plasma exosome-derived sentrin SUMO-specific protease 1: a prognostic biomarker in patients with osteosarcoma. Front Oncol. 2021;11:625109. doi: 10.3389/fonc.2021.625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshida A, Fujiwara T, Uotani K, Morita T, Kiyono M, Yokoo S, et al. Clinical and functional significance of intracellular and extracellular microRNA-25-3p in osteosarcoma. Acta Med Okayama. 2018;72:165–174. doi: 10.18926/AMO/55857. [DOI] [PubMed] [Google Scholar]
- 94.Weinman MA, Ramsey SA, Leeper HJ, Brady JV, Schlueter A, Stanisheuski S, et al. Exosomal proteomic signatures correlate with drug resistance and carboplatin treatment outcome in a spontaneous model of canine osteosarcoma. Cancer Cell Int. 2021;21:245. doi: 10.1186/s12935-021-01943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu X, Wen Y, Tan LY, Wang J, Tang F, Wang YT, et al. Exosomal long non-coding RNA ANCR mediates drug resistance in osteosarcoma. Front Oncol. 2021;11:735254. doi: 10.3389/fonc.2021.735254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torreggiani E, Roncuzzi L, Perut F, Zini N, Baldini N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int J Oncol. 2016;49:189–196. doi: 10.3892/ijo.2016.3509. [DOI] [PubMed] [Google Scholar]
- 97.Shiozawa K, Shuting J, Yoshioka Y, Ochiya T, Kondo T. Extracellular vesicle-encapsulated microRNA-761 enhances pazopanib resistance in synovial sarcoma. Biochem Biophys Res Commun. 2018;495:1322–1327. doi: 10.1016/j.bbrc.2017.11.164. [DOI] [PubMed] [Google Scholar]
- 98.Wei H, Chen J, Wang S, Fu F, Zhu X, Wu C, et al. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int J Nanomed. 2019;14:8603–8610. doi: 10.2147/IJN.S218988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shimbo K, Miyaki S, Ishitobi H, Kato Y, Kubo T, Shimose S, et al. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem Biophys Res Commun. 2014;445:381–387. doi: 10.1016/j.bbrc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 100.Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, et al. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J Control Release. 2022;343:107–117. doi: 10.1016/j.jconrel.2022.01.026. [DOI] [PubMed] [Google Scholar]
- 101.Behzadi E, Hosseini HM, Halabian R, Fooladi A. Macrophage cell-derived exosomes/staphylococcal enterotoxin B against fibrosarcoma tumor. Microb Pathog. 2017;111:132–138. doi: 10.1016/j.micpath.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 102.Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–155. doi: 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T, et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165:77–84. doi: 10.1016/j.jbiotec.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 104.Yang Z, Shi J, Xie J, Wang Y, Sun J, Liu T, et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat Biomed Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parada N, Romero-Trujillo A, Georges N, Alcayaga-Miranda F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J Adv Res. 2021;31:61–74. doi: 10.1016/j.jare.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen YS, Lin EY, Chiou TW, Harn HJ. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Ci Ji Yi Xue Za Zhi. 2020;32:113–120. doi: 10.4103/tcmj.tcmj_182_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang K, Dong C, Chen M, Yang T, Wang X, Gao Y, et al. Extracellular vesicle-mediated delivery of miR-101 inhibits lung metastasis in osteosarcoma. Theranostics. 2020;10:411–425. doi: 10.7150/thno.33482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xue Q, Yang Y, Yang L, Yan X, Shen Z, Liu J, et al. miR-371b-5p-engineered exosomes enhances tumor inhibitory effect. Front Cell Dev Biol. 2021;9:750171. doi: 10.3389/fcell.2021.750171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.