Abstract
Background
The human mouth is a natural laboratory for studying how bacterial communities differ across habitats. Different bacteria colonize different surfaces in the mouth—teeth, tongue dorsum, and keratinized and non-keratinized epithelia—despite the short physical distance between these habitats and their connection through saliva. We sought to determine whether more tightly defined microhabitats might have more tightly defined sets of resident bacteria. A microhabitat may be characterized, for example, as the space adjacent to a particular species of bacterium. Corncob structures of dental plaque, consisting of coccoid bacteria bound to filaments of Corynebacterium cells, present an opportunity to analyze the community structure of one such well-defined microhabitat within a complex natural biofilm. Here, we investigate by fluorescence in situ hybridization and spectral imaging the composition of the cocci decorating the filaments.
Results
The range of taxa observed in corncobs was limited to a small subset of the taxa present in dental plaque. Among four major groups of dental plaque streptococci, two were the major constituents of corncobs, including one that was the most abundant Streptococcus species in corncobs despite being relatively rare in dental plaque overall. Images showed both Streptococcus types in corncobs in all individual donors, suggesting that the taxa have different ecological roles or that mechanisms exist for stabilizing the persistence of functionally redundant taxa in the population. Direct taxon-taxon interactions were observed not only between the Streptococcus cells and the central corncob filament but also between Streptococcus cells and the limited subset of other plaque bacteria detected in the corncobs, indicating species ensembles involving these taxa as well.
Conclusions
The spatial organization we observed in corncobs suggests that each of the microbial participants can interact with multiple, albeit limited, potential partners, a feature that may encourage the long-term stability of the community. Additionally, our results suggest the general principle that a precisely defined microhabitat will be inhabited by a small and well-defined set of microbial taxa. Thus, our results are important for understanding the structure and organizing principles of natural biofilms and lay the groundwork for future work to modulate and control biofilms for human health.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-022-01323-x.
Keywords: Oral microbiome, Biogeography, Fluorescence in situ hybridization, FISH, Microscopy, Imaging, Corncob, Hedgehog, Microbial ecology
Background
Microbial community complexity in the human mouth depends on the scale on which it is assessed. The mouth as a whole has some 700 resident microbial taxa [1–3]. This large set of microbes is subdivided into smaller sets specialized for the different habitats within the mouth, such as dental plaque, tongue dorsum, and buccal mucosa [4–9]. Some of the microbes within dental plaque are specialized for subgingival rather than supragingival habitats [8, 10]; others are rare in healthy plaque but abundant in disease states such as caries or periodontal disease [11]. Investigating spatial organization at sub-millimeter scales using imaging, we have discovered organized consortia tens to hundreds of micrometers in diameter both in supragingival dental plaque [12] and on the tongue dorsum [13]. Each of these consortia contained a subset of the taxa that were found at the site overall. These findings raise the question: is it possible that the apparent enormous complexity of microbial communities is a consequence of combining many distinct habitats in a single sample, and that the more precisely a habitat can be defined, the smaller the number of microbes that grow there?
The habitat for a microbe is defined in large part by the other microbes located within a radius of a few micrometers to tens of micrometers. Short-range interactions between taxa shape the physiology of individual microbes and of microbial communities as a whole. Microbes exude metabolites that stimulate or inhibit growth of neighboring microbes [14–16] or cause them to alter their metabolism [16–18]. These interactions are strongest at distances of only a few micrometers, particularly in situations where fluid flow can rapidly attenuate the concentration of a metabolite [19] or within dense aggregations of microbes in which the distance over which a metabolite is available depends on the rates at which it is secreted and taken up by neighboring microbes [20]. Microbial surfaces also present binding sites to which other microbes may adhere and which thereby enable direct taxon-taxon interaction [21] and permit the localization of a microbe into a favorable habitat. For these reasons, the local neighborhood and nearest-neighbor relationships of a microbe play a major role in defining its habitat.
Corncob structures of dental plaque present an opportunity to analyze a well-defined microhabitat within the full complexity of a natural microbial community. In an otherwise amorphous mass of plaque bacteria, corncobs are discrete, readily recognizable structures characterized by direct physical interaction between filaments and cocci, as shown first by light microscopy [22, 23] and subsequently by electron microscopy [24–26]. Microdissection of corncobs followed by cultivation [27] identified the filament as C. matruchotii and the cocci as Streptococcus sanguis (subsequently renamed S. crista [28], then S. cristatus [29]). The potential involvement of additional partners was suggested by reconstruction experiments showing that S. sanguis cocci could associate with Fusobacterium nucleatum to form corncob-like structures in vitro [30]. However, the relationship of these corncob-like, cocci-filament associations to the structures previously identified as corncobs in dental plaque was not established. Our imaging confirmed the presence in natural plaque of a filamentous Corynebacterium core decorated by cocci of genus Streptococcus but also revealed additional participants in these corncobs, including members of the genus Porphyromonas and the family Pasteurellaceae [12]. These observations suggest that the species composition of corncobs is simple enough to be tractable but complex enough to offer insight into the rules governing community assembly within a natural microbiome.
To investigate the degree of selectivity of the corncob microhabitat, and the site-specificity of its component taxa, we focused on the healthy human mouth and on species of the genus Streptococcus. Among the genera of oral bacteria, Streptococcus stands out for its high abundance throughout the mouth, with multiple species that are abundant and prevalent in healthy dental plaque. Here, we investigate whether corncob structures in dental plaque represent a species ensemble involving a single species of Streptococcus, or whether more than one Streptococcus species can associate with Corynebacterium and with the other cocci in corncobs. Our results indicate that the corncob microhabitat can be occupied apparently interchangeably by more than one species but not all species of plaque Streptococcus. To our knowledge, this is the first report identifying at the species level direct spatial interaction involving more than two partners in a natural biofilm. Our results are thus important for learning how such biofilms are constructed and eventually how to manipulate their composition, particularly in the human microbiome. Notwithstanding their apparently interchangeable positions, two types of corncob-forming Streptococcus coexisted in all donors sampled, suggesting either that they occupy different ecological niches or that mechanisms exist that maintain redundancy and diversity in this host-microbiome system.
Methods
Sample collection and preparation
Samples of supragingival dental plaque from 14 healthy donors older than 18 years old were collected using toothpicks. All donors provided written informed consent. Donors were asked to provide information on their diet (vegetarian or meat-eating) and current tobacco use; all 14 donors reported a meat-eating or non-vegetarian diet and no current tobacco use. Donors were asked not to perform oral hygiene for at least 12 to 24 h prior to sample collection. Samples were fixed in 2% paraformaldehyde (Electron Microscopy Sciences) in 1× phosphate buffered saline (PBS), with 4 h of incubation on ice. Samples were then washed 3 times with 10 mM Tris-HCl pH 7.5 allowing settling by gravity rather than centrifugation between each wash to minimize disruption of structures in the dental plaque. Samples were stored in a 1:1 mixture of 96% ethanol and 10 mM Tris-HCl pH 7.5 at −20 °C until use.
Probe design and testing
We designed FISH probes targeting species of Streptococcus that are abundant in supragingival plaque and potentially involved in the formation of corncobs; one probe targeted S. gordonii, one targeted S. cristatus, one targeted S. mitis and its close relatives S. infantis and S. oralis, and one targeted both S. cristatus and the S. mitis/oralis/infantis group (Additional file 1). Probe sites were selected by inspecting an alignment of 16S rRNA sequences of oral Streptococcus species extracted from sequences deposited in the expanded Human Oral Microbiome Database (eHOMD) [31]. Candidate oligonucleotide probes were tested for specificity in silico using mathFISH [32] to calculate predicted free energy of hybridization and predicted hybridization efficiency on probe sites of target taxa and non-target taxa. If the calculated hybridization efficiency on the target taxon in 20–30% formamide was low, the probes were lengthened by several nucleotides.
The abbreviated probe names indicate the major targeted taxon and the position of the probe target site along the 16S rRNA; probe Scri995 targets S. cristatus, Sgor63 targets S. gordonii, Smit651 targets the S. mitis/oralis/infantis group, and Smit371 targets the S. mitis/oralis/infantis group and S. cristatus. Target sequences are shown in Fig. 1 together with an alignment of the corresponding sequences from four major supragingival plaque Streptococcus spp. The alignments also indicate some potential off-target interactions: Scri995 and Smit371 are expected to hybridize with S. sinensis, and Sgor63 with S. anginosus. However, oligotyping has shown that both species are rare in healthy supragingival plaque [7, 9] and thus unlikely to be a source of ambiguity. The S. mitis group, which we define as species nearly identical to S. mitis in 16S rRNA gene sequence, includes S. infantis, S. oralis, and S. pneumoniae. A complete description of the expected specificity of these probes on all Streptococcus spp. in eHOMD is given in Additional file 1.
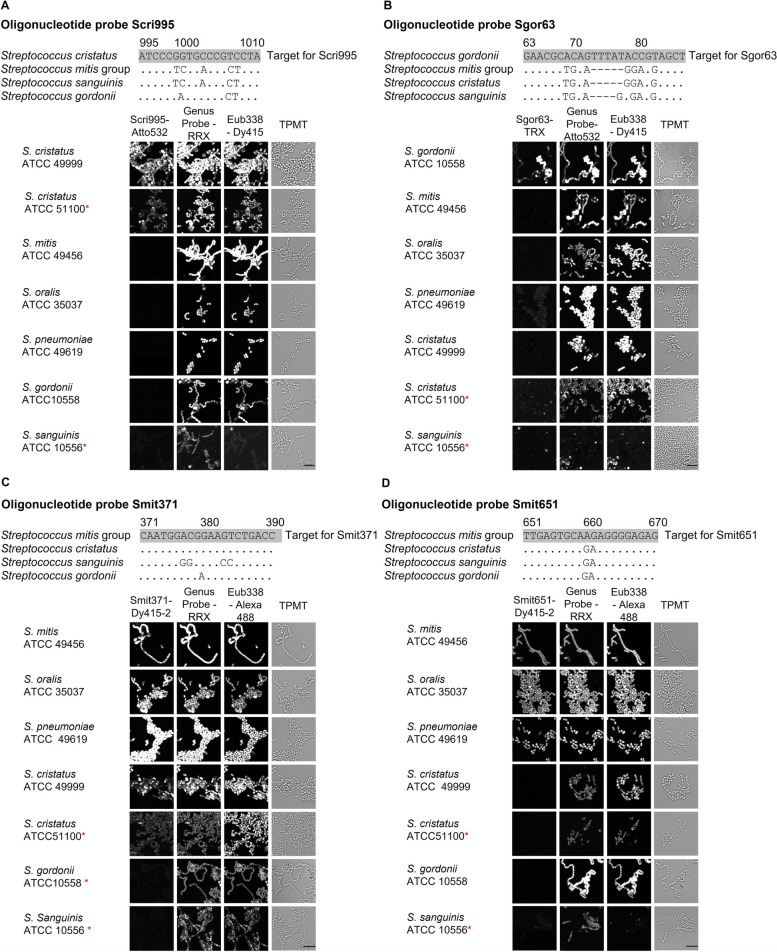
Fig. 1.
Validation of new oligonucleotide probes targeting subsets of the genus Streptococcus. Each newly designed probe was hybridized to pure cultures simultaneously with existing probes targeting genus Streptococcus and most Bacteria. 15 pure cultures were hybridized; 7 are shown here and the remaining 8 are shown in Additional file 1. A Probe Scri995 targeting S. cristatus. B Probe Sgor63 targeting S. gordonii. C Probe Smit371 targeting S. mitis and its close relatives and S. cristatus. D probe Smit651 targeting S. mitis and its close relatives. For each probe set, image acquisition and linear unmixing were carried out under the same conditions using Zeiss ZEN software. Images were imported into FIJI and the range of display intensities was kept constant for each fluorophore (each column in the figure); for cultures where the fluorescence was dim in all channels, the display range was then additionally adjusted by a constant factor for all images in the row in order to improve visibility of cells; these rows are marked (*). All probes hybridized with their expected targets and showed negligible cross-hybridization to unexpected targets. A full list of oral Streptococcus species and their matches and mismatches to each probe is shown in Additional file 1. (RRX: Rhodamine Red X; TRX: Texas Red X). Scale bars = 5 micrometers
Fluorophore-labeled oligonucleotide probes were custom-synthesized (Biomers.net, Ulm/Donau, Germany) and tested experimentally for specificity and sensitivity on 15 pure cultures of streptococci as well as additional taxa. FISH was carried out on S. cristatus (3 strains), S. mitis (2 strains), S. gordonii, S. vestibularis, S. sanguinis, S. parasanguinis, Porphyromonas gingivalis, Aggregatibacter aphrophilus, Corynebacterium durum, and C. matruchotii to validate the specificity and sensitivity of each set of probes. A complete list of strains used to test the specificity and sensitivity of the probes is shown in Additional file 2.
Probe sets
To image corncobs, we used 3 sets of probes (Additional file 3) targeting bacteria representing some of the most abundant taxa in the supragingival plaque: family Pasteurellaceae (genera Haemophilus and Aggregatibacter), genera Streptococcus, Corynebacterium, and Porphyromonas, and species S. cristatus, S. gordonii, and S. mitis/oralis/infantis and close relatives [7, 12]. Probe sequences and the composition of probe sets are shown in Additional file 3. The Pasteurellaceae, Corynebacterium, and Porphyromonas probes are described in Mark Welch et al. 2016 [12]; the Corynebacterium genus probe targets both major oral Corynebacterium species, C. matruchotii and C. durum, and the Porphyromonas probe targets the P. gingivalis group including P. catoniae and P. pasteri but not P. endodontalis.
FISH
Approximately 100 μl of fixed dental plaque in 50% ethanol/10 mM Tris pH 7.5 was spread onto Gold Seal UltraStick adhesion slides (ThermoFisher) and allowed to air-dry immediately before FISH. One hundred microliters of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.5, 0.01% SDS, 20% Hi-Di formamide (ThermoFisher)) containing 2 pmol/μl of each probe were added. Each slide was covered with a 22 × 40 mm cover slip and incubated in a humid chamber at 46 oC for 4 h. Slides were washed once with wash buffer (215 mM NaCl, 20 mM Tris-HCl pH 7.5, 5 mM EDTA), incubated 15 min at 48 oC, and rinsed with cold water. Slides were mounted in ProLong Gold antifade and covered with a 22 × 50 mm #1.5 coverslip. The same protocol was used for hybridization on pure cultures except that 10 μl of a fixed culture was used.
Image acquisition
Spectral images were acquired using either a Zeiss 780 or Zeiss 880 laser scanning confocal microscope equipped with a 32-anode spectral detector and a 40×, 1.4 NA Plan-Apochromat objective. Samples were imaged using 633, 561, 488, and 405 nm excitation wavelengths. Images of pure cultures were acquired using the same imaging conditions as the plaque samples. Images were acquired at a resolution of 9.64 pixels/μm (2048 × 2048 pixels and 212.55 × 212.55 μm).
Image analysis
Reference spectra for each fluorophore used in this study were measured on Leptotrichia buccalis cells labeled with the Eub338 probe conjugated to the appropriate fluorophore. The acquired images were processed by applying a median filter with a 3 × 3 kernel, followed by linear unmixing in the Zeiss ZEN Black software using the respective reference spectra. Unmixed images were imported into FIJI [33] to generate maximum intensity projections of z-stack images and to select and false-color unmixed channels for overlay images using the Image5D plug-in.
Results
Dental plaque hedgehogs contain both single-taxon and mixed corncobs
The genus Streptococcus is species-rich, with 36 oral or potentially oral species recognized in the expanded Human Oral Microbiome Database (eHOMD) [31]. Of these, four subgroups are abundant in dental plaque: the S. mitis/oralis/infantis group, S. sanguinis, S. gordonii, and S. cristatus [7]. To investigate spatial organization of Streptococcus species in corncobs, we designed FISH probes targeting subgroups of Streptococcus species and applied them to supragingival plaque sampled from healthy volunteers. We designed probes to differentiate among groups of species so that collectively the probes could generate a distinctive hybridization pattern for S. cristatus, S. gordonii, and the S. mitis group including S. mitis, S. oralis, and S. infantis. We tested each probe for effectiveness and specificity by hybridizing it with pure cultures representative of target and non-target taxa. For comparison, each culture was also hybridized simultaneously with a universal bacterial probe and a probe for the genus Streptococcus. The target taxa showed the expected probe signals (Fig. 1, Additional file 1).
Having established the specificity of these new species and subgroup-level probes, we combined them with existing probes targeting genus- and family-level taxa to create probe sets to illuminate corncob structure. In addition to Streptococcus at the genus and species level, the probe sets targeted the other taxa previously demonstrated to participate in corncobs: the genera Corynebacterium and Porphyromonas and the family Pasteurellaceae [12]. We employed probes in different combinations, using different fluorophores, to ensure robustness of results to the details of the probe set composition. A detailed description of each probe set and its validation on pure cultures is presented in Additional files 3 and 4.
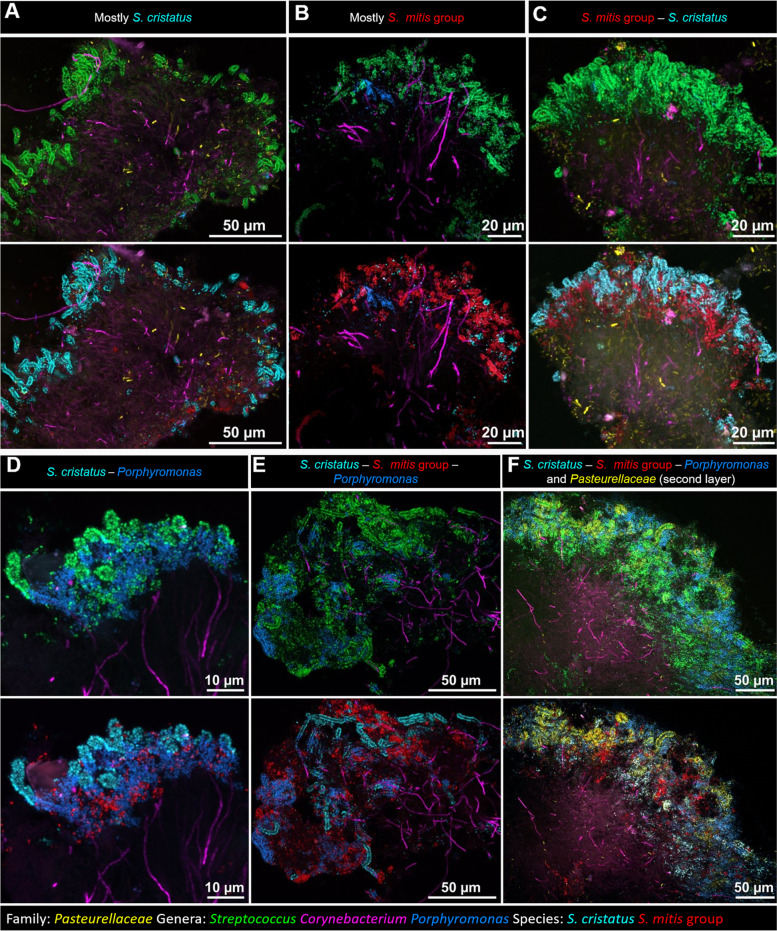
A characteristic feature of dental plaque seen in our previous work was the ‘hedgehog’ structure [12]. Operationally, we define a hedgehog as a cluster of Corynebacterium filaments with corncobs at their tips. Applying the FISH probe sets to samples from 14 healthy subjects revealed that individual hedgehogs have heterogeneous sets of corncobs (Fig. 2). In some of the hedgehogs, most of the corncob “kernels” (the cocci surrounding the tips of the Corynebacterium filaments) were of the same species (Fig. 2A, B). In some cases the species was S. cristatus (Fig. 2A) and in other cases it was S. mitis/oralis/infantis (Fig. 2B). Other hedgehogs contained corncobs with a mixture of S. mitis/oralis/infantis and S. cristatus (Fig. 2C). Some hedgehogs had corncobs containing the additional taxa Porphyromonas (Fig. 2D–F) and Pasteurellaceae (Fig. 2F). Qualitatively, our results show that dental plaque hedgehogs can be composed of corncobs of varying composition, from either or both of two subgroups of Streptococcus and with the presence or absence of members of two additional taxa, Porphyromonas and Pasteurellaceae. Thus the plaque hedgehog is not a structure of consistent composition but a category of organization in which the filament is constant but the taxonomic composition of the kernels is variable.
Fig. 2.
Dental plaque hedgehogs contain corncobs of mixed composition. Hedgehog structures are identified in plaque by the presence of a cluster of Corynebacterium filaments with corncobs at the periphery. Family- and genus-level probes (top in each pair of images) show corncobs composed of cells of genera Streptococcus (green), Porphyromonas (blue), and Corynebacterium (magenta) and family Pasteurellaceae (yellow). Species-level probes (bottom in each pair of images) show that the Streptococcus population in a given hedgehog can contain mostly S. cristatus (cyan) (A), mostly S. mitis/oralis/infantis (red) (B), or a mixture of both S. cristatus and S. mitis/oralis/infantis (C). Mixed corncob communities in hedgehogs can also contain Porphyromonas together with both S. cristatus and S. mitis/oralis/infantis (D, E), and may also contain cells of family Pasteurellaceae as an additional, outer layer on the corncobs (F)
Composition of individual corncobs
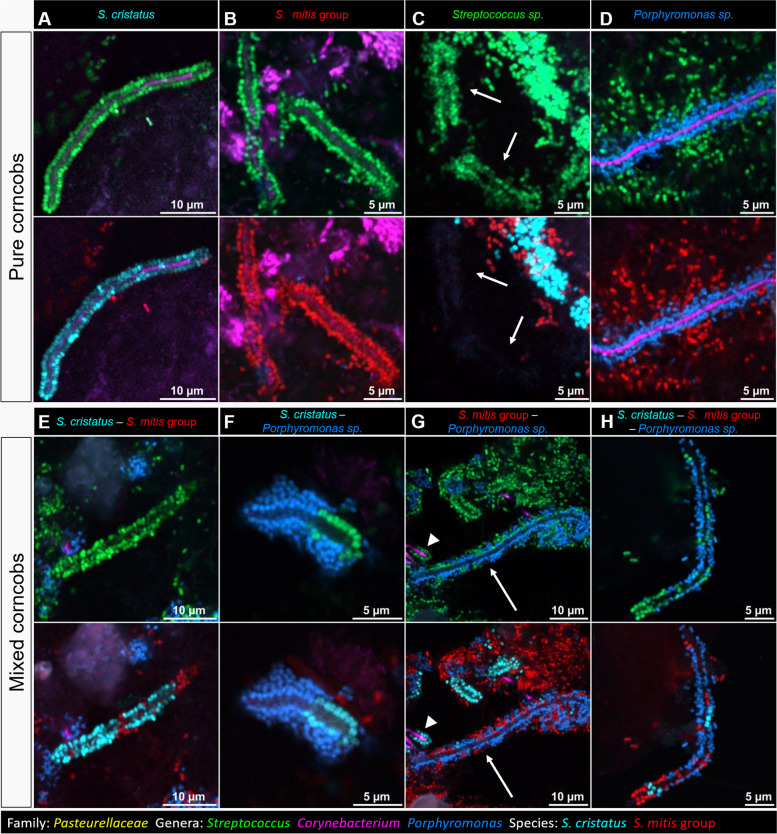
Visualizing individual corncobs at higher magnification, we observed that the kernels of a corncob can be composed of a single species or contain mixtures of different species (Fig. 3). The species that were frequently observed were S. cristatus, S. mitis/oralis/infantis, and Porphyromonas sp., each being observed individually (Fig. 3A–D) and in combinations (Fig. 3 E–H). Interestingly, although we visualized cells of S. gordonii in the vicinity of corncobs (Additional file 5), we never observed S. gordonii as part of a corncob. We did occasionally observe cells in corncobs that hybridized with the Streptococcus genus probe but not with any of the species probes we employed (Fig. 3C). Thus, the individual corncob, like the hedgehog, is not a structure of consistent composition, but is a category of structure in which the taxonomic composition is variable—but the range of variability appears to be limited to a subset of the cocci present in dental plaque.
Fig. 3.
Individual corncobs can contain single or multiple species of Streptococcus. Genus probes (top in each pair of images) show the overall structure of corncobs and distinguish between the central filament (Corynebacterium) and the surrounding Streptococcus (green) or Porphyromonas (blue). Staining of the central filament is sometimes absent and in these cases its identity is not confirmed. Species probes (bottom in each pair) distinguish between S. cristatus (cyan) and S. mitis/oralis/infantis (red). “Pure” corncobs were those in which all the imaged cells around the central filament hybridized to the same probe, targeting S. cristatus (A), S. mitis/oralis/infantis (B), a third species of Streptococcus not identified with the set of probes used (C), or Porphyromonas (D). “Mixed” corncobs contained more than one type of cells surrounding the central filament: S. cristatus and S. mitis/oralis/infantis (E), S. cristatus and Porphyromonas sp. (F), S. mitis/oralis/infantis and Porphyromonas sp. (G), S. cristatus, S. mitis/oralis/infantis and Porphyromonas sp. (H). Different types are sometimes near each other in the same field of view, e.g., in G a mixed corncob of S. mitis/oralis/infantis and Porphyromonas sp. (arrow), and a pure corncob of S. cristatus (△) are observed
Corncobs presented themselves in variable conformations within the plaque biofilm. They occurred in groups at the periphery of hedgehog structures, consistent with earlier results showing corncobs forming an outer layer on filament-rich plaque [26]. They also occurred as isolated corncobs embedded in filamentous or non-filamentous plaque. Their length was variable, generally 10–20 μm and as long as 50 μm in a single image. The central filament was sometimes stained with the Corynebacterium genus probe (Fig. 3A, D, G) but at other times no staining of the central filament was observed. Variation in intensity of staining of Corynebacterium cells is frequently observed (e.g., [12]) and likely results from variation in permeability of the cells to the probe or from variation in ribosome content. Staining is frequently stronger in the center of hedgehog structures than in the corncobs at the periphery (e.g., Fig. 2) and is often variable even within a single corncob (Fig. 3A, B).
Quantification of corncob types
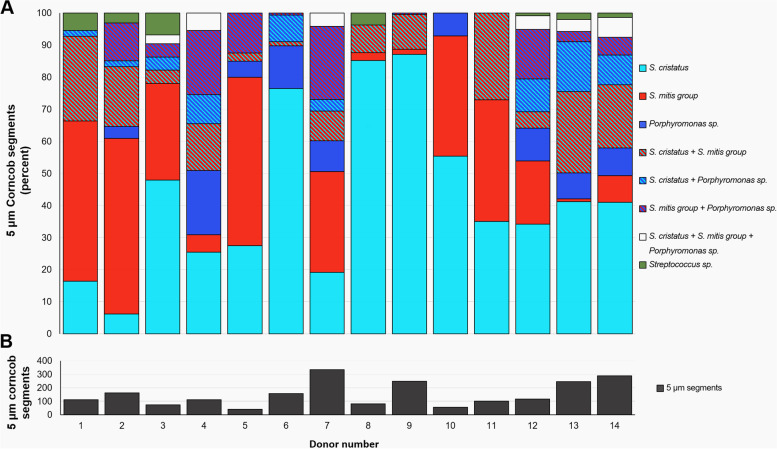
To quantify the relative abundance of different corncob types, we analyzed images of corncobs by dividing the corncobs into segments of 5 μm in length, and classifying the 5 μm segments according to the identity of the layer of cells immediately adjacent to the filament (Fig. 4). For purposes of this quantification, a corncob in longitudinal section was defined operationally as 2 continuous rows of cocci that were at least 5 μm in length on both sides of a core filament—either a visible filament or a gap that was presumed to contain an unstained filament. A corncob in cross section or oblique section was defined as a continuous circle or oval of cocci surrounding a filament or a space presumed to contain an unstained filament. We imaged 10 fields of view (FOV) per probe set and donor, for each of two probe sets and 14 donors, selecting fields of view to image where corncobs were visible through the eyepieces. Not all of these images contained corncobs meeting the criteria for quantification; therefore the total number of FOV in which corncobs were counted averaged 8.3 FOV per probe set and donor (range 3 to 11). The total FOV per donor in which corncobs were counted averaged 17 (range 9–20). From this dataset, we counted a total of 2122 corncob segments with a mean of 152 per donor (range 40–334).
Fig. 4.
Each donor has a mixed population of corncobs. For each of 14 donors, the imaged corncobs were divided into 5 μm segments and the presence of S. mitis/oralis/infantis, S. cristatus, other Streptococcus spp., and Porphyromonas spp. in each segment was recorded. The relative abundance of each type of corncob in each donor is shown (A). Black bars indicate the total number of 5 μm corncob segments counted in each donor (B). Corncob segments consisting of S. cristatus alone constituted 42.3% of the total and a mean of 42.7% (range 6% to 87%) across the 14 individuals. Segments consisting of pure S. mitis/oralis/infantis were 19.4% of the total, mean 23.8% (range 0 to 55%) and pure Porphyromonas sp. segments were 6.8% of the total, mean 6.1% (range 0–20%). Mixed segments and segments including unidentified Streptococcus spp. were 31.6% of the total and mean 27.4% (range 0–50%)
The results of this analysis show that all donors had corncobs of diverse composition (Fig. 4). The imaged corncobs from every donor included both S. cristatus and S. mitis/oralis/infantis, and images from most donors also included Porphyromonas. In about half of the donors, we also detected a small number of corncobs containing cells hybridizing with the Streptococcus genus probe but with none of the species probes. About two thirds of the corncob segments contained only a single taxon, either S. cristatus, S. mitis/oralis/infantis, or Porphyromonas, while the remaining one third of segments were mixed (Fig. 4 and Additional file 6). Thus, while the majority of corncob segments were composed of a single taxon, the overall corncob community within each donor was complex.
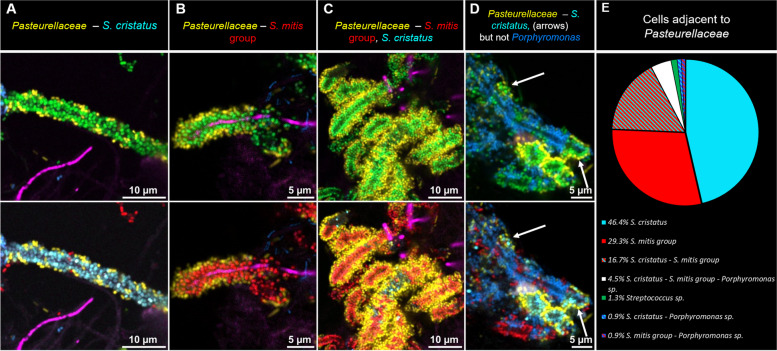
Taxon adjacency relationships in corncobs include not only the relationship between the central Corynebacterium filament and the surrounding cocci but also the relationship between this first layer of cocci and the outer layer of cells belonging to the Pasteurellaceae [12], of which the representatives abundant in the human mouth are Haemophilus and Aggregatibacter [1, 7]. To determine whether these cells were found adjacent to all types of inner-layer cocci or only a subset of them, we included a probe for Pasteurellaceae in one probe set and detected Pasteurellaceae in corncobs from 7 of the 14 donors and in a total of 21.3% of the 1053 corncob segments counted using this probe set (Fig. 5). Results showed that the Pasteurellaceae in corncobs associated with both major types of corncob streptococci and also with the unidentified streptococci, in the approximate ratios in which these Streptococcus spp. were present in the corncobs overall, but were not found adjacent to corncob Porphyromonas (Fig. 5). Thus, our results suggest the possibility of metabolic or binding interactions between Pasteurellaceae and both of the two major Streptococcus types in corncobs, but not with corncob Porphyromonas.
Fig. 5.
Cells of family Pasteurellaceae adhere to S. mitis/oralis/infantis and S. cristatus but not Porphyromonas. In addition to the layer of cells of Streptococcus sp. or Porphyromonas sp. directly adjacent to the central filament, a second layer of cells from the family Pasteurellaceae is sometimes observed on corncobs. We observed these cells attached to corncobs in which the inner layer of cells was S. cristatus (A), S. mitis/oralis/infantis (B), or a mixture of S. cristatus and S. mitis/oralis/infantis (C). We also observed a second layer on mixed corncobs that included Porphyromonas sp.; in these corncobs the Pasteurellaceae cells were adjacent to Streptococcus cells but not to the nearby Porphyromonas cells (D). In the samples analyzed, we did not observe any layer of Pasteurellaceae attached to pure Porphyromonas corncobs. Among the 5 μm corncob segments counted on the samples hybridized with probe set 1, 21.1% presented a second layer formed by Pasteurellaceae (E). Of these, close to half (47%) were 5 μm segments in which all the Streptococcus cells were S. cristatus; 30% of the segments consisted of pure S. mitis/oralis/infantis; and the Streptococcus population of the remaining 23% was mixed or unidentified
Discussion
Our findings show that the corncob microhabitat is selective for a subset of the Streptococcus species in dental plaque. The most abundant streptococci in dental plaque are members of the S. mitis/oralis/infantis cluster and S. sanguinis, which together make up about 90% of plaque streptococci. The species S. cristatus is a minor component of the genus in plaque as a whole, making up less than 4% of supragingival plaque Streptococcus in Human Microbiome Project samples across 148 individuals [7]. Nonetheless, in the 14 individuals studied here, S. cristatus was the most abundant Streptococcus species in corncobs. Thus, the site-specificity displayed by S. cristatus is for a well-defined microhabitat within dental plaque: adhered to Corynebacterium filaments as corncobs. The association is not exclusive, however, in that a different Streptococcus species, identified by FISH as a member of the S. mitis/oralis/infantis group, was almost as abundant as S. cristatus in corncobs. Detailed comparison of staining from the Streptococcus genus vs. species probes indicated that S. cristatus and the S. mitis/oralis/infantis group together comprised almost all the Streptococcus cells in corncobs; other unidentified Streptococcus were rarely present, detected in 1.4% of corncob segments. In particular, S. gordonii, a species with overall abundance in supragingival plaque similar to that of S. cristatus, was detected in surrounding plaque but not in corncobs. Thus, the corncob represents an interaction between Corynebacterium and a limited subset of the pool of plaque Streptococcus species. Filament-rich plaque including corncobs makes up only a portion of dental plaque, and in some samples only a modest number of corncobs could be detected. Additional studies with a larger cohort will be needed to make more precise estimates of relative abundance and to determine the identity of the additional streptococci in corncobs.
Members of other genera present in corncobs likewise were adjacent to multiple partners but not all potential partners in plaque. Cells hybridizing with the Porphyromonas probe were present in corncobs, either as the only cocci surrounding a filament or sharing a central filament with S. mitis/oralis/infantis, S. cristatus, or both. The additional outer layer of Pasteurellaceae was found adjacent to cells of both S. mitis/oralis/infantis and S. cristatus, but not Porphyromonas. This distribution indicates that the Pasteurellaceae-Streptococcus relationship in corncobs, like the Corynebacterium-Streptococcus relationship, is a selective interaction. Although Pasteurellaceae spp. associated with two different Streptococcus spp. partners, it did not associate with Porphyromonas spp. Further study will be needed to determine the mechanistic underpinnings of this spatial selectivity: whether it results from differential binding or differential reproductive success of Pasteurellaceae spp. when bound to Streptococcus spp. rather than Porphyromonas spp., or both.
Although corncob-like structures have been reported to form around other taxa, several lines of evidence suggest that in supragingival plaque the corncob filament is generally Corynebacterium spp. In vitro studies [30] have shown that Fusobacterium nucleatum, when mixed with Streptococcus, can form the central filament of corncob-like structures. However, we have not seen an association of cocci with Fusobacterium spp. in natural dental plaque. Our previous results with a probe set targeting different filamentous bacteria in plaque, including Fusobacterium, indicated that the corncob association was highly specific to Corynebacterium spp. [12]. Although staining of Corynebacterium was variable in intensity, the central filament of corncobs, when staining was evident, was always Corynebacterium. Other filamentous or elongated taxa such as Fusobacterium, Leptotrichia, and Capnocytophaga were not detected as the central filament even when they were detected in the immediate surroundings of the corncob. A previous study [34] showed associations of streptococci with hyphae of Candida albicans in natural plaque. However, Candida generally has low abundance in the healthy mouth. In the present study, to focus on species-level identification of streptococci, we omitted probes for the filamentous taxa that our previous study did not detect in corncobs. The quantification results in this study apply to the full population of corncob cocci that we visualized in healthy subjects, whether or not the identity of the central filament could be confirmed.
Our finding of complex but limited taxon composition in corncobs bears on an important question in microbial ecology, namely the question of how a stable, healthy interaction is maintained between a host and its microbiome [35]. Theoretical work predicts that mutualistic interactions tend to fall apart over time, for example because the loss of one of the partners results in the loss of the other, or because one partner ceases to behave as a mutualist and instead becomes a parasite [36]. Such a shift from mutualism to parasitism is more likely if the interaction is highly specific, so that an organism is dependent on a single partner [37]. Bacteria within the densely packed dental plaque biofilm depend on one another for metabolites and signals [19, 38, 39], but the composition of oral microbial communities is characterized by wide fluctuations in the relative abundance of taxa even as the overall community membership remains stable, a pattern known as stationary dynamics [40, 41]. Although the consistent composition and direct cell-cell attachment in corncobs suggests a degree of metabolic dependency of the partners on one another, we observed flexibility in the taxon relationships involved in corncobs, in the sense that several partners were capable of interacting with the central filament and several streptococci could interact with the outer layer of Pasteurellaceae. Thus the spatial relationships we observe in corncobs suggest that each of the microbial participants is capable of interacting with multiple, albeit limited, potential partners, a feature that may encourage the long-term stability of the community.
A related open question in microbial ecology is whether microbial communities assemble with a consistent species composition or, alternatively, with a consistent set of functional genes that can be contributed by a range of different species [42, 43]. It has been proposed, for example, that under conditions common in the mouth (horizontal gene transfer and migration), species identity can be insignificant because genes, rather than species, inhabit niches [44]. Despite the flexibility we observed in the composition of individual corncobs, however, both S. cristatus and S. mitis/oralis/infantis were observed in corncobs in every donor. At the scale of individual corncobs or corncob segments, these distinct Streptococcus species were apparently interchangeable in their ability to bind to the central Corynebacterium filament and the exterior shell of Pasteurellaceae, yet both types persisted in the plaque community. This persistence suggests that the different taxa possess distinct ecological roles, or that mechanisms exist that stabilize the continued persistence of multiple, functionally redundant taxa within the same microbiome ecosystem. Our data thus indicate that in the corncob microhabitat within the dental plaque biofilm, species composition remains consistent from mouth to mouth.
The heterogeneity of corncob structures has important implications for mechanistic studies such as in vitro co-culture or multi-taxon metabolic modeling of plaque bacteria as a model microbial community. In addition to the Corynebacterium-S. cristatus relationship, our results show numerous pairs of taxa directly adjacent to one another in corncobs, including all combinations of S. cristatus, S. mitis/oralis/infantis, Porphyromonas spp., and Corynebacterium spp. as well as Pasteurellaceae with both S. mitis/oralis/infantis and S. cristatus. Thus, a number of potentially significant taxon-taxon relationships have been identified in this study, and our results suggest that a natural corncob may be modeled not only as a two-taxon relationship but also as a relationship containing three, four, or five partners. The mechanistic underpinnings of the corncob association likely are founded on adhesion of taxa to one another. C. matruchotii itself adheres not directly to the tooth surface, but to early colonizers such as Actinomyces naeslundii [45]. Among oral streptococci, S. cristatus has tufts of fibrils that enable its adhesion to C. matruchotii [46] and S. oralis subsp. dentisani forms fibrils distributed asymmetrically on its surface that likely play a role in adhesion [47]. Whether this subspecies of S. oralis is the taxon identified in corncobs by our S. mitis/oralis/infantis probe is an important question that could be resolved by the development of in situ sequencing approaches. Although the metabolism of the species visualized here in corncobs has not yet been the subject of extensive in vitro investigation, other oral species within these same genera have been investigated and their taxon-taxon interactions have been shown to change the gene expression and biology of the partners [14, 48, 49]. For example, co-culture of C. durum and S. sanguinis results in interspecies interactions involving fatty acid metabolism of both partners [45]. Our results enable the selection of taxa for in vitro co-culture studies that are grounded in the frequently adjacent taxa of natural plaque; these are the taxa that are likely to engage in metabolic interactions with physiologically relevant consequences.
Corncobs bear some resemblance to another tight spatial relationship in the oral microbiome, in which ultrasmall Saccharibacteria spp. live epibiotically on filamentous Actinobacteria such as Schaalia odontolytica, but there are important differences between the two consortia. Oral Saccharibacteria are obligate epibionts; they have genome sizes under 0.9 Mb, lack numerous genes essential for a free-living lifestyle, and consequently cannot be cultivated except in co-culture with their host [50–52]. By contrast, corncob taxa (not only C. matruchotii but also S. cristatus, S. oralis, S. mitis, and the Porphyromonas species most likely to be in corncobs, P. catoniae and P. pasteri) have genome sizes of approximately 2 Mb and are free-living, capable of growth in pure culture in standard media. In ecological terms, corncob cocci have a fundamental niche (capable of independent growth in standard nutrient-rich conditions) that is broader than their realized niche (generally found adjacent to Corynebacterium spp. in corncobs). Thus while the Saccharibacteria-host relationship is that of a potentially parasitic epibiont, the corncob represents a consortium of organisms that may prosper in each other’s company but are capable of independent growth.
Several lines of evidence suggest that the taxa participating in corncobs are associated with human health. Recent studies have found C. matruchotii and C. durum associated with health rather than caries [53–55]. Metabolites produced by C. durum have also been found to extend lifespan in the model organism Caenorhabditis elegans [56] and C. durum elicited no inflammatory response from human gingival and oral mucosal cells, suggesting it is a commensal [57]. S. cristatus has been shown to inhibit biofilm formation of the periodontal pathogen P. gingivalis by repression of virulence genes [58, 59]. Because of their location towards the outside of plaque, corncob taxa may represent the first organisms that a microbe would encounter when landing on the tooth biofilm; corncobs composed of S. cristatus might therefore inhibit colonization of the mouth by this potential pathogen. On the other hand, some mitis group streptococci potentiate the virulence of C. albicans [60, 61]. Under what circumstances the species that participate in corncob structures inhibit or enhance the colonization and virulence of pathogens, and the mechanisms by which they do so, is an important question for further research. In studies of the development of plaque on epoxy resin crowns worn by volunteers, the initial plaque was coccus-rich, corncobs were first observed after 3 days, and filament-rich plaque did not occur until approximately 1 week of incubation [25, 26]. These observations might suggest that hedgehog structures and corncobs would be rare in people engaging in daily dental hygiene. In the present study, however, we detected corncobs in all donors and hedgehog structures in most, even though donors were instructed to refrain from oral hygiene for only 24 h and no donor reported going longer than 26 h without tooth brushing. We conclude that hedgehogs and corncobs can be formed in less than 24 h in the dental plaque of healthy individuals, perhaps growing from already-established patches of filament-rich plaque, and that corncob structure and function may play an important role in normal oral microbial ecology and in maintenance of human health.
The unique benefit of the imaging approach we took here is that taxon-taxon spatial relationships can be directly visualized at micrometer scales. The results we report here represent substantial effort in imaging and analysis; to analyze 2122 corncob segments we quantified 232 fields of view, each of which was composed of 5 to 8 unmixed fluorophore channels, for a total of more than 1400 images. Each field of view in our study represented an area of 212 × 212 micrometers and a volume of 4 × 10−5 mm3. By comparison, each data point in a sequencing study may represent a homogenized sample from a cubic millimeter of biofilm or a swab over several cm of area. Thus, sequencing generates abundant data that can be subjected to sophisticated analysis, but at the cost of losing fine-scale spatial information, and quantitative comparison of our results with the results of sequencing studies is not straightforward because the scale of sampling is so different. Future studies with increasingly complex probe sets and automated image processing may be able to identify hundreds of thousands of cells with micrometer spatial resolution, enabling analysis of the full complexity and micron-scale arrangement of microbes in biofilms.
Conclusions
Within the complex dental plaque biofilm, corncob structures represent a well-defined microhabitat that is inhabited by a specific subset of the bacteria found in dental plaque as a whole. The spatial adjacency relationships in corncobs indicate that each taxon associates with a limited number of potential partners, but always with more than one potential partner, a feature that may encourage the long-term stability of the community. Our results suggest the general principle that the more precisely a microhabitat can be defined, the more well-defined will be the set of microbial taxa that grow in this habitat. Further work elucidating bacterial nearest-neighbor relationships in oral biofilms may identify additional taxon-taxon associations which could be exploited to enable targeted modulation of these communities for the maintenance of health and the treatment of disease.
Supplementary Information
Additional file 1. Validation of new oligonucleotide probes targeting subsets of the genus Streptococcus. For each probe, a full list of oral Streptococcus species and their matches and mismatches to the probe is shown (top), along with images showing the intensity of signal imaged after hybridization of pure cultures of cells to the probes. Each newly designed probe was hybridized simultaneously with existing probes targeting genus Streptococcus and most Bacteria as controls. 15 pure cultures were hybridized; 8 are shown here and the remaining 7 are shown in Fig. 1. A) Probe Scri995 targeting S. cristatus; B) probe Sgor63 targeting S. gordonii; C) probe Smit371 targeting S. mitis and its close relatives and S. cristatus; D) probe Smit651 targeting S. mitis and its close relatives. For each probe set, image acquisition and linear unmixing of all cultures were carried out under the same conditions using Zeiss ZEN software. Images were imported into FIJI and the range of display intensities was kept constant for each fluorophore (each column in the figure); for cultures where the fluorescence was dim in all channels, the display range was then additionally adjusted by a constant factor for all images in the row to improve visibility of cells; these rows are marked (*). All probes hybridized with their expected targets and showed negligible cross-hybridization to unexpected targets. (RRX: Rhodamine Red X; TRX: Texas Red X).
Additional file 2. List of strains used for FISH on pure cultures.
Additional file 3. Ribosomal RNA-targeted oligonucleotide probes used in this study and their combination into probe sets [62–64].
Additional file 4. Probe set validation matrices showing hybridization of pure cultures with complete probe sets. For each probe set, image acquisition and linear unmixing of all cultures were carried out under consistent conditions using Zeiss ZEN software. Images were imported into FIJI and the range of display intensities was kept constant for each fluorophore (each column in the figure). In addition to the species probes shown in Fig. 1 and Additional File 1, probes and their targets and fluorophores are as follows: Str405 [62] targeting genus Streptococcus, labeled with Rhodamine Red X (RRX); Por1160 [64] targeting the gingivalis group of genus Porphyromonas, labeled with Alexa 555; Pas111 [64] targeting family Pasteurellaceae, labeled with Dy615; Cor633 [12] targeting genus Corynebacterium, labeled with Dy490 (part A) or Atto 620 (part B); probe Cmat175 [12] targeting species C. matruchotii, labeled with Atto 655. All probes hybridized with their expected targets and showed negligible cross-hybridization to unexpected targets. (At532: Atto 532; TRX: Texas Red X).
Additional file 5. Cells of Streptococcus gordonii visualized in supragingival dental plaque. Top panel shows Streptococcus spp. at the genus level; bottom panel shows Streptococcus at the species level. (A): Cells of S. gordonii (arrows) are visualized in the vicinity of corncobs but not attached to them. (B): Cells of S. gordonii are also observed in hedgehog structures but not as part of corncobs. (C): a large clump of S. mitis group cells with scattered S. gordonii cells.
Additional file 6. Taxon composition of corncob segments by donor. This is the numerical data underlying the bar chart in Fig. 4.
Acknowledgments
We thank Floyd Dewhirst, Matthew Ramsey, Samantha King, Molly Bennett, Janina Schumann, and Louie Kerr and the staff of the Marine Biological Laboratory Central Microscopy Facility for helpful discussions and technical help; Anthony McLean for a close reading of the manuscript; Jack Gilbert for support of A.S.’s summer research at the Marine Biological Laboratory; the Woods Hole Partnership Education Program for support of I.I.; and Jim McIlvain and Zeiss Corp. for use of the LSM 880 confocal microscope. We thank the plaque donors for their participation.
Abbreviations
- FISH
Fluorescence in situ hybridization
- FOV
Field of view
- NA
Numerical aperture
Authors’ contributions
JMW and GGB conceived and designed the research; VM-L, AS, II, and JMW participated in data collection; JMW, GGB, and VM-L wrote the manuscript; and all authors read and approved the final manuscript.
Funding
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers DE027958 and DE022586. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. V.M-L. was supported in part by a G. Unger Vetlesen Foundation grant to the Marine Biological Laboratory. A.S. was partially supported by the Jeff Metcalf Internship Program of the University of Chicago. I.I. was supported by the Woods Hole Partnership Education Program.
Availability of data and materials
The datasets supporting the conclusions of this article are available in Zenodo under doi:10.5281/zenodo.6426109, doi:10.5281/zenodo.6529696, and doi:10.5281/zenodo.6529719.
Declarations
Ethics approval and consent to participate
Sampling from human subjects for this study was carried out under protocol 120160354 approved by the New England Institutional Review Board. All subjects provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Viviana Morillo-Lopez, Email: vivikamorillo@gmail.com.
Alexandra Sjaarda, Email: alisjaarda@gmail.com.
Imon Islam, Email: iislam@ucsc.edu.
Gary G. Borisy, Email: gborisy@forsyth.org
Jessica L. Mark Welch, Email: jmarkwelch@forsyth.org
References
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 2018;3(6). https://msystems.asm.org/content/3/6/e00187-18. [DOI] [PMC free article] [PubMed]
- 3.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol. 2005;38(1):135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 5.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason MR, Chambers S, Dabdoub SM, Thikkurissy S, Kumar PS. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome. 2018;6(1):67. doi: 10.1186/s40168-018-0443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mark Welch JL, Dewhirst FE, Borisy GG. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol. 2019;73:335–358. doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, Haffajee AD. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2011;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, Ellwood R, Giacaman RA, Herrera D, Herzberg MC, Könönen E, Marsh PD, Meyle J, Mira A, Molina A, Mombelli A, Quirynen M, Reynolds EC, Shapira L, Zaura E. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J Clin Periodontol. 2017;44 Suppl 18:S5–S11. doi: 10.1111/jcpe.12682. [DOI] [PubMed] [Google Scholar]
- 12.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilbert SA, Mark Welch JL, Borisy GG. Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 2020;30:4003–4015.e3. doi: 10.1016/j.celrep.2020.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7(3):e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of Synergy in Polymicrobial Infections. J Microbiol. 2014;52(3):188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pande S, Kost C. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 2017;25:349–361. doi: 10.1016/j.tim.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Seyedsayamdost MR, Traxler MF, Clardy J, et al. Old meets new: using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 2012;517:89–109. doi: 10.1016/B978-0-12-404634-4.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies Interactions Stimulate Diversification of the Streptomyces coelicolor Secreted Metabolome. mBio. 2013;4(4):e00459–e00413. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 20.Dal Co A, van Vliet S, Kiviet DJ, et al. Short-range interactions govern the dynamics and functions of microbial communities. Nat Ecol Evol. 2020;4:366–375. doi: 10.1038/s41559-019-1080-2. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175(11):3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vicentini R. Bacteria of the sputa and cryptogamic flora of the mouth. London: Bailliere, Tindall & Cox; 1897. [Google Scholar]
- 23.Williams JL. A contribution to the bacteriology of the human mouth. Dent Cosmos. 1899;41:317–349. [Google Scholar]
- 24.Jones SJ. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch Oral Biol. 1972;17:613–616. doi: 10.1016/0003-9969(72)90081-7. [DOI] [PubMed] [Google Scholar]
- 25.Listgarten MA, Mayo H, Amsterdam M. Ultrastructure of the attachment device between coccal and filamentous micro-organisms in “corn cob” formations of dental plaque. Arch Oral Biol. 1973;18:651–656. doi: 10.1016/0003-9969(73)90105-2. [DOI] [PubMed] [Google Scholar]
- 26.Listgarten MA, Mayo HE, Tremblay R. Development of dental plaque on epoxy resin crowns in man: a light and electron microscopic study. J Periodontol. 1975;46(1):10–26.8. doi: 10.1902/jop.1975.46.1.10. [DOI] [PubMed] [Google Scholar]
- 27.Mouton C, Reynolds HS, Genco RJ. Characterization of tufted streptococci isolated from the “corn cob” configuration of human dental plaque. Infect Immun. 1980;27(1):235–245. doi: 10.1128/iai.27.1.235-245.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handley P, Coykendall A, Beighton D, Hardie JM, Whiley RA. Streptococcus crista sp. nov., a Viridans Streptococcus with Tufted Fibrils, Isolated from the Human Oral Cavity and Throat. Int J Syst Bacteriol. 1991;41(1):543–547. doi: 10.1099/00207713-41-4-543. [DOI] [PubMed] [Google Scholar]
- 29.Trüper H, De’clari L. Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) “in apposition”. Int J Syst Bacteriol. 1997;47(3):908–909. doi: 10.1099/00207713-47-3-908. [DOI] [Google Scholar]
- 30.Lancy P, Jr, Dirienzo JM, Appelbaum B, Rosan B, Holt SC. Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect Immun. 1983;40(1):303–309. doi: 10.1128/iai.40.1.303-309.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 2018;3(6). 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed]
- 32.Yilmaz LS, Parnerkar S, Noguera DR. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol. 2011;77(3):1118–1122. doi: 10.1128/AEM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zijnge V, Leeuwen VMB, Degener E, Abbas F, Thurnheer T, Gmür R, Harmsen HJ. Oral biofilm architecture on natural teeth. PLoS One. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar PS. Microbial dysbiosis: the root cause of periodontal disease. J Periodontol. 2021. 10.1002/JPER.21-0245. [DOI] [PubMed]
- 36.Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21(10):585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 37.Chomicki G, Kiers ET, Renner SS. The evolution of mutualistic dependence. Annu Rev Ecol Evol Syst. 2020;51:409–432. doi: 10.1146/annurev-ecolsys-110218-024629. [DOI] [Google Scholar]
- 38.Stacy A, Fleming D, Lamont RJ, Rumbaugh KP, Whiteley M. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. mBio. 2016;7(3). 10.1128/mBio.00782-16. [DOI] [PMC free article] [PubMed]
- 39.Hoare A, Wang H, Meethil A, Abusleme L, Hong B-Y, Moutsopoulos NM, Marsh PD, Hajishengallis G, Diaz PI. A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J. 2021;15:1490–1504. doi: 10.1038/s41396-020-00865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbons SM, Kearney SM, Smillie CS, Alm EJ. Two dynamic regimes in the human gut microbiome. PLoS Comput Biol. 2017;13(2):e1005364. doi: 10.1371/journal.pcbi.1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burke C, Steinberg P, Rusch D, Kjellberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. PNAS. 2011;108(34):14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niehus R, Mitri S, Fletcher AG, Foster KR. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun. 2015;6:8924. doi: 10.1038/ncomms9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esberg A, Barone A, Eriksson L, Lif Holgerson P, Teneberg S, Johansson I. Corynebacterium matruchotii Demography and Adhesion Determinants in the Oral Cavity of Healthy Individuals. Microorganisms. 2020;8:1780. doi: 10.3390/microorganisms8111780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handley PS, Correia FF, Russell K, Rosan B, DiRienzo JM. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol Immunol. 2005;20:131–140. doi: 10.1111/j.1399-302X.2004.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronis A, Brockman K, Singh AK, Gaytán MO, Wong A, McGrath S, Owen CD, Magrini V, Wilson RK, van der Linden M, King SJ. Streptococcus oralis subsp. dentisani produces monolateral serine-rich repeat protein fibrils, one of which contributes to saliva binding via sialic acid. Infect Immun. 2019;87(10):e00406–e00419. doi: 10.1128/IAI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treerat P, Redanz U, Redanz S, Giacaman RA, Merritt J, Kreth J. Synergism between Corynebacterium and Streptococcus sanguinis reveals new interactions between oral commensals. ISME J. 2020;14(5):1154–1169. doi: 10.1038/s41396-020-0598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perera D, McLean A, Morillo-Lopez V, Cloutier-Leblanc K, Almeida E, Cabana K, Mark Welch J, Ramsey M. Mechanisms underlying interactions between two abundant oral commensal bacteria. ISME J. 2022;16(4):948–957. doi: 10.1038/s41396-021-01141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 2015;112(1):244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bor B, Collins AJ, Murugkar PP, Balasubramanian S, To TT, Hendrickson EL, Bedree JK, Bidlack FB, Johnston CD, Shi W, McLean JS, He X, Dewhirst FE. Insights obtained by culturing Saccharibacteria with their bacterial hosts. J Dent Res. 2020;99(6):685–694. doi: 10.1177/0022034520905792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murugkar PP, Collins AJ, Chen T, Dewhirst FE. Isolation and cultivation of candidate phyla radiation Saccharibacteria (TM7) bacteria in coculture with bacterial hosts. J Oral Microbiol. 2020;12(1):1814666. doi: 10.1080/20002297.2020.1814666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fakhruddin KS, Ngo HC, Samaranayake LP. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 2019;25:982–995. doi: 10.1111/odi.12932. [DOI] [PubMed] [Google Scholar]
- 54.Schoilew K, Ueffing H, Dalpke A, Wolff B, Frese C, Wolff D, Boutin S. Bacterial biofilm composition in healthy subjects with and without caries experience. J Oral Microbiol. 2019;11:1. doi: 10.1080/20002297.2019.1633194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qudeimat MA, Alyahya A, Karched M, Behbehani J, Salako NO. Dental plaque microbiota profiles of children with caries-free and caries-active dentition. J Dent. 2021;104:103539. doi: 10.1016/j.jdent.2020.103539. [DOI] [PubMed] [Google Scholar]
- 56.Kim JH, Bang IH, Noh YJ, Kim DK, Bae EJ, Hwang IH. Metabolites Produced by the Oral Commensal Bacterium Corynebacterium durum Extend the Lifespan of Caenorhabditis elegans via SIR-2.1 Overexpression. Int J Mol Sci. 2020;21(6):2212. doi: 10.3390/ijms21062212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redanz U, Redanz S, Treerat P, Prakasam S, Lin LJ, Merritt J, Kreth J. Differential Response of Oral Mucosal and Gingival Cells to Corynebacterium durum, Streptococcus sanguinis, and Porphyromonas gingivalis Multispecies Biofilms. Front Cell Infect Microbiol. 2021;11:686479. doi: 10.3389/fcimb.2021.686479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho MH, Lamont RJ, Xie H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci Rep. 2017;7(1):1413. doi: 10.1038/s41598-017-01551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47(12):3902–3906. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paster BJ, Bartoszyk IM, Dewhirst FE. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 1998;20:223–231. doi: 10.1023/A:1009715710555. [DOI] [Google Scholar]
- 63.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-Targeted Oligonucleotide Probes with Flow Cytometry for Analyzing Mixed Microbial Populations. Appl Environ Microbiol. 1990;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108(10):4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Validation of new oligonucleotide probes targeting subsets of the genus Streptococcus. For each probe, a full list of oral Streptococcus species and their matches and mismatches to the probe is shown (top), along with images showing the intensity of signal imaged after hybridization of pure cultures of cells to the probes. Each newly designed probe was hybridized simultaneously with existing probes targeting genus Streptococcus and most Bacteria as controls. 15 pure cultures were hybridized; 8 are shown here and the remaining 7 are shown in Fig. 1. A) Probe Scri995 targeting S. cristatus; B) probe Sgor63 targeting S. gordonii; C) probe Smit371 targeting S. mitis and its close relatives and S. cristatus; D) probe Smit651 targeting S. mitis and its close relatives. For each probe set, image acquisition and linear unmixing of all cultures were carried out under the same conditions using Zeiss ZEN software. Images were imported into FIJI and the range of display intensities was kept constant for each fluorophore (each column in the figure); for cultures where the fluorescence was dim in all channels, the display range was then additionally adjusted by a constant factor for all images in the row to improve visibility of cells; these rows are marked (*). All probes hybridized with their expected targets and showed negligible cross-hybridization to unexpected targets. (RRX: Rhodamine Red X; TRX: Texas Red X).
Additional file 2. List of strains used for FISH on pure cultures.
Additional file 3. Ribosomal RNA-targeted oligonucleotide probes used in this study and their combination into probe sets [62–64].
Additional file 4. Probe set validation matrices showing hybridization of pure cultures with complete probe sets. For each probe set, image acquisition and linear unmixing of all cultures were carried out under consistent conditions using Zeiss ZEN software. Images were imported into FIJI and the range of display intensities was kept constant for each fluorophore (each column in the figure). In addition to the species probes shown in Fig. 1 and Additional File 1, probes and their targets and fluorophores are as follows: Str405 [62] targeting genus Streptococcus, labeled with Rhodamine Red X (RRX); Por1160 [64] targeting the gingivalis group of genus Porphyromonas, labeled with Alexa 555; Pas111 [64] targeting family Pasteurellaceae, labeled with Dy615; Cor633 [12] targeting genus Corynebacterium, labeled with Dy490 (part A) or Atto 620 (part B); probe Cmat175 [12] targeting species C. matruchotii, labeled with Atto 655. All probes hybridized with their expected targets and showed negligible cross-hybridization to unexpected targets. (At532: Atto 532; TRX: Texas Red X).
Additional file 5. Cells of Streptococcus gordonii visualized in supragingival dental plaque. Top panel shows Streptococcus spp. at the genus level; bottom panel shows Streptococcus at the species level. (A): Cells of S. gordonii (arrows) are visualized in the vicinity of corncobs but not attached to them. (B): Cells of S. gordonii are also observed in hedgehog structures but not as part of corncobs. (C): a large clump of S. mitis group cells with scattered S. gordonii cells.
Additional file 6. Taxon composition of corncob segments by donor. This is the numerical data underlying the bar chart in Fig. 4.
Data Availability Statement
The datasets supporting the conclusions of this article are available in Zenodo under doi:10.5281/zenodo.6426109, doi:10.5281/zenodo.6529696, and doi:10.5281/zenodo.6529719.